Abstract
The SMRT (silencing mediator of retinoic acid and thyroid hormone receptor) and N-CoR (nuclear receptor corepressor) corepressors are important mediators of transcriptional repression by nuclear hormone receptors. SMRT is regulated by MAPK kinase kinase (MAPKKK) cascades that induce its release from its receptor partners, its export from nucleus to cytoplasm, and derepression of target gene expression. Intriguingly, the otherwise closely related N-CoR is refractory to MAPKKK signaling under the same conditions. However, both SMRT and N-CoR are expressed as a series of alternatively spliced protein variants differing in structure and function. We have now characterized the impact of this alternative mRNA splicing on the corepressor response to MAPKKK signaling. Whereas the SMRTα, SMRTτ, and SMRTsp2 splice variants are released from their nuclear receptor partners in response to MAPKKK activation, the SMRTsp18 variant, which resembles N-CoR in its overall molecular architecture, is relatively refractory to this kinase-induced release. Alternative splicing of N-CoR, in contrast, had only minimal effects on the resistance of this corepressor to MAPKKK inhibition. Notably, all of the SMRT splice variants examined redistributed from nucleus to cytoplasm in response to MAPKKK cascade signaling, but none of the N-CoR splice variants did so. Different tiers of the MAPKKK cascade hierarchy contributed to these different aspects of corepressor regulation, with MAP/ERK kinase kinase 1 and MAP/ERK kinase 1 regulating subcellular redistribution and ERK2 regulating nuclear receptorcorepressor interaction. We conclude that cells can customize their transcriptional response to MAPKKK cascade signaling by selective expression of the SMRT or N-CoR locus, by selective utilization of a specific corepressor splice variant, and by selective exploitation of specific tiers of the MAPK cascade.
Many Eukaryotic Transcription factors possess bidirectional regulatory properties and can either repress or activate target gene expression by alternatively recruiting either corepressors or coactivators (1–7). It is these auxiliary coregulator proteins that mediate, in turn, the actual molecular events necessary for transcriptional regulation. Corepressors typically place repressive marks in chromatin and mediate inhibitory interactions with the general transcriptional machinery, whereas coactivators do the opposite (8–17). This type of bipolar transcriptional regulation is particularly evident in the actions of the nuclear receptors, a large family of hormone-regulated transcription factors that play key roles in metazoan reproduction, development, and homeostasis (18–22). Nuclear receptors such as the thyroid hormone receptors (TRs) and retinoic acid receptors (RARs) can bind to corepressors and repress target gene expression in the absence of hormone ligand but release from corepressors, recruit coactivators, and become transcriptional activators after binding to hormone agonist (23–25).
The most extensively characterized corepressors for the nuclear receptors are SMRT (silencing mediator of retinoic acid and thyroid hormone receptor) and N-CoR (nuclear receptor corepressor) (26–34). Although expressed from two different loci, these two corepressors closely resemble one another in sequence and in overall structural organization and likely diverged from a common genetic ancestor near the beginning of the vertebrate radiation (6, 20, 35). Both SMRT and N-CoR bind to their nuclear receptor partners through C-terminal receptor interaction [corepressor nuclear receptor (CoRNR) box] motifs and nucleate the assembly of still larger, multisubunit corepressor complexes (36–42). SMRT and N-CoR interact with overlapping sets of nuclear receptor partners, display similar biochemical properties in vitro, and assemble into similar corepressor complexes that can include histone deacetylase 3, transducin β-like protein 1, transducin β-like protein-related protein 1, G protein pathway suppressor 2, and a variety of additional protein subunits (43–51).
Despite their otherwise close interrelatedness, we reported previously one significant difference between these two corepressor paralogs: SMRT function is negatively regulated by a MAPK kinase kinase (MAPKKK) cascade that phosphorylates SMRT in response to growth factor or stress signals, leading to release of SMRT from its nuclear receptor partners and SMRT export from the nucleus to the cytoplasm (52–54). Derepression through this pathway contributes to the prodifferentiation, antineoplastic effects of arsenic in treatment of acute promyelocytic leukemia (55). N-CoR, in contrast, is refractory to MAPKKK signals under the same conditions but is instead reported to be inhibited by Akt signaling (54, 56).
It has recently become appreciated that both SMRT and N-CoR are subject to extensive alternative mRNA splicing events that generate a diverse series of distinct corepressor variants from each locus (30, 31, 33, 57–61). These corepressor splice variants differ in their molecular architecture, are expressed at different abundances in different tissues, and display distinct affinities for different nuclear receptor partners (28, 57–63). At least several of the alternative mRNA splicing events impact known sites of phosphorylation within these corepressors (58); we therefore examined the effect of these alternative mRNA splicing events on the response of SMRT and N-CoR to MAPKKK signaling. We report here that the receptor interaction properties of the previously characterized SMRTτ and SMRTα variants, as well as a newly identified SMRTsp2 variant, are strongly inhibited by MAPK signaling; in contrast, an additional, newly recognized splicing variant of SMRT, denoted SMRTsp18, retains nuclear receptor interaction even in the presence of MAPKKK signaling. We also demonstrate that both the originally studied N-CoR isoform and retinoid X receptor interacting protein 13Δ1 (RIP13Δ1) (an N-CoR splice variant) are relatively resistant to MAPK cascade signaling in the same assay. All forms of SMRT studied here relocalize to the cytoplasm in response to activation of the cascade, whereas all forms of N-CoR characterized are resistant to this redistribution. Interestingly, whereas ERK kinases operating at the bottom of the kinase cascade are responsible for the release of SMRTα, SMRTτ, and SMRTsp2 from their nuclear receptor partners, MAP/ERK kinase kinase 1 (MEKK1) and MAP/ERK kinase 1 (MEK1), operating higher in the kinase hierarchy, play a separate role in mediating SMRT nuclear to cytoplasmic relocalization. We conclude that alternative mRNA splicing can generate forms of SMRT that are either sensitive or resistant to inhibition by MAPK cascades, that N-CoR variants are generically resistant to this regulatory network, and that the interaction properties vs. subcellular distribution properties of these corepressors are independently regulated by distinct elements in the MAPKKK hierarchy.
RESULTS
Alternative Splicing of SMRT Generates Corepressor Variants that Differ in Their Interactions with Nuclear Receptors
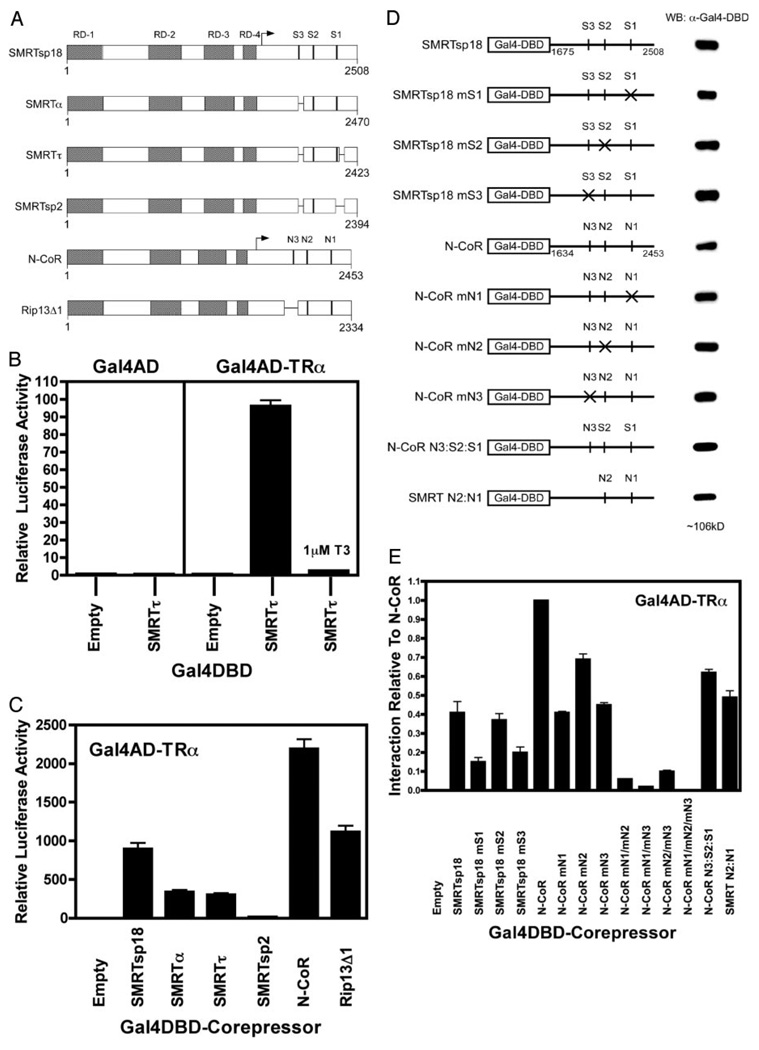
Our previous MAPKKK studies focused primarily on SMRTα and N-CoR, which are the splice variants of these corepressors first characterized in detail (26, 29–32). SMRTα contains two interaction sites for nuclear receptors (S2+S1), whereas N-CoR contains three (N3+N2+N1) (Fig. 1A). A newly recognized splice version of SMRT, denoted SMRTsp18, was subsequently identified that contains a third receptor interaction domain (Fig. 1A, S3) and therefore more closely resembles the N-CoR variant than does SMRTα (59, 61). Conversely, there is an alternatively spliced form of N-CoR, denoted RIP13Δ1, that deletes N3 from N-CoR and consequently more closely resembles SMRTα (28, 33) (Fig. 1A). Additional corepressor splice forms have also been identified, including SMRTτ (which alters the sequences flanking the S1 domain) and SMRTsp2 (which deletes the S1 domain entirely) (Fig. 1A) (33, 61). We therefore used a mammalian two-hybrid assay to determine the ability of these various corepressor variants to interact with nuclear receptors in the absence or presence of MAPKKK cascade signaling, using GAL4 DNA binding domain (DBD)-corepressor fusions (Fig. 1D), GAL4-activation domain (AD)-TRα fusions, and a GAL4–17mer-luciferase reporter. In this manner, a physical interaction between the corepressor and the TRα reconstitutes GAL4 transcription factor function and can be measured as an induction of luciferase activity in transfected CV-1 cells. A β-galactosidase reporter driven by a constitutive promoter was included in all transfections as an internal normalization control.
Fig. 1. Different Corepressor Splice Variants Differ in Their Ability to Interact with TRα.
A, Schematic representations of the SMRT and N-CoR alternative splice variants are shown. The locations of the repression domains (RD) and the CoRNR box motifs (S3, S2, S1, N3, N2, N1) are indicated. Overall length of each variant is expressed in codons. The receptor interaction domains of these corepressors, indicated by an arrow, were fused to GAL4DBD for use in the two-hybrid studies (see also D). B, SMRTτ and TRα interact strongly in a mammalian two-hybrid assay. A GAL4DBD-SMRTτ construct and a GAL4AD-TRα construct were cotransfected into CV-1 cells together with a GAL-17mer luciferase reporter. Relative luciferase activity was calculated as the absolute luciferase activity normalized to the expression of a constitutive β-galactosidase construct used as an internal transfection control. Negative controls included the use of empty GAL4DBD and/or empty GAL4AD constructs, as indicated, or performing the transfection in the presence of T3, which releases corepressor from TRα. C, Different corepressor splice variants differ in their interaction with TRα. The receptor interaction domains of the various corepressor splice variants in A were introduced into the GAL4DBD construct, as noted below the panel, and tested for their ability to interact with the GAL4AD-TRα construct in the mammalian two-hybrid assay. D, Schematic representations of the SMRT and N-CoR mutants used and their expression levels are shown. Mutations were introduced into individual CoRNR boxes in the GAL4DBD-SMRTsp18 (codons 1675–2508) and GAL4DBD-N-CoR (codons 1634–2453) constructs as indicated by the × symbols and by the mS2, mS3, etc. nomenclature. The relative levels of expression of each of these constructs, determined by Western blotting, are depicted to the right. Alternatively, specific CoRNR boxes were exchanged between SMRT and N-CoR (e.g. N-CoR N3:S2:S1 contains the N3 motif of N-CoR linked to the S2 and S1 motifs of SMRT). E, Both the iteration and the identity of the CoRNR motifs in SMRT and N-CoR determine the ability of the corepressors to interact with TRα. The mammalian two-hybrid assay in C was repeated using the GAL4DBD-SMRTsp18 and GAL4DBD-N-CoR constructs bearing disruptions in specific CoRNR boxes. Single mutants are as depicted in D; double mutants (e.g. mN1/mN2) bear disruptive mutations in two CoRNR boxes. The mammalian two-hybrid interaction of TRα with the N-CoR construct was defined as 1.
We first examined the interaction of the various corepressor variants with their nuclear receptor partners in the absence of MAPKKK signaling. As established previously (54), cointroduction of a GAL4DBDSMRT τ construct and a GAL4AD-TRα construct strongly induced luciferase activity in these cells (Fig. 1B). This induction of luciferase in the mammalian two-hybrid assay appeared to be an authentic reflection of the physical interaction between corepressor and nuclear receptor: 1) little or no luciferase activity was detected if either or both constructs were replaced with corresponding empty GAL4DBD or empty GAL4AD constructs, 2) the GAL4DBD-corepressor/ GAL4AD-TRα interaction was strongly inhibited by thyroid hormone, 3) no interaction was observed when using corepressor domains lacking the known receptor interaction domains or when using mutant TRs that do not interact with corepressor, 4) the β-galactosidase internal control was not affected by the GAL4DBD-corepressor or GAL4AD-TRα constructs, and 5) results from the mammalian two-hybrid assay closely paralleled those from protein pull-down, coimmunoprecipitation, and electrophoretic mobility supershift techniques (Fig. 1B) (52–55, 64).
The SMRTα variant yielded a very slightly higher two-hybrid interaction with TRα than did SMRTτ, whereas a much greater two-hybrid interaction with TRα was observed for the SMRTsp18 variant, which contains three receptor interaction motifs (S3+S2+S1) (Fig. 1C). Conversely, the SMRTsp2 splice variant, which lacks both the S3 and the S1 motifs, severely reduced the two-hybrid interaction to levels just above background (Fig. 1, A and C). Paralleling these SMRT results, the full-length N-CoR variant (containing N3+N2+N1) displayed a stronger two-hybrid interaction with TRα than did the N-CoR-derived RIP13Δ1 variant (which contains only N2+N1) (Fig. 1, A and C). Interestingly, both of the NCoR variants displayed a 2- to 3-fold higher two-hybrid interaction with TRα than did the “corresponding” SMRT version: compare the (N3+N2+N1) N-CoR with the (S3+S2+S1) SMRTsp18, and the (N2+N1) RIP13Δ1 with the (S2+S1) SMRTα or SMRTτ. The corepressor constructs were expressed at approximately comparable levels with the exception of GAL4DBD-N-CoR, which was somewhat underexpressed (Fig. 1D and data not shown); therefore, the true interaction of N-CoR with TRα might be somewhat stronger than the luciferase level suggests.
We next defined the specific CoRNR boxes that mediate the interaction of each corepressor variant with TRα. Disrupting any one of the three CoRNR boxes in N-CoR or in SMRTsp18 by site-directed mutagenesis reduced, but did not abolish, interaction with TRα in our assay, demonstrating that no one specific CoRNR box is indispensable (Fig. 1, D and E, compare the mS1, mS2, mS3, mN1, etc. mutants with the wild-type corepressor). However, any additional reduction in CoRNR box number from two to one severely attenuated two-hybrid interaction with TRα, consistent with the weak interaction properties of SMRTsp2 above (Fig. 1E, e.g. N-CoR mN1/mN3). For all splice variants tested, the central N2 or S2 CoRNR box contributed less to the two-hybrid interaction with TRα than did the flanking N3/S3 or N1/S1 CoRNR boxes (Fig. 1E). As noted for the naturally spliced variants, the N-CoR-derived mutants displayed a stronger interaction with TRα than did the corresponding SMRT-derived mutants (Fig. 1E). Similarly, adding the N-CoR sequences containing the N3 box to the SMRTα S2 and S1 domains results in a stronger interaction than either SMRTα itself or SMRTsp18 (Fig. 1, D and E, compare SMRTα and SMRTsp18 with N-CoR N3:S2:S1); conversely, replacing the N3 sequence in N-CoR with sequences derived from SMRTα (i.e. lacking a third CoRNR box) reduced the interaction (Fig. 1, D and E, compare N-CoR with SMRT N2:N1). The various GAL4DBD-corepressor mutant constructs were expressed at comparable levels by Western blot analysis (Fig. 1D and data not shown). We conclude that a minimum of two receptor interaction domains are required to obtain significant two-hybrid interaction between any of the corepressor splice variants and TRα, with the preferred pairing being N3 and N1 or S3 and S1, but with a variety of alternative pairings able to replace these optimal ones in corepressor variants lacking this optimal configuration. Furthermore, the CoRNR boxes in N-CoR interact intrinsically more strongly with TRα than do the corresponding CoRNR boxes in SMRT.
A Distinct Set of Corepressor Variants Interact Preferentially with RARα vs. TRα
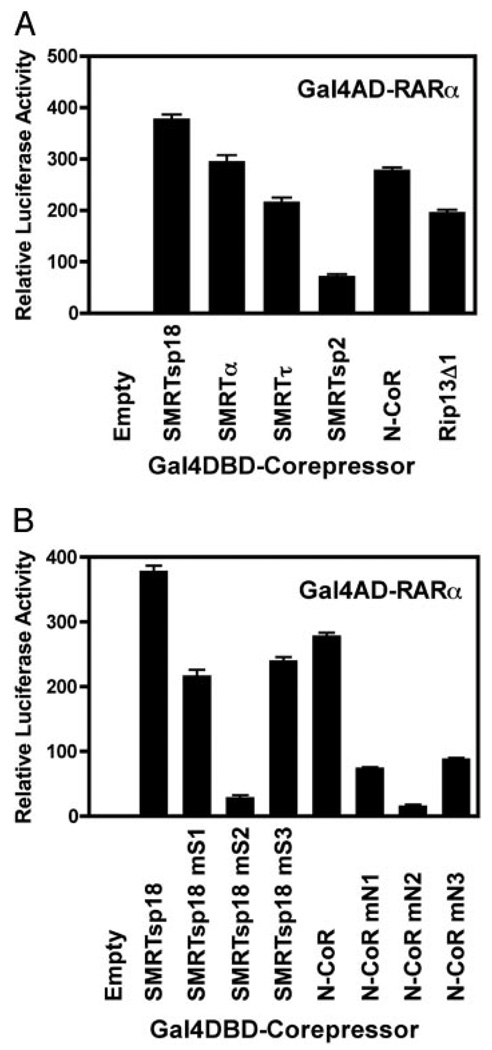
We next repeated the two-hybrid interaction analysis with RARα in place of TRα. Notably the N-CoR vs. SMRT specificity was reversed from that of TRα, with RARiα preferentially interacting with any given SMRT variant more strongly than with the corresponding form of N-CoR (e.g. compare SMRTsp18 with N-CoR or SMRTα with RIP13Δ1) (Fig. 2A). For any given corepressor locus, splice variants encoding three receptor interaction motifs (e.g. SMRTsp18 or N-CoR itself) interacted more strongly with RARα than did splice variants encoding two receptor interaction domains (e.g. SMRTα, SMRTτ, or RIP13Δ1) (Fig. 2A). Similarly, artificial disruption of any one CoRNR box motif in either SMRTsp18 or N-CoR also significantly reduced the interaction with RARα, with the effect being more pronounced on the RARα/N-CoR interaction than on the RARα/SMRTsp18 interaction (Fig. 2B). Unlike the case with TRα, the interaction of RARα with corepressor was stabilized primarily through the central S2/N2 interaction domain rather than the N3/S3 or N1/S1 domains, and natural or artificial SMRT variants retaining only the middle S2 CoRNR box motif (e.g. SMRTsp2) retained the ability to interact detectably with RARα (Fig. 2B). Despite the generic preference of RARα for SMRT over N-CoR when comparing comparable splice variants, alternative mRNA splicing nonetheless plays a crucial role, with certain splice variants of N-CoR able to interact with RARα at levels equal to or greater than do other alternative splice variants of SMRT (Fig. 2A). Similarly, the generic preference of TRα for N-CoR over SMRT is also dependent on which splice variants are compared (Fig. 1C).
Fig. 2. The Ability of the Various Corepressor Splice Variants to Interact with RARα Differs from Their Ability to Interact with TRα.
A, Different corepressor splice variants differ in their interaction with RARα. The mammalian two-hybrid assays were performed as in Fig. 1C, except a GAL4AD-RARα construct was used instead of the GAL4AD-TRα fusion. B, Both the identity and the iteration of the CoRNR motifs in SMRT and N-CoR determine the ability of the corepressors to interact with RARα. The mammalian two-hybrid assay in A was repeated using the GAL4AD-RARα construct and the GAL4DBD-SMRTsp18 or GAL4DBD-N-CoR CoRNR motif mutants described in Fig. 1D.
Alternative mRNA Splicing of SMRT Defines whether the Corepressor/Nuclear Receptor Interaction Responds to MAPKKK Cascade Signaling or Not
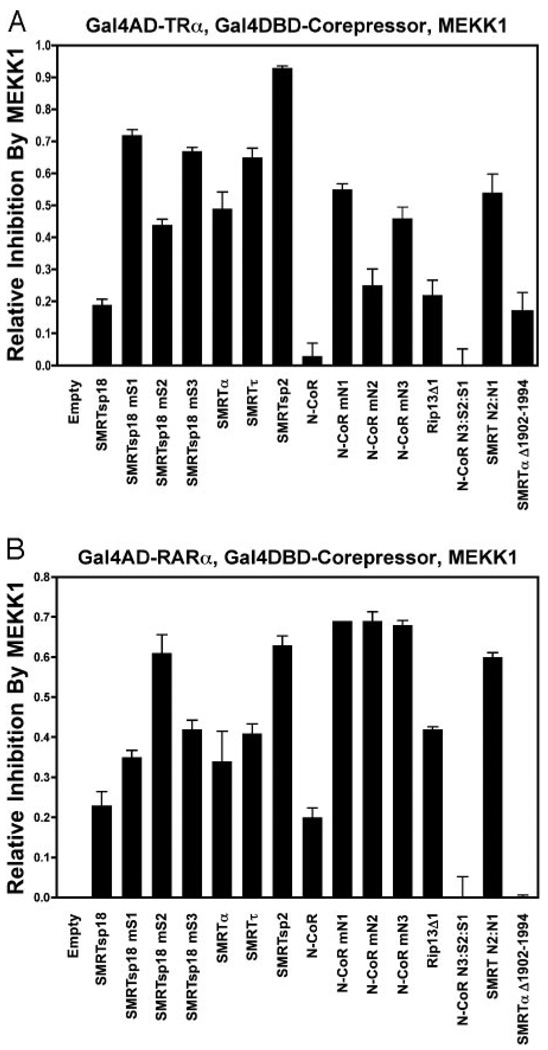
We next examined the effect of cointroducing an activated MAPKKK (MEKK1) to the mammalian two-hybrid interaction assay. This construct initiates a kinase activation cascade that strongly inhibits the interaction between SMRTα and a variety of nuclear receptors, including TRα (52–54). Extending these previous studies, we found that the two-hybrid interaction of TRα with either SMRTα or SMRTτ was potently inhibited by activated MEKK1 (Fig. 3A). This MAPKKK-mediated inhibition of the mammalian two-hybrid assay reflected a true inhibition of the physical interaction between SMRT and TRα and was also observed in coimmunoprecipitation assays (52–55). MEKK1 did not inhibit expression of the GALDBD-fusion proteins themselves, nor did MEKK1 significantly alter the expression of the luciferase reporter itself in the absence of the GAL4DBD-corepressor or GAL4AD-TRα constructs (54) (data not shown).
Fig. 3. The Ability of MAPKKK Signaling to Inhibit the Core-pressor Interaction with TRα or RARα depends on both the Class and the Splice Form of the Corepressor.
A, Certain corepressor splice variants are refractory, whereas others are sensitive to inhibition by MEKK1 signaling/ interaction assay with TRα. The mammalian TRα/corepressor two-hybrid assays in Fig. 1, C and D, were repeated in the absence or presence of a cotransfected, constitutively active MEKK1 construct. A GAL4DBD-SMRTα construct lacking SMRTα codons 1902 to 1994 was also included. “Relative-inhibition by MEKK1” represents the portion of the relative luciferase activity observed for each splice variant in the absence of MEKK1 signaling that is lost in response to MEKK1 signaling, e.g. 1.0 would represent a complete loss of interaction in response to MEKK1, whereas 0 would represent no inhibition. B, Certain corepressor splice variants are refractory, whereas others are sensitive to inhibition by MEKK1 signaling/interaction assay with RARα. The mammalian two-hybrid assays in Fig. 2, A and B, measuring the interaction of GAL4AD-RARα with the various GAL4DBD-corepressor constructs, was repeated in the absence or presence of a cotransfected, constitutively active MEKK1 construct. The overall experiments and analysis was as in A above.
Notably, SMRTsp18 differed from SMRTα and SMRTτ by being much more resistant to inhibition by MAPKKK cascade signaling in the two-hybrid assay (Fig. 3A). N-CoR was fully resistant in the same assay. Both SMRTsp18 and N-CoR possess a third receptor interaction motif (Fig. 1A, denoted N3 or S3). However, this third interaction site was not itself sufficient to confer resistance to MAPK cascade inhibition on either SMRT or N-CoR. For example, the two-hybrid interaction of TRα with SMRTsp18 mutants retaining S3, but with either S2 or S1 disrupted (SMRTsp18 mS2 or mS1), was sensitive to MAPK cascade inhibition; the same was true of N-CoR mutants retaining N3 but lacking either N2 or N1 (N-CoR mN2 or mN1) (Fig. 3A). Instead, it was total number of CoRNR boxes that defined MEKK1 resistance vs. sensitivity. Addition of the N3 CoRNR box sequences to SMRTα (yielding an N3+S2+S1 corepressor, denoted N-CoR N3:S2:S1) conferred resistance to MAPKKK signaling, whereas replacement of the N3 motif of N-CoR with the CoRNR-box lacking sequences from SMRTα (yielding an N2+N1 corepressor, denoted SMRT N2:N1) conferred sensitivity to MAPKKK inhibition (Fig. 3A). Corepressors containing (N3+N1) or (S3+S1) pairings (i.e. N-CoR mN2 or SMRTsp18 mS2) were less sensitive to MEKK1 disruption than were corepressors containing other pairings, and the relatively weak two-hybrid interaction mediated by any individual interaction motif was particularly sensitive to disruption by activated MEKK1 signaling (Fig. 3A). We conclude that the presence of three interaction domains on a corepressor variant confers resistance to disruption of the interaction of SMRT with TRα in the two-hybrid assay, whereas corepressor mutants or splice variants that possess two (or fewer) interaction domains are typically sensitive to MEKK1 inhibition in this assay.
There was one partial exception to this general rule. Although the RIP13Δ1 splice variant of N-CoR contains only two receptor interaction domains (N2+N1), it was nonetheless reproducibly more resistant to MEKK1 inhibition than were the corresponding SMRT variants (α or τ) or an N-CoR mutant artificially lacking the N3 CoRNR box (Fig. 3A). This higher resistance of RIP13Δ1 to inhibition by MAPKKK signaling mapped to a region upstream of the N2 CoRNR box in RIP13Δ1. Replacement of the RIP13Δ1 sequences in this region with corresponding sequences originating from SMRT counteracted this resistance and enhanced sensitivity to MAPKKK cascade inhibition (Fig. 3A, compare RIP13Δ1 with SMRT N2:N1), whereas deletion of the equivalent region in SMRTα conferred resistance (Fig. 3A, compare SMRTα with SMRTαΔ1902–1994). A likely explanation for this phenomenon, the presence of a kinase docking site present in this region of N-CoR and SMRTα but lost from RIP13Δ1, is described further below.
We also examined the effect of MEKK1 signaling on the interaction of RARα with the different corepressor variants and observed both similarities and differences relative to the TRα data (Fig. 3B). Both N-CoR and RIP13Δ1 were somewhat more sensitive to inhibition by MEKK1 when assayed with RARα than with TRα (Fig. 3, compare A and B). Nonetheless, as observed for TRα, the interaction of RARα with corepressor splice variants containing three CoRNR motifs (SMRTsp18, N-CoR, and N-CoR N3: S2:S1) was relatively resistant to inhibition by coin-troduction of an activated MEKK1 (Fig. 3B), whereas corepressor forms containing only two CoRNR boxes (SMRTα, SMRTτ, RIP13Δ1, or the artificial CoRNR box mutants) were more sensitive to inhibition (Fig. 3B). The SMRTsp2 splice variant, which encodes only the S2 motif, was among the most sensitive to inhibition by MEKK1 signaling (Fig. 3B). Taken as a whole, these results implicate similar but not identical mechanisms operating in the MAPKKK cascade inhibition of the interaction of corepressors with TRα and RARα.
The Effect of MEKK1 Signaling on the Subcellular Distribution of SMRT and N-CoR Is Independent of the Nature of the Splice Variant
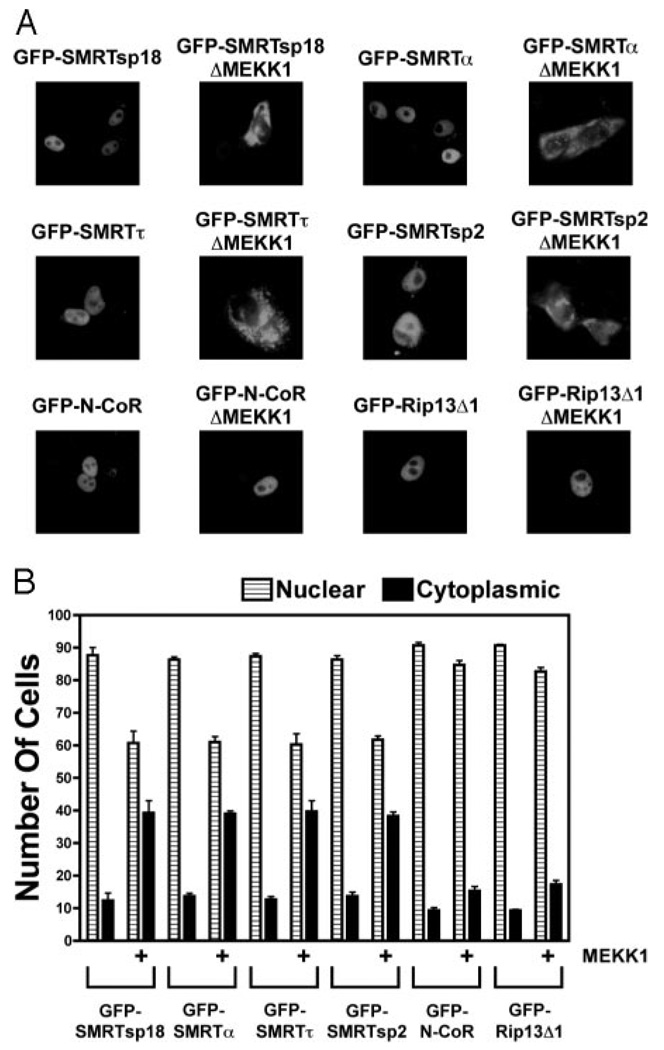
We previously reported that SMRTτ, but not N-CoR, relocalizes from the nucleus to the cytoplasm in response to MAPKKK cascade signaling (53, 54).We revisited this issue to determine whether the nature of the corepressor splice variant influenced the relocalization response. In the absence of MEKK1 signaling, all SMRT and N-CoR variants tested displayed a primarily nuclear localization (Fig. 4, A and B). In agreement with our previous work, the subcellular localization of both green fluorescent protein (GFP)-SMRTτ and GFP-SMRTα was responsive to introduction of an activated MEKK1: only 12–14% of untreated cells displayed a non-nuclear localization of GFP-SMRTτ or SMRTα, whereas introduction of an activated MEKK1 resulted in a cytoplasmic GFP-SMRT localization in up to 40% of these cells (Fig. 4, A and B). Similar results were observed using immunolocalization of native SMRT constructs or biochemical subcellular fractionations (53, 54). Intriguingly, GFP-SMRTsp18 displayed a nuclear to cytoplasmic relocalization indistinguishable from that of GFP-SMRT α or GFP-SMRTτ in response to MEKK1 signaling, despite SMRTsp18 being essentially resistant to MEKK1 signaling in the two-hybrid nuclear receptor interaction assay (Fig. 4B). In contrast to the SMRT variants, the subcellular localization of either N-CoR or RIP13Δ1 was relatively unaffected by cointroduction of MEKK1 (~10% non-nuclear in the absence of MEKK1 vs. 15% in the presence of MEKK1) (Fig. 4, A and B). We conclude that the identity of the locus, SMRT vs. N-CoR, determines the ability of the corepressor to undergo subcellular relocalization in response to MAPK cascade signaling, whereas the nature of the mRNA splice variant determines the ability of the corepressor to release from its nuclear receptor partners under the same circumstances.
Fig. 4. SMRT Splice Variants Display a Nuclear to Cytoplasmic Relocalization in Response to MEKK1, whereas N-CoR Splice Variants Do Not.
A, Representative fluorescent microscope photographs display a nuclear to cytoplasmic relocalization of GFP-SMRT constructs, but not of GFP-N-CoR constructs, in response to MEKK1 signaling. CV-1 cells were transiently transfected with the GFP-SMRT or GFP-N-CoR splice variants indicated in each panel, together with a control vector or a vector expressing a constitutively active form of MEKK1. The cells were subsequently fixed and visualized by fluorescent light microscopy as noted in Materials and Methods. Representative micrographs are shown. B, Quantification confirms that SMRT but not N-CoR splice variants respond to MEKK1 signaling by redistribution from the nuclear to the cytoplasmic compartment. The same assay was performed as in A for each of the GFP-SMRT and GFP-N-CoR splice variants noted and was quantified by counting the percentage of the cells displaying a predominantly nuclear vs. a predominantly cytoplasmic localization of each corepressor.
Corepressor Redistribution from Nucleus to Cytoplasm Is Regulated by Distinct Steps in the MAPKKK Cascade Hierarchy from Those Governing Nuclear Receptor Release
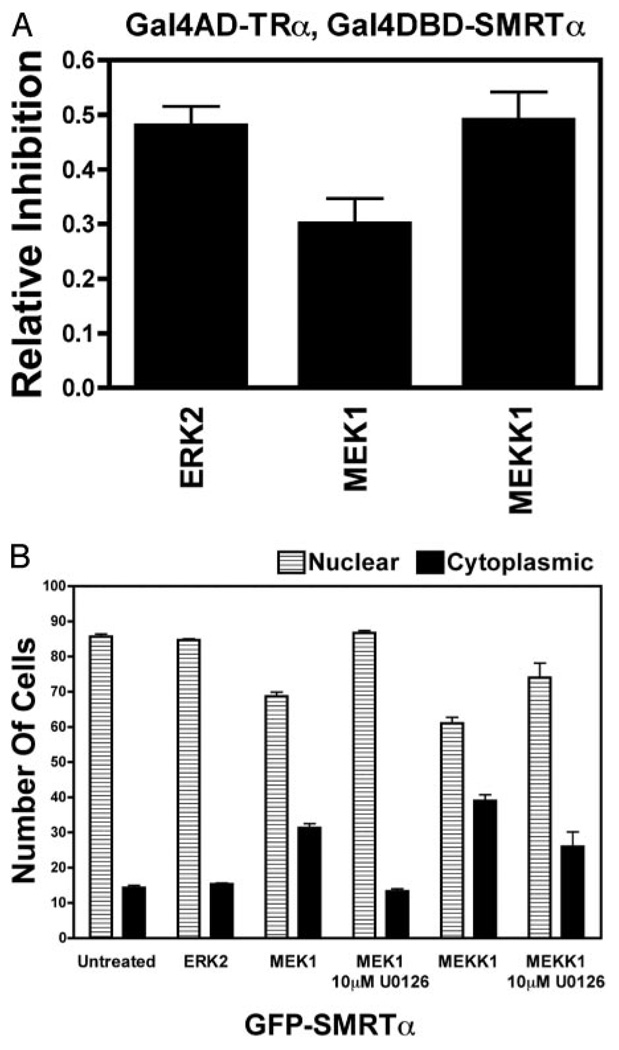
Our observation that all SMRT variants tested redistributed from nucleus to cytoplasm in response to MEKK1 signaling whereas only certain SMRT variants released from TRα or RARα under the same conditions suggested that these events are controlled by distinct regulatory mechanisms. We next explored whether these distinct regulatory mechanisms represented the actions of different tiers of the MAPKKK cascade. Introduction of either an activated MEKK1 (representing the top tier of the kinase cascade) or an activated MEK1 (operating at the second tier down) resulted in strong inhibition of the two-hybrid interaction between SMRTα and TRα (Fig. 5A) and the relocalization of these corepressors from nucleus to cytoplasm (Fig. 5B). In contrast, introduction of an activated ERK2, representing the third tier of the cascade, strongly inhibited the SMRTα/TRα interaction (Fig. 5A) but conferred no significant change in SMRTα subcellular distribution (Fig. 5B). N-CoR or SMRT constructs that were resistant to inhibition by MEKK1 in the two-hybrid interaction assay were similarly resistant to inhibition by ERK2, and constructs sensitive to MEKK1 inhibition in this assay were sensitive to ERK2 inhibition (data not shown). Interestingly, although the ability of MEK1 to redistribute SMRT constructs in this assay was completely blocked by a cognate chemical inhibitor, U0126 [1,4-diamino-2,3-dicyano-1,4-bis(oaminophenylmercapto) butadiene], the ability of MEKK1 to do so was only partially blocked by U0126 (Fig. 5B). This suggests that MEKK1 induces nuclear export of SMRT both directly and through its ability to activate MEK1.
Fig. 5. ERK2 Alone Is Sufficient to Inhibit the Interaction of SMRT with TRα, whereas MEKK1 and MEK1 Are Responsible for the SMRT Subcellular Redistribution.
A, ERK2, MEK1, or MEKK1 display comparable abilities to inhibit the mammalian two-hybrid interaction between TRα and SMRTα. A mammalian two-hybrid interaction assay was performed as in Fig. 3A using the GAL4DBD-SMRTα and GAL4AD-TRα constructs but cotransfecting activated MEKK1, MEK1, or ERK2 constructs as described in Materials and Methods. Relative luciferase and relative inhibition was calculated as in Fig. 3A. It should be noted that the activity and expression levels of the various kinases may not be identical, and this may contribute to any differences observed in their impact on the two-hybrid interaction. B, In contrast to MEKK1 or MEK1, ERK2 alone has little or no effect on the localization of SMRT. The subcellular distribution assay described in Fig. 4 was repeated using GFP-SMRTα in a transient transfection of CV-1 cells but using constitutively active ERK2, MEK1, or MEKK1 constructs as indicated. The experiments with MEKK1 and MEK1 were performed in the absence or presence of U0126, a specific inhibitor of MEK1 activity. Quantification was as in Fig. 4B.
These results indicate that ERK2, when activated either autonomously or naturally by its MEKK1-MEK1 upstream regulatory cascade, induces release of SMRT (α, τ, or sp2) from TRα but has little or no effect on the subcellular localization of any SMRT variant tested. MEKK1 or MEK1, in contrast, each have the ability to induce the relocalization of any form of SMRT from nucleus to cytoplasm but mediate the release of corepressor from nuclear receptors indirectly through their ability to activate ERK2.
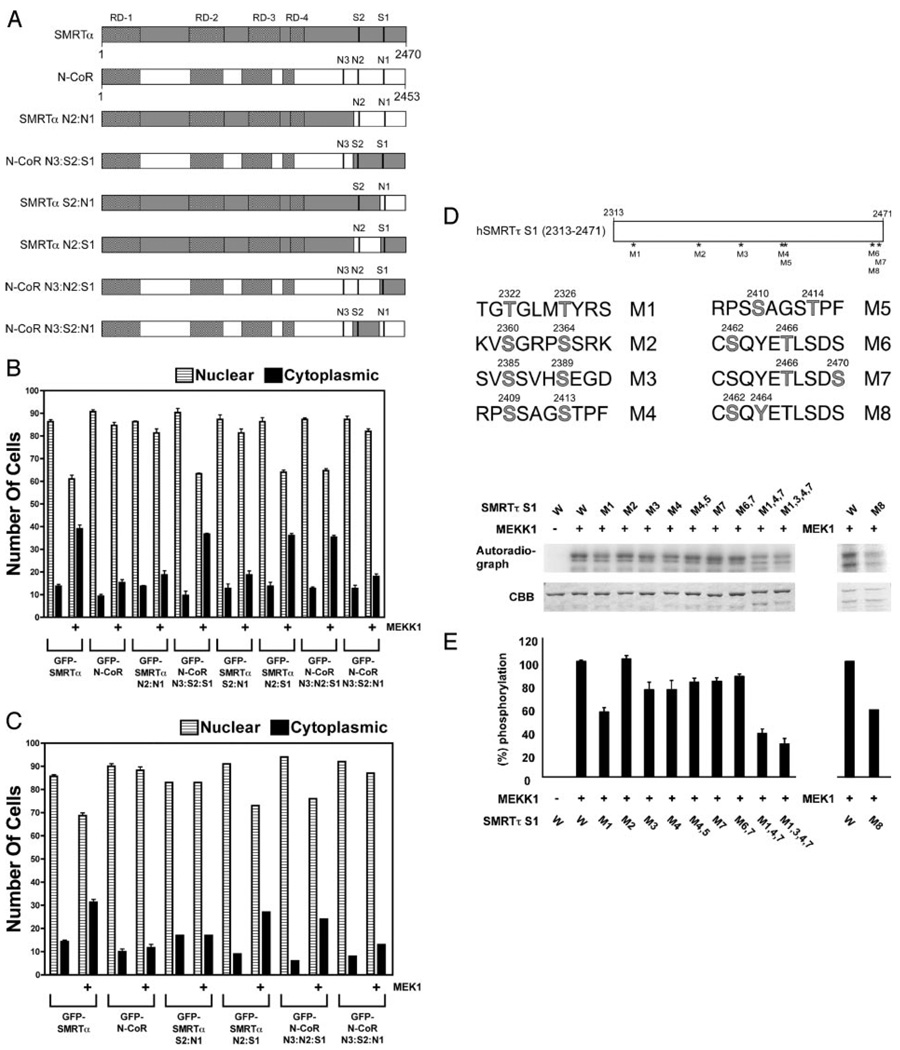
All three kinases, MEKK1, MEK1, and ERK2, can efficiently phosphorylate SMRT (and N-CoR) in vitro and in vivo at high stoichiometries (53). For example, up to 8 mol phosphate per mole protein were incorporated into the SMRTα S1 fragment by MEKK1 in vitro (data not shown). Presumably, the different consequences of these phosphorylations on SMRT and N-CoR function reflect differences in the sites or in the effects of these phosphorylations in the two different corepressor contexts. We therefore localized the phosphorylation site(s) in SMRT responsible for mediating its subcellular redistribution in response to MEKK1/MEK1 by analyzing a series of SMRT/N-CoR chimeras. Corepressor chimeras containing sequences derived from the SMRT S1 domain (e.g. SMRTsp18, SMRTα, SMRTτ, SMRTα N2:S1, N-CoR N3:N2:S1, and N-CoR N3:S2:S1) redistributed in response to MEKK1 or MEK1, whereas corepressor chimeras containing the corresponding region derived from N-CoR (N-CoR, RIP13Δ1, SMRTα N2:N1, SMRTα N3:S2:N1, and SMRTα S2:N1) did not; these chimeras therefore implicated the S1 region (Fig. 6, B and C). Consistent with this conclusion, the C-terminal 1449 amino acids of SMRT were sufficient to mediate a nuclear to cytoplasmic redistribution in response to either MEKK1 or MEK1 signaling (data not shown). Inspection of the SMRT sequence revealed seven potential phosphorylation sites for MEKK1 (S/T-X-X-XS/T) and one potential phosphorylation site for MEK1 (Thr-X-Tyr) within this S1 region (Fig. 6D) (65); site-directed mutagenesis demonstrated that many of these potential sites were, in fact, phosphorylated by MEKK1 or MEK1 in vitro (Fig. 6E). Although no single mutation reduced total phosphorylation by more than 50%, stronger reductions in overall phosphorylation were observed when introducing simultaneous mutations at multiple sites (Fig. 6E). Assay of these SMRT mutants in the form of a GFP-SMRT construct revealed that no individual serine or threonine was fully responsible for the MEKK1- or MEK1-mediated relocalization phenotype; instead, our results suggested that multiple phosphorylation events contribute combinatorially to this phenomenon (data not shown).
Fig. 6. Subcellular Relocalization of SMRT in Response to MEKK1 or MEK1 Signaling Requires a C-Terminal Corepressor Domain near the S1 Motif.
A, Schematics of the various SMRT and N-CoR constructs used in this experiment are shown. B, SMRT determinants required for nuclear to cytoplasmic relocalization in response to MEKK1 map near the S1 region. The GFP-SMRT transient transfection experiments described in Fig. 4 were repeated using the GFP-SMRT and GFP-N-CoR constructs indicated below the panel, in the presence or absence of a constitutively active MEKK1 construct. C, The same experiment as in B was repeated but using an activated MEK1 construct in place of the activated MEKK1 construct. D, Potential sites of MEKK1 or MEK1 phosphorylation in S1 region of SMRT are indicated in light gray. Specific S to A or T to A substitutions at these sites are denoted as M1, M2, etc. E, Effect of alanine substitutions at putative MEKK1 sites in SMRTα on phosphorylation by MEKK1 in vitro. GST-SMRTα constructs were incubated with [γ-32P]rATP in vitro and analyzed by SDS-PAGE (54, 84). Phosphor imager imaging of the radiolabel incorporated into each GST-SMRT protein (W, wild-type; mutants as in E) is shown in the top and quantified in the bottom; Coomassie brilliant blue staining is shown in the middle.
To localize the SMRT sequences responsible for ERK2 inhibition of the corepressor/receptor interaction, we used a combination of strategies. SMRTα constructs containing amino acids 1745–2470 were sensitive to ERK2 inhibition, whereas SMRT constructs limited to amino acids 2050–2470 of SMRTα were more resistant (Fig. 7A), implicating the 1745–2049 domain as contributing to this phenomenon. The relative resistance of RIP13Δ1 and SMRTαΔ1902–1994 to ERK2 inhibition (Fig. 3) further identified a subregion of this domain as crucial for efficient ERK2-mediated inhibition of the twohybrid interaction. This subdomain includes a series of potential ERK phosphorylation sites and encompasses a putative D element of a larger bipartite ERK docking site (Fig. 7B) (65, 66). Supporting the concept that this domain is recognized by ERK2, a glutathione S-transferase (GST) fusion of this region of SMRT physically interacts with ERK2 in vitro (Fig. 7C). We suggest that the presence of these ERK sites in most splice variants of SMRT and N-CoR allows them to be efficiently phosphorylated by ERK2. For SMRT or N-CoR variants containing two or less CoRNR boxes, this phosphorylation inhibits their interaction with nuclear receptors. Either loss of the crucial ERK recognition sites (as in RIP13Δ1) or introduction of a third CoRNR box (as in SMRTsp18 or N-CoR) confers resistance to this ERK2 inhibition. There are 12 potential sites of ERK phosphorylation (S/T-P) within the 1745–2049 region, many of which are phosphorylated by ERK2 in vitro; because of the multiplicity of these (and other) ERK phosphorylation sites in SMRT, we were unable to further identify the specific phospho-amino acids responsible for the inhibition of the SMRT/TRα interaction (data not shown).
Fig. 7. Efficient ERK2 Inhibition of the SMRT Interaction with TRα Requires the Integrity of a Putative ERK Docking Site.
A, Loss of codons 1745–2050 reduces the ability of ERK2 to inhibit the interaction between SMRTα and TRα. The mammalian two-hybrid interaction assay described in Fig. 5A was repeated using GAL4DBD constructs containing the SMRTα sequences indicated below the panel. B, The sequence of a possible bipartite ERK docking site that maps to this region of SMRT is presented, and numbering refers to codon position in SMRTα sequence. C, ERK2 interacts with this region of SMRT in vitro. A GST fusion of SMRTα codons 1878–1974, immobilized on glutathione agarose, was incubated with soluble, 35S-radiolabeled constitutively active ERK2 using a GST-pull-down protocol (80); radiolabeled ERK2 remaining bound to the GST-SMRT construct after repeated washings was eluted and quantified (relative to input) by SDS-PAGE/ phosphor imager analysis (80).
MEKK1 Reverses Repression by SMRT Variants But Not by N-CoR
To determine the effect of MEKK1 on transcriptional repression by SMRT and N-CoR, we performed transient transfections in CV-1 cells using a GAL4DBD-RARα fusion and a GAL17mer-luciferase reporter gene. Basal luciferase expression in this system was repressed by introduction of the GAL4DBD-RARα construct, and this repression was further enhanced by ectopic introduction of SMRTsp18, SMRTα, SMRTτ, or N-CoR (Fig. 8A). Cointroduction of an activated MEKK1 construct had no detectable effect on the N-CoR enhanced repression but reversed the repression mediated by all three SMRT variants tested. The ability of MEKK1 to interfere with repression mediated by SMRTτ and SMRTα, but not N-CoR, is fully anticipated from both our two-hybrid and our subcellular distribution studies. However, the ability of MEKK1 to also interfere with repression mediated by SMRTsp18 suggests that the MEKK1-mediated export of this splice variant out of the nucleus may be sufficient to interfere with RARα-mediated repression, despite the SMRTsp18/RARα interaction being relatively resistant to MEKK1 disruption in our two-hybrid assays. Alternatively, it is possible that MEKK1 has multiple disruptive effects on SMRT function, and the inhibition of repression by SMRTsp18 seen in Fig. 8A is through a mechanism unrelated to either subcellular distribution or corepressor-receptor interaction.
Fig. 8. MAPKKK Signaling Can Reverse Nuclear Receptor Repression in a Corepressor-Specific Fashion.
A, MEKK1 can reverse repression mediated by SMRTα, SMRTτ, or SMTRsp18 but not by N-CoR. GAL4DBD-RARα constructs were transfected into CV-1 cells together with a GAL17mer-luciferase reporter and a pCH110 β-galactosidase internal control, either alone (no exogenous corepressor) or with expression vectors for N-CoR, SMRTα, SMRTτ, or SMRT-sp18 as indicated. Parallel experiments were performed with an empty GAL4DBD vector. Relative luciferase activity was determined after 48 h. Relative repression was calculated by comparing the effect of MEKK1 on each GAL4DBD-RARα construct relative with the effect of MEKK1 on the empty GAL4DBD construct under the same conditions. The average and SD from three experiments is presented. B, Model of the impact of MAPKKK signaling on SMRT and N-CoR corepressor function. MEKK1 and MEK1 are proposed to induce redistribution of SMRT isoforms from nucleus to cytoplasm, whereas N-CoR isoforms are resistant to this effect. Conversely, the interaction of SMRT or N-CoR with nuclear receptors is primarily inhibited by ERK2 operating at the “bottom” of the kinase cascade; the presence of three CoRNR box motifs (RIDS) or loss of an ERK docking site in either SMRT or N-CoR stabilizes the corepressor/TRα interaction against disruption by ERK2.
DISCUSSION
A lternative mRNA Splicing Determines the Affinity of the SMRT and N-CoR Corepressors for Their Nuclear Receptor Partners
Our goal was to understand how corepressor splicing affects the response to MAPKKK cascades. Not unexpectedly, we also observed that corepressor splicing influences the basal interaction of N-CoR and SMRT with their nuclear receptor partners. Several generalities were noted. First, corepressor variants that encode three CoRNR box motifs (e.g. N-CoR and SMRTsp18) display a substantially higher interaction with either TRα or RARα in the mammalian two-hybrid assay than do splice versions of the same corepressors with only two CoRNR boxes. Similarly, splice variants having two CoRNR boxes interact with these nuclear receptors significantly more strongly than do variants possessing only one. Although representing novel observations for SMRTsp18, our results are consistent with previous studies comparing N-CoR and RIP13Δ1 (28, 36, 41, 67, 68). Our data generally support the model that nuclear receptors can simultaneously contact two CoRNR boxes within a single corepressor molecule, most likely by forming receptor dimers (69), but also indicate that receptors can interact with a given corepressor through a variety of alternative CoRNR boxes and iterations. Intriguingly, even corepressor variants containing a single CoRNR box (such as SMRTsp2) may potentially be able to interact sufficiently with certain nuclear receptors (such as RARα) so as to exert biological effects.
Despite these modifying effects of mRNA splicing, any given form of N-CoR displayed a stronger interaction with TRα than did the corresponding form of SMRT (e.g. N-CoR vs. SMRTsp18, RIP13Δ1 vs. SMRTα or SMRTτ). Conversely, any given form of SMRT displayed a stronger interaction with RARα than did the corresponding form of N-CoR. Both N-CoR and SMRTsp18 preferentially used the flanking CoRNR boxes (N3+N1 or S3+S1) to interact with TRα but the internal CoRNR box (N2 or S2) to interact with RARα. Therefore, the preference of TRα for N-CoR vs. SMRTsp18 reflects an intrinsically higher affinity of TRα for N3 and N1 vs. S3 and S1, rather than a use of different CoRNR box parings by TRα in N-CoR vs. SMRTsp18.
Alternative mRNA Splicing Determines the SMRT Corepressor Response to MAPKKK Cascade Signaling
We reported previously that growth factor signaling initiates an MAPKKK cascade resulting in SMRT phosphorylation, release of SMRT from its nuclear receptor partners, redistribution of SMRT from nucleus to cytoplasm, and derepression of associated target genes; N-CoR, in contrast, was insensitive to MAPK cascade signaling under the same conditions (53, 54). Our previous studies focused on the prototypic isolate of each corepressor: N-CoR itself (which encodes three receptor interaction domains, N3+N2+N1) and SMRTα or SMRTτ (which contain only two receptor interaction domains, S2+S1). However, alternative mRNA splicing generates a series of additional SMRT and N-CoR variants, leading us to examine the impact of this corepressor diversity on regulation by MAPKKK. Our experiments reveal that splice variants generated from a single corepressor locus can differ significantly in their sensitivity to MAPKKK signals. The presence of three CoRNR boxes in either SMRTsp18 or N-CoR stabilizes the interaction of these corepressor variants with their nuclear receptor partners, making the interaction relatively refractory to inhibition by MEKK1 signaling. Conversely, splice forms of SMRT lacking the S3 CoRNR box (SMRTα, SMRTτ, or SMRTsp2) are sensitive to MEKK1-mediated disruption of their interactions with nuclear receptors. We explored this question further by artificially disrupting specific CoRNR box motifs or exchanging CoRNR boxes between SMRT and N-CoR. Unexpectedly, the response of the corepressor/nuclear receptor interaction to kinase inhibition was not dependent on the presence or absence of any given CoRNR box. Instead, corepressors that contained three CoRNR boxes were generally refractory to inhibition by MEKK1 in these assays, whereas corepressors that contained two or fewer CoRNR boxes were generally sensitive to MEKK1 inhibition. We conclude that mRNA splicing and CoRNR box numeration, not the identity of the corepressor paralog per se, is a primary determinant of whether a given corepressor variant can be released from nuclear receptors by these kinase cascades (Fig. 8).
The naturally occurring RIP13Δ1 variant of N-CoR presented an exception to this overall rule. RIP13Δ1 contains only two CoRNR boxes (N2+N1) and, in keeping with our model, was slightly more sensitive to MAPKKK signaling compared with the virtually fully refractory N-CoR (N3+N2+N1). However, RIP13Δ1 was clearly much more resistant to MAPKKK inhibition than were other natural or artificial corepressors containing two CoRNR boxes (e.g. SMRTα, SMRTτ, or an N-CoR construct with an N3 disrupted by site-directed mutagenesis). This relative resistance of RIP13Δ1 appeared to reflect the splice-mediated deletion of an ERK docking site in exon 37b present in N-CoR and the SMRT variants but absent in the RIP13Δ1 variant.
All SMRT Splice Variants Tested Redistribute from Nucleus to Cytoplasm in Response to MEKK1 Signaling, whereas all N-CoR Variants Tested Are Resistant to this Kinase-Mediated Redistribution
Although alternative mRNA splicing modified the ability of SMRT to respond to MAPKKK signaling in the corepressor/nuclear receptor interaction assay, alternative mRNA splicing had no observable effect in the nuclear export assay. Both N-CoR itself and RIP13Δ1 remained primarily nuclear in the presence of an activated MEKK1 or MEK1, whereas SMRTα, SMRTτ, SMRTsp18, and SMRTsp2 were able to relocalize from nucleus to cytoplasm in response to MEKK1 or MEK1. One implication from these results is that kinase-mediated nuclear export is regulated by a mechanism distinct from that regulating nuclear receptor interaction, as described in more detail below. Intriguing to contemplate in this regard is the behavior of the SMRTsp18 variant, the bulk of which is exported from the nucleus in response to MEKK1 signaling yet retains an apparent interaction with TRα or RARα in the two-hybrid assay under the same conditions. It is possible that SMRTsp18 redistribution into the cytoplasm in response to MEKK1 may be accompanied by a parallel codistribution of the associated nuclear receptors. Alternatively, the bulk redistribution of SMRTsp18 to the cytoplasm under these circumstances may leave behind a smaller, nuclear subpopulation of SMRTsp18 that remains tethered to target DNA through its stable interaction with TRα or RARα. The latter model is consistent with the activity of SMRTsp18 in the mammalian two-hybrid assay, which presumably requires a nuclear localization to activate the luciferase reporter. However, a variety of intermediate scenarios are possible in which SMRTsp18 is retained on certain target genes but not on others based on the affinity of the associated transcription factor partner for its cognate DNA binding sites and on the strength of the SMRTsp18/transcription factor interaction itself. This latter interpretation is consistent with our observation that the ability of SMRTsp18 to mediate repression by an RARα construct can be inhibited by MEKK1 in at least certain contexts.
The Subcellular Distribution of SMRT and the Release of this Corepressor from Its Nuclear Receptor Partners Are Regulated by Distinct Tiers of the MAPKKK Cascade
All three tiers of the MAPKKK cascade, represented by MEKK1, MEK1, and ERK1/2, are able to independently phosphorylate SMRT in vitro (53). Our current study implicates the highest two tiers of the MAPKKK cascade, MEKK1 and MEK1, as the primary mediators of the SMRT nuclear to cytoplasmic relocalization, whereas the bottom tier kinase, represented by the ERK kinases, is the principal effector by which the SMRT/receptor interaction is inhibited (Fig. 8); these results raise the possibility that, in certain contexts, the subcellular distribution of SMRT might be regulated separately from its interaction with its nuclear receptor partners. The effects of MEKK1 and MEK1 on SMRT subcellular localization are strongest in the presence of both MEKK1 and MEK1 but can be observed with either kinase alone, whereas ERK2 is neither required nor sufficient to induce redistribution of SMRT. Conversely, ERK2 alone inhibits the interaction of SMRT with TRα and with RARα, whereas the ability of MEKK1 and MEK1 to interfere with the SMRT/receptor interaction is likely indirect and reflects, at least in part, the ability of these higher-order kinases to induce ERK2 activity (Fig. 8). A candidate ERK docking site near the S2/N2 CoRNR box appears likely to be required for this ERK-mediated inhibition of corepressor/ receptor interaction; its absence from RIP13Δ1 may account for the relative insensitivity of this splice variant to ERK2 inhibition.
We previously reported identification of an MEKK1 site in SMRTα that interfered with binding of a short, corepressor peptide to TRα in vitro (54). These results, obtained by use of an electrophoretic mobility shift/super-shift assay, are reproducible. However, we did not observe an effect of this MEKK1 phosphorylation on the interaction of any of the longer versions of SMRT tested here, either in vitro or in vivo; instead, ERK2 was the key mediator of inhibition of the SMRT/TRα and SMRT/ RARα interaction in the experiments reported here. It remains possible that direct MEKK1 phosphorylation of SMRT may contribute to the regulation of the corepressor/receptor interaction in other contexts or in a manner not detectable by the methods used here. It is also possible that the effects of MEKK1 on the subcellular localization of SMRT include actions of this kinase on proteins other than SMRT itself, such as components of the nuclear translocation machinery.
Notably, both SMRT and N-CoR are responsive to a wider network of additional kinases that modulate their function in response to cellular signaling (56, 70–77). Some signals are shared, whereas others differ for SMRT and for N-CoR. For example, N-CoR but not SMRT has been reported to be inhibited by Akt phosphorylation (56), the reciprocal of the response of these corepressors to MAPKKK signaling as reported here. Conversely, the interaction of SMRT (and probably N-CoR) with many nuclear receptors is enhanced by casein kinase 2 phosphorylation (76). It remains to be determined whether the response of these corepressors to Akt or casein kinase 2 is altered by alternative mRNA splicing. It should also be noted that the transcription factor partner can itself influence the ability of these corepressor variants to respond to their kinase regulators. As noted here, the interaction of RARα with SMRTα or SMRTτ is less sensitive to MEKK1 inhibition than is the interaction of TRα with these corepressors. Similarly, other investigators have reported that, in contrast to the thyroid hormone and retinoid nuclear receptors studied here, certain steroid receptors can render N-CoR responsive to MEKK1 inhibition indirectly by recruiting TAB2 (TGF-β-activated kinase 1 binding domain 2), an MEKK1 target protein that, on phosphorylation, results in an ubiquitin- mediated disassembly of the corepressor complex and derepression (70, 77). The prevailing theme in eukaryotic repression therefore appears to be one of great diversity, by which the transcriptional program of a cell is adapted by the combined impact of hormone ligand, kinase signaling, transcription factor partnership, and alternative mRNA splicing to yield the evolutionarily most adaptive outcome for a given set of circumstances.
MATERIALS AND METHODS
Plasmid Constructs
The origins of the pSG5-GAL4DBD-RARα, pUC18, pCH110, pADH-Gal4–17mer, pSG5-GAL4AD, pSG5-GAL4AD-T3Rα, pSG5-GAL4AD-RARα, pSG5-GAL4DBD, pSG5-GAL4DBD-SMRT τ (1773–2471), pCMV5-FLAG-ΔMEKK1 (817–1493), and pCMV-HA-MEK1 (R4F) plasmids were described previously (52, 53, 78–82). The pSG6-Gal4DBD vector was created by inserting a synthetic oligonucleotide (Biosource International, Camarillo, CA) encoding an expanded multiple cloning site into pSG5-Gal4DBD. The pSG6-Gal4DBDSMRT α (1675–2470) expression vector was created by inserting an EcoRI-SalI (blunt) fragment from a parental full-length SMRTα (1–2470) cloning vector into EcoRI-SmaI digested pSG6-Gal4DBD. pSG6-Gal4DBD expression vectors containing sequences from SMRTsp18 (1675–2508), SMRTτ (1675–2423), and SMRTsp2 (1675–2394) were subsequently created by inserting SgrAI-PstI fragments from appropriate parental full-length SMRT cloning vectors into SgrAI-PstI digested pSG6-Gal4DBD-SMRTα (1675–2470). The pSG6-Gal4DBD-SMRTα (1675–2470) Δ1902–1994 vector was created from the equivalent SMRTα vector using QuikChange-mediated mutagenesis and the protocol recommended by the manufacturer (Stratagene, La Jolla, CA). The pSG6-Gal4DBD-N-CoR (1634–2453) expression vector, corresponding to the equivalent sequence in the SMRT vectors, was created by inserting an EcoRI-SalI (blunt) fragment from a parental full-length N-CoR (1–2453) cloning vector into E coRI-SmaI digested pSG6-Gal4DBD. The pSG6-Gal4DBDRIP13 Δ1 (1675–2334) vector was generated from the equivalent N-CoR vector using site-directed mutagenesis. Corepressor constructs with mutant receptor interaction domains were first generated in parental vectors using standard site-directed mutagenesis protocols and then shuttled into the appropriate vectors. In the case of SMRT, mS1 corresponds to I2335A/I2336A, mS2 to V2133A/I2134A, and mS3 to I2007A/I2008A, whereas for N-CoR, mN1 corresponds to I2280A/ I2281A, mN2 to I2076A/I2077A, and mN3 to I1952A/I1953A (54). pSG6-Gal4DBD-N-CoR N3:S2:S1 (N-CoR 1634–2031/ SMRTα 2049–2470) and pSG6-Gal4DBD-SMRTα N2:N1 (SMRTα 1675–2048/N-CoR 2032–2453) expression vectors were created using a combination of standard PCR, site-directed mutagenesis, and restriction endonuclease protocols.
The pCMV-sGFPc vector was created by inserting a synthetic oligonucleotide (Biosource International) encoding an expanded multiple cloning site into a restriction endonuclease site-depleted pCMV-sGFPBH vector (53). The pCMV-sGFPc-SMRTsp18 (1–2508), SMRTα (1–2470), SMRTτ (1– 2423), and SMRTsp2 (1–2394) expression vectors were created by inserting HinDIII-XhoI fragments from appropriate parental full-length SMRT cloning vectors into HinDIII-XhoI digested pCMV-sGFPc. The pCMV-sGFPc-N-CoR (1–2453) expression vector was created by inserting an EcoRI/SalI fragment from the parental full-length N-CoR cloning vector into EcoRI/SalI digested pCMV-sGFPc. The pCMV-sGFPc-RIP13Δ1 (1–2334) expression vector was created by inserting a BstXI/SalI fragment from an RIP13Δ1 intermediate cloning vector into BstXI/SalI digested pCMV-sGFPc-N-CoR (1– 2453). pCMV-sGFPc-N-CoR N3:S2:S1 (N-CoR 1–2031/ SMRTα 2049–2470), pCMV-sGFPc-N-CoR N3:N2:S1 (NCoR 1–2248/SMRTα 2265–2470), pCMV-sGFPc-N-CoR N3: S2:N1 (N-CoR 1–2031/SMRTα 2049–2264/N-CoR 2249– 2453), pCMV-sGFPc-SMRTα N2:N1 (SMRTα 1–2048/N-CoR 2032–2453), pCMV-sGFPc-SMRTα S2:N1 (SMRTα 1–2264/N-CoR 2249–2453), and pCMV-sGFPc-SMR0054α N2:S1 (SMRTα 1–2048/N-CoR 2032–2248/SMRTα 2265–2470) expression vectors were created using a combination of PCR, site-directed mutagenesis, and restriction endonuclease protocols. The pSG5-Myc-SMRTsp18 (1–2508) expression vector was created by inserting the HinDIII-XhoI fragment from a parental vector into an equivalently cleaved pSG5-Myc vector. The pSG5-HA-ERK2 (L75P/S153D) vector, which encodes a constitutively active form of ERK2 (83), was created by QuikChange mutagenesis of a wild-type ERK2 molecular clone.
Mammalian Two-Hybrid Analysis
CV-1 cells were propagated in DMEM formulated with high glucose, L-glutamine, and pyridoxine hydrochloride (Invitrogen, Carlsbad, CA) and supplemented with 5% heat-inactivated fetal bovine serum (Hyclone, Logan, UT); cells were maintained at 37 C in a humidified 5% CO2 atmosphere. Transient transfections were performed using 3.0 × 104 CV−1 cells per well in a 24-well plate, Effectene reagent, and the protocol of the manufacturer (Qiagen, Valencia, CA). Transfection mixtures included 50 ng of the appropriate pSG5-GAL4AD vector, 12.5 ng of the appropriate pSG5-GAL4DBD vector, 50 ng of the pADH-GAL4–17mer luciferase reporter, 50 ng of pCH110 as an internal transfection control, appropriate expression vectors for the indicated signal transducers, and/or an empty vector as appropriate to bring to total DNA to 250 ng/well. Twenty-four hours after transfection, the medium was replaced with fresh medium with or without 1 µM T3 as indicated. Cells were collected 48 h after transfection and were lysed in 100 µl/well of Triton lysis buffer (0.2% Triton X−100, 91 mM K2HPO4, and 9.2 mM KH2PO4). Luciferase and β-galactosidase activity were determined as described previously (53, 81).
Fluorescent Microscopy
CV-1 cells (1.0 × 105 cells per well in a six-well plate) were allowed to attach to 22 × 22 mm coverslips and were transfected using the Effectene protocol described above, using 500 ng of the appropriate pCMV-sGFPc-corepressor vector, 100 ng of the appropriate expression vector for the indicated signal transducers, and empty vector for a total of 750 ng of DNA per well. Twenty-four hours after transfection, the medium was replaced with fresh medium with or without 10 µM U0126 (Cell Signaling Technology, Danvers, MA) as indicated. Cells were fixed 48 h after transfection for 10 min at room temperature in 4% formalin. After aspiration of the fixing agent, cells were washed three times in PBS and incubated for 5 min at room temperature in PBS containing 0.5 µg/ml 4′,6-diamidino-2-phenylindole. The coverslips were again washed three times in PBS and once in distilled water, and the excess moisture was removed by aspiration. The coverslips were mounted on slides using 25 µl Vectashield (Vector Laboratories, Burlingame, CA) and were sealed with fingernail polish. The slides were visualized using a Nikon (Tokyo, Japan) Microphot Epifluorescence microscope and a Nikon Coll Pix 450 digital camera. For quantification of the fluorescent microscopic data, 100 transfected cells were counted at random from each slide and scored for the following GFP-corepressor subcellular localizations: predominantly nuclear or predominantly cytoplasmic.
Repression Assay
CV-1 cells were propagated as described above. Transient transfections were performed using 3.0 × 104 CV−1 cells per well in a 24-well plate, Effectene reagent, and the protocol of the manufacturer (Qiagen). Transfection mixtures included 12.5 ng of either the pSG5-GAL4DBD or pSG5-GAL4DBDRAR α vector, 50 ng of the appropriate expression vectors for the indicated Myc-tagged corepressors (54, 57), 50 ng of the pADH-GAL4–17mer luciferase reporter, 50 ng of pCH110 as an internal transfection control, 50 ng of the pCMV5-FLAG-ΔMEKK1 expression vector, and/or an empty vector, as appropriate to bring to total DNA to 250 ng/well. Twenty-four hours after transfection, the medium was replaced with fresh medium. Cells were collected 48 h after transfection and were lysed in 100 µl/well of Triton lysis buffer (0.2% Triton X-100, 91 mM K2HPO4, and 9.2 mM KH2PO4). Luciferase and β-galactosidase activity were determined as described previously (53, 81).
Acknowledgments
We gratefully acknowledge the superb technical assistance of Liming Liu. We also sincerely thank Meghan Dukerich for excellent assistance in characterization of the subcellular distribution properties of specific SMRT mutants.
This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases/National Institutes of Health Grant R01DK53528 and National Institute of General Medical Sciences/National Institutes of Health/University of California Davis Physician Scientist Training Program Predoctoral Training Award T32GM07377.
Abbreviations
- AD
Activation domain
- CoRNR
corepressor nuclear receptor
- DBD
DNA binding domain
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- MAPKKK
MAPK kinase kinase
- MEK1
MAP/ERK kinase 1
- MEKK1
MAP/ERK kinase kinase 1
- RAR
retinoic acid receptor
- RIP13Δ1
retinoid X receptor interacting protein 13Δ1
- TR
thyroid hormone receptor.
Footnotes
Molecular Endocrinologyis published monthly by The Endocrine Society (http://www.endo-society.org), the foremost professional society serving the endocrine community.
Disclosure Statement: The authors have nothing to disclose.
REFERENCES
- 1.Baniahmad A. Nuclear hormone receptor co-repressors. J Steroid Biochem Mol Biol. 2005;93:89–97. doi: 10.1016/j.jsbmb.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 2.Edwards DP. The role of coactivators and corepressors in the biology and mechanism of action of steroid hormone receptors. J Mammary Gland Biol Neoplasia. 2000;5:307–324. doi: 10.1023/a:1009503029176. [DOI] [PubMed] [Google Scholar]
- 3.Lee JW, Lee YC, Na SY, Jung DJ, Lee SK. Transcriptional coregulators of the nuclear receptor superfamily: coactivators and corepressors. Cell Mol Life Sci. 2001;58:289–297. doi: 10.1007/PL00000856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McKenna NJ, Xu J, Nawaz Z, Tsai SY, Tsai MJ, O’Malley BW. Nuclear receptor coactivators: multiple enzymes, multiple complexes, multiple functions. J Steroid Biochem Mol Biol. 1999;69:3–12. doi: 10.1016/s0960-0760(98)00144-7. [DOI] [PubMed] [Google Scholar]
- 5.Hu X, Lazar MA. Transcriptional repression by nuclear hormone receptors. Trends Endocrinol Metab. 2000;11:6–10. doi: 10.1016/s1043-2760(99)00215-5. [DOI] [PubMed] [Google Scholar]
- 6.Privalsky ML. The role of corepressors in transcriptional regulation by nuclear hormone receptors. Annu Rev Physiol. 2004;66:315–360. doi: 10.1146/annurev.physiol.66.032802.155556. [DOI] [PubMed] [Google Scholar]
- 7.Xu L, Glass CK, Rosenfeld MG. Coactivator and corepressor complexes in nuclear receptor function. Cur Opin Genetics Develop. 1999;9:140–147. doi: 10.1016/S0959-437X(99)80021-5. [DOI] [PubMed] [Google Scholar]
- 8.Berger SL. Molecular biology. The histone modification circus. Science. 2001;292:64–65. [PubMed] [Google Scholar]
- 9.Chen H, Tini M, Evans RM. HATs on and beyond chromatin. Cur Opin Cell Biol. 2001;13:218–224. doi: 10.1016/s0955-0674(00)00200-3. [DOI] [PubMed] [Google Scholar]
- 10.Ishizuka T, Lazar MA. The N-CoR/histone deacetylase 3 complex is required for repression by thyroid hormone receptor. Mol Cell Biol. 2003;23:5122–5131. doi: 10.1128/MCB.23.15.5122-5131.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 12.Jones PL, Shi YB. N-CoR-HDAC corepressor complexes: roles in transcriptional regulation by nuclear hormone receptors. Curr Top Microbiol Immunol. 2003;274:237–268. doi: 10.1007/978-3-642-55747-7_9. [DOI] [PubMed] [Google Scholar]
- 13.Kuzmichev A, Reinberg D. Role of histone deacetylase complexes in the regulation of chromatin metabolism. Cur Top Microbiol Immunol. 2001;254:35–58. doi: 10.1007/978-3-662-10595-5_2. [DOI] [PubMed] [Google Scholar]
- 14.Rice JC, Allis CD. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Cur Opin Cell Biol. 2001;13:263–273. doi: 10.1016/s0955-0674(00)00208-8. [DOI] [PubMed] [Google Scholar]
- 15.Rosenfeld MG, Glass CK. Coregulator codes of transcriptional regulation by nuclear receptors. J Biol Chem. 2001;276:36865–36868. doi: 10.1074/jbc.R100041200. [DOI] [PubMed] [Google Scholar]
- 16.Urnov FD, Wolffe AP, Guschin D. Molecular mechanisms of corepressor function. Curr Top Microbiol Immunol. 2001;254:1–34. doi: 10.1007/978-3-662-10595-5_1. [DOI] [PubMed] [Google Scholar]
- 17.Workman JL, Kingston RE. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Ann Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 18.Beato M, Klug J. Steroid hormone receptors: an update. Human Reprod Update. 2000;6:225–236. doi: 10.1093/humupd/6.3.225. [DOI] [PubMed] [Google Scholar]
- 19.Li X, Lonard DM, O’Malley BW. A contemporary understanding of progesterone receptor function. Mech Ageing Dev. 2004;125:669–678. doi: 10.1016/j.mad.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 20.Moehren U, Eckey M, Baniahmad A. Gene repression by nuclear hormone receptors. Essays Biochem. 2004;40:89–104. doi: 10.1042/bse0400089. [DOI] [PubMed] [Google Scholar]
- 21.Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J, Lazar MA. The mechanism of action of thyroid hormones. Annu Rev Physiol. 2000;62:439–466. doi: 10.1146/annurev.physiol.62.1.439. [DOI] [PubMed] [Google Scholar]
- 23.Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 24.Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell. 2004;116:511–526. doi: 10.1016/s0092-8674(04)00133-3. [DOI] [PubMed] [Google Scholar]
- 25.Privalsky ML. Regulation of SMRT and N-CoR corepressor function. Curr Top Microbiol Immunol. 2001;254:117–136. doi: 10.1007/978-3-662-10595-5_6. [DOI] [PubMed] [Google Scholar]
- 26.Chen JD, Evans RM. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 27.Chen JD, Umesono K, Evans RM. SMRT isoforms mediate repression and anti-repression of nuclear receptor heterodimers. Proc Natl Acad Sci USA. 1996;93:7567–7571. doi: 10.1073/pnas.93.15.7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Downes M, Burke LJ, Bailey PJ, Muscat GE. Two receptor interaction domains in the corepressor, N-CoR/RIP13, are required for an efficient interaction with ReverbA α and RVR: physical association is dependent on the E region of the orphan receptors. Nucleic Acids Res. 1996;24:4379–4386. doi: 10.1093/nar/24.22.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hörlein AJ, Näär AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Söderström M, Glass CK, Rosenfeld MG. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 30.Ordentlich P, Downes M, Xie W, Genin A, Spinner NB, Evans RM. Unique forms of human and mouse nuclear receptor corepressor SMRT. Proc Natl Acad Sci USA. 1999;96:2639–2644. doi: 10.1073/pnas.96.6.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park EJ, Schroen DJ, Yang M, Li H, Li L, Chen JD. SMRTe, a silencing mediator for retinoid and thyroid hormone receptors-extended isoform that is more related to the nuclear receptor corepressor. Proc Natl Acad Sci USA. 1999;96:3519–3524. doi: 10.1073/pnas.96.7.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sande S, Privalsky ML. Identification of TRACs (T3 receptor-associating cofactors), a family of cofactors that associate with, and modulate the activity of, nuclear hormone receptors. Mol Endocrinol. 1996;10:813–825. doi: 10.1210/mend.10.7.8813722. [DOI] [PubMed] [Google Scholar]
- 33.Seol W, Mahon MJ, Lee YK, Moore DD. Two receptor interacting domains in the nuclear hormone receptor corepressor RIP13/N-CoR. Mol Endocrinol. 1996;10:1646–1655. doi: 10.1210/mend.10.12.8961273. [DOI] [PubMed] [Google Scholar]
- 34.Zamir I, Harding HP, Atkins GB, Hörlein A, Glass CK, Rosenfeld MG, Lazar MA. A nuclear hormone receptor corepressor mediates transcriptional silencing by receptors with distinct repression domains. Mol Cell Biol. 1996;16:5458–5465. doi: 10.1128/mcb.16.10.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ordentlich R, Downes M, Evans RM. Corepressors and nuclear hormone receptor function. Curr Top Microbiol Immunol. 2001;254:101–116. doi: 10.1007/978-3-662-10595-5_5. [DOI] [PubMed] [Google Scholar]
- 36.Cohen RN, Brzostek S, Kim B, Chorev M, Wondisford FE, Hollenberg AN. The specificity of interactions between nuclear hormone receptors and corepressors is mediated by distinct amino acid sequences within the interacting domains. Mol Endocrinol. 2001;15:1049–1061. doi: 10.1210/mend.15.7.0669. [DOI] [PubMed] [Google Scholar]
- 37.Hu X, Lazar MA. The CoRNR motif controls the recruitment of corepressors by nuclear hormone receptors. Nature. 1999;402:93–96. doi: 10.1038/47069. [DOI] [PubMed] [Google Scholar]
- 38.Marimuthu A, Feng W, Tagami T, Nguyen H, Jameson JL, Fletterick RJ, Baxter JD, West BL. TR surfaces and conformations required to bind nuclear receptor corepressor. Mol Endocrinol. 2002;16:271–286. doi: 10.1210/mend.16.2.0777. [DOI] [PubMed] [Google Scholar]
- 39.Nagy L, Kao HY, Love JD, Li C, Banayo E, Gooch JT, Krishna V, Chatterjee K, Evans RM, Schwabe JWR. Mechanism of corepressor binding and release from nuclear hormone receptors. Genes Dev. 1999;13:3209–3216. doi: 10.1101/gad.13.24.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perissi V, Staszewski LM, McInerney EM, Kurokawa R, Krones A, Rose DW, Lambert MH, Milburn MV, Glass CK, Rosenfeld MG. Molecular determinants of nuclear receptor-corepressor interaction. Genes Dev. 1999;13:3198–3208. doi: 10.1101/gad.13.24.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Webb P, Anderson CM, Valentine C, Nguyen P, Marimuthu A, West BL, Baxter JD, Kushner PJ. The nuclear receptor corepressor (N-CoR) contains three isoleucine motifs (I/LXXII) that serve as receptor interaction domains (IDs) Mol Endocrinol. 2000;14:1976–1985. doi: 10.1210/mend.14.12.0566. [DOI] [PubMed] [Google Scholar]
- 42.Xu HE, Stanley TB, Montana VG, Lambert MH, Shearer BG, Cobb JE, McKee DD, Galardi CM, Plunket KD, Nolte RT, Parks DJ, Moore JT, Kliewer SA, Willson TM, Stimmel JB. Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARα. Nature. 2002;415:813–817. doi: 10.1038/415813a. [DOI] [PubMed] [Google Scholar]
- 43.Fischle W, Dequiedt F, Fillion M, Hendzel MJ, Voelter W, Verdin E. Human HDAC7 histone deacetylase activity is associated with HDAC3 in vivo. J Biol Chem. 2001;276:35826–35835. doi: 10.1074/jbc.M104935200. [DOI] [PubMed] [Google Scholar]
- 44.Guenther MG, Barak O, Lazar MA. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol. 2001;21:6091–6101. doi: 10.1128/MCB.21.18.6091-6101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones PL, Sachs LM, Rouse N, Wade PA, Shi YB. Multiple N-CoR complexes contain distinct histone deacetylases. J Biol Chem. 2001;276:8807–8811. doi: 10.1074/jbc.C000879200. [DOI] [PubMed] [Google Scholar]
- 46.Kao HY, Downes M, Ordentlich P, Evans RM. Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes Dev. 2000;14:55–66. [PMC free article] [PubMed] [Google Scholar]
- 47.Pascual G, Fong AL, Ogawa S, Gamliel A, Li AC, Perissi V, Rose DW, Willson TM, Rosenfeld MG, Glass CK. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-γ. Nature. 2005;437:759–763. doi: 10.1038/nature03988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoon HG, Chan DW, Huang ZQ, Li J, Fondell JD, Qin J, Wong J. Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. EMBO J. 2003;22:1336–1346. doi: 10.1093/emboj/cdg120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Underhill C, Qutob MS, Yee SP, Torchia J. A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J Biol Chem. 2000;275:40463–40470. doi: 10.1074/jbc.M007864200. [DOI] [PubMed] [Google Scholar]
- 50.Zhang J, Kalkum M, Chait BT, Roeder RG. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol Cell. 2002;9:611–623. doi: 10.1016/s1097-2765(02)00468-9. [DOI] [PubMed] [Google Scholar]
- 51.Zhang D, Yoon HG, Wong J. JMJD2A is a novel N-CoR-interacting protein and is involved in repression of the human transcription factor achaete scute-like homologue 2 (ASCL2/Hash2) Mol Cell Biol. 2005;25:6404–6414. doi: 10.1128/MCB.25.15.6404-6414.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hong SH, Wong CW, Privalsky ML. Signaling by tyrosine kinases negatively regulates the interaction between transcription factors and SMRT (silencing mediator of retinoic acid and thyroid hormone receptor) corepressor. Mol Endocrinol. 1998;12:1161–1171. doi: 10.1210/mend.12.8.0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hong SH, Privalsky ML. The SMRT corepressor is regulated by a MEK-1 kinase pathway: inhibition of corepressor function is associated with SMRT phosphorylation and nuclear export. Mol Cell Biol. 2000;20:6612–6625. doi: 10.1128/mcb.20.17.6612-6625.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jonas BA, Privalsky ML. SMRT and N-CoR corepressors are regulated by distinct kinase signaling pathways. J Biol Chem. 2004;279:54676–54686. doi: 10.1074/jbc.M410128200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hong SH, Yang Z, Privalsky ML. Arsenic trioxide is a potent inhibitor of the interaction of SMRT corepressor with its transcription factor partners, including the PML-RAR α oncoprotein found in human acute promyelocytic leukemia. Mol Cell Biol. 2001;21:7172–7182. doi: 10.1128/MCB.21.21.7172-7182.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hermanson O, Jepsen K, Rosenfeld MG. N-CoR controls differentiation of neural stem cells into astrocytes. Nature. 2002;419:934–939. doi: 10.1038/nature01156. [DOI] [PubMed] [Google Scholar]
- 57.Goodson ML, Jonas BA, Privalsky ML. Alternative mRNA splicing of SMRT creates functional diversity by generating corepressor isoforms with different affinities for different nuclear receptors. J Biol Chem. 2005;280:7493–7503. doi: 10.1074/jbc.M411514200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goodson M, Jonas BA, Privalsky MA. Corepressors: custom tailoring and alterations while you wait. Nucl Recept Signal. 2005;3:e003. doi: 10.1621/nrs.03003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Malartre M, Short S, Sharpe C. Alternative splicing generates multiple SMRT transcripts encoding conserved repressor domains linked to variable transcription factor interaction domains. Nucleic Acids Res. 2004;32:4676–4686. doi: 10.1093/nar/gkh786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Malartre M, Short S, Sharpe C. Xenopus embryos lacking specific isoforms of the corepressor SMRT develop abnormal heads. Dev Biol. 2006;292:333–343. doi: 10.1016/j.ydbio.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 61.Short S, Malartre M, Sharpe C. SMRT has tissue-specific isoform profiles that include a form containing one CoRNR box. Biochem Biophys Res Commun. 2005;334:845–852. doi: 10.1016/j.bbrc.2005.06.175. [DOI] [PubMed] [Google Scholar]
- 62.Bailey PJ, Dowhan DH, Franke K, Burke LJ, Downes M, Muscat GE. Transcriptional repression by COUP-TF II is dependent on the C-terminal domain and involves the N-CoR variant, RIP13Δ1. J Steroid Biochem Mol Biol. 1997;63:165–174. doi: 10.1016/s0960-0760(97)00079-4. [DOI] [PubMed] [Google Scholar]
- 63.Dwivedi PP, Muscat GE, Bailey PJ, Omdahl JL, May BK. Repression of basal transcription by vitamin D receptor: evidence for interaction of unliganded vitamin D receptor with two receptor interaction domains in RIP13Δ1. J Mol Endocrinol. 1998;20:327–335. doi: 10.1677/jme.0.0200327. [DOI] [PubMed] [Google Scholar]
- 64.Wong CW, Privalsky ML. Transcriptional silencing is defined by isoform- and heterodimer-specific interactions between nuclear hormone receptors and corepressors. Mol Cell Biol. 1998;18:5724–5733. doi: 10.1128/mcb.18.10.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Masuda K, Shima H, Katagiri C, Kikuchi K. Activation of ERK induces phosphorylation of MAPK phosphatase-7, a JNK specific phosphatase, at Ser-446. J Biol Chem. 2003;278:32448–32456. doi: 10.1074/jbc.M213254200. [DOI] [PubMed] [Google Scholar]
- 67.Hu X, Li Y, Lazar MA. Determinants of CoRNR-dependent repression complex assembly on nuclear hormone receptors. Mol Cell Biol. 2001;21:1747–1758. doi: 10.1128/MCB.21.5.1747-1758.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Makowski A, Brzostek S, Cohen RN, Hollenberg AN. Determination of nuclear receptor corepressor interactions with the thyroid hormone receptor. Mol Endocrinol. 2003;17:273–286. doi: 10.1210/me.2002-0310. [DOI] [PubMed] [Google Scholar]
- 69.Zamir I, Zhang J, Lazar MA. Stoichiometric and steric principles governing repression by nuclear hormone receptors. Genes Dev. 1997;11:835–846. doi: 10.1101/gad.11.7.835. [DOI] [PubMed] [Google Scholar]
- 70.Baek SH, Ohgi KA, Rose DW, Koo EH, Glass CK, Rosenfeld MG. Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-κB and β-amyloid precursor protein. Cell. 2002;110:55–67. doi: 10.1016/s0092-8674(02)00809-7. [DOI] [PubMed] [Google Scholar]
- 71.Hoberg JE, Yeung F, Mayo MW. SMRT derepression by the IκB kinase α: a prerequisite to NF-κB transcription and survival. Mol Cell. 2004;16:245–255. doi: 10.1016/j.molcel.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 72.Lavinsky RM, Jepsen K, Heinzel T, Torchia J, Mullen TM, Schiff R, Del-Rio AL, Ricote M, Ngo S, Gemsch J, Hilsen-beck SG, Osborne CK, Glass CK, Rosenfeld MG, Rose DW. Diverse signaling pathways modulate nuclear receptor recruitment of N-CoR and SMRT complexes. Proc Natl Acad Sci USA. 1998;95:2920–2925. doi: 10.1073/pnas.95.6.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McKenzie GJ, Stevenson P, Ward G, Papadia S, Bading H, Chawla S, Privalsky M, Hardingham GE. Nuclear Ca2+ and CaM kinase IV specify hormonal- and Notch-responsiveness. J Neurochem. 2005;93:171–185. doi: 10.1111/j.1471-4159.2005.03010.x. [DOI] [PubMed] [Google Scholar]
- 74.Ogawa S, Lozach J, Jepsen K, Sawka-Verhelle D, Perissi V, Sasik R, Rose DW, Johnson RS, Rosenfeld MG, Glass CK. A nuclear receptor corepressor transcriptional checkpoint controlling activator protein 1-dependent gene networks required for macrophage activation. Proc Natl Acad Sci USA. 2004;101:14461–14466. doi: 10.1073/pnas.0405786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wagner BL, Norris JD, Knotts TA, Weigel NL, McDonnell DP. The nuclear corepressors NCoR and SMRT are key regulators of both ligand- and 8-bromo-cyclic AMP-dependent transcriptional activity of the human progesterone receptor. Mol Cell Biol. 1998;18:1369–1378. doi: 10.1128/mcb.18.3.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou Y, Gross W, Hong SH, Privalsky ML. The SMRT corepressor is a target of phosphorylation by protein kinase CK2 (casein kinase II) Mol Cell Biochem. 2001;220:1–13. doi: 10.1023/a:1011087910699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhu P, Baek SH, Bourk EM, Ohgi KA, Garcia-Bassets I, Sanjo H, Akira S, Kotol PF, Glass CK, Rosenfeld MG, Rose DW. Macrophage/cancer cell interactions mediate hormone resistance by a nuclear receptor derepression pathway. Cell. 2006;124:615–629. doi: 10.1016/j.cell.2005.12.032. [DOI] [PubMed] [Google Scholar]
- 78.Hadzic E, Desaiyajnik V, Helmer E, Guo S, Wu SJ, Koudinova N, Casanova J, Raaka BM, Samuels HH. A 10-amino-acid sequence in the N-terminal A/B domain of thyroid hormone receptor α is essential for transcriptional activation and interaction with the general transcription factor TFIIB. Mol Cell Biol. 1995;15:4507–4517. doi: 10.1128/mcb.15.8.4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hong SH, David G, Wong CW, Dejean A, Privalsky ML. SMRT corepressor interacts with PLZF and with the PML-retinoic acid receptor α (RARα) and PLZF-RARα oncoproteins associated with acute promyelocytic leukemia. Proc Natl Acad Sci USA. 1997;94:9028–9033. doi: 10.1073/pnas.94.17.9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wong CW, Privalsky ML. Transcriptional repression by the SMRT-mSin3 corepressor: multiple interactions, multiple mechanisms, and a potential role for TFIIB. Mol Cell Biol. 1998;18:5500–5510. doi: 10.1128/mcb.18.9.5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hauksdottir H, Farboud B, Privalsky ML. Retinoic acid receptors β and γ do not repress, but instead activate target gene transcription in both the absence and presence of hormone ligand. Mol Endocrinol. 2003;17:373–385. doi: 10.1210/me.2002-0340. [DOI] [PubMed] [Google Scholar]
- 82.Chen HW, Privalsky ML. The erbA oncogene represses the actions of both retinoid X and retinoid A receptors but does so by distinct mechanisms. Mol Cell Biol. 1993;13:5970–5980. doi: 10.1128/mcb.13.10.5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Emrick MA, Hoofnagle AN, Miller AS, Ten Eyck LF, Ahn NG. Constitutive activation of extracellular signal-regulated kinase 2 by synergistic point mutations. J Biol Chem. 2001;276:46469–46479. doi: 10.1074/jbc.M107708200. [DOI] [PubMed] [Google Scholar]
- 84.Hayakawa F, Privalsky ML. Phosphorylation of PML by mitogen-activated protein kinases plays a key role in arsenic trioxide-mediated apoptosis. Cancer Cell. 2004;5:389–401. doi: 10.1016/s1535-6108(04)00082-0. [DOI] [PubMed] [Google Scholar]