Abstract
Numerous studies have demonstrated that β1- and β2-adrenergic receptor gene (ADRB1 and ADRB2) variants influence cardiovascular risk and β-blocker responses in hypertension and heart failure. We evaluated the relationship between ADRB1 and ADRB2 haplotypes, cardiovascular risk (death, nonfatal myocardial infarction (MI ), and nonfatal stroke), and atenolol-based vs. verapamil sustained-release (SR )-based antihypertensive therapy in 5,895 coronary artery disease (CAD) patients. After an average of 2.8 years, death rates were higher in patients carrying the ADRB1 Ser49-Arg389 haplotype (hazard ratio (HR ) 3.66, 95% confidence interval (95% CI) 1.68–7.99). This mortality risk was significant in patients randomly assigned to verapamil SR (HR 8.58, 95% CI 2.06–35.8) but not atenolol (HR 2.31, 95% CI 0.82–6.55), suggesting a protective role for the β-blocker. ADRB2 haplotype associations were divergent within the treatment groups but did not remain significant after adjustment for multiple comparisons. ADRB1 haplotype variation is associated with mortality risk, and β-blockers may be preferred in subgroups of patients defined by ADRB1 or ADRB2 polymorphisms.
Cardiovascular disease is the leading cause of morbidity and mortality in the United States.1 Evidence is accumulating that genetic polymorphisms may be predictive of cardiovascular risk.2,3 Moreover, several studies have identified genetic factors that influence blood pressure and metabolic responses to β-blockers, thiazide diuretics, and renin–angiotensin system antagonists.4 Whether such pharmacogenetic differences translate to differences in the clinical outcome of antihypertensive therapy is less clear, particularly when patients receive multiple drugs that are titrated to a target blood pressure.5 A pharmacogenetic approach to treating hypertension could not only reduce the number and cost of medications but also reduce morbidity and mortality if the outcome of drug treatment differs by genotype.
Single-nucleotide polymorphisms (SNPs) in the genes encoding the adrenergic receptors have functional and physiological consequences.6 β1-adrenergic receptors are important in regulating heart rate and contractility as well as renin release in the kidney. Nonsynonymous SNPs in the β1-adrenergic receptor gene (ADRB1; Ser49Gly and Arg389Gly) result in differences in agonist-mediated downregulation and signaling activity.7 Similarly, β2-adrenergic receptors play an important role in cardiac function, metabolism, and vascular tone. Nonsynonymous SNPs in the β2-adrenergic receptor gene (ADRB2; Arg16Gly and Gln27Glu) affect expression, agonist responsiveness, and desensitization.8,9 Previous epidemiologic studies have suggested that ADRB1 and ADRB2 SNPs correlate with a variety of intermediate cardiovascular phenotypes,10 cardiovascular risk,11,12 and β-blocker responses in hypertension13,14 and heart failure.15,16
Taken together, genetically determined differences in β-adrenergic receptor function may influence cardiovascular risk as well as the degree of risk reduction conferred by drugs that inhibit adrenergic activity. Therefore, we investigated the influence of ADRB1 and ADRB2 haplotype variation on the incidence of death, nonfatal myocardial infarction (MI), and nonfatal stroke as well as the pharmacogenetics of β-blocker– and calcium channel blocker–based antihypertensive therapy in the INternational VErapamil SR/Trandolapril STudy— GENEtic Substudy (INVEST-GENES). Based on the literature, we hypothesized that patients with the ADRB1 Ser49-Arg389 haplotype or the ADRB2 haplotype containing the Arg16 and Gln27 alleles would be at relatively higher risk for cardiovascular events11,12 and that atenolol would be beneficial as compared with sustained-release verapamil (verapamil SR) in patients with the ADRB1 Ser49-Arg389 haplotype.
RESULTS
Study population and baseline characteristics
INVEST-GENES consisted of an ethnically diverse, elderly population of patients with hypertension and documented coronary artery disease (CAD), including a large proportion of women and individuals with diabetes. Patients were randomly assigned to receive atenolol or verapamil SR. Trandolapril and hydrochlorothiazide were added as needed to control blood pressure. Demographic and clinical characteristics did not differ significantly by treatment strategy (Table 1). Haplotypes for at least one gene were inferred for a total of 5,895 patients, including 5,817 who were successfully genotyped at both ADRB1 loci (Ser49Gly and Arg389Gly) and 5,877 who were successfully genotyped at two of three ADRB2 loci (Gly16Arg, Gln27Glu, and 523C>A). Genotype concordance was >97% at all loci. All loci were in Hardy–Weinberg equilibrium except ADRB2 Gly16Arg in Hispanic patients (χ2 = 6.12, P < 0.05). The minor deviation from Hardy–Weinberg equilibrium in Hispanics appeared to be due to admixture, as this was the most admixed population in our sample. In Hispanics with >80% European ancestry, there was no evidence of deviation from Hardy–Weinberg equilibrium (P = 0.47), which was selected as the cutoff because the black and white subjects in this sample have, on average, >80% ancestry from a single continental population. Low to moderate levels of linkage disequilibrium were noted between the SNPs in ADRB1 (r2 = 0.04–0.19) and ADRB2 (r2 = 0.07–0.45). The two variant loci in ADRB1 formed three common haplotypes, and the three variant loci in ADRB2 also formed three common haplotypes (Table 2). The genotype and haplotype distributions differed significantly by race/ ethnicity (Table 2).
Table 1.
Baseline characteristics
| Atenolol strategy (n = 2,973) |
Verapamil SR strategy (n = 2,922) |
|
|---|---|---|
| Demographic | ||
| Age—mean (SD) (years) | 66 (9.7) | 66 (9.6) |
| Age > 70 | 1,010 (34.0) | 971 (33.4) |
| Female, no. (%) | 1,660 (55.8) | 1,637 (56.0) |
| Race/ethnicity, no. (%) | ||
| White | 1,188 (40.0) | 1,221 (41.8) |
| Hispanic | 1,438 (48.4) | 1,363 (46.7) |
| Black | 347 (11.7) | 338 (11.6) |
| BMI—mean (SD) (kg/m2) | 29.5 (5.6) | 29.3 (5.6) |
| Medical history, no. (%) | ||
| History of MI | 673 (22.6) | 689 (23.6) |
| Heart failure (class I–III) | 106 (3.6) | 95 (3.3) |
| Chronic stable angina | 2,219 (74.6) | 2,168 (74.2) |
| Unstable angina | 277 (9.3) | 293 (10.1) |
| Dyslipidemiaa | 1,779 (59.8) | 1,743 (59.7) |
| LVH | 463 (15.6) | 423 (14.5) |
| Arrhythmia | 191 (6.4) | 211 (7.2) |
| Stroke or TIAb | 183 (6.2) | 228 (7.8) |
| PVD | 327 (11.0) | 328 (11.2) |
| Renal insufficiencyc | 38 (1.3) | 54 (1.9) |
| Diabetesa | 851 (28.6) | 806 (27.6) |
| Obese | 1,226 (41.2) | 1,158 (39.6) |
| Cancer | 108 (3.6) | 131 (4.5) |
| Ever-smoker | 1,220 (41.0) | 1,213 (41.5) |
| Medications, no. (%) | ||
| Aspirin/antiplatelet | 1,355 (45.6) | 1,339 (45.8) |
| Other NSAIDs | 718 (24.2) | 682 (23.3) |
| Antidiabetic | 736 (24.7) | 661 (22.6) |
| Lipid lowering | 1,049 (35.3) | 1,072 (36.7) |
| Nitrates | 848 (28.5) | 812 (27.8) |
| Potassium | 172 (5.8) | 175 (6.0) |
| HRT | 393 (13.2) | 366 (12.5) |
| Blood pressure | ||
| Systolic—mean (SD) (mm Hg) | 148 (18) | 149 (19) |
| Diastolic—mean (SD) (mm Hg) | 86 (11) | 85 (11) |
| Controlledd—no. (%) | 718 (24.1) | 780 (26.7) |
BMI, body mass index; HRT, hormone replacement therapy; LVH, left ventricular hypertrophy; NSAIDs, nonsteroidal anti-inflammatory drugs; PVD, peripheral vascular disease; TIA, transient ischemic attack.
History of and/or currently taking lipid-lowering or antidiabetic medications.
P < 0.05 for verapamil sustained-release (SR) vs. atenolol.
History of or currently have elevated serum creatinine but less than 4 mg/dl.
Blood pressure control defined as ≤140/90 or ≤130/80 in patients with diabetes or a history of renal insufficiency.
Table 2.
Minor allele and haplotype frequencies by race/ethnicity
| All | White | Hispanic | Black | |
|---|---|---|---|---|
| ADRB1 (%)a | n = 5,817 | n = 2,375 | n = 2,766 | n = 676 |
| Gly49 | 17.8 | 12.2 | 20.8 | 23.2 |
| Gly389 | 29.0 | 27.2 | 28.4 | 39.1 |
| Ser49-Arg389 | 53.4 | 60.8 | 51.0 | 37.6 |
| Ser49-Gly389 | 28.9 | 27.0 | 28.3 | 39.2 |
| Gly49-Arg389 | 17.6 | 12.1 | 20.6 | 23.2 |
| ADRB2 (%)b | n = 5,877 | n = 2,400 | n = 2,795 | n = 682 |
| Arg16 | 43.4 | 38.7 | 45.3 | 51.0 |
| Glu27 | 32.2 | 41.8 | 28.7 | 16.3 |
| 523A | 25.2 | 19.3 | 27.4 | 34.8 |
| Arg16-Gln27-523C | 41.1 | 35.9 | 43.3 | 48.2 |
| Gly16-Glu27-523C | 29.7 | 39.9 | 25.7 | 14.3 |
| Gly16-Gln27-523A | 22.2 | 16.8 | 23.8 | 32.4 |
| Other | 7.0 | 7.4 | 7.2 | 5.1 |
Gly49-Gly389 haplotype observed in 0.1%.
Rare haplotypes accounted for 6.2% of haplotype diversity.
The average follow-up period for the primary outcome was 2.8 ± 0.7 years. The treatment strategies did not differ in terms of the primary outcome (defined as the first occurrence of death, nonfatal MI, or nonfatal stroke; hazard ratio (HR) 0.81, 95% confidence interval (95% CI) 0.63–1.05, P = 0.12). The median doses of atenolol and verapamil SR at the end of the study were 100 and 240 mg/day, respectively. Hydrochlorothiazide was used in 74% of atenolol-treated patients and 61% of verapamil SR–treated patients, whereas trandolapril was used in 70% of atenolol-treated patients and 78% of verapamil SR–treated patients. Mean on-treatment systolic blood pressure values were not significantly different across treatment strategies and haplotype groups (data not shown). Mean on-treatment diastolic blood pressure was statistically lower in the verapamil SR strategy group (77.7 mm Hg vs. 78.2 mm Hg, P = 0.003), patients with the ADRB1 Ser49-Arg389 haplotype (0 copies, 78.4 mm Hg; 1 copy, 77.9 mm Hg; 2 copies, 77.6 mm Hg; P = 0.003), and patients with the ADRB2 Gly16-Glu27-523C haplotype (0 copies, 78.4 mm Hg; 1 copy, 77.8 mm Hg; 2 copies, 76.6 mm Hg; P < 0.0001). In all cases the differences were <2 mm Hg and therefore not likely to be clinically important. Mean on-treatment heart rate did not differ according to ADRB1 haplotype and was statistically, but not clinically, lower in patients with the ADRB2 Gly16-Glu27-523C haplotype (0 copies, 72.2 beats/min; 1 copy, 71.8 beats/min; 2 copies, 71.7 beats/min; P = 0.005).
Associations of ADRB1 with primary and secondary outcomes
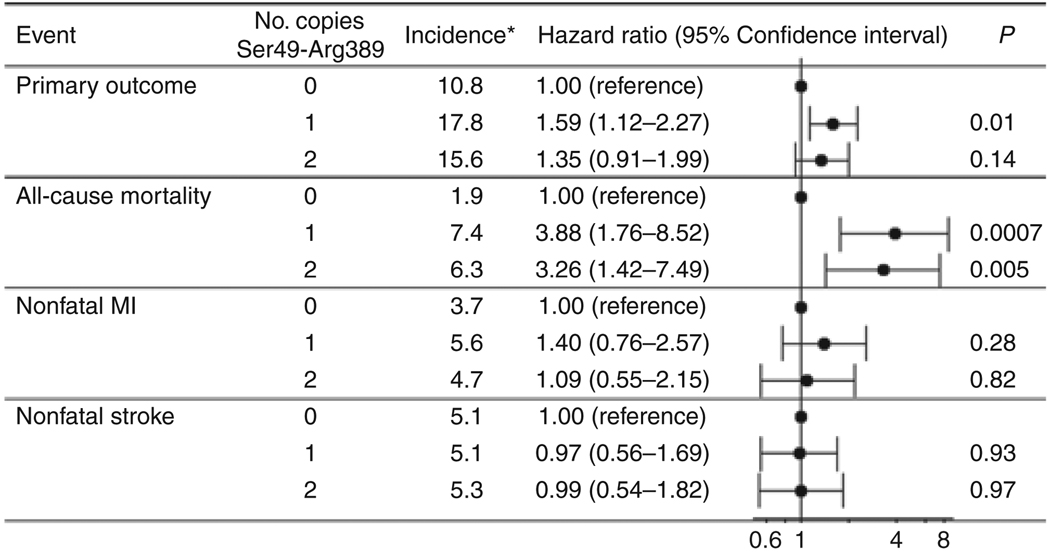
The model based on the Ser49-Arg389 haplotype best characterized the risk for the primary outcome (log-rank P = 0.02), whereas the other two common haplotypes, Ser49-Gly389 and Gly49-Arg389, were not associated with the primary outcome (Supplementary Table S1 online). The haplotype risk was similar between patients with one or two copies of the Ser49-Arg389 (Figure 1), and these patients had a relatively higher risk for the primary outcome relative to non-carriers (Ser49-Arg389 carriers vs. noncarriers, HR 1.51, 95% CI 1.07–2.12, P = 0.02). This association was driven entirely by mortality (Ser49-Arg389 carriers vs. noncarriers, HR 3.66, 95% CI 1.68–7.99, P = 0.001), whereas no association was noted for nonfatal MI or nonfatal stroke (Figure 1). The cause of death was adjudicated, although the exact cause of noncardiovascular death was not specified. The Ser49-Arg389 haplotype was significantly associated with both cardiovascular and noncardiovascular mortality (data not shown). The increased risk of death among carriers of the Ser49-Arg389 haplotype was consistent across racial/ethnic groups and after adjustment for ancestry (Ser49-Arg389 carriers vs. noncarriers: whites, HR 2.32, 95% CI 0.84–6.44, P = 0.1; Hispanics, HR 2.95, 95% CI 0.89–9.78, P = 0.08; blacks, HR not calculated because all events occurred in carriers). Given the similarity in effect across racial/ethnic groups and the lack of apparent population stratification, all subsequent analyses are presented for the combined population to maintain power, controlling for race/ethnicity. In genotype-based analyses, the mortality association was detectable for Arg389Gly under a recessive model (P = 0.03); the data suggest that ADRB1 haplotype is more informative than the individual SNPs.
Figure 1.
Associations of the ADRB1 Ser49-Arg389 haplotype with primary and secondary outcomes. Hazard ratios are based on reduced model adjusted for age, sex, and race/ethnicity. *Crude incidence per 1,000 patient years. MI, myocardial infarction.
ADRB2 associations with primary outcomes
None of the ADRB2 haplotype models revealed differential risk for the primary outcome in the overall population, with the lowest P value being 0.39 (Supplementary Table S1 online). Given the lack of statistical trends for a main effect for the primary outcome, secondary outcomes were not tested.
Pharmacogenetic associations with ADRB1 and ADRB2
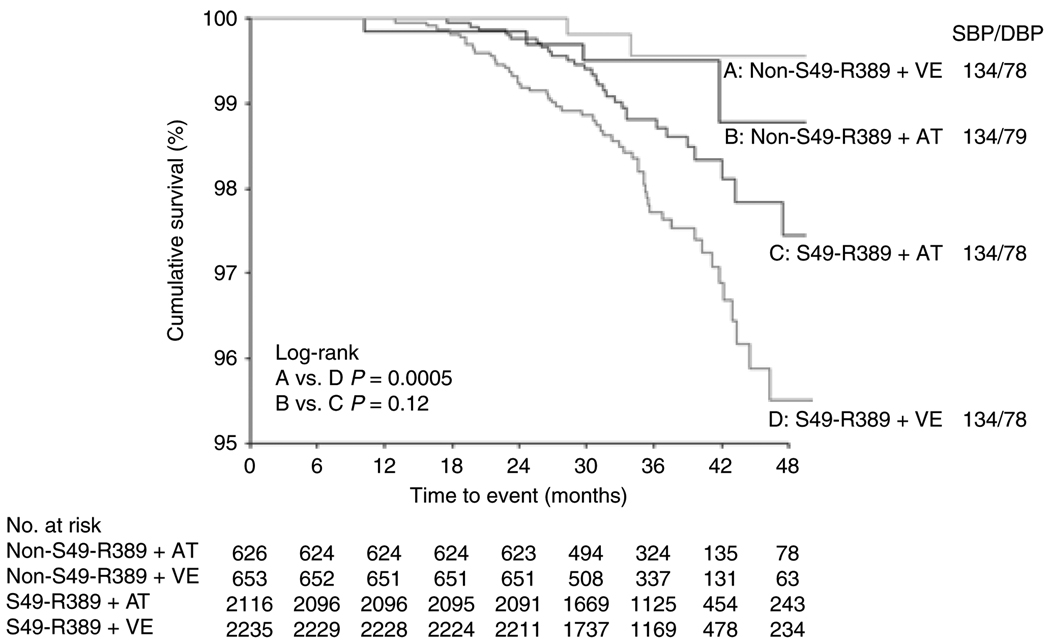
Because genetic associations for ADRB1 with the primary outcome were driven by differences in death, pharmacogenetic analyses focused on this outcome. The increase in mortality risk among patients with one or two copies of the Ser49-Arg389 haplotype was significant in patients randomly assigned to verapamil SR (HR 8.58, 95% CI 2.06–35.8, P = 0.003) but not in patients assigned to atenolol (HR 2.31, 95% CI 0.82–6.55, P = 0.11). These data suggest that atenolol offsets the risk associated with the ADRB1 haplotype (Pinteraction = 0.19; Figure 2). The point estimates were minimally affected when adjusted for clinical covariates and secondary drug use (data not shown). Adjustment for ancestry did not change the magnitude or direction of any of these associations (data not shown). These associations remained significant even when using the Bonferroni-corrected P value. Blood pressure and heart rate values were similar across haplotype and drug use strata (Figure 2). The median atenolol and verapamil doses did not differ according to ADRB1 haplotype, although Ser49-Arg389 carriers were less likely than noncarriers to receive a second-line drug in either strategy (atenolol + hydrochlorothiazide 73% vs. 78%, P = 0.007; verapamil SR + trandolapril 69% vs. 75%, P = 0.006).
Figure 2.
All-cause mortality and mean on-treatment blood pressure by ADRB1 Ser49-Arg389 haplotype and atenolol/verapamil sustained-release (SR) therapy. HR, hazard ratio; 95% CI, 95% confidence interval; S49-R389, Ser49-Arg389 haplotype; AT, atenolol; VE, verapamil SR; SBP, systolic blood pressure; DBP, diastolic blood pressure.
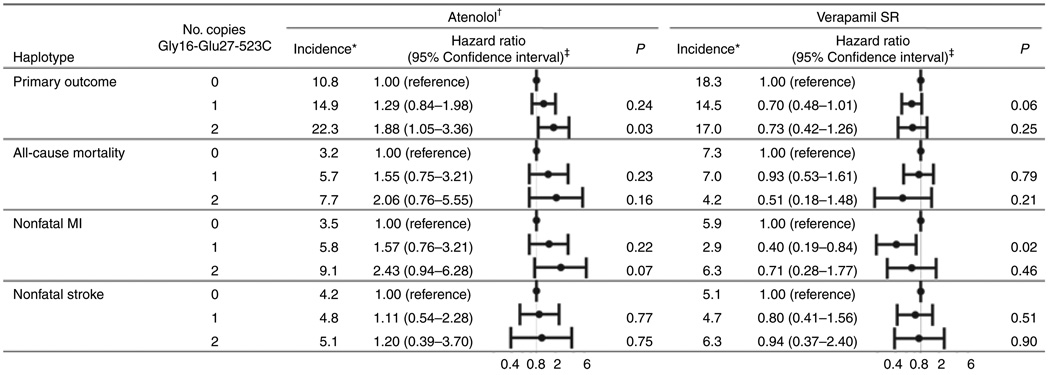
Pharmacogenetic analysis of common ADRB2 haplotypes revealed that the risk for the primary outcome differed significantly across the Gly16-Glu27-523C haplotype in verapamil SR–treated patients but not in atenolol-treated patients (Pinteraction = 0.05; Figure 3). None of the other haplotype models was associated with the primary outcome within the treatment strategies. The haplotype-associated risks were driven largely by mortality (Pinteraction = 0.11) and nonfatal MI (Pinteraction = 0.06; Figure 3). On the basis of the Bonferroni-corrected P value, none of the subgroup associations would be defined as significant, despite the significant P values for the interaction term. Genotype-based analysis revealed that this was driven largely by the 523C>A genotype, but similar trends that were consistent with the haplotype association were noted for the Gly16Arg and Gln27Glu SNPs (data not shown), again supporting the view that the haplotype analysis is the more powerful one. The median atenolol and verapamil SR doses did not differ by ADRB2 haplotype; nor did the rates of hydrochlorothiazide or trandolapril use (data not shown).
Figure 3.
Primary and secondary outcomes by ADRB2 Gly16-Glu27-523C haplotype and antihypertensive drug therapy. *Crude incidence per 1,000 patient years. †Includes only patients ever exposed to atenolol (94% in atenolol strategy). ‡Hazard ratios based on reduced model adjusted for age, sex, and race/ ethnicity. MI, myocardial infarction; SR, sustained release.
As an exploratory analysis, mortality was modeled for the randomized drugs on the basis of both ADRB1 and ADRB2 haplotype information. The analysis revealed that patients with at least one copy of the Ser49-Arg389 haplotype and zero copies of the Gly16-Glu27-523C haplotype (representing 42% of the study population) had better outcomes when treated with atenolol than with verapamil SR (HR 0.42, 95% CI 0.21–0.82, P = 0.01). Comparing this result to the HR of 0.64 when considering the ADRB1 gene alone suggests that a consideration of both genes may be even more informative for identifying those most likely to benefit from β-blocker therapy.
DISCUSSION
Amino-acid changing SNPs in the adrenergic receptors have previously been shown to be associated with cardiovascular and drug response phenotypes. In treated hypertensive patients with CAD, we identified a significant association between SNPs in ADRB1 and the incidence of all-cause death, showing that patients with haplotypes bearing the common alleles (Ser49 and Arg389) were at relatively higher risk. The risk associated with this haplotype was significantly reduced by β-blocker therapy, but not by calcium channel blocker therapy, a finding consistent with the understanding of the functional consequences of these genetic polymorphisms. Specifically, the Arg389 form of the receptor has been documented in several in vitro and ex vivo studies to be associated with increased coupling of the β1-adrenergic receptor to G protein, leading to greater adenylyl cyclase activation.17,18 The Ser49 form of the receptor has most consistently been associated with resistance to receptor downregulation.19,20 Therefore, the Ser49-Arg389 haplotype would be expected to be most responsive to activation by catecholamines, and consequently a greater response to β-blockade with this haplotype would also be expected.
Consistent with our primary hypothesis, patients with at least one copy of the haplotype containing the wild-type Ser49 and Arg389 alleles were at relatively higher risk for the primary outcome than patients with variant alleles. The main haplotype association was driven by more than a threefold difference in the rate of all-cause mortality.21 Our findings are in line with those of Iwai et al.,22 who reported an association between the Arg389 allele and MI, but other observational studies have not supported a major influence of ADRB1 variants on cardiovascular risk.12,23 The lack of consistency may be attributable to differences in patient populations studied (i.e., those with or without overt CAD)23 or prevalent use of β-blockers.12 Our data support the use of β-blockers as being a potential confounder for detecting the genetic association with outcomes, given that there were no differences in outcomes by genotype among the atenolol-treated patients. Overall, the results of the current investigation are in line with the known functionality of these variants and the widely recognized adverse consequences of chronic sympathetic activation.
The pharmacogenetic findings regarding the Ser49-Arg389 haplotype are consistent not only with the in vitro and ex vivo studies documenting the functional basis of these polymorphisms but also with the existing β-blocker pharmacogenetics literature. 24,25 This study extends those findings to include outcomes of antihypertensive therapy. In our investigation, patients carrying the Ser49-Arg389 haplotype derived a significant survival benefit from the use of a β-blocker. We previously demonstrated that hypertensive patients who were Ser49-Arg389 homozygotes experienced a significantly greater blood pressure response to β-blockers than those with haplotypes containing a variant allele.13 With variations on this theme, four other studies have corroborated the finding that patients with the Arg389 allele or Ser49-Arg389 haplotype show a greater blood pressure response to β-blockers.8,14,26,27 It follows that, in certain subpopulations, hypertension may have a strong adrenergic component that is particularly amenable to β-blocker therapy. Similarly, studies in heart failure patients have also suggested the Arg389 allele or Ser49-Arg389 haplotype is associated with the greatest improvement in ejection fraction after initiation of β-blocker therapy.15,28,29 Liggett et al. recently demonstrated that Arg389 homozygous heart failure patients derived a significant survival benefit from bucindolol as compared with placebo, whereas in Gly389 carriers the outcomes with bucindolol and placebo were similar.30
Taken together, these studies suggest that the ADRB1 gene is an important pharmacogenetic target for β-blocker response. The literature suggests that polymorphisms in the gene influence intermediate-response phenotypes (e.g., blood pressure reduction and ejection fraction improvement) along with mortality outcomes in hypertension and heart failure. The specific findings from this study suggest that β-blockers may be the preferred antihypertensive therapy in hypertensive CAD patients who are Ser49-Arg389 carriers.
It should be noted that carriers of the major alleles (Ser49 and Arg389) were at relatively increased risk and benefited from β-blocker therapy, and, conversely, those with a variant on both chromosomes were at lower risk. This is in contrast to the apparent inheritance patterns of some of the other β-blocker pharmacogenetics studies for ADRB1 described above, in which a dominant model was typically assumed, with Ser49-Arg389 homozygotes exhibiting the greatest β-blocker response and carriers of at least one variant allele/haplotype having a lesser response. However, very few studies have been sufficiently powered to test for mode of inheritance, given that variant homozygotes, typically, have been minimally represented. In these cases a dominant model (whereby variant carriers were often collapsed to a single group) was typically pursued out of statistical necessity. Thus, although the mode of inheritance of this association seems to be in contrast to some of the β-blocker pharmacogenetics literature, it can also be concluded that the literature has not revealed a clear pattern of inheritance for this gene and the resulting phenotypes. This is the largest ADRB1 pharmacogenetic study to date and, from this perspective, may have been the best powered to assess for mode of inheritance. Additionally, with complex phenotypes, it is possible that different phenotypes will exhibit different inheritance patterns.15,30
We also identified a statistical interaction between atenolol and verapamil SR and ADRB2 haplotypes. However, this did not meet our threshold for significance after adjusting for multiple comparisons. This association was driven primarily by divergent risks for both death and nonfatal MI, and none of the associations appeared to be driven by differences in blood pressure. If the findings for ADRB2 are validated, knowledge of this haplotype may further enhance the ability to identify patients who might benefit from β-blocker therapy.
Limitations
As a cohort study nested in a randomized trial with adjudicated end points, INVEST-GENES has several advantages over population-based studies, and the results are generalizable to other CAD populations managed with contemporary interventions. However, the current investigation also has limitations that deserve consideration. First, despite the large size of the study population, the event rate was low, and this study may have been underpowered to evaluate gene–drug interactions, particularly for the individual end points and within certain subgroups of patients (i.e., individual end point by drug by gene by race). Second, the INVEST-GENES population is racially/ ethnically diverse. To control for potential confounding by population stratification, we considered analyses separately by race and by inclusion of ancestry informative marker data. These various analyses suggest that our findings are not confounded by population stratification. Third, the use of trandolapril and hydrochlorothiazide was, by design, different in the two treatment strategies and may have influenced the results. However, the findings were similar in the expanded model that adjusted for exposure to these drugs. Last, replication in independent cohorts is desirable. In the absence of another study with randomized drug therapy and comparably rigorous follow-up and phenotype definition, we must rely on the existing evidence from the laboratory and endophenotype studies.31
Conclusion
Identifying genetic markers for cardiovascular risk has the potential to improve cardiovascular risk stratification and identify those requiring more aggressive management of hypertension and related chronic diseases. Common SNPs in the genes encoding the β1- and β2-adrenergic receptors alter receptor activity and have physiological consequences. Consistent with the known functionality of the β1-adrenergic receptor variants, we identified an association between ADRB1 haplotypes and the risk of death. More importantly, our data suggest that β-blockers offset this mortality risk, in keeping with observations that patients bearing the wild-type alleles are more responsive to β-blocker therapy in settings of blood pressure lowering, improvement in ejection fraction, and survival in heart failure. ADRB2 variants were similarly associated with treatment outcomes, but given the inconsistencies in the literature, these findings require independent replication. The pharmacogenetic evidence for β-blockers and adrenergic receptor genes is highly convincing, particularly for ADRB1, and our data suggest that a patient’s genotype could influence the antihypertensive drug choice independent of blood pressure responses.
METHODS
INVEST-GENES design and participants
INVEST was a prospective, randomized, open-label, blinded-end point (PROBE) trial designed to compare antihypertensive treatment outcomes in 22,576 patients. The INVEST-GENES cohort consisted of 5,979 patients from 184 sites in the United States and Puerto Rico who provided DNA samples and additional written informed consent for genetic studies. The details of the INVEST methods and main outcomes have been previously reported.32 Briefly, INVEST enrolled hypertensive patients over the age of 50 years with documented CAD. Patients were randomly assigned to a verapamil SR- or an atenolol-based treatment strategy. Trandolapril was recommended for all patients with heart failure, renal dysfunction, or diabetes. Hydrochlorothiazide and/or trandolapril were added as needed to achieve blood pressure targets defined in the sixth report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure. Details on the addition of study drugs and dose titration may be found in the original INVEST publication. 32 Patients were followed every 6 weeks for the first 6 months and every 6 months thereafter until 2 years after the last patient was enrolled. Blood pressure control and cardiovascular outcomes were similar between the treatment strategies in the main trial.32
Outcomes
The primary outcome was a composite of the first occurrence of all-cause mortality, nonfatal MI, or nonfatal stroke. Secondary outcomes included the individual components of the primary outcome. Events were adjudicated by an independent committee that was blinded to treatment strategy.
Genotyping
Buccal tissue samples were obtained by mouthwash, and genomic DNA was isolated using the Gentra Systems PureGene kit. Patients were genotyped for two variants in ADRB1 (Ser49Gly (145A>G), rs1801252; and Arg389Gly (1165C>G), rs1801253) and three variants in ADRB2 (Gly16Arg (46G>A), rs1042713; Gln27Glu (79A>G), rs1042714; and Arg175Arg (523C>A), rs1042718) using pyrosequencing (Biotage, Uppsala, Sweden) and TaqMan allelic discrimination (Applied Biosystems, Foster City, CA). Genotype accuracy was verified by genotyping 5–10% randomly selected duplicate samples for each SNP on the alternate platform. Ancestry informative markers (87 total) were genotyped using either allele-specific PCR with universal energy transfer labeled primers or competitive allele-specific PCR at Prevention Genetics (Marshfield, WI).
Statistics
Hardy–Weinberg equilibrium was tested for each racial/ethnic group using χ2 analysis. Haplotypes were reconstructed separately for each racial/ethnic group from raw genotype data using PHASE software (version 2.1)33 for patients who were successfully genotyped at both ADRB1 loci and at least two ADRB2 loci. Each haplotype was coded according to the number of copies (zero, one, or two). Linkage disequilibrium (r2) between the SNPs in each racial/ethnic group was estimated using Haploview.34 Demographic, baseline clinical characteristics, and mean on-treatment (from 6 months to the last follow-up) blood pressure levels and heart rates were compared by haplotype to identify potentially confounding factors, using χ2 tests for categorical data and t-tests, analysis of variance, or a nonparametric equivalent for continuous data.
Given that INVEST-GENES was a cohort of patients from a randomized clinical trial, that the candidate genes encode the drug targets, and that the hypotheses are pharmacogenetic in nature (in an effort to validate published associations), we screened for primary outcome associations both in the overall population and within the randomized treatment arms. The main effects of each haplotype were first evaluated in the intention-to-treat population for the primary outcome based on haplotype copy number (zero, one, or two) using Kaplan–Meier analysis with pooled log-rank tests. Cox proportional hazards regression was performed to estimate HR and 95% CI for each copy of the haplotype relative to zero copies of the haplotype. The regression model was initially adjusted for race/ethnicity, age, sex, and treatment strategy (reduced model). The following covariates were subsequently entered into the model using the stepwise procedure if P < 0.1 and retained if P < 0.05 (expanded model): history of heart failure, MI, diabetes, stroke or transient ischemic attack, renal insufficiency, dyslipidemia, left ventricular hypertrophy, peripheral vascular disease, stable angina, unstable angina, arrhythmia, cancer, or ever having smoked; body mass index; and baseline systolic and diastolic blood pressure values. The reduced model is presented unless the point estimates differ from the expanded model by more than 0.1. The threshold for significance in the screening analysis was set at P < 0.05 based on the a priori genetic and pharmacogenetic hypotheses that follow the existing clinical and experimental data in the literature for both genes and the studied polymorphisms. The proportionality of haplotype effects was evaluated by examining Schoenfeld residuals.
To reduce multiple comparisons, only significant haplotype associations with the primary outcome were followed by analysis of the secondary end points and pharmacogenetic relationships. To account for comparisons incurred in the stepwise analysis approach, the final significant P value for log-rank tests was defined as P ≤ 0.003, which was arrived at on the basis of a Bonferroni correction for 15 comparisons as follows: ADRB1: one gene test for primary outcome (one in overall population), one haplotype tested for three secondary outcomes, one haplotype tested for one secondary outcome in two treatment strategies (six tests for ADRB1 in total), and ADRB2: three gene tests for primary outcome (one in overall population and two within treatment strategies), one haplotype tested for three secondary outcomes in two treatment strategies (nine tests for ADRB2 in total). This correction might be viewed as being overly conservative, given that an assumption of the Bonferroni correction is independence of all the tests, and these tests were not all independent. Additionally, the most appropriate inheritance model (i.e., additive, dominant, or recessive) was selected on the basis of visual inspection of per-allele point estimates in an effort to maintain power in these analyses. Genotype-based analyses were considered exploratory.
Verapamil SR and atenolol were started at baseline in 100 and 94% of patients in the respective strategies. Consequently, pharmacogenetic analyses examined haplotype risks in the intention-to-treat population for patients in whom verapamil SR or atenolol was ever prescribed. Associations were evaluated by Kaplan–Meier analysis with pairwise log-rank tests and by Cox proportional hazard regression stratified by exposure or with haplotype–drug interaction terms. Given the differences in time to initiation and the overlapping use of trandolopril and hydrochlorothiazide, the expanded model for pharmacogenetic analyses also adjusted for average trandolapril and hydrochlorothiazide doses as time-varying covariates.
To control for the potential of population stratification in our racially and ethnically diverse population, we used a total panel of 87 ancestry informative markers, selected to show large allele-frequency differences across three parental populations (West Africans, Native Americans, and Europeans) from a large panel of more than 10,000 SNPs.35 The maximum-likelihood criterion was then used to estimate each patient’s individual genomic ancestry proportions on these three axes, and these terms were included in the statistical models for patients with data for at least 50% of the ancestry informative markers. All analyses were performed with and without stratification by race/ethnicity because of reduced power in the racial/ethnic subgroup analysis. Data are presented for the combined population unless the results differed across racial/ ethnic strata.
All statistical analyses were performed using SAS version 9.1 (SAS Institute, Cary, NC).
Supplementary Material
is linked to the online version of the paper at http://www.nature.com/cpt
ACKNOWLEDGMENTS
We thank Lynda Stauffer and Kathy Eberst (posthumously) for processing and genotyping the samples, Robert Kolb for coordinating INVEST sites for INVEST-GENES participation, and all the INVEST patients who also agreed to participate in the genetic substudy. The phenotype and genotype data are available at http://www.PharmGKB.org; PS203971, PS205525, PS205530, PS203972, PS203974, PS205526, PS205527, PS205529, PS206277, PS207726, PS207727, PS207729, PS207730, PS207731, PS207732, PS207733, PS207734, and PS207735. This project was funded by National Institutes of Health grants HL074730, HL69758, and RR017568; a grant from Abbott Pharmaceuticals; and American Heart Association Postdoctoral Fellowship 0625619B. Clinical trials registration: http://clinicaltrials.gov, identifier NCT00133692.
Footnotes
Conflict of interest
Drs Johnson, Pepine, Cooper-DeHoff, and Langaee received grant funding from Abbott Laboratories. Drs Cooper-DeHoff and Pepine along with the University of Florida hold U.S. patent 5,991,731 related to INVEST. Dr Pepine has been a consultant for Abbott Laboratories. The other authors declared no conflict of interest.
References
- 1.Rosamond W, et al. Heart disease and stroke statistics—2007 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2006;115:e69–e171. doi: 10.1161/CIRCULATIONAHA.106.179918. [DOI] [PubMed] [Google Scholar]
- 2.Ginsburg GS, Donahue MP, Newby LK. Prospects for personalized cardiovascular medicine: the impact of genomics. J. Am. Coll. Cardiol. 2005;46:1615–1627. doi: 10.1016/j.jacc.2005.06.075. [DOI] [PubMed] [Google Scholar]
- 3.Samani NJ, et al. Genomewide association analysis of coronary artery disease. N. Engl. J. Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson JA, Turner ST. Hypertension pharmacogenomics: current status and future directions. Curr. Opin. Mol. Ther. 2005;7:218–225. [PubMed] [Google Scholar]
- 5.Davis BR, et al. Antihypertensive therapy, the alpha-adducin polymorphism, and cardiovascular disease in high-risk hypertensive persons: the Genetics of Hypertension-Associated Treatment Study. Pharmacogenomics J. 2007;7:112–122. doi: 10.1038/sj.tpj.6500395. [DOI] [PubMed] [Google Scholar]
- 6.Kirstein SL, Insel PA. Autonomic nervous system pharmacogenomics: a progress report. Pharmacol. Rev. 2004;56:31–52. doi: 10.1124/pr.56.1.2. [DOI] [PubMed] [Google Scholar]
- 7.Sandilands A, Yeo G, Brown MJ, O’Shaughnessy KM. Functional responses of human beta1 adrenoceptors with defined haplotypes for the common 389R>G and 49S>G polymorphisms. Pharmacogenetics. 2004;14:343–349. doi: 10.1097/00008571-200406000-00003. [DOI] [PubMed] [Google Scholar]
- 8.Dishy V, et al. The effect of common polymorphisms of the beta2-adrenergic receptor on agonist-mediated vascular desensitization. N. Engl. J. Med. 2001;345:1030–1035. doi: 10.1056/NEJMoa010819. [DOI] [PubMed] [Google Scholar]
- 9.Drysdale CM, et al. Complex promoter and coding region beta 2-adrenergic receptor haplotypes alter receptor expression and predict in vivo responsiveness. Proc. Natl. Acad. Sci. USA. 2000;97:10483–10488. doi: 10.1073/pnas.97.19.10483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bengtsson K, et al. Polymorphism in the beta(1)-adrenergic receptor gene and hypertension. Circulation. 2001;104:187–190. doi: 10.1161/01.cir.104.2.187. [DOI] [PubMed] [Google Scholar]
- 11.Heckbert SR, et al. Beta2-adrenergic receptor polymorphisms and risk of incident cardiovascular events in the elderly. Circulation. 2003;107:2021–2024. doi: 10.1161/01.CIR.0000065231.07729.92. [DOI] [PubMed] [Google Scholar]
- 12.Lanfear DE, Jones PG, Marsh S, Cresci S, McLeod HL, Spertus JA. Beta2-adrenergic receptor genotype and survival among patients receiving beta-blocker therapy after an acute coronary syndrome. JAMA. 2005;294:1526–1533. doi: 10.1001/jama.294.12.1526. [DOI] [PubMed] [Google Scholar]
- 13.Johnson JA, Zineh I, Puckett BJ, McGorray SP, Yarandi HN, Pauly DF. Beta 1-adrenergic receptor polymorphisms and antihypertensive response to metoprolol. Clin. Pharmacol. Ther. 2003;74:44–52. doi: 10.1016/S0009-9236(03)00068-7. [DOI] [PubMed] [Google Scholar]
- 14.Liu J, et al. Beta1-Adrenergic receptor polymorphisms influence the response to metoprolol monotherapy in patients with essential hypertension. Clin. Pharmacol. Ther. 2006;80:23–32. doi: 10.1016/j.clpt.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 15.Mialet Perez J, et al. Beta 1-adrenergic receptor polymorphisms confer differential function and predisposition to heart failure. Nat. Med. 2003;9:1300–1305. doi: 10.1038/nm930. [DOI] [PubMed] [Google Scholar]
- 16.Terra SG, et al. Beta1-adrenergic receptor polymorphisms and left ventricular remodeling changes in response to beta-blocker therapy. Pharmacogenet. Genomics. 2005;15:227–234. doi: 10.1097/01213011-200504000-00006. [DOI] [PubMed] [Google Scholar]
- 17.Liggett SB. Beta2-adrenergic receptor polymorphisms and sudden cardiac death: a signal to follow. Circulation. 2006;113:1818–1820. doi: 10.1161/CIRCULATIONAHA.105.618967. [DOI] [PubMed] [Google Scholar]
- 18.Mason DA, Moore JD, Green SA, Liggett SB. A gain-of-function polymorphism in a G-protein coupling domain of the human beta 1-adrenergic receptor. J. Biol. Chem. 1999;274:12670–12674. doi: 10.1074/jbc.274.18.12670. [DOI] [PubMed] [Google Scholar]
- 19.Levin MC, Marullo S, Muntaner O, Andersson B, Magnusson Y. The myocardium-protective Gly-49 variant of the beta 1-adrenergic receptor exhibits constitutive activity and increased desensitization and downregulation. J. Biol. Chem. 2002;277:30429–30435. doi: 10.1074/jbc.M200681200. [DOI] [PubMed] [Google Scholar]
- 20.Rathz DA, Brown KM, Kramer LA, Liggett SB. Amino acid 49 polymorphisms of the human beta1-adrenergic receptor affect agonistpromoted trafficking. J. Cardiovasc. Pharmacol. 2002;39:155–160. doi: 10.1097/00005344-200202000-00001. [DOI] [PubMed] [Google Scholar]
- 21.A randomized trial of propranolol in patients with acute myocardial infarction. I. Mortality results. JAMA. 1982;247:1707–1714. doi: 10.1001/jama.1982.03320370021023. [DOI] [PubMed] [Google Scholar]
- 22.Iwai C, et al. Arg389Gly polymorphism of the human beta1-adrenergic receptor in patients with nonfatal acute myocardial infarction. Am. Heart. J. 2003;146:106–109. doi: 10.1016/S0002-8703(03)00110-8. [DOI] [PubMed] [Google Scholar]
- 23.White HL, et al. An evaluation of the beta-1 adrenergic receptor Arg389Gly polymorphism in individuals at risk of coronary events. A WOSCOPS substudy. Eur. Heart. J. 2002;23:1087–1092. doi: 10.1053/euhj.2001.3037. [DOI] [PubMed] [Google Scholar]
- 24.Pacanowski MA, Johnson JA. PharmGKB submission update: IX. ADRB1 gene summary. Pharmacol. Rev. 2007;59:2–4. doi: 10.1124/pr.59.1.6. [DOI] [PubMed] [Google Scholar]
- 25.Shin J, Johnson JA. Pharmacogenetics of beta-blockers. Pharmacotherapy. 2007;27:874–887. doi: 10.1592/phco.27.6.874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J, et al. Gly389Arg polymorphism of beta1-adrenergic receptor is associated with the cardiovascular response to metoprolol. Clin. Pharmacol. Ther. 2003;74:372–379. doi: 10.1016/S0009-9236(03)00224-8. [DOI] [PubMed] [Google Scholar]
- 27.Karlsson J, et al. Beta1-adrenergic receptor gene polymorphisms and response to beta1-adrenergic receptor blockade in patients with essential hypertension. Clin. Cardiol. 2004;27:347–350. doi: 10.1002/clc.4960270610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Terra SG, et al. Association between beta-adrenergic receptor polymorphisms and their G-protein-coupled receptors with body mass index and obesity in women: a report from the NHLBI-sponsored WISE study. Int. J. Obes. (Lond) 2005;29:746–754. doi: 10.1038/sj.ijo.0802978. [DOI] [PubMed] [Google Scholar]
- 29.Yu WP, Lou M, Deng B, Song HM, Wang HB. [Beta1-adrenergic receptor (Arg389Gly) polymorphism and response to bisoprolol in patients with chronic heart failure] Zhonghua Xin Xue Guan Bing Za Zhi. 2006;34:776–780. [PubMed] [Google Scholar]
- 30.Liggett SB, et al. A polymorphism within a conserved beta(1)-adrenergic receptor motif alters cardiac function and beta-blocker response in human heart failure. Proc. Natl. Acad. Sci. USA. 2006;103:11288–11293. doi: 10.1073/pnas.0509937103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chanock SJ, et al. Replicating genotype-phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- 32.Pepine CJ, et al. A calcium antagonist vs a non-calcium antagonist hypertension treatment strategy for patients with coronary artery disease. The International Verapamil-Trandolapril Study (INVEST): a randomized controlled trial. JAMA. 2003;290:2805–2816. doi: 10.1001/jama.290.21.2805. [DOI] [PubMed] [Google Scholar]
- 33.Pritchard JK, Stephens M, Rosenberg NA, Donnelly P. Association mapping in structured populations. Am. J. Hum. Genet. 2000;67:170–181. doi: 10.1086/302959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 35.Shriver MD, et al. Large-scale SNP analysis reveals clustered and continuous patterns of human genetic variation. Hum. Genomics. 2005;2:81–89. doi: 10.1186/1479-7364-2-2-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
is linked to the online version of the paper at http://www.nature.com/cpt