Abstract
In this review, we take a survey of bioinformatics databases and quantitative structure-activity relationship studies reported in published literature. Databases from the most general to special cancer-related ones have been included. Most commonly used methods of structure-based analysis of molecules have been reviewed, along with some case studies where they have been used in cancer research. This article is expected to be of use for general bioinformatics researchers interested in cancer and will also provide an update to those who have been actively pursuing this field of research.
Introduction
Bioinformatics has played a crucial role in structure based drug and target discovery, diagnosis and analysis of various diseases and their diversity. In particular there is enormous potential of its application in cancer research, which has only been partially exploited so far. Essentially all bioinformatics starts with a database and proceeds to some kind of knowledge discovery and prediction. In this article, we review bioinformatics databases and different types of quantitative structure-activity relationship (QSAR) studies, which have either been used in cancer research or have the potential of such application.
Bioinformatics databases
Biological experiments result in useful information. This information has remained scattered in published literature, technical lab reports and patent files until not very long ago. However, there has been a tremendous effort during last couple of decades to compile, share, standardize and model biological information (e.g. Wu et al. 2003; Bairoch and Boeckmann 1991; Benson et al. 2005; Hamosh et al. 2005; Bateman et al. 2004; Boguski et al. 1993; Bauer et al. 2005; Smigielski et al. 2000; Wu et al. 2001; Berman 2000; Hulo et al. 2006; Attwood et al. 2000; Gromiha et al. 1999; Mulder et al. 2002; Pongor et al. 1992; Kanehisa and Goto 2000; Dowell et al. 2001;). There has also been relatively recent interest in improving the quality of databases, developing web-interfaces and integration of databases (Achard et al. 2001; He et al. 2005; Hanisch et al. 2002; Westbrook et al. 2002; Arauzo-Bravo and Ahmad 2005). These efforts have made it possible to know the state of the art in a given area of biology and provide a basis for what is sometimes called in-silico biology, as opposed to in-vivo and in-vitro biology. Some of the most widely used databases have been listed in Table 1.
Table 1.
General Bioinformatics Databases.
|
Major sequence repositories | ||
| DNA Data Bank of Japan (DDBJ) | http://www.ddbj.nig.ac.jp | All known nucleotide and protein sequences; International Nucleotide Sequence Database Collaboration |
| EMBL Nucleotide Sequence Database | http://www.ebi.ac.uk/embl.html | All known nucleotide and protein sequences; International Nucleotide Sequence Database Collaboration |
| GenBank | http://www.ncbi.nlm.nih.gov/ | All known nucleotide and protein sequences; International Nucleotide Sequence Database Collaboration |
| NCBI Reference Sequence Project | http://www.ncbi.nlm.nih.gov/RefSeq/ | Non-redundant collection of naturally-occurring biological molecules |
| Ensembl | http://www.ensembl.org/ | Annotated information on eukaryotic genomes |
| UCSC Genome Browser | http://genome.ucsc.edu/ | Genome assemblies and annotation |
| UniGene | http://www.ncbi.nlm.nih.gov/UniGene/ | Non-redundant, gene-oriented clusters |
|
Protein Databases | ||
| CSDBase | http://www.chemie.uni-marburg.de/~csdbase/ | Cold shock domain-containing proteins |
| DExH/D Family Database | http://www.helicase.net/dexhd/dbhome.htm | DEAD-box, DEAH-box and DExH-box proteins |
| Endogenous GPCR List | http://www.tumor-gene.org/GPCR/gpcr.html | G protein-coupled receptors; expression in cell lines |
| EXProt | http://www.cmbi.Kun.nl/EXProt/ | Proteins with experimentally-verified function |
| GenProtEC | http://genprotec.mbl.edu | E. coli K-12 genome, gene products and homologs |
| Histone Database | http://research.nhgri.nih.gov/histones/ | Histone and histone fold sequences and structures |
| HIV Molecular Immunology Database | http://hiv-web.lanl.gov/content/immunology/index | HIV epitopes |
| HIV RT and Protease Sequence Database | http://hivdb.stanford.edu | HIV reverse transcriptase and protease sequences |
| Homeodomain Resource genomic | http://genome.nhgri.nih.gov/homeodomain/ | Homeodomain sequences, structures and related genetic and genomic information |
| HUGE | http://www.kazusa.or.jp/huge/ | Large (>50 kDa) human proteins and cDNA sequences |
| IMGT | http://imgt.cines.fr | Immunoglobulin, T cell receptor and MHC sequences from human and other vertebrates |
| IMGT/HLA | http://www.ebi.ac.uk/imgt/hla/ | Polymorphic sequences of human MHC and related genes |
| IMGT/MHC Database | http://www.ebi.ac.uk/ipd/mhc/index.html | Major histocompatibility complex sequences |
| InBase | http://www.neb.com/neb/inteins.html | All known inteins (protein splicing elements): properties, sequences, bibliography |
| InterPro | http://www.ebi.ac.uk/interpro | Protein families and domains |
| LGICdb | http://www.ebi.ac.uk/compneur-srv/LGICdb/LGICdb.php | Ligand-gated ion channel subunit sequences |
| Nuclear Protein Database (NPD) | http://npd.hgu.mrc.ac.uk | Proteins localized in the nucleus |
| NRMD | http://www.receptors.org/NR/ | Nuclear receptor superfamily |
| NUREBASE | http://www.ens-lyon.fr/LBMC/laudet/nurebase.html | Nuclear hormone receptors |
| ooTFD | http://www.ifti.org/ootfd | Transcription factors and gene expression |
| PANTHER | http://www.pantherdb.org/ | Gene products organized by biological function |
| Peptaibol | http://www.cryst.bbk.ac.uk/peptaibol/home.shtml | Peptaibol (antibiotic peptide) sequences |
| Phospho.ELM | http://phospho.elm.eu.org/ | Protein phosphorylation sites |
| PKR | http://www.kinasenet.org/pkr/Welcome.do | Protein kinase sequences, enzymology, genetics and molecular and structural properties |
| Prolysis | http://delphi.phys.univ-tours.fr/Prolysis/ | Proteases and natural or synthetic protease inhibitors |
| Protein Information Resource (PIR) | http://pir.georgetown.edu | Comprehensive, annotated, non-redundant protein sequence databases |
| ProtoNet | http://www.protonet.cs.huji.ac.il/ | Hierarchical clustering of protein sequences |
| RTKdb | http://pbil.univ-lyon1.fr/RTKdb/ | Receptor tyrosine kinase sequences |
| SEVENS | http://sevens.cbrc.jp | 7-transmembrane helix receptors |
| SWISS-PROT/TrEMBL | http://www.expasy.org/sprot | Curated protein sequences |
| TIGRFAMs | http://www.tigr.org/TIGRFAMs | Functional identification of proteins |
| trEST, trGEN, Hits | http://hits.isb-sib.ch | Hypothetical protein sequences |
|
Structure | ||
| ASTRAL | http://astral.stanford.edu/ | Sequences of domains of known structure, selected subsets and sequence-structure correspondences |
| BioMagResBank acids | http://www.bmrb.wisc.edu/ | NMR spectroscopic data from proteins, peptides, and nucleic acids |
| CATH | http://www.biochem.ucl.ac.uk/bsm/cath_new | Protein domain structures |
| CKAAPs DB | http://ckaaps.sdsc.edu/perl/browser.pl | Structurally-similar proteins with dissimilar sequences |
| CSD | http://www.ccdc.cam.ac.uk/products/csd/ | Crystal structure information for organic and metal organic compounds |
| Database of Macromolecular Movements | http://bioinfo.mbb.yale.edu/Mol-MovDB/ | Descriptions of protein and macromolecular motions, including movies |
| Decoys ‘R’ Us | http://dd.stanford.edu/ | Computer-generated protein conformations based on sequence data |
| DSMM | http://projects.eml.org/mcm/database/dsmm | Database of Simulated Molecular Motions |
| Gene3D | http://cathwww.biochem.ucl.ac.uk:8080/Gene3D/ | Precalculated structural assignments for genes within whole genomes |
| GTOP | http://spock.genes.nig.ac.jp/~genome/gtop.html | Protein fold predictions from genome sequences |
| HIC-Up | http://alpha2.bmc.uu.se/hicup/ | Structures of small molecules (‘hetero-compounds’) |
| HSSP | http://www.sander.ebi.ac.uk/hssp/ | Structural families and alignments; structurally-conserved regions and domain architecture |
| LPFC | http://smi-web.stanford.edu/projects/helix/LPFC/ | Library of protein family core structures |
| MMDB linked | http://www.ncbi.nlm.nih.gov/Structure/ | All experimentally-determined three-dimensional structures, linked to NCBI Entrez |
| ModBase | http://modbase.compbio.ucsf.edu/modbase-cgi-new/index.cgi | Annotated comparative protein structure models |
| NDB | http://ndbserver.rutgers.edu/ | Nucleic acid-containing structures |
| NTDB | http://ntdb.chem.cuhk.edu.hk¶ | Thermodynamic data for nucleic acids |
| PALI | http://pauling.mbu.iisc.ernet.in/~pali | Phylogeny and alignment of homologous protein structures |
| PASS2 | http://caps.ncbs.res.in/campass/pass.html | Structural motifs of protein superfamilies |
| PDB | http://www.pdb.org/ | Structure data determined by X-ray crystallography and NMR |
| PDB-REPRDB | http://mbs.cbrc.jp/pdbreprdb-cgi/reprdb_menu.pl | Representative protein chains, based on PDB entries |
| PDBsum | http://www.ebi.ac.uk/thornton-srv/databases/pdbsum/ | Summaries and analyses of PDB structures |
| ProTherm | http://gibk26.bse.kyutech.ac.jp/jouhou/therm/protherm.html | Thermodynamic data for Pro-wild-type and mutant proteins |
| PSSH | http://srs3d.ebi.ac.uk/ | Alignments between protein sequences and tertiary structures |
| RNABase | http://www.rnabase.org | RNA-containing structures from PDB and NDB |
| SCOP | http://scop.mrc-lmb.cam.ac.uk/scop | Familial and structural protein relationships |
| SCOR | http://scor.lbl.gov | RNA structural relationships |
| Sloop | http://www-cryst.bioc.cam.ac.uk/~sloop/ | Classification of protein loops |
| Structure-Superposition Database | http://ssd.rbvi.ucsf.edu | Pairwise superposition of TIM-barrel structures |
| SUPERFAMILY | http://supfam.org | Assignments of proteins to structural superfamilies |
|
Retrieval Systems and Database Structure | ||
| TESS | http://www.cbil.upenn.edu/cgi-bin/tess/tess | Transcription element search system |
| Virgil | http://www.infobiogen.fr/services/virgil¶ | Database interconnectivity |
The cancer research community has not remained indifferent to the importance of databases. From the big organizations such as National Cancer Institute (NCI; http://www.cancer.gov) to smaller research groups, scientists have developed databases relating to the genetics, molecular biology, microarray clinical reports and several other aspects of cancer. Table 2 lists some of the most prominent databases, which have emerged in respect of cancer research. Some of these databases are discussed below:
Table 2.
Cancer related bioinformatics databases.
| Database Name | URL | Description |
|---|---|---|
| Atlas of Genetics and Cytogenetics in Oncology and Haematology | http://www.infobiogen.fr/services/chromcancer/ | Cancer-related genes, chromosomal abnormalities in oncology and haematology, and cancer-prone diseases |
| Cancer Chromosomes | http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=cancerchromosomes | Cytogenetic, clinical and reference information on cancer-related aberrations |
| CGED | http://cged.hgc.jp/cgi-bin/input.cgi | Cancer gene expression database |
| COSMIC | http://www.sanger.ac.uk/genetics/CGP/cosmic/ | Catalogue of somatic mutations in cancer: sequence data, samples and publications |
| Germline p53 Mutations | http://www.lf2.cuni.cz/win/projects/p53.htm | Mutations in germline_mut_ human tumor and cell line p53 gene |
| IARC TP53 Database | http://www.p53.iarc.fr/index.html | Human TP53 somatic and germline mutations |
| MTB | http://tumor.informatics.jax.org/mtbwi/index.do | Mouse tumor biology database: tumor types, genes, classification, incidence, pathology |
| OncoMine | http://www.oncomine.org/ | Cancer microarray data by gene or cancer type |
| Oral Cancer Gene Database | http://www.tumor-gene.org/Oral/oral.html | Cellular and molecular data for genes involved in oral cancer |
| RB1 Gene Mutation DB | http://www.verandi.de/joomla/ | Mutations in the human retinoblastoma (RB1) gene |
| RTCGD | http://rtcgd.ncifcrf.gov/ | Mouse retroviral tagged cancer gene database |
| SNP500Cancer | http://snp500cancer.nci.nih.gov | Re-sequenced SNPs from 102 reference samples |
| SV40 Large T-Antigen Mutants | http://www.pitt.edu/pipaslab/¶ | Mutations in SV40 large tumor antigen gene |
| Tumor Gene Family Databases | http://www.tumor-gene.org/tgdf.html | Cellular, molecular and biological data about genes involved in various cancers |
These sites could not be opened at the time of revising the manuscript.
Cancer Chromosomes database
(http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=cancerchromosomes) Cancer Chromosomes integrates data from three sources: the NCI/NCBI SKY/M-FISH & CGH Database, the NCI Mitelman Database of Chromosome Aberrations in Cancer, and the NCI Recurrent Aberrations in Cancer (Knutsen et al. 2005). This is a publicly available database and can be searched for cytogenetic, clinical, and/or reference information. Similarity reports demonstrating cytogenetic and clinical relatedness at varying levels of specificity are also returned on querying this database.
CGED (Cancer Gene Expression Database)
(http://cged.hgc.jp/cgi-bin/input.cgi) CGED is a database containing expression profiles and accompanying clinical information of breast, colorectal, and hepatocellular cancer related genes (Kato et al. 2005). The data in CGED have been obtained through collaborative efforts made at the Nara Institute of Science and Technology and Osaka University School of Medicine to identify genes of clinical importance. The expression data have been obtained by a high-throughput RT-PCR technique (adaptor-tagged competitive PCR). The data can be retrieved either using gene identifiers or by functional categories defined by Gene Ontology terms or the SwissProt annotation. Gene expression data are displayed in mosaic plots. This database also provides for the expression patterns of multiple genes, selected by names or similarity search of the patterns. The sorting function enables users for easy recognition of relationships between gene expression and clinical parameters.
The Atlas of Genetics and Cytogenetics in Oncology and Haematology
(http://www.infobiogen.fr/services/chromcancer) The Atlas of Genetics and Cytogenetics in Oncology and Haematology is a database containing information about genes related to cancer (Huret et al. 2000). This database contains information in the form of cards on cancer related genes, chromosomal abnormalities, cancers, and cancer-prone diseases. These cards are well-structured papers, which represent the body of the Atlas. Cards on genes include data on DNA/RNA, protein, mutations, and diseases. Cards on leukemias and solid tumours include data on: clinics, cytogenetics, genes, hybrid gene and fusion protein. Cards on cancer-prone diseases include data on: inheritance mode, clinics, neoplastic risk, cytogenetics, genes and proteins, mutations. These Cards are linked to NCBI published literature database PubMed, and to other major databases (nomenclature, cartography, gene structure, transcripts, proteins, domain families, diseases, mutations, probes). This database has another component called Deep Insights and Case Reports. Deep insights are review articles related to special topics and the Case Reports section is dedicated to rare cytogenetic entities of leukemia including the associated prognosis. This database also referred to as The Atlas is part of the genome project and participates in the research in cancer epidemiology.
Database of germline p53 mutations
(http://www.lf2.cuni.cz/projects/germline_mut_p53.htm) Somatic mutations in the p53 tumor suppressor gene are found in many human cancers (Le Roux et al. 2005). In addition, germline p53 mutations have been identified in individuals from cancer-prone families and in isolated cancer patients affected at a young age or suffering from multiple tumours (Harris 1996; Hollstein et al. 1991). A large fraction of the cancer-prone families with germline p53 mutation follow the criteria of Li-Fraumeni syndrome (LFS) (Li et al. 1988;). This syndrome is a rare familial autosomal dominant cancer syndrome characterised by early-onset sarcomas, brain tumours, premenopausal breast cancer, leukaemias and adrenocortical tumours. It is with this view that a database dedicated to p53 mutations has been developed. Genotype-phenotype correlations, compiled in this data may improve the counseling and preventive approaches in the affected families. This is a comprehensive database of those cases of germline p53 mutations for which sufficient detail is given in the literature. In addition to listing all mutations, the database includes detailed information about the families, affected individuals and their tumours. It therefore provides a powerful means for drawing correlations between various aspects of germline p53 mutations. Each p53 mutation (type of the mutation, exon and codon affected by the mutation, nucleotide and amino acid change), have been explained. In addition, it has the information on the family history of cancer, diagnosis of LFS, each affected individual (sex, generation, p53 status, from which parent the mutation was inherited) and each tumour (type, age of onset, p53 status (loss of heterozygosity and immunostaining). Each entry contains the original research article as reference(s).
COSMIC database
(http://www.sanger.ac.uk/genetics/CGP/cosmic/) COSMIC is a database designed to store and display somatic mutation information-relevant for cancer (Forbes et al. 2006). In particular, it contains information relating to human cancers. COSMIC contains information on publications, samples and mutations implicated in cancer. It also includes samples, which have been found to be negative for mutations during screening. This allows the calculation of frequency data normalized by control frequencies for mutations in different genes in different cancer types. Samples entered include benign neoplasms and other benign proliferations, in situ and invasive tumors, recurrences, metastases and cancer cell lines. Histology and tissue ontology has also been created in this database. All mutations are mapped to a single version of each gene. The data can be queried by tissue, histology or gene and displayed as a graph, as a table or exported to other formats.
EHCO database
(http://ehco.nchc.org.tw) EHCO (Encyclopedia of Hepatocellular Carcinoma genes Online) is an integrative database for HCC (hepatocellular carcinoma) research. It carries gene annotations collected by computer-assisted mining, manual curation, and extraction from public databases. Currently EHCO contains information for about 3500 HCC-related genes. Various entries in this database can be compared online. Detailed annotations for particular genes, including sequence, ontology, cited literature, and expression profiles are also available.
Human p53 database
(http://metalab.unc.edu/dnam/mainpage.html) A collection of databases relating to p53 gene mutations, lacI and lacZ is available on this website (Cariello et al. 1994; Cariello et al. 1996). There are nearly 6000 entries corresponding to p53, 200 for lacZ and 1500 of lacI. In addition 1500 transgenic and 8000 bacterial entries are also included. A software for analysis of the databases is also included. Each database has a separate software analysis program. All these databases include information about mutations such as base position, the nature of the mutation, amino acid position, molecular weight and the name of mutant amino acid, the local sequence around a mutation and literature citation as the source of listed information. Information specific to the p53 database includes cancer type, cell origin, loss of heterozygosity.
IARC TP53 Database
(http://www.p53.iarc.fr/index.html) The IARC TP53 Database compiles data on human somatic and germline TP53 genetic variations that are reported in the published literature. (Olivier et al. 2002; Hernandez-Boussard 1999; Hainaut et al. 1997; Hainaut et al. 1998 ; Hollstein et al. 1994, Hollstein et al. 1996). With over 18,500 somatic and 225 germline mutations and 1,000 citations in the world literature, this database is now recognized as a major source of information on TP53 mutation patterns in human cancer. It can be searched and analyzed online and is useful to draw hypotheses on the nature of the molecular events involved in TP53 mutagenesis and on the natural history of cancer.
ITTACA Gene expression and clinical database
(http://bioinfo-out.curie.fr/ittaca/) ITTACA is a database of microarray experimental results and clinical information retrieved form published papers (Elfilali et al. 2006). It contains information on breast carcinoma, bladder carcinoma, and uveal melanoma. Online service also allows some basic statistical analysis of the database such as the comparison of expression distribution profiles, tests for differential expression, and patient survival analyses.
The Mouse Tumor Biology Database (MTB)
(http://www.informatics.jax.org) MTB database compiles and shares information about tumor frequency, genetics, and pathology in genetically denned mice (i.e., transgenics, targeted mutations, and inbred strains) (Bult et al. 2001). The database collects crucial information about incidence of different types of tumors in different strains, mutations relating to specific genes and tumors corresponding to them, which have been reported in medical journals. Existing standards for anatomy, tumor names, gene names, and strain names are well enforced, enabling direct links to information across MTB entries and to other relevant databases.
The Tumor Gene Family Databases (TGDBs)
(http://condor.bcm.tmc.edu/ermb/tgdb/tgdf.html) TGDB is made up of two databases viz. Oral Cancer Gene Database (OrCGDB) and Breast Cancer Gene Database (BCGD). Both these databases contain information on a mechanism of oncogenic activation, regulation, frequency of involvement in various tumor types, and chromosomal location for the genes involved in cancer (e.g. proto-oncogenes and tumor supressor genes). Data about the encoded proteins includes the cell type in which they are found, subcellular location, DNA, protein, and ligand binding, role in development, and normal biochemical function.
QSAR and in-silico analysis of molecular recognition
Once the molecular mechanism and the chemistry of a disease is understood, the next crucial task is to find a suitable cure for it. Atypical requirement is to find a suitable drug target and the drug itself (Brooijmans and Kuntz 2003). Target discovery draws much on bioinformatics tools today and in case of cancer the DNA and protein molecules both can be potential targets for drugs (Choudhary et al. 2005; Bandyopadhyaya et al. 2005; Bhongade et al. 2004; Asseffa et al. 2003; Gellert et al. 2005; Khaleque et al. 2006; Yao et al. 2005; McColl et al. 2005).
Drug discovery is a complex, expensive and very time-consuming exercise, as there is no single systematic way to automatically discover a drug even when the disease and targets have been well understood (Dixit and Mitra 2002).
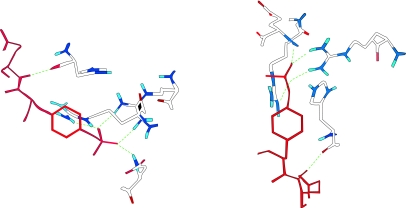
There may be millions of candidate molecules if in-silico filtering is not performed. Experiments cannot be performed on such large number of drug candidates due to prohibitive costs both in terms of time and money. Quantitative structure-activity relationship (QSAR) studies form the center stage when a protein (typically an enzyme) is the target and there is a need to find a suitable molecule, which can control (inhibit) the activity of its target. The basic principle of such a study is the structure-dependence of chemical activity. QSAR has existed much longer than the first popularity of computers, because chemical structure has always been able to explain at least some aspects of chemical properties. However, with the availability of powerful computers and high quality databases of molecular libraries and interactions have made QSAR an essential component of drug discovery today. Role of structure in determining the activity of a chemical compound is illustrated in an example of protein-ligand complex in Fig 1.
Figure 1.
Identical A & B Chain Residues of 1A1E in complex with its ligand (ACE-PTR-GLU-DIY). Ligand in red.
QSAR based (in-silico) analysis may be better regarded as an exercise to screen or filter drug candidates, before they are subjected to more intensive calculations such as docking or an experimental measurement of activity (in-vitro) and finally under real conditions (in-vivo). Many times this step will pick up a dozen of drug candidate from a library of millions of well-studied molecules. Traditional QSAR is specific to a particular target or enzyme and all the screening is performed on drug candidates (ligand molecules). These ligand molecules are very diverse and in order to screen them suitably, we need to describe their structure as well as chemical nature. This leads to the issue of finding descriptors of molecular properties of ligands and drugs. Hundreds of molecular properties or descriptors are used to represent molecules (Labute 2000; Xue and Bajorath 2000; Wildman and Crippen 2002; Gozalbes et al. 2002).
These properties may be purely geometric, topological, electromagnetic, classical and quantum-mechanical. Often, predicting activity of a protein-ligand combination if the descriptors of the ligand are known carries out this screening. Regression techniques such as Principal Component Analysis (PCA), Neural Network and Multi-variate correlation are the major techniques used for this purpose. In the following we review some of these techniques and special reference will be wherever a successful application to cancer has been reported.
A large number of molecular descriptors are available and used (Todeschini and Consonni; Labute 2000; Wildman and Crippen 2002; Hansch et al. 1995; Basak et al. 1980; Gozalbes et al. 2002; Pirard and Picket 2000; Basak et al. 1981; Basak et al. 1982; Kier and Hall 1999; Raevsky 1999; Xue and Bajorath 2000). Molecular descriptors used in QSAR for a unique representation and identification of ligand molecules, which are likely to be drug candidates, may be classified as follows:
Constitutional descriptors such as molecular weight, van der Waals volume, electronegativities, polarizability, number of atoms, non-H atoms, number of H bonds, multiple bonds, bond orders, aromatic ratio, number of rings, number of double and triple bonds, aromatic bonds, 3 different types of (n-membered) rings, benzene-like rings.
Topological descriptors such as total structure connectivity index, Pogliani index, ramification index, polarity number, average vertex distance degree, mean square distance index (Balaban), Schultz Molecular Topological Index (MTI), square reciprocal distance sum index, quasi-Wiener index (Kirchhoff number), spanning tree number, hyper-distance-path index, reciprocal hyper-distance-path index, detour index, hyper-detour index, reciprocal hyper-detour index, distance/detour index, all-path Wiener index, Wiener-type index from Z weighted distance matrix (Barysz matrix), molecular electrotopological variation, E-state topological parameter, Kier symmetry index eccentricity, mean distance degree deviation, unipolarity, centralization, variation.
Walk and path counts such as molecular walk counts, total walk count, self-returning walk counts, molecular path counts, molecular multiple path counts, total path count, conventional bond-order ID number, Randic ID number, Balaban ID number, ratio of multiple path count over path count, difference between multiple path count and path count.
Connectivity indices such as connectivity indices, average connectivity indices, valence connectivity indices, average valence connectivity indices, solvation connectivity indices, modified, reciprocal distance Randic-type index, reciprocal distance squared Randic-type index.
Information indices such as information index on molecular size, total information index of atomic composition, mean information index on atomic composition, mean information content on the distance equality, mean information content on the distance magnitude, mean information content on the distance degree equality, mean information content on the distance degree magnitude, total information content on the distance equality, total information content on the distance magnitude, mean information content on the vertex degree equality, mean information content on the vertex degree magnitude, graph vertex complexity index, graph distance complexity index (log), Balaban U index, Balaban V index, Balaban X index, Balaban Y index Basak indices of neighborhood symmetry.
2D autocorrelations Broto-Moreau autocorrelations of a topological structure, Moran autocorrelations, Geary autocorrelations.
Edge adjacency indices edge connectivity index of order 0, edge connectivity index of order 1 eigenvalues from edge adj. matrix weighted by edge degrees, eigenvalues from edge adj. matrix weighted by dipole moments, eigenvalues from edge adj. matrix weighted by resonance integrals spectral moments from edge adj. matrix, spectral moments from edge adj. matrix weighted by edge degrees, spectral moments from edge adj. matrix weighted by dipole moments, spectral moments from edge adj. matrix weighted by resonance integrals.
Eigenvalue-based indices Lovasz-Pelikan index (leading eigenvalue), leading eigenvalue from Z weighted distance matrix (Barysz matrix), leading eigenvalue from mass weighted distance matrix, leading eigenvalue from van der Waals weighted distance matrix, leading eigenvalue from electro-negativity weighted distance matrix, leading eigenvalue from polarizability weighted distance matrix.
Geometrical descriptors 3D-Wiener index, 3D-Balaban index, 3D-Harary index average geometric distance degree, D/D index, average distance/distance degree gravitational index G1, gravitational index G2 (bond-restricted), radius of gyration (mass weighted), span R, average span R.
Functional group counts terminal primary C(sp3), total secondary C(sp3), total tertiary C(sp3), total quaternary C(sp3), ring secondary C(sp3), ring tertiary C(sp3), ring quaternary C(sp3) aromatic C(sp2), unsubstituted benzene C(sp2), substituted benzene C(sp2), non-aromatic conjugated C(sp2), terminal primary C(sp2), aliphatic secondary C(sp2), aliphatic tertiary C(sp2), allenes groups, terminal C(sp), non-terminal C(sp) cyanates (aliphatic), cyanates (aromatic), isocyanates (aliphatic), isocyanates (aromatic), thiocyanates (aliphatic), thiocyanates (aromatic), isothiocyanates (aliphatic), isothiocyanates (aromatic).
Charge descriptors maximum positive charge, maximum negative charge, total positive charge, total negative charge, total absolute charge (electronic charge index – ECI), mean absolute charge (charge polarization), total squared charge, relative positive charge, relative negative charge, submolecular polarity parameter, topological electronic descriptor, topological electronic descriptor (bond resctricted), partial charge weighted topological electronic descriptor, local dipole index.
Molecular properties unsaturation index hydrophilic factor Ghose-Crippen molar refractivity topological polar and non-polar surface area.
Many more descriptors may be calculated and comprehensive lists can be found. A comprehensive review of molecular descriptors is presented by Karelson (2000). Many free and commercial software also provide a current list of descriptors (e.g. http://www.talete.mi.it/products/dragon_molecular_descriptors.htm and http://preadmet.bm-drc.org/preadmet/query/query1.php, from where, list of many of the above descriptors is compiled.). An excellent coverage of issues and topics related to QSAR is also provided in a text book by Gasteiger and Engel (2003).
After the descriptors of molecules have been calculated, redundant descriptors are removed using Principal Component Analysis or Multivariate analysis (Jolliffe 1986: Xue and Bajorath 2000). Many commercial and some free software programs are now available which may be used to calculate some of the descriptors and/or develop a QSAR model using them. Some of these programs are listed in Table 3. These softwares can give few key descriptors (such as 5 descriptors in Molinspiration) or a very large number of them (e.g. DRAGON gives more than 1500 descriptors), which will need to be reduced by some analysis.
Table 3.
Available QSAR and molecular descriptor programs.
| Name of the software | Brief Description | URL/Reference |
|---|---|---|
| PEST | Shape properties, Wavelet decomposition properties, Electrostatic potential, electronic kinetic energy density etc. | C. Matthew Sundling, N. Sukumar and Curt Breneman Rensselaer Polytechnic Institute http://www.chem.rpi.edu/chemweb/recondocpest.html |
| Pharma Algorithm’s QSAR Builder | QSAR and QSPR modeling; Excess molar refraction, H-bond acidity, H-donor capability, H-bond basicity, H-acceptor capability Hexadecane/gas partition coefficient, LogP partition coefficient, TPSA - topological polar surface area | Hugo Kubinyi, Professor of Pharmaceutical Chemistry at the University of Heidelberg, Germany http://apalgorithms.com/qsar_builder.htm |
| Bioreason | QSAR, QPSR | Commercial by Bioreason |
| ClassPharmer | http://www.bioreason.com | |
| ChemTK | Molecule design, descriptors and modeling | http://www.sageinformatics.com/chemtk.html |
| Molinspiration toolkit | Java Based software and free online calculations of fragments and basic properties/descriptors. | http://www.molinspiration.com |
| ShapeSig | Shape desciptors, and statistical analysis | http://histidine.umdnj.edu/~shape/index.php |
| Cerius (QSAR module) | Modeling and QSAR, includes MOPAC Quantum mechanical calculations, alignments etc. | http://www.accelrys.com |
| CODESSA | QSAR program | http://www.semichem.com/codessa/default.php |
| HASL | 3D QSAR | http://www.bio.com/store/product.jhtml?id=prod300024 |
| QTRFIT | Rigid body superposition | http://www.osc.edu/PET/CCM/skeleton/software/tested/source/qtrfit/qtrfit.html |
| DRAGON | 1664 molecular descriptors | http://www.talete.mi.it/main_net.htm |
Cancer researchers have frequently used these methods for a systematic filtering of potential drug candidates or for generalizing principles governing the choice of ligands that prefer to bind to a particular family of proteins in a selective and competitive way. Several aspects of cancer have been studied using QSAR techniques. Classical efforts at using QSAR for cancer drug research date back to 1970s (e.g. Hansch 1979). Antitumour drugs have remained a regular subject of investigation using QSAR (Ren and Lien 2004). During that time, focus was to discover drugs for chemotherapy. As cases of multidrug resistance were observed, a need to have alternative medicine for the same action were felt. Thus, a large number of researchers have focused on multidrug resistance in regards to chemotherapy and employed QSAR as a means to solve this problem. For example Breier et al. (2000) have studied multidrug resistance (MDR) for L1210/VCR-1 and L1210/VCR-2 cell lines in regards to leukemia treatment. They related the developed adaptation and drug resistance to structure descriptors of drugs viz. binding energy, molecular weight, pKa, log P etc. Klopman et al. (1997) have studied 609 diverse compounds to understand the drug resistance in P388/ADR resistant cell lines. In this study they identified several structural characteristics of MDR such as log P and graph index. More advanced techniques of QSAR such as Comparative Molecular Simillarity Index Analysis (CoMSIA) have been used to study antiviral and anticancer drugs targeting Thymidine Kinase (e.g. Bandyopadhyaya et al. 2005, Bhongade and Gadad 2004). Principle of CoMSIA is the alignment and comparison of drug molecules by comparing their similarity indices (selected descriptors). A similar approach, called Comparative Molecular Field Analysis (CoMFA) focuses on molecular field descriptors for this purpose (Cramer et al. 1988). Epidermal Growth Factor Receptors (EGFR) are one of the most popular class of proteins studied by QSAR method. Assefa et al. (2003) have used CoMFA for such a study and concluded that electrostatics and hydrophobicity descriptors play the most important role in EPGR target binding. Similarly, electrotopological state atom (ETSA) indices have been shown to play the most important role in anti tumour effect of pyridoacridine ascididemin analagues (Debnath et al. 2003). Thus, if a drug is available for chemotherapy and more such drugs are required to have redundancy against drug resistance, previously known successful drug/inhibitor is compared with a large data set of diverse molecules and those having their molecular indices (CoMSIA), or molecular fields (CoMFA) similar to that drug are picked up for potential use. Most recent QSAR related cancer studies have focused on genomic aspects of cancer related drug discovery (Workman 2001, Jung et al. 2003). This allows for individual prescriptions based on the genetic makeup of the patient. Thus, the possibility of having a large number of drugs having similar inhibitory ability but diverse genetic response opens a myriad of possibilities for cancer related research for peoples and individuals.
Summary
A number of databases directly and indirectly useful for cancer research have been reviewed. QSAR techniques and its application to cancer research have been outlined.
References
- Achard F, Vaysseix G, Barillot E. XML, bioinformatics and data integration. Bioinformatics. 2001;17(2):115–25. doi: 10.1093/bioinformatics/17.2.115. [DOI] [PubMed] [Google Scholar]
- Arauzo-Bravo, Marcos J, Ahmad S. Protein Sequence and Structure Databases: A Review. Current Analytical Chemistry. 2005;vol. 1(3):355–371. (17) [Google Scholar]
- Assefa H, Kamath S, Buolamwini JK. 3D-QSAR and docking studies on 4-anilinoquinazoline and 4-anilinoquinoline epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors. J Comput Aided Mol Des. 2003;17(8):475–93. doi: 10.1023/b:jcam.0000004622.13865.4f. [DOI] [PubMed] [Google Scholar]
- Attwood TK, Croning MDR, Flower DR, Lewis AP, Mabey JE, Scordis P, Selley JN, Wright W. PRINTS-S: the database formerly known as PRINTS. Nucleic Acids Res. 2000;28(1):225–227. doi: 10.1093/nar/28.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A, Boeckmann B. The SWISS-PROT protein sequence data bank. Nucleic Acids Res. 1991;19(Suppl):2247–2249. doi: 10.1093/nar/19.suppl.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyaya AK, Johnsamel J, Al-Madhoun AS, Eriksson S, Tjarks W. Comparative molecular field analysis and comparative molecular similarity indices analysis of human thymidine kinase 1 substrates. Bioorg Med Chem. 2005;13(5):1681–9. doi: 10.1016/j.bmc.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Basak SC. Bindingofbarbituratesto cytochromeP450: AQSARstudy using log P and topological indices. Med. Sci. Res. 1988;16:281. [Google Scholar]
- Basak SC, Gieschen DP, Magnuson VR, Harriss DK. Structure-activity relationships and pharmacokinetics: a comparative study of hydrophobicity, van der Waals’ volume and topological parameters. IRCS Med. Sci. 1982;10:619. [Google Scholar]
- Basak SC, Raychaudhury C, Roy AB, Ghosh JJ. Quantitative Structure-Activity Relationship (QSAR) studies of bioactive agents using structural information indices. Ind. J. Pharmacol. 1981;13:112. [Google Scholar]
- Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer ELL, Studholme DJ, Yeats C, Eddy SR. The Pfam protein families’ database. Nucleic Acids Res. 2004;32(Database issue):D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2005;33(Database Issue):D34–D38. doi: 10.1093/nar/gki063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhongade BA, Gadad AK. 3D-QSAR CoMFA/CoMSIA studies on Urokinase plasminogen activator (uPA) inhibitors: a strategic design in novel anticancer agents. Bioorg Med Chem. 2004;12(10):2797–805. doi: 10.1016/j.bmc.2004.02.019. [DOI] [PubMed] [Google Scholar]
- Boguski MS, Lowe TMJ, Carolyn M. Tolstoshev. dbEST: a database for “expressed sequence tags”. Nature Genetics. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- Breier A, Drobna Z, Docolomansky P, Barancik M, Neoplasma “Cytotoxic activity of several unrelated drugs on L1210 mouse leukemic cell sublines with P-glycoprotein (PGP) mediated multidrug resistance (MDR) phenotype. A QSAR study.”. 2000;47:100–106. [PubMed] [Google Scholar]
- Brooijmans N, Kuntz ID. MOLECULAR RECOGNITION AND DOCKING ALGORITHMS. Annual Review of Biophysics and Biomolecular Structure. 2003;32:335–373. doi: 10.1146/annurev.biophys.32.110601.142532. [DOI] [PubMed] [Google Scholar]
- Bult CJ, Krupke DM, Naf D, Sundberg JP, Eppig JT. Web-based access to mouse models of human cancers: the Mouse Tumor Biology (MTB) Database. Nucleic Acids Res. 2001;29(1):95–7. doi: 10.1093/nar/29.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood SK, Khaleque MA, Sawyer DB, Ciocca DR. Heat shock proteins in cancer: chaperones of tumorigenesis. Trends Biochem Sci. 2006 doi: 10.1016/j.tibs.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Cariello NF, et al. Database and software for the analysis of mutations in the human p53 gene. Cancer Research. 1994;54:4454–4460. [PubMed] [Google Scholar]
- Cariello NF, et al. Database and software for the analysis of mutations at the human p53 gene. Nucleic Acids Research. 1994;22:3549–3550. [PMC free article] [PubMed] [Google Scholar]
- Cariello NF, Douglas GR, Soussi T. Databases and software for the analysis of mutations in the human p53 gene, the human hprt gene and the lacZ gene intransgenic rodents. Nucl. Acids Res. 1996;24:119–120. doi: 10.1093/nar/24.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary G, Karthikeyan C, Hari Narayana Moorthy NS, Sharma SK, Trivedi P. QSAR Analysis of Some Cytotoxic Thiadiazino-acridines. Internet Electron. J. Mol. Des. 2005;4:793–802. [Google Scholar]
- Cramer RD, Patterson DE, Bunce JD. Comparative molecular field analysis (CoMFA) 1. Effect of shape on binding of steroids to carrier proteins. J. Am. Chem. Soc. 1988;110(18):5959–67. doi: 10.1021/ja00226a005. [DOI] [PubMed] [Google Scholar]
- Debnath B, Gayen S, Bhattacharya S, Samanta S, Jha T. “QSAR study on some pyridoacridine ascididemin analogues as antitumor agents.”. Bioorg Med Chem. 2003;11(24):5493–9. doi: 10.1016/j.bmc.2003.09.022. [DOI] [PubMed] [Google Scholar]
- Dixit KS, Mitra SN. Bioinformatics in Drug Discovery. CRIPS. 2002;3:2–6. [Google Scholar]
- Dowell RD, Jokerst RM, Day A, Eddy SR, Stein L. The Distributed Annotation System. BMC Bioinformatics. 2001;2:7. doi: 10.1186/1471-2105-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfilali A, Lair S, Verbeke C, La Rosa P, Radvanyi F, Barillot E. ITTACA: a new database for integrated tumor transcriptome array and clinical data analysis. Nucleic Acids Research. 2006;34(Database issue):D613–D616. doi: 10.1093/nar/gkj022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes S, Clements J, Dawson E, Bamford S, Webb T, Dogan A, Flanagan A, Teague J, Wooster R, Futreal PA, Stratton MR. COSMIC 2005. Br J Cancer. 2006;94(2):318–22. doi: 10.1038/sj.bjc.6602928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger J, Engel T. John Wiley & Sons; New York: 2003. Chemoinformatics. [Google Scholar]
- Gellert GC, Jackson SR, Dikmen ZG, Wright WE, Shay JW. Telomerase as a therapeutic target in Cancer. Drug Discovery Today: Disease Mechanisms. 2005;2(2) [Google Scholar]
- Gonzales-Diaz H, Gia O, Uriarte E, Hernadez I, Ramos R, Chaviano M, Seijo S, Castillo JA, Morales L, Santana L, Akpaloo D, Molina E, Cruz M, Torres LA, Cabrera MA. “Markovian chemicals “in silico” design (MARCH-INSIDE), apromising approach for computer-aided molecular design I: discovery of anticancer compounds”. J Mol Model. 2003;9(6):395–407. doi: 10.1007/s00894-003-0148-7. [DOI] [PubMed] [Google Scholar]
- Gozalbes R, Doucet JP, Derouin F. Application of topological descriptors in QSAR and drug design: history and new trends. Curr Drug Targets Infect Disord. 2002;2(1):93–102. doi: 10.2174/1568005024605909. [DOI] [PubMed] [Google Scholar]
- Gromiha MM, An J, Kono H, Oobatake M, Uedaira H, Sarai A. ProTherm: Thermodynamic Database for Proteins and Mutants. Nucleic Acids Res. 1999;27:286–288. doi: 10.1093/nar/27.1.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainaut P, Hernandez T, Robinson A, Rodriguez-Tome P, Flores T, Hollstein M, Harris CC, Montesano R. IARC Database of p53 gene mutations in human tumors and cell lines: updated compilation, revised formats and new visualisation tools. Nucleic Acids Res. 1998;26(1):205–13. doi: 10.1093/nar/26.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainaut P, Soussi T, Shomer B, Hollstein M, Greenblatt M, Hovig E, Harris CC, Montesano R. Database of p53 gene somatic mutations in human tumors and cell lines: updated compilation and future prospects. Nucleic Acids Res. 1997;25(1):151–7. doi: 10.1093/nar/25.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33(Database Issue):D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handbook of Molecular Descriptors Roberto Todeschini and Viviana Consonni. WILEY - VCH, pp. 667, 2000 in the Series of Methods and Principles in Medicinal Chemistry - Volume 11 Mannhold R, Kubinyi H, Timmerman H.ISBN 3-52-29913-0 (DM 490).
- Hanisch D, Zimmer R, Lengauer T. ProML - the Protein Markup Language for specification of protein sequences, structures and families. In Silico Biol. 2002;2:313–324. [PubMed] [Google Scholar]
- Hansch C. “QSAR in cancer chemotherapy.”. Farmaco Sci. 1979;34(1):89–104. [PubMed] [Google Scholar]
- Hansch C, Leo L, Hoekman D. Monograph: ‘Exploring the QSAR. Hydrophobic, Electronic, and Steric Constants’. In: Heller SR, editor. ACS; Washington, DC: 1995. [Google Scholar]
- Harris CC. p53 tumor suppressor gene: at the crossroads of molecular carcinogenesis, molecular epidemiology, and cancer risk assessment. Environ Health Per spect. 1996;104(Suppl 3):435–9. doi: 10.1289/ehp.96104s3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Vines RR, Wattam AR, Abramochkin GV, Dickerman AW, Eckart JD, Sobral BWS. PIML: the Pathogen Information Markup Language. Bioinformatics. 2005;21(1):116–121. doi: 10.1093/bioinformatics/bth462. [DOI] [PubMed] [Google Scholar]
- Hernandez-Boussard T, Montesano R, Hainaut P. Sources of bias in the detection and reporting of p53 mutations in human cancer: analysis of the IARC p53 mutation database. Genet Anal. 1999;14(5–6):229–33. doi: 10.1016/s1050-3862(98)00030-8. [DOI] [PubMed] [Google Scholar]
- Hernandez-Boussard T, Rodriguez-Tome P, Montesano R, Hainaut P. IARC p53 mutation database: a relational database to compile and analyze p53 mutations in human tumors and cell lines. International Agency for Research on Cancer. Hum Mutat. 1999;14(1):1–8. doi: 10.1002/(SICI)1098-1004(1999)14:1<1::AID-HUMU1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253(5015):49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- Hollstein M, Shomer B, Greenblatt M, Soussi T, Hovig E, Montesano R, Harris CC. Somatic point mutations inthep53 gene of human tumors and cell lines: updated compilation. Nucleic Acids Res. 1996;24(1):141–6. doi: 10.1093/nar/24.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollstein M, Rice K, Greenblatt MS, Soussi T, Fuchs R, Sorlie T, Hovig E, Smith-Sorensen B, Montesano R, Harris CC. Database of p53 gene somatic mutations in human tumors and cell lines. Nucleic Acids Res. 1994;22(17):3551–5. [PMC free article] [PubMed] [Google Scholar]
- Hulo N, Bairoch A, Bulliard V, Cerutti L, De Castro E, Langendijk-Genevaux PS, Pagni M, Sigrist CJA. The PROSITE database. Nucleic Acids Research. 2006;34:D227–D230. doi: 10.1093/nar/gkj063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huret J, Le Minor S, Dorkeld F, Dessen P, Bernheim A. Atlas of Genetics and Cytogenetics in Oncology and Haematology, an Interactive Database Nucleic Acids Research. 2000;28(1):349–351. doi: 10.1093/nar/28.1.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolliffe LT. Principal Component Analysis. Springer; New york: 1986. [Google Scholar]
- Jung M, Kim H, Kim M. Chemical genomics strategy for the discovery of new anticancer agents. Curr Med Chem. 2003;10(9):757–62. doi: 10.2174/0929867033457782. [DOI] [PubMed] [Google Scholar]
- Kamal A, Laxman E, Khanna GB, Reddy PS, Rehana T, Arifuddin M, Neelima K, Kondapi AK, Dastidar SG. “Design, synthesis, biological evaluation and QSAR studies of novel bisepipodophyllotoxins as cytotoxic agents.”. Bioorg Med Chem. 2004;(15):4197–209. doi: 10.1016/j.bmc.2004.05.026. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karelson M. John Wiley & Sons; New York: 2000. Molecular Descriptors in QSAR/QSPR. [Google Scholar]
- Kato K, Yamashita R, Matoba R, Monden M, Noguchi S, Takagi T, Nakai K. Cancer gene expression database (CGED): a database for gene expression profiling with accompanying clinical information of human cancer tissues. Nucleic Acids Research. 2005;33:D533–D536. doi: 10.1093/nar/gki117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kier LB, Hall LH. Molecular Structure Descriptors: The Electrotopological State. Academic Press; San Diego, CA: 1999. [Google Scholar]
- Klopman G, Shi LM, Ramu A. “Quantitative structure-activity relationship of multidrug resistance reversal agents”. Mol Pharmacol. 1997;52(2):323–34. doi: 10.1124/mol.52.2.323. [DOI] [PubMed] [Google Scholar]
- Knutsen T, Gobu V, Knaus R, Padilla-Nash H, Augustus M, Strausberg RL, Kirsch IR, Sirotkin K, Ried T. The interactive online SKY/M-FISH and CGH database and the Entrez cancer chromosomes search database: linkage of chromosomal aberrations with the genome sequence. Genes Chromosomes Cancer. 2005;44(1):52–64. doi: 10.1002/gcc.20224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutsen T, Gobu V, Knaus R, Padilla-Nash H, Augustus M, Strausberg RL, Kirsch IR, Sirotkin K, Ried T. Chromosomes Search Database: Linkage of Chromosomal Aberrations with the Genome Sequence Genes. Chromosomes Cancer. 2005;44(1):52–64. doi: 10.1002/gcc.20224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuz’min VE, Artemenko AG, Lozytska RN, Fedtchouk AS, Lozitsky VP, Muratov EN, Mescheriakov AK. “Investigation of anticancer activity of macrocyclic Schiff bases by means of 4D-QSAR based on simplex representation of molecular structure”. SAR QSAR Environ Res. 2005;16(3):219–30. doi: 10.1080/10659360500037206. [DOI] [PubMed] [Google Scholar]
- Kuz’min VE, Artemenko AG, Lozitsky VP, Muratov EN, Fedtchouk AS, Dyachenko NS, Nosach LN, Gridina TL, Shitikova LI, Mudrik LM, Mescheriakov AK, Chelombitko VA, Zheltvay AI, Vanden Eynde JJ. “The analysis of structure-anticancer and antiviral activity relationships for macrocyclic pyridinophanes and their analogues on the basis of 4D QSAR models (simplex representation of molecular structure).”. Acta Biochim Pol. 2002;49(1):157–68. [PubMed] [Google Scholar]
- Labute P. Awidely applicable set of descriptors. JMol Graph Model. 2000;18(4–5):464–77. doi: 10.1016/s1093-3263(00)00068-1. [DOI] [PubMed] [Google Scholar]
- Le Roux E, Gormally E, Hainaut P. Somatic mutations in human cancer: applications in molecular epidemiology. Rev Epidemiol Sante Publique. 2005;53(3):257–66. doi: 10.1016/s0398-7620(05)84603-6. Review. [DOI] [PubMed] [Google Scholar]
- Li FP, Fraumeni JF., Jr Soft-tissue sarcomas, breast cancer and other neoplasms: a familial syndrome? Ann Intern Med. 1969;71:747–52. doi: 10.7326/0003-4819-71-4-747. [DOI] [PubMed] [Google Scholar]
- Li FP, Fraumeni JF, Jr, Mulvihill JJ, Blattner WA, Dreyfus MG, Tucker MA, Miller RW. A cancer family syndrome in twentyfour kindreds. Cancer Res. 1988;48(18):5358–62. [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Cherukuri PF, DeWeese-Scott C, Geer LY, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Liebert CA, Liu C, Lu F, Marchler GH, Mullokandov M, Shoemaker BA, Simonyan V, Song JS, Thiessen PA, Yamashita RA, Yin JJ, Zhang D, Bryant SH. CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 2005;33(Database Issue):D192–D196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McColl BK, Loughran SJ, Davydova N, Stacker SA, Achen MG. Mechanisms of lymphangiogenesis: targets for blocking the metastatic spread of cancer. Curr Cancer Drug Targets. 2005;5(8):561–71. doi: 10.2174/156800905774932833. [DOI] [PubMed] [Google Scholar]
- Mewes HW, Frishman D, Guldener U, Mannhaupt G, Mayer K, Mokrejs M, Morgenstern B, Munsterkotter M, Rudd S, Weil B. MIPS: a database for genomes and protein sequences. Nucleic Acids Res. 2002;30:31–34. doi: 10.1093/nar/30.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Biswas M, Bradley P, Bork P, Bucher P, Copley R, Courcelle E, Durbin R, Falquet L, Fleischmann W, Gouzy J, Griffith-Jones S, Haft D, Hermjakob H, Hulo N, Kahn D, Kanapin A, Krestyaninova M, Lopez R, Letunic I, Orchard S, Pagni M, Peyruc D, Ponting CP, Servant F, Sigrist CJA. InterPro: An integrated documentation resource for protein families, domains and functional sites. Briefings in Bioinformatics. 2002;3(3):225–235. doi: 10.1093/bib/3.3.225. [DOI] [PubMed] [Google Scholar]
- Olivier M, Eeles R, Hollstein M, Khan MA, Harris CC, Hainaut P. New online mutation analysis and recommendations to users. Hum Mutat. 2002;19(6):607–14. doi: 10.1002/humu.10081. [DOI] [PubMed] [Google Scholar]
- Pirard B, Pickett SD. Classification of kinase inhibitors using BCUT descriptors. J Chem Inf Comput Sci. 2000;40(6):1431–40. doi: 10.1021/ci000386x. [DOI] [PubMed] [Google Scholar]
- Pongor S, Sker V, Cserzo M, Hatsagi Z, Simon G, Bevilacqua V. The SBASE domain library: a collection of annotated protein sequence segments. Protein Eng. 1992;6:391–395. doi: 10.1093/protein/6.4.391. [DOI] [PubMed] [Google Scholar]
- Raevsky OA. “Molecular structure descriptors in the computer-aided design of biologically active compounds”. RUSS CHEM REV. 1999;68(6):505–524. [Google Scholar]
- Raychaudhury C, Basak SC, Roy AB, Ghosh JJ. Quantitative Structure-Activity Relationship (QSAR) studies of pharmacological agents using topological information content. Indian Drugs. 1980;18:97. [Google Scholar]
- Ray SK, Basak SC, Raychaudhury C, Roy AB, Ghosh JJ. The utility of information content (IC), structural information content (SIC), hydrophobicity (log P) and van der Waals’ volume (VW) in the design of barbiturates and tumor-inhibitory triazenes: a comparative study. Arzneim.-Forsch./Drug Res. 1983;33:352. [PubMed] [Google Scholar]
- Ren SS, Lien EJ. “Anticancer agents: tumor cell growth inhibitory activity and binary QSAR analysis”. Curr Pharm Des. 2004;10(12):1399–415. doi: 10.2174/1381612043384925. [DOI] [PubMed] [Google Scholar]
- Smigielski EM, Sirotkin K, Ward M, Sherry ST. dbSNP: a database of single nucleotide polymorphisms. Nucleic Acids Research. 2000;28(1):352–355. doi: 10.1093/nar/28.1.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westbrook J, Feng Z, Jain S, Bhat TN, Thanki N, Ravichandran V, Gilliland GL, Bluhm WF, Weissig H, Greer DS, Bourne PE, Berman HM. The Protein Data Bank: unifying the archive. Nucleic Acids Res. 2002;30(1):245–248. doi: 10.1093/nar/30.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildman SA, Crippen GM. Three-dimensional molecular descriptors and a novel QSAR method. J Mol Graph Model. 2002;21(3):161–70. doi: 10.1016/s1093-3263(02)00147-x. [DOI] [PubMed] [Google Scholar]
- Winkler DA. The Role of Quantitative Structure-activity relationships (QSAR) in Biomolecular Discovery. Briefings in Bioinformatics. 2002;3(1):73–86. doi: 10.1093/bib/3.1.73. [DOI] [PubMed] [Google Scholar]
- Workman P. New drug targets for genomic cancer therapy: successes, limitations, opportunities and future challenges. Curr Cancer Drug Targets. 2001;1(1):33–7. doi: 10.2174/1568009013334269. Review. [DOI] [PubMed] [Google Scholar]
- Wu CH, Yeh LL, Huang H, Arminski L, Castro-Alvear J, Chen Y, Hu Z, Kourtesis P, Ledley RS, Suzek BE, Vinayaka CR, Zhang J, Barker WC. The Protein Information Resource. Nucleic Acids Res. 2003;31(1):345–347. doi: 10.1093/nar/gkg040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Xiao C, Hou Z, Huang H, Barker WC. iProClass: an integrated, comprehensive and annotated protein classification database. Nucleic Acids Res. 2001;29(1):52–54. doi: 10.1093/nar/29.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Xiao YD, Feng J, Golbraikh A, Tropsha A, Lee KH. “Anti-tumor agents. 213. Modeling of epipodophyllotoxin derivatives using variable selection k nearest neighbor QSAR method”. J Med Chem. 2002;45(11):2294–309. doi: 10.1021/jm0105427. [DOI] [PubMed] [Google Scholar]
- Xue L, Bajorath J. “The analysis of structure-anticancer and antiviral activity relationships for macrocyclic pyridinophanes and their analogues on the basis of 4D QSAR models (simplex representation of molecular structure)”. J Chem Inf Comput Sci. 2000;40(3):801–809. [Google Scholar]
- Xue L, Bajorath J. Molecular Descriptors in Chemoinformatics, Computational Combinatorial Chemistry, and Virtual Screening. J. Combinatorial Chemistry & High Throughput Screening. 2000;3(5):363–372. doi: 10.2174/1386207003331454. [DOI] [PubMed] [Google Scholar]
- Yao SW, Lopes VH, Fernandez F, Garcia-Mera X, Morales M, Rodriguez-Borges JE, Cordeiro MN. “Synthesis and QSAR study of the anticancer activity of some novel indane carbocyclic nucleosides”. Bioorg Med Chem. 2003;11(23):4999–5006. doi: 10.1016/j.bmc.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Yao YL, Yang WM. Nuclear proteins: promising targets for cancer drugs. Curr Cancer Drug Targets. 2005;5(8):595–610. doi: 10.2174/156800905774932815. [DOI] [PubMed] [Google Scholar]
- Zheng S, Luo X, Chen G, Zhu W, Shen J, Chen K, Jiang H. “A new rapid and effective chemistry space filter in recognizing a drug like database”. J Chem Inf Model. 2005;45(4):856–62. doi: 10.1021/ci050031j. [DOI] [PubMed] [Google Scholar]