The crystal structures of TTHA1623 from T. thermophilus HB8 in an iron-bound and a zinc-bound form have been determined to 2.8 and 2.2 Å resolution, respectively.
Keywords: TTHA1623, Thermus thermophilus HB8, metallo-β-lactamase superfamily
Abstract
TTHA1623 is a metallo-β-lactamase superfamily protein from the extremely thermophilic bacterium Thermus thermophilus HB8. Homologues of TTHA1623 exist in a wide range of bacteria and archaea and one eukaryote, Giardia lamblia, but their function remains unknown. To analyze the structural properties of TTHA1623, the crystal structures of its iron-bound and zinc-bound forms have been determined to 2.8 and 2.2 Å resolution, respectively. TTHA1623 possesses an αββα-fold similar to that of other metallo-β-lactamase superfamily proteins with glyoxalase II-type metal coordination. However, TTHA1623 exhibits a putative substrate-binding pocket with a unique shape.
1. Introduction
TTHA1623 is a metallo-β-lactamase superfamily protein from the extremely thermophilic bacterium Thermus thermophilus HB8. The metallo-β-lactamases, which were first identified as class B β-lactamases, possess an αββα-fold and a di-metal-binding site (Carfi et al., 1995 ▶; Concha et al., 1996 ▶). Other enzymes with this fold include glyoxalase II, rubredoxin oxygen:oxidoreductase, phosphorylcholine esterase and tRNA maturase. The function of TTHA1623 is unknown, since it possesses neither β-lactamase nor glyoxalase II activity (unpublished data). However, a BLAST search has revealed that homologues of TTHA1623 are present in a wide range of bacteria and archaea and one eukaryote (Giardia lamblia). Therefore, TTHA1623 and its homologues are likely to play an important role in these organisms. To analyze the structural properties of TTHA1623, we have determined the crystal structures of TTHA1623 in its di-iron-bound and di-zinc-bound forms to 2.8 and 2.2 Å resolution, respectively. TTHA1623 exhibits a uniquely shaped putative substrate-binding pocket with glyoxalase II-type metal coordination (Bebrone, 2007 ▶).
2. Materials and methods
2.1. Cloning, expression and purification
The gene encoding TTHA1623 from T. thermophilus HB8 (gi:55981592) was amplified by PCR using T. thermophilus HB8 genomic DNA as the template. The PCR primers used were 5′-GGAATTCCATATGAGGGTCTTTCCCGTC-3′ (including an NdeI site, shown in bold) and 5′-GAAGATCTTATCACGCTTCCCATTCTAAC-3′ (including a BglII site, shown in bold). 30 cycles of PCR were performed using KOD-plus (Toyobo, Japan) with a melting phase of 367 K for 30 s, an annealing phase of 326 K for 30 s and a polymerization phase of 341 K for 60 s. The PCR product and the vector plasmid pET-11a(+) (Novagen) were digested by NdeI and BglII and were ligated with Ligation High (Toyobo). The DNA sequence of the TTHA1623-encoding region of the resulting plasmid was verified. The protein without any tags was overexpressed in Escherichia coli BL21 (DE3) (Novagen). Harvested cells were resuspended in 20 mM Tris–HCl pH 8.0 and 50 mM NaCl and then disrupted by sonication. The lysate was centrifuged at 40 000g at 277 K for 30 min. The supernatant was incubated at 343 K for 10 min and then centrifuged at 40 000g at 277 K for 30 min. Ammonium sulfate was added to the supernatant to a final concentration of 1.5 M, which was followed by centrifugation at 40 000g at 277 K for 30 min. The supernatant solution was loaded onto a Resource ISO (GE Healthcare) column equilibrated with 50 mM sodium phosphate buffer pH 7.0 and 1.5 M ammonium sulfate. Proteins were eluted with a linear gradient of 1.5–0 M ammonium sulfate. The fractions containing TTHA1623 were then loaded onto a hydroxyapatite CHT10 (Bio-Rad) column equilibrated with 10 mM sodium phosphate buffer pH 8.0 and eluted with a linear gradient of 10–100 mM sodium phosphate pH 8.0. In the final purification step, the fractions containing TTHA1623 were applied onto a HiLoad 16/60 Superdex 75 pg (GE Healthcare) column equilibrated with 20 mM Tris–HCl pH 8.0 and 150 mM NaCl. The purified TTHA1623 was applied onto a HiPrep 26/10 Desalting (GE Healthcare) column equilibrated with 20 mM Tris–HCl pH 8.0 to remove NaCl.
2.2. Crystallization and data collection
The sitting-drop vapour-diffusion method was used for crystallization. 1 µl purified TTHA1623 solution (9.7 mg ml−1) and 1 µl reservoir solution were mixed to prepare a crystallization drop and the drop was equilibrated against 200 µl reservoir solution. Cocrystallization of TTHA1623 with zinc was performed at 293 K with a reservoir solution containing 100 mM sodium cacodylate buffer pH 7.3, 30% PEG 200 (Hampton Research) and 200 mM zinc acetate. The crystals were picked up in mounting loops and directly frozen in liquid nitrogen without using any cryoprotectant. X-ray diffraction data for the crystals were obtained on beamline BL26B1 at SPring-8 (Harima, Japan). The data for the first crystal were obtained at three wavelengths, the zinc peak (1.2822 Å), edge (1.2829 Å) and remote (1.0000 Å), for use in the zinc multiple-wavelength anomalous dispersion (MAD) method. The data for the second crystal were obtained at 1.0000 Å. Crystallization without zinc was performed at 293 K with a reservoir solution containing 100 mM sodium cacodylate pH 6.5 and 1.4 M sodium acetate trihydrate. The crystals were picked up in mounting loops and directly frozen in liquid nitrogen using 20% glycerol as the cryoprotectant. The X-ray diffraction data for the zinc-free TTHA1623 crystal were obtained at a wavelength of 1.0000 Å on beamline BL26B1 at SPring-8 (Harima, Japan).
2.3. Structure solution and refinement
The data were processed with the HKL-2000 program package (Otwinowski & Minor, 1997 ▶) and the CCP4 suite (Collaborative Computational Project, Number 4, 1994 ▶). The data for the first zinc-bound crystal of TTHA1623 were processed with the ‘scale anomalous’ flag of HKL-2000 to keep Bijvoet pairs separate both in scaling and in the output file; thus, the number of unique reflections for these data sets are double the values for the other data sets (Table 1 ▶). Zinc sites were determined with the program SOLVE (Terwilliger & Berendzen, 1999 ▶) using the MAD data sets of the first crystal and the resulting phases were improved with the program RESOLVE (Terwilliger & Berendzen, 1999 ▶). Molecular replacement with the program MOLREP (Vagin & Teplyakov, 2000 ▶) was carried out against the data set obtained from the second crystal using the initial model obtained from the MAD data sets as a search model. The model was refined using Coot (Emsley & Cowtan, 2004 ▶) and REFMAC5 (Murshudov et al., 1997 ▶). Using the refined structure of zinc-bound TTHA1623 as the search model, molecular replacement with the program MOLREP (Vagin & Teplyakov, 2000 ▶) was carried out against the data set obtained from the zinc-free TTHA1623 crystal. The model was refined using Coot (Emsley & Cowtan, 2004 ▶) and REFMAC5 (Murshudov et al., 1997 ▶). Noncrystallographic symmetry (NCS) restraints were applied throughout the refinement. The stereochemistry of the structure was checked by the program PROCHECK (Laskowski et al., 1993 ▶).
Table 1. Summary of data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell.
| Zinc-bound crystal | |||||
|---|---|---|---|---|---|
| 1st crystal | |||||
| Zn edge | Zn peak | Zn remote | 2nd crystal | Zinc-free iron-bound crystal | |
| Data collection | |||||
| Wavelength (Å) | 1.2829 | 1.2822 | 1.0000 | 1.0000 | 1.0000 |
| Resolution range (Å) | 50.0–2.40 (2.49–2.40) | 50.0–2.20 (2.28–2.20) | 50.0–2.80 (2.90–2.80) | ||
| No. of observed reflections | 125496 | 136427 | 139258 | 125281 | 69903 |
| No. of unique reflections | 36071 | 37010 | 37531 | 26551 | 12243 |
| Data completeness (%) | 96.6 (74.3) | 99.9 (99.3) | 100.0 (100.0) | 99.3 (98.4) | 99.8 (100.0) |
| Rmerge† | 0.040 (0.384) | 0.041 (0.256) | 0.040 (0.239) | 0.063 (0.272) | 0.096 (0.148) |
| 〈I〉/〈〈σ(I)〉 | 15.5 (1.3) | 19.0 (2.7) | 20.8 (2.9) | 9.1 (2.1) | 51.1 (23.3) |
| Space group | I222 | P31 | |||
| Unit-cell parameters (Å, °) | a = 79.1, b = 114.1, c = 114.7 | a = b = 78.6, c = 71.9, α = β = 90, γ = 120 | |||
| Refinement | |||||
| Resolution range used for refinement (Å) | 50.0–2.20 | 20.0–2.80 | |||
| Rfactor‡ (%) | 18.8 | 26.7 | |||
| Rfree‡ (%) | 22.9 | 29.9 | |||
| No. of reflections used for refinement | 24819 | 12171 | |||
| Protein residues modelled | 406 of 414 | 397 of 414 | |||
| No. of protein atoms modelled | 3061 | 3019 | |||
| No. of water molecules modelled | 158 | 16 | |||
| Mean overall B value (Å2) | 16.3 | 42.0 | |||
| R.m.s.d. bond angle (°) | 1.225 | 0.895 | |||
| R.m.s.d. bond length (Å) | 0.010 | 0.005 | |||
| Ramachandran plot | |||||
| Residues in most favoured regions (%) | 85.5 | 82.4 | |||
| Residues in additionally allowed regions (%) | 14.5 | 17.3 | |||
| Residues in generously allowed regions (%) | 0.0 | 0.3 | |||
| Residues in disallowed regions (%) | 0.0 | 0.0 | |||
R
merge = 
 .
.
R
factor = 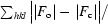
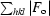 . R
free was calculated using 5% of the data that were excluded from refinement.
. R
free was calculated using 5% of the data that were excluded from refinement.
2.4. Analytical gel filtration
The purified TTHA1623 was loaded onto a Superdex 75 HR 10/30 column (GE Healthcare) equilibrated with 50 mM Tris–HCl buffer pH 8.0 and 150 mM NaCl and eluted with the same buffer at a flow rate of 0.5 ml min−1 at room temperature. The molecular weight of TTHA1623 was estimated by comparing its elution volume with those of the following standard proteins: transferrin (MW 81 000), ovalbumin (MW 43 000), myoglobin (MW 17 600), ribonuclease A (MW 13 700) and aprotinin (MW 6500).
2.5. Metal content analysis
Metal-content analysis of TTHA1623 was carried out with an SPS-1200VR inductively coupled plasma atomic emission spectrometer (ICP-AES; Seiko Instruments) that covered eight types of element (Mg, Al, Ca, Mn, Fe, Ni, Cu and Zn). The purified TTHA1623 was dialyzed against 10 mM Tris–HCl pH 8.0 at 277 K overnight to remove loosely bound metal ions and then diluted to 0.5 mg ml−1. The ICP-AES measurements were carried out as described previously (Crowder et al., 1997 ▶).
2.6. Structure-similarity search
The structures of TTHA1623 and its homologues were superposed using DaliLite (Holm & Park, 2000 ▶). The r.m.s.d. value was calculated using the same program.
3. Results and discussion
3.1. Overall structure of zinc-bound TTHA1623
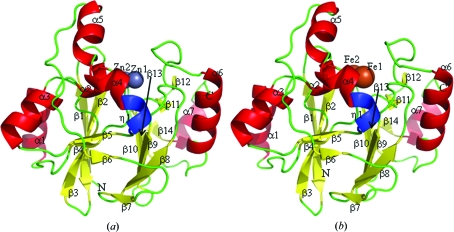
The crystal structure of zinc-bound TTHA1623 was determined at 2.2 Å resolution by a combination of zinc MAD and molecular-replacement methods. The final structure included two protein molecules (residues 1–200 for chain A and 1–206 for chain B of the 207 residues of the protein), 160 water molecules and 14 zinc ions in the asymmetric unit. No electron density was observed for the remaining residues. Data-collection and refinement statistics are summarized in Table 1 ▶. The interface of two TTHA1623 molecules in the asymmetric unit was 290 Å2, corresponding to only 3% of the total surface area of TTHA1623 and indicating that TTHA1623 was monomeric in the crystal, as shown in solution by gel-filtration chromatography. The TTHA1623 monomer contained 14 β-strands, seven α-helices and one 310-helix (Fig. 1 ▶ a), and formed a ββ-sandwich with helices on each external face, as in other metallo-β-lactamase superfamily proteins (Carfi et al., 1995 ▶; Yamamura et al., 2008 ▶). The di-metal-binding site is located on one edge of the ββ-sandwich, with the two zinc ions located 3.3 Å apart.
Figure 1.
Overall structure of TTHA1623 in di-zinc-bound (a) and di-iron-bound (b) forms. The N- and C-termini are labelled N and C, respectively. β-strands (β1–14), α-helices (α1–7) and 310-helix (η1) are shown in yellow, red and blue, respectively. Zinc and iron ions in the putative active site are presented as grey and brown spheres, respectively. This figure was drawn with PyMOL (DeLano, 2002 ▶).
3.2. Overall structure and metal content of iron-bound TTHA1623
The crystal structure of TTHA1623 crystallized without the addition of any metal ion was determined at 2.8 Å resolution by the molecular-replacement method. Metal ions were observed in the crystal structure and were determined to be iron using ICP-AES (Table 2 ▶). The final structure thus included two TTHA1623 monomers (residues 3–200 for chain A and 3–201 for chain B), 16 water molecules, four iron ions and two acetate ions in the asymmetric unit. Data-collection and refinement statistics are summarized in Table 1 ▶. The interface of two TTHA1623 molecules in the asymmetric unit was 406 Å2, corresponding to only 5% of the total surface area of TTHA1623. The TTHA1623 monomer contained 14 β-strands, seven α-helices and one 310-helix (Fig. 1 ▶ b). The di-metal-binding site is located on one edge of the ββ-sandwich, with the two iron ions located 3.1 Å apart. The overall structures of di-iron-bound and di-zinc-bound TTHA1623 are almost identical, with an r.m.s.d. of 0.7 Å for the corresponding Cα atoms.
Table 2. Metal content of TTHA1623.
Each value represents the mean ± standard deviation (n = 3).
| Metal ion | Moles of metal per mole of TTHA1623 |
|---|---|
| Fe | 1.7 ± 0.0 |
| Mn | 0.068 ± 0.001 |
| Zn | 0.043 ± 0.002 |
| Cu | 0.011 ± 0.000 |
| Mg | <0.010 |
| Al | <0.010 |
| Ca | <0.010 |
| Ni | <0.010 |
3.3. Metal-binding modes of di-zinc-bound and di-iron-bound TTHA1623
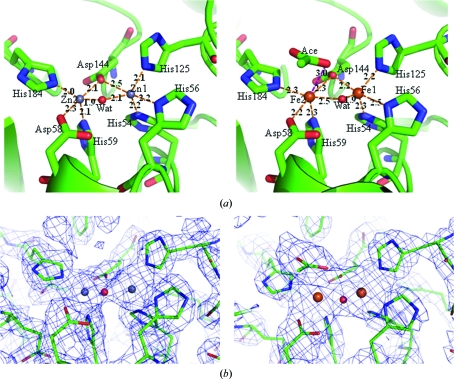
In di-zinc-bound TTHA1623, Zn1 was recognized by His54, His56, His125, Asp144 and a water molecule and Zn2 was recognized by Asp58, His59, Asp144, His184 and a water molecule; both formed square-pyramidal coordinations (Fig. 2 ▶ a, left). In di-iron-bound TTHA1623, Fe1 and Fe2 were recognized by the same residues and in the same coordination modes as in di-zinc-bound TTHA1623 (Fig. 2 ▶ a, right). The residues involved in the metal coordination are identical to those in glyoxalase IIs. Interestingly, an acetate was observed 3.0 Å away from Fe2 in the metal-binding site of di-iron-bound TTHA1623. As a result, Fe2 is in a distorted octahedral coordination (Fig. 2 ▶ a, right). It is presumed that the acetate mimics the hydrolyzed product of an amide or an ester, which are the putative substrates of TTHA1623. The acetate was not observed in di-zinc-bound TTHA1623, despite the presence of 200 mM zinc acetate in the reservoir solution (Fig. 2 ▶ b).
Figure 2.
(a) The putative active sites of TTHA1623 in di-zinc-bound (left) and di-iron-bound (right) forms. Zinc ions are labelled Zn1 and Zn2 and iron ions are labelled Fe1 and Fe2. The ligand water molecules are shown in red spheres and labelled Wat. The acetate ion is shown in a stick model and labeled as Ace. Dotted lines show the coordination of metal ions by their ligands. The distances between atoms in Å are labeled on the dotted lines. (b) σA-weighted 2F o − F c OMIT maps of the putative active sites of TTHA1623 in di-zinc-bound (left) and di-iron-bound (right) forms. The maps are contoured at 1σ. This figure was drawn with PyMOL (DeLano, 2002 ▶).
3.4. Structural similarity search
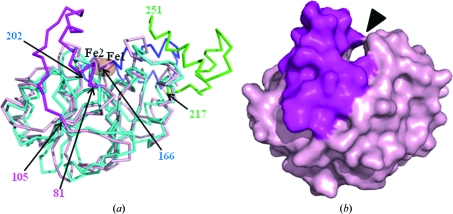
A structural similarity search was performed with the atomic coordinates of di-iron-bound TTHA1623 using the DALI server (Holm & Sander, 1995 ▶), as the native TTHA1623 is presumed to bind iron ions. The top five hits were Arabidopsis thaliana glyoxalase II (PDB code 2q42; Levin et al., 2007 ▶), A. thaliana ETHE1-like protein (PDB code 2gcu; McCoy et al., 2006 ▶), Salmonella typhimurium glyoxalase II (PDB code 2qed; Campos-Bermudez et al., 2007 ▶), human glyoxalase II (PDB code 1qh5; Cameron et al., 1999 ▶) and Bacillus cereus metallo-β-lactamase (PDB code 2bfz; Davies et al., 2005 ▶) (Table 3 ▶). The superimposition of TTHA1623 with 2qed, the only di-iron-binding enzyme among the DALI hits, was performed using DaliLite (Holm & Park, 2000 ▶). The αββα-folds of these metallo-β-lactamase superfamily proteins are similar to each other, with an r.m.s.d. of 1.8 Å between 170 pairs of Cα atoms. However, their putative substrate-binding pockets were dissimilar. TTHA1623 lacks residues corresponding to 166–202 and 217–251 of 2qed, which form part of the substrate-binding pocket of 2qed, while 2qed lacks residues corresponding to 81–105 of TTHA1623, which form part of the putative substrate-binding pocket of TTHA1623 (Fig. 3 ▶).
Table 3. Summarization of DALI results.
| Z score | PDB code | Source | Protein name | Residues involved in coordination | Metal ions |
|---|---|---|---|---|---|
| 2zzi | T. thermophilus | TTHA1623 | Site 1: His54, His56, His125, Asp144 | Site 1: Fe | |
| Site 2: Asp58, His59, Asp144, His184 | Site 2: Fe | ||||
| 21.6 | 2q42 | A. thaliana | Glyoxalase II | Site 1: His54, His56, His112 | Site 1: Zn |
| Site 2: Asp58, His59, Asp131, His169 | Site 2: Fe | ||||
| 21.6 | 2gcu | A. thaliana | ETHE1-like protein | Site 1: His72, His128, Asp153 | Site 1: Fe |
| Site 2: None | |||||
| 20.2 | 2qed | S. typhimurium | Glyoxalase II | Site 1: His53, His55, His110, Asp127 | Site 1: Fe |
| Site 2: Asp57, His58, Asp127, His165 | Site 2: Fe | ||||
| 19.7 | 1qh5 | Homo sapiens | Glyoxalase II | Site 1: His54, His56, His110, Asp134 | Site 1: Zn |
| Site 2: Asp58, His59, Asp134, His173 | Site 2: Zn | ||||
| 19.2 | 2bfz | B. cereus | Metallo-β-lactamase | Site 1: His116, His118, His196 | Site 1: Zn |
| Site 2: None |
Figure 3.
(a) Superposed diagram of TTHA1623 (shown in pink) and 2qed (light blue). The region unique to TTHA1623, residues 81–105, is shown in magenta. The regions unique to 2qed, residues 166–202 and 217–251, are shown in blue and green, respectively. The residue numbers of TTHA1623 and 2qed are labelled in the diagram. (b) Surface diagram of TTHA1623. The region unique to TTHA1623 (residues 81–105) is shown in magenta and the putative substrate-binding pocket is indicated by an arrowhead. The viewing points of (a) and (b) are the same. This figure was drawn with PyMOL (DeLano, 2002 ▶).
4. Conclusion
The present results reveal that (i) TTHA1623 exhibits a uniquely shaped putative substrate-binding pocket with a glyoxalase II-type metal-coordination mode, (ii) the recombinant TTHA1623 protein was in the di-iron-bound form and (iii) the bound iron ions were replaced by zinc ions in the presence of 200 mM zinc ions in the environment.
Supplementary Material
PDB reference: zinc-bound TTHA1623, 2zwr, r2zwrsf
PDB reference: iron-bound TTHA1623, 2zzi, r2zzisf
Acknowledgments
This work was performed under the Structural-Biological Whole Cell Project led by Dr Seiki Kuramitsu at the RIKEN SPring-8 Center. The synchrotron-radiation experiments were performed at BL26B1 at SPring-8 (Harima, Japan). This work was supported by the RIKEN Structural Genomics/Proteomics Initiative (RSGI) and by the National Project on Protein Structural and Functional Analyses and Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- Bebrone, C. (2007). Biochem. Pharmacol.74, 1686–1701. [DOI] [PubMed]
- Cameron, A. D., Ridderström, M., Olin, B. & Mannervik, B. (1999). Structure, 7, 1067–1078. [DOI] [PubMed]
- Campos-Bermudez, V. A., Leite, N. R., Krog, R., Costa-Filho, A. J., Soncini, F. C., Oliva, G. & Vila, A. J. (2007). Biochemistry, 46, 11069–11079. [DOI] [PubMed]
- Carfi, A., Pares, S., Duée, E., Galleni, M., Duez, C., Frère, J. M. & Dideberg, O. (1995). EMBO J.14, 4914–4921. [DOI] [PMC free article] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Concha, N. O., Rasmussen, B. A., Bush, K. & Herzberg, O. (1996). Structure, 4, 823–836. [DOI] [PubMed]
- Crowder, M. W., Maiti, M. K., Banovic, L. & Makaroff, C. A. (1997). FEBS Lett.418, 351–354. [DOI] [PubMed]
- Davies, A. M., Rasia, R. M., Vila, A. J., Sutton, B. J. & Fabiane, S. M. (2005). Biochemistry, 44, 4841–4849. [DOI] [PubMed]
- DeLano, W. L. (2002). The PyMOL Molecular Graphics System. http://www.pymol.org.
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Holm, L. & Park, J. (2000). Bioinformatics, 16, 566–567. [DOI] [PubMed]
- Holm, L. & Sander, C. (1995). Trends Biochem. Sci.20, 478–480. [DOI] [PubMed]
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst.26, 283–291.
- Levin, E. J., Kondrashov, D. A., Wesenberg, G. E. & Phillips, G. N. Jr (2007). Structure, 15, 1040–1052. [DOI] [PMC free article] [PubMed]
- McCoy, J. G., Bingman, C. A., Bitto, E., Holdorf, M. M., Makaroff, C. A. & Phillips, G. N. (2006). Acta Cryst. D62, 964–970. [DOI] [PubMed]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Terwilliger, T. C. & Berendzen, J. (1999). Acta Cryst. D55, 849–861. [DOI] [PMC free article] [PubMed]
- Vagin, A. & Teplyakov, A. (2000). Acta Cryst. D56, 1622–1624. [DOI] [PubMed]
- Yamamura, A., Ohtsuka, J., Kubota, K., Agari, Y., Ebihara, A., Nakagawa, N., Nagata, K. & Tanokura, M. (2008). Proteins, 73, 1053–1057. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: zinc-bound TTHA1623, 2zwr, r2zwrsf
PDB reference: iron-bound TTHA1623, 2zzi, r2zzisf
PDB reference: zinc-bound TTHA1623, 2zwr, r2zwrsf
PDB reference: iron-bound TTHA1623, 2zzi, r2zzisf