The Nac1 POZ domain resembles the POZ-domain dimers of the POZ-zinc finger transcription factors; the structure will have relevance for the design of therapeutics that target Nac1 function in ovarian cancer.
Keywords: BTB, ovarian cancer, stem cells, Miz-1
Abstract
Nac1 is a POZ-domain transcription factor that is involved in the self-renewal of embryonic stem cells. It is overexpressed in ovarian serous carcinoma and targeting the interactions of its POZ domain is a potential therapeutic strategy. Nac1 lacks a zinc-finger DNA-binding domain and thereby differs from most other POZ-domain transcription factors. Here, the crystal structure of the Nac1 POZ domain at 2.1 Å resolution is reported. The Nac1 POZ domain crystallized as a dimer in which the interaction interfaces between subunits resemble those found in the POZ-zinc finger transcription factors. The organization of the Nac1 POZ-domain core resembles reported POZ-domain structures, whereas the C-terminus differs markedly. The C-terminal α-helix of the Nac1 POZ domain is shorter than that observed in most other POZ-domain transcription factors; variation in the organization of this region may be a general feature of POZ-domain structures.
1. Introduction
Nac1 is a POZ-domain protein that was originally characterized as a cocaine-inducible gene product in the nucleus accumbens of rat brain (Cha et al., 1997 ▶). It was initially shown to function in the behavioural responses to psychostimulants (Mackler et al., 2000 ▶), although has subsequently been implicated in diverse physiological processes. Nac1 mediates the translocation of the ubiquitin–proteosome (UPS) complex from the nucleus into neuronal dendritic spines, thereby regulating proteolysis during synaptic remodelling (Shen et al., 2007 ▶). Nac1 also acts as a transcriptional repressor in both neuronal and non-neuronal cells (Korutla et al., 2002 ▶, 2005 ▶; Nakayama et al., 2007 ▶); it interacts with the homeodomain protein Nanog and is thus a central component of the transcription-factor network that maintains the pluripotency of embryonic stem cells (Wang et al., 2006 ▶). Levels of Nac1 are elevated in advanced ovarian serous carcinoma and cervical carcinoma (Nakayama et al., 2006 ▶; Yeasmin et al., 2008 ▶); in ovarian cancer, expression is particularly high in recurrent disease and in effusions and is thought to contribute to the development of resistance to chemotherapy (Davidson et al., 2007 ▶; Ishibashi et al., 2008 ▶). Artificial knock-down of Nac1 resulted in the apoptosis of ovarian cancer cell lines and rescued their sensitivity to chemotherapy, suggesting that this protein may be a potential therapeutic target.
POZ (poxvirus and zinc finger; also known as BTB, bric-à-brac, tramtrack and broad complex) domains are protein–protein interaction motifs that are found in over 200 human proteins (reviewed in Stogios et al., 2005 ▶). POZ-domain proteins function in a wide range of biological activities, including transcription, ion-channel gating, cytoskeletal dynamics and protein degradation via the ubiquitin–proteasome (UPS) system. Approximately 40 transcription factors contain N-terminal POZ domains (POZ-TFs); many of these play roles in development and the deregulation of POZ-TF expression has been observed in several human cancers (reviewed in Kelly & Daniel, 2006 ▶). Most POZ-TFs contain C2H2 zinc-finger or basic leucine-zipper DNA-binding domains; however, Nac1 is distinct and does not contain any characterized DNA-binding motifs. The mechanism by which Nac1 interacts with DNA is not known, although target genes have been identified by chromatin immunoprecipitation and microarray approaches (Kim et al., 2008 ▶). POZ domains mediate the homodimerization of POZ-zinc finger transcription factors and also direct the formation of higher order oligomers and heteromeric POZ–POZ complexes; the oligomeric organization of Nac1 has not been characterized. Crystal structures of the POZ domains from the zinc-finger proteins PLZF (promyelocytic leukaemia zinc finger; Ahmad et al., 1998 ▶; Li et al., 1999 ▶), BCL6 (B-cell lymphoma 6; Ahmad et al., 2003 ▶; Ghetu et al., 2008 ▶; Stead et al., 2008 ▶) and LRF (leukaemia/lymphoma-related factor; Schubot et al., 2006 ▶; Stogios et al., 2007 ▶) revealed domain-swapped homodimers that are thought to represent the biologically active unit. In contrast, the Miz-1 (Myc-interacting zinc-finger protein 1) POZ domain crystallized as a tetramer in which two POZ dimers interact via a β-sheet region (Stead et al., 2007 ▶); this tetrameric organization was also observed in solution, suggesting that this interface may be important in the higher order homo- and hetero-oligomerization of the POZ-TFs in vivo.
POZ domains interact with non-POZ partners in a highly specific manner. The POZ-TF POZ domains recruit transcriptional co-regulators: for example, BCL6 binds the co-repressors BCoR (BCL6-interacting co-repressor), SMRT (silencing mediator for retinoid and thyroid hormone receptors) and NCoR (nuclear receptor co-repressor), whereas Nac1 recruits CoREST (co-repressor of RE1-silencing transcription factor, also known as RCoR; Korutla et al., 2007 ▶). The Nac1 POZ domain also interacts with cullin 3, consistent with its role in the UPS system (Shen et al., 2007 ▶). Targeting the interactions of POZ domains with their transcriptional co-repressors is a strategy for the treatment of tumours associated with deregulated POZ-TF expression. Crystal structures of BCL6 in association with BCoR (Ghetu et al., 2008 ▶) and SMRT (Ahmad et al., 2003 ▶) peptides led to the design of inhibitors that interfere with co-repressor recruitment; these inhibitors induced the apoptosis of diffuse large B-cell lymphoma (DLBCL) tumour cells that overexpress BCL6 and are promising therapeutic agents (Parekh et al., 2008 ▶). Targeting the interactions of the Nac1 POZ domain is a potential strategy for the treatment of ovarian cancer; here, we report the crystal structure of the Nac1 POZ domain to 2.1 Å resolution.
2. Materials and methods
2.1. Cloning
A DNA fragment encoding the Nac1 POZ domain (Nac1 residues 2–125) was amplified by PCR from a human placental cDNA library and inserted into the expression vector pGEX-6P-1 (GE Healthcare). The POZ-domain F98D mutant was generated by PCR with Phusion High-Fidelity DNA polymerase (Finnzymes) using the oligonucleotides ATGAACGTGGGCGACCAGGACCTGCTCATGTACACGGCT and GCTCAGCCGGCCCGTGTAGCAGAAGCTGAGGTACTG.
2.2. Protein expression and purification
GST-fusion proteins were expressed in Escherichia coli BL21 (DE3) pLysS. Bacteria were cultured in 2TY at 310 K to an OD600nm of 0.8. Expression of recombinant protein was then induced with 0.1 mM IPTG at 289 K for 16 h. Cells were lysed and fusion proteins were bound to glutathione-Sepharose 4B (GE Healthcare). The N-terminal GST tag was removed by cleavage with PreScission protease in 20 mM Tris–HCl, 75 mM NaCl, 5 mM DTT pH 7.5. The Nac1 POZ domain was further purified by gel-filtration chromatography in 20 mM Tris–HCl, 150 mM NaCl, 5 mM DTT, 5% glycerol pH 8.6 and concentrated to 14 mg ml−1 using Amicon centrifugal concentrators (Millipore).
2.3. Crystallization
Crystals of the Nac1 POZ domain were grown by sitting-drop vapour diffusion at 291 K by mixing 2 µl protein solution (14 mg ml−1) with 2 µl reservoir solution (6 M ammonium nitrate, 0.1 M bis-tris propane pH 7.5). Large crystals were typically obtained after 48 h. Crystals were mounted in a nylon cryoloop (Hampton Research) and transferred to mother liquor supplemented with 20% glycerol for 30 s before being flash-frozen in liquid nitrogen.
2.4. Data collection, structure determination and refinement
X-ray diffraction data were collected under a nitrogen-gas stream at 100 K on beamline I03 at the Diamond Light Source (Didcot, England). Data reduction was performed using iMOSFLM and SCALA (Leslie, 1992 ▶; Collaborative Computational Project, Number 4, 1994 ▶). The structure was solved using the MrBUMP molecular-replacement pipeline (Keegan & Winn, 2007 ▶); the MrBUMP solution was obtained using a MOLREP-generated search model from the LRF POZ domain (PDB code 2if5) with Phaser as the molecular-replacement program (McCoy et al., 2005 ▶). A model was built using ARP/wARP (Perrakis et al., 1999 ▶); this was followed by iterative model building and refinement in Coot (Emsley & Cowtan, 2004 ▶) and REFMAC (Murshudov et al., 1997 ▶), including TLS refinement with three TLS groups per POZ-domain monomer. The stereochemistry was analysed with PROCHECK (Laskowski et al., 1993 ▶) and MolProbity (Davis et al., 2007 ▶) and structure superpositions were calculated using the SUPERPOSE server (Maiti et al., 2004 ▶); the PDB entries used for superpositions were Miz-1, 2q81; BCL6, 1r28; PLZF, 1buo; LRF, 2nn2. Images of protein structures were prepared using PyMOL (DeLano, 2002 ▶).
3. Results and discussion
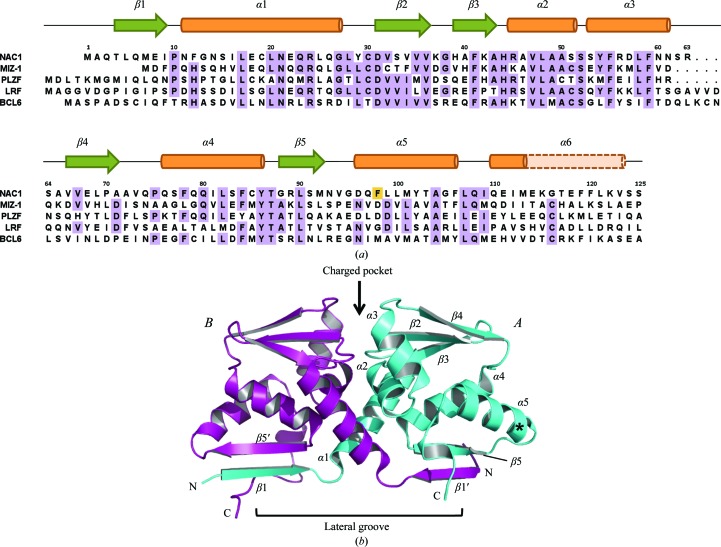
Expression of the wild-type human Nac1 POZ domain in E. coli produced highly insoluble recombinant protein, even upon attempted optimization of the expression and purification protocols. The Nac1 POZ-domain sequence is 38% identical to that of Miz-1; since recombinant Miz1 POZ domain is highly soluble, we used the crystal structure of the Miz1 POZ domain to predict surface residues that might contribute to the insolubility of Nac1. Mutation of a single surface hydrophobic residue (F98D; Fig. 1 ▶ a) enabled the purification of highly soluble recombinant Nac1 POZ domain. This mutation does not reside near residues that have been implicated either in POZ-domain oligomerization or in the recruitment of transcriptional co-repressors. The F98D Nac1 POZ domain was crystallized and the structure was solved by molecular replacement and refined to R = 20.63%, R free = 24.09% at 2.1 Å resolution (Table 1 ▶; Fig. 1 ▶ b).
Figure 1.
Structure of the Nac1 POZ domain. (a) Sequence alignment of the Nac1, Miz-1, PLZF, BCL6 and LRF POZ domains. Sequences were aligned using ClustalW. The observed secondary-structure elements of the Nac1 POZ domain are indicated, with α-helices in orange and β-sheets in green. The residues of α6 in the Miz-1, PLZF, BCL6 and LRF POZ domains are shown in pale orange. (b) Ribbon representation of the Nac1 POZ-domain dimer. The position of the F98D mutation is shown with an asterisk. The secondary-structure elements of chain A are indicated, together with β1′ and β5′ of chain B.
Table 1. Data-collection and processing statistics.
Values in parentheses are for the highest resolution shell (2.12.21).
| Crystal parameters | |
| Space group | P41212 |
| Unit-cell parameters (, ) | a = 57.69, b = 57.69, c = 172.60, = 90, = 90, = 90 |
| Data collection | |
| Resolution () | 47.962.10 (2.212.10) |
| Wavelength () | 0.9763 |
| R merge † (%) | 10.3 (50.6) |
| R p.i.m. ‡ (%) | 4.3 (20.5) |
| I/(I) | 13.5 (4.0) |
| No. of unique reflections | 17928 |
| Multiplicity | 6.7 (7.0) |
| Completeness (%) | 100 (100) |
| Refinement | |
| Resolution () | 2.1 |
| R § (%) | 0.206 |
| R free ¶ (%) | 0.241 |
| R.m.s.d. stereochemistry†† | |
| Bond lengths () | 0.114 |
| Bond angles () | 1.294 |
| No. of protein atoms | 1857 |
| No. of water molecules | 107 |
| Average B factor (2) | 18.09 |
| Ramachandran analysis‡‡ (%) | |
| Favoured | 96.2 |
| Allowed | 100.0 |
| Disallowed | 0 |
R
merge = 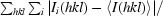
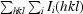 , where I
i(hkl) is the integrated intensity of a given reflection and I(hkl) is the mean intensity of multiple corresponding symmetry-related reflections.
, where I
i(hkl) is the integrated intensity of a given reflection and I(hkl) is the mean intensity of multiple corresponding symmetry-related reflections.
R
p.i.m. = 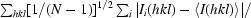
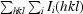 , where I
i(hkl) is the integrated intensity of a given reflection, I(hkl) is the mean intensity of multiple corresponding symmetry-related reflections and N is the multiplicity of a given reflection.
, where I
i(hkl) is the integrated intensity of a given reflection, I(hkl) is the mean intensity of multiple corresponding symmetry-related reflections and N is the multiplicity of a given reflection.
R = 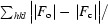
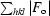 , where F
o and F
c are the observed and calculated structure factors, respectively.
, where F
o and F
c are the observed and calculated structure factors, respectively.
R free is the same as R but calculated using 5% random data excluded from the refinement.
R.m.s.d. stereochemistry is the deviation from ideal values.
Ramachandran analysis was carried out using MolProbity (Davis et al., 2007 ▶).
Nac1 is a distinct POZ-domain transcription factor that does not contain a characteristic zinc-finger or basic leucine-zipper DNA-binding domain. The POZ domains of the individual POZ-zinc finger transcription factors have approximately the same degree of homology to each other as they do to Nac1, suggesting similar timescales of evolutionary divergence (Fig. 1 ▶ a). The Nac1 POZ domain crystallized as a domain-swapped dimer whose interfaces closely resemble those reported in the POZ domains of the POZ-zinc finger proteins BCL6, PLZF and LRF (Fig. 1 ▶ b). A central hydrophobic interface that buries 1018 Å2 surface area per monomer is formed by the tight packing of helices α1, α2 and α3, whereas two β-strand interfaces that bury an additional 947 Å2 per monomer are formed by the interaction of β1 of one subunit with β5 of the other (Fig. 1 ▶ b). It is therefore likely that Nac1 functions biologically as an obligate homodimer in a manner resembling the POZ-zinc finger proteins. The organization of secondary-structure elements in the centre of the Nac1 POZ dimer (β1–α5) is the same as in reported POZ structures; compared with Nac1, the r.m.s.d. values of Cα-atom positions are 1.62 Å for BCL6, 2.19 Å for Miz-1, 2.31 Å for PLZF and 2.37 Å for LRF. Conserved residues that constitute a charged pocket in the POZ-TF POZ domains are also present in Nac1 (Asp31 and Arg45); this region is thought to be important in transcriptional repression in BCL6 and PLZF, although the mechanism involved is unknown (Melnick et al., 2002 ▶).
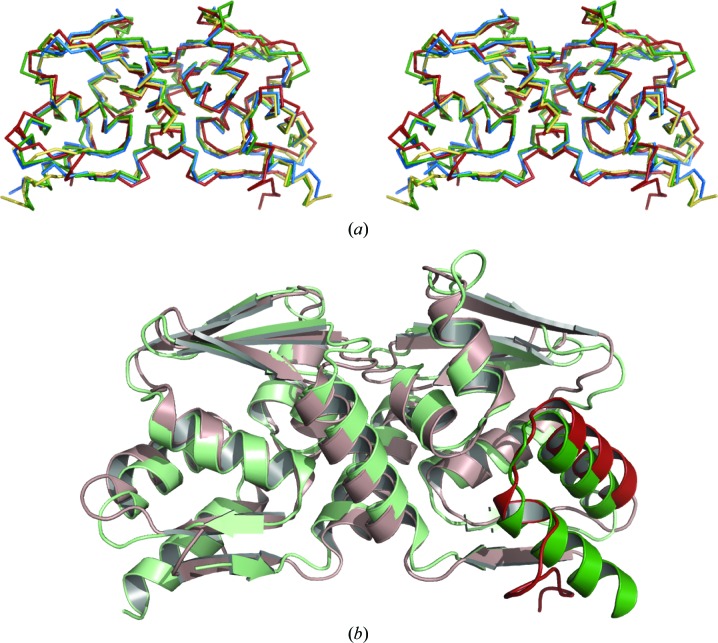
The organization of the C-terminus in the Nac1 POZ domain differs from that in the reported BCL6, LRF, PLZF and Miz-1 POZ-domain structures (Fig. 2 ▶). The C-terminal residues of the latter POZ domains form a 12-residue α-helix (α6) that lies adjacent to α5 on the outside of the molecule. The corresponding helix in Nac1 is extremely short and the C-terminal residues are unstructured and diverge away from α5 to interact with the lower β1–β5′ sheet. These C-terminal residues have the highest B factors of the Nac1 POZ-domain crystal structure in chain B and are disordered in chain A. The residues corresponding to α6 constitute the least conserved sequences among the POZ domains of the POZ-TFs and the α-helical propensities of the amino acids in the middle of this region differ greatly. The POZ-domain α6 helix of the zinc-finger transcriptional repressor Kaiso (also known as ZBTB33; PDB code 3fkc) is also extremely short; the low α-helical propensity of the Nac1 and Kaiso residues in the middle of α6 (glycine in Nac1; glycine and proline in Kaiso) may contribute to this structural feature. The elongin C proteins contain a POZ domain that lacks an entire α6 helix; interestingly, the α5 helix of elongin C is involved in its interaction with the VHL protein in a manner that would not be compatible with the presence of a POZ-domain α6 helix in its classical location (Stebbins et al., 1999 ▶). Variation in the length of the C-terminal α6 helix may therefore be a general feature of POZ domains, although its biological relevance in the POZ-TFs is unknown.
Figure 2.
Superposition of POZ-domain structures. (a) Stereo-image of POZ-domain Cα-atom superposition for Nac1 (red), BCL6 (green), PLZF (yellow) and LRF (blue). The accession codes of the PDB entries used for superposition were BCL6, 1r28; PLZF, 1buo; LRF, 2nn2. (b) Superposition of the BCL6 (pale green) and Nac1 (pale pink) POZ domains. The α5 and α6 regions of chain B are highlighted in green (BCL6) and red (Nac1).
The recruitment of transcriptional co-repressors to the POZ-TF POZ domains is highly specific: for example, BCL6 interacts with SMRT and BCoR in a mutually exclusive manner, whereas neither of these co-repressors bind to LRF or Nac1. The residues of SMRT and BCoR that interact with BCL6 share no sequence homology, although both bind to the POZ-domain lateral grooves that run across β1 and up through the dimerization interface. The POZ-domain dimer recruits two co-repressor molecules symmetrically; contacts are mediated by residues in β1, αl, α2, α3 and α6 of both POZ subunits and involve the rearrangement of some amino-acid side chains. Conserved interactions of the BCL6 POZ domain with SMRT and BCoR involve the main-chain atoms of the co-repressor, whereas nonconserved interactions are mediated by the side chains. The most important BCL6 residues that interact with SMRT and BCoR are notably not conserved in the Nac1 POZ domain: BCL6 residues Gln10, Arg13, Arg24, His116, Arg28 and Tyr58 are replaced by amino acids of different charge in Nac1, with the equivalent positions being Glu8, Asn11, Glu22, Glu114, Gln26 and Arg56, respectively. It will now be relevant to determine the residues that direct the specific interactions of the Nac1 POZ domain with the transcriptional co-repressor CoREST.
Supplementary Material
PDB reference: Nac1 POZ domain, 3ga1, r3ga1sf
Acknowledgments
This work was funded by Yorkshire Cancer Research and by a BBSRC studentship to MAS. We thank the staff at the Diamond Light Source (Didcot, UK) for assistance during X-ray data collection.
References
- Ahmad, K. F., Engel, C. K. & Privé, G. G. (1998). Proc. Natl Acad. Sci. USA, 95, 12123–12128. [DOI] [PMC free article] [PubMed]
- Ahmad, K. F., Melnick, A., Lax, S., Bouchard, D., Liu, J., Kiang, C. L., Mayer, S., Takahashi, S., Licht, J. D. & Privé, G. G. (2003). Mol. Cell, 12, 1551–1564. [DOI] [PubMed]
- Cha, X. Y., Pierce, R. C., Kalivas, P. W. & Mackler, S. A. (1997). J. Neurosci. 17, 6864–6871. [DOI] [PMC free article] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Davidson, B., Berner, A., Trope, C. G., Wang, T. L. & Shih, I.-M. (2007). Hum. Pathol. 38, 1030–1036. [DOI] [PubMed]
- Davis, I. W., Leaver-Fay, A., Chen, V. B., Block, J. N., Kapral, G. J., Wang, X., Murray, L. W., Arendall, W. B. III, Snoeyink, J., Richardson, J. S. & Richardson, D. C. (2007). Nucleic Acids Res. 35, W375–W383. [DOI] [PMC free article] [PubMed]
- DeLano, W. L. (2002). The PyMol Molecular Graphics System. http://www.pymol.org.
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Ghetu, A. F., Corcoran, C. M., Cerchietti, L., Bardwell, V. J., Melnick, A. & Privé, G. G. (2008). Mol. Cell, 29, 384–391. [DOI] [PMC free article] [PubMed]
- Ishibashi, M., Nakayama, K., Yeasmin, S., Katagiri, A., Iida, K., Nakayama, N., Fukumoto, M. & Miyazaki, K. (2008). Clin. Cancer Res. 14, 3149–3155. [DOI] [PubMed]
- Keegan, R. M. & Winn, M. D. (2007). Acta Cryst. D63, 447–457. [DOI] [PubMed]
- Kelly, K. F. & Daniel, J. M. (2006). Trends Cell Biol. 16, 578–587. [DOI] [PubMed]
- Kim, J., Chu, J., Shen, X., Wang, J. & Orkin, S. H. (2008). Cell, 132, 1049–1061. [DOI] [PMC free article] [PubMed]
- Korutla, L., Degnan, R., Wang, P. & Mackler, S. A. (2007). J. Neurochem. 101, 611–618. [DOI] [PubMed]
- Korutla, L., Wang, P. J., Lewis, D. M., Neustadter, J. H., Stromberg, M. F. & Mackler, S. A. (2002). Neuroscience, 110, 421–429. [DOI] [PubMed]
- Korutla, L., Wang, P. J. & Mackler, S. A. (2005). J. Neurochem. 94, 786–793. [DOI] [PubMed]
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst. 26, 283–291.
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr. 26
- Li, X., Peng, H., Schultz, D. C., Lopez-Guisa, J. M., Rauscher, F. J. III & Marmorstein, R. (1999). Cancer Res. 59, 5275–5282. [PubMed]
- Mackler, S. A., Korutla, L., Cha, X. Y., Koebbe, M. J., Fournier, K. M., Bowers, M. S. & Kalivas, P. W. (2000). J. Neurosci. 20, 6210–6217. [DOI] [PMC free article] [PubMed]
- Maiti, R., Van Domselaar, G. H., Zhang, H. & Wishart, D. S. (2004). Nucleic Acids Res. 32, W590–W594. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Storoni, L. C. & Read, R. J. (2005). Acta Cryst. D61, 458–464. [DOI] [PubMed]
- Melnick, A., Carlile, G., Ahmad, K. F., Kiang, C. L., Corcoran, C., Bardwell, V., Privé, G. G. & Licht, J. D. (2002). Mol. Cell. Biol. 22, 1804–1818. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Nakayama, K., Nakayama, N., Davidson, B., Sheu, J. J., Jinawath, N., Santillan, A., Salani, R., Bristow, R. E., Morin, P. J., Kurman, R. J., Wang, T. L. & Shih, I.-M. (2006). Proc. Natl Acad. Sci. USA, 103, 18739–18744. [DOI] [PMC free article] [PubMed]
- Nakayama, K., Nakayama, N., Wang, T. L. & Shih, I.-M. (2007). Cancer Res. 67, 8058–8064. [DOI] [PubMed]
- Parekh, S., Privé, G. & Melnick, A. (2008). Leuk. Lymphoma, 49, 874–882. [DOI] [PMC free article] [PubMed]
- Perrakis, A., Morris, R. & Lamzin, V. S. (1999). Nature Struct. Biol. 6, 458–463. [DOI] [PubMed]
- Schubot, F. D., Tropea, J. E. & Waugh, D. S. (2006). Biochem. Biophys. Res. Commun. 351, 1–6. [DOI] [PubMed]
- Shen, H., Korutla, L., Champtiaux, N., Toda, S., LaLumiere, R., Vallone, J., Klugmann, M., Blendy, J. A., Mackler, S. A. & Kalivas, P. W. (2007). J. Neurosci. 27, 8903–8913. [DOI] [PMC free article] [PubMed]
- Stead, M. A., Rosbrook, G. O., Hadden, J. M., Trinh, C. H., Carr, S. B. & Wright, S. C. (2008). Acta Cryst. F64, 1101–1104. [DOI] [PMC free article] [PubMed]
- Stead, M. A., Trinh, C. H., Garnett, J. A., Carr, S. B., Baron, A. J., Edwards, T. A. & Wright, S. C. (2007). J. Mol. Biol. 373, 820–826. [DOI] [PubMed]
- Stebbins, C. E., Kaelin, W. G. Jr & Pavletich, N. P. (1999). Science, 284, 455–461. [DOI] [PubMed]
- Stogios, P. J., Chen, L. & Privé, G. G. (2007). Protein Sci. 16, 336–342. [DOI] [PMC free article] [PubMed]
- Stogios, P. J., Downs, G. S., Jauhal, J. J., Nandra, S. K. & Privé, G. G. (2005). Genome Biol. 6, R82. [DOI] [PMC free article] [PubMed]
- Wang, J., Rao, S., Chu, J., Shen, X., Levasseur, D. N., Theunissen, T. W. & Orkin, S. H. (2006). Nature (London), 444, 364–368. [DOI] [PubMed]
- Yeasmin, S., Nakayama, K., Ishibashi, M., Katagiri, A., Iida, K., Purwana, I. N., Nakayama, N. & Miyazaki, K. (2008). Clin. Cancer Res. 14, 1686–1691. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: Nac1 POZ domain, 3ga1, r3ga1sf