Glutathione S-transferases (GSTs) are a group of detoxifying enzymes that are found in animals, plants and microorganisms. Here, the crystallizations of two cyanobacterial GSTs are reported with the aim of determining their atomic structures.
Keywords: cyanobacteria, detoxification, glutathione transferases, isothiocyanates, toxins
Abstract
Glutathione S-transferases (GSTs) are a group of multifunctional enzymes that are found in animals, plants and microorganisms. Their primary function is to remove toxins derived from exogenous sources or the products of metabolism from the cell. Mammalian GSTs have been extensively studied, in contrast to bacterial GSTs which have received relatively scant attention. A new class of GSTs called Chi has recently been identified in cyanobacteria. Chi GSTs exhibit a high glutathionylation activity towards isothiocyanates, compounds that are normally found in plants. Here, the crystallization of two GSTs are presented: TeGST produced by Thermosynechococcus elongates BP-1 and SeGST from Synechococcus elongates PCC 6301. Both enzymes formed crystals that diffracted to high resolution and appeared to be suitable for further X-ray diffraction studies. The structures of these GSTs may shed further light on the evolution of GST catalytic activity and in particular why these enzymes possess catalytic activity towards plant antimicrobial compounds.
1. Introduction
Glutathione S-transferases (GSTs; EC 2.5.1.18) belong to a multifunctional family of proteins that are produced by most, if not all, aerobic organisms, in which they play a major role in protecting cells against exogenous and endogeous small-molecule toxins. They catalyze the conjugation of glutathione (GSH) to a range of electrophilic compounds, making the target molecules more water-soluble and thus facilitating their excretion from the cell. In mammals, GSTs can be divided into three major families based on structural similarities and differences. The major cytosolic family, comprising the Alpha, Mu, Omega, Pi, Sigma, Theta and Zeta classes, have been the most extensively studied, in part because of their involvement in disease. This family of GSTs assemble as dimers, with each monomer being composed of two domains. The N-terminal domain contains most of the residues that form the GSH-binding site, whereas the C-terminal domain includes residues that are involved in recognizing the target toxin molecule (Wilce & Parker, 1994 ▶; Armstrong, 1997 ▶; Sheehan et al., 2001 ▶; Hayes et al., 2005 ▶). The N-terminal domain of GSTs possesses a thioredoxin fold, suggesting that GSTs may have evolved from the thioredoxin superfamily (Martin, 1995 ▶). Mitochondrial GSTs, called Kappa class GSTs, form a distinct family. The third family includes membrane-associated proteins denoted MAPEG (membrane-associated proteins in eicosanoid and glutathione metabolism).
The best described bacterial GSTs (Allocati et al., 2009 ▶) are the enzymes from Proteus mirabilis (Perito et al., 1996 ▶) and Escherichia coli (Nishida et al., 1994 ▶). Structural studies suggested that these GSTs exhibited novel features, justifying their classification into a separate class termed Beta (Nishida et al., 1998 ▶; Rossjohn et al., 1998 ▶). Subsequently, crystal structures have been described for bacterial GSTs from Burkholderia xenovorans (Tocheva et al., 2006 ▶), Ochrobactrum anthropi (Federici et al., 2007 ▶) and Shewanella oneidensis (Remmerie et al., 2008 ▶). Interestingly, S. oneidensis GST is better classified as a Theta-class enzyme, whereas the others are clearly Beta-class enzymes. Very recently, a novel class of bacterial GSTs called the Chi class has been described (Wiktelius & Stenberg, 2007 ▶). These GSTs were found in cyanobacteria and represent a new class of GST that appears to be specific to cyanobacteria. The GSTs from Thermosynechococcus elongates (TeGST) and Synechococcus elongates (SeGST) have a shorter amino-acid sequence than most GSTs described to date. Their catalytic activities and substrate specificities were generally comparable with those of other bacterial GSTs. A surprising finding of the study was that both enzymes showed an exceptionally high activity towards structurally different isothiocyanates (ITCs), which are antimicrobial toxins that are found in plants. The structures of these enzymes might provide insight into the evolution of this surprising catalytic activity.
2. Cloning, expression and purification
2.1. TeGST and SeGST
The cloning, expression and purification protocols for TeGST and SeGST have been reported elsewhere (Wiktelius & Stenberg, 2007 ▶). Briefly, amplification of GST-encoding genes was achieved by PCR. DNA was subcloned into pGEM3-Z (Promega) and used for transformation of competent E. coli cells. The amplified DNA was ligated into expression vector pKK-D. Expression was induced by the addition of isopropyl β-d-1-thiogalactopyranoside. The cells were harvested by centrifugation, resuspended in buffer containing lysozyme and further disrupted by ultrasonication. The lysates were mixed with Glutathione Sepharose (Amersham Biosciences). The gel was washed with 10 mM Tris–HCl buffer pH 7.8, 1 mM EDTA and proteins were eluted with 50 mM glycine pH 10. The samples were passed through a PD-10 Sephadex G25 column and dialyzed against 10 mM Tris–HCl buffer pH 7.8 and 1 mM EDTA. TeGST and SeGST were concentrated to 18 and 3 mg ml−1, respectively.
3. Protein crystallization
3.1. TeGST
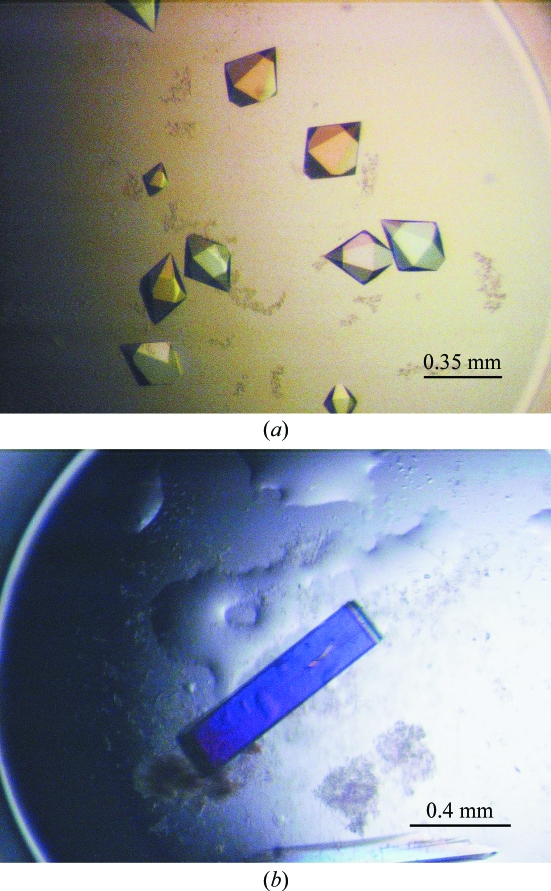
Screening for crystallization conditions of TeGST was performed using the hanging-drop vapour-diffusion method in 24-well plates (Linbro, ICN Biochemicals Inc., Ohio, USA) at 295 K. For crystallization, the protein solution was diluted to 6 mg ml−1. 2 µl protein solution was mixed with 2 µl reservoir solution on a cover slip and equilibrated over 1 ml well solution. The initial crystallization conditions were established using Index Screen from Hampton Research (California, USA). After 1 d, crystals appeared in a wide range of different conditions. A few days later, 85 drops out of 96 conditions contained crystals. Most crystals were hexagonal bipyramids (Fig. 1 ▶ a), but their detailed morphology and quantity varied depending on the condition. It was noted that in certain drops the crystals lost their well defined edges within a few days. The condition that gave the best-looking and largest crystals was optimized. The reservoir solution of this condition was 35%(v/v) pentaerythritol propoxylate, 0.2 M potassium chloride and 50 mM HEPES buffer pH 7.5. After optimization, the best-looking crystals appeared in 25–35%(v/v) pentaerythritol propoxylate, 0.2 M potassium chloride and 50 mM HEPES buffer pH 7.5. The largest crystals reached maximum dimensions of 0.35 × 0.25 × 0.25 mm within 3 d (Fig. 1 ▶ a).
Figure 1.
Crystals of cyanobacterium GSTs. Crystal dimensions are detailed in the text. (a) TeGST, (b) SeGST.
3.2. SeGST
Initial crystallization trials used Index Screen from Hampton Research (California, USA). After one week, small spines grew in 2.0 M sodium malonate. The crystals grew up to 1 mm in length but were very thin. In an attempt to increase the crystal thickness, the crystallization conditions were varied by using different sodium malonate concentrations and by the use of additives from Additive Screens I and II (Hampton Research, California, USA), without success. All trials were carried out at a constant temperature of 295 K. In order to find a new crystal form, crystallization trials were set up in a nanolitre format with a Phoenix crystallization robot (Art Robbins Instruments, Sunnyvale, California, USA) located at the Bio21 Collaborative Crystallization Centre (Bio21-C3), Parkville, Melbourne (http://www.csiro.au/c3/). Screening experiments were performed using 768 crystallization conditions from eight commercial kits including Com1 (Crystal Screen HT from Hampton Research), Com2 (Wizard Full from Emerald Biosystems, Bainbridge Island, Washington, USA), Com3 (Index Screen from Hampton Reseach), Com4 (PEG/Ion, Quik and Grid MPD from Hampton Research), Com5 (Precipitant Synergy Screen Primary and conditions 1–32 of Precipitant Synergy Screen 67% from Emerald Biosystems), Com6 (conditions 33–64 of Precipitant Synergy Screen 67% and Precipitant Synergy Screen 33% from Emerald Biosystems), Com7 (The PACT Suite from Qiagen, Doncaster, Australia) and Com8 (The Anions Suite from Qiagen). The nanolitre crystallization experiments were set up using the sitting-drop method in Innovadyne SD-2 plates with the reservoir containing 50 µl precipitant solution. 100 nl protein solution was mixed with 100 nl reservoir solution. Crystals appeared after 5 d in 20%(w/v) PEG 6000, 0.2 M CaCl2 and 100 mM HEPES buffer pH 7.5 or 100 mM MES buffer pH 6.5. Crystallization conditions were scaled up using the hanging-drop vapour-diffusion method in 24-well plates (Linbro, ICN Biochemicals Inc., Ohio, USA). 2 µl droplets were mixed with an equal volume of reservoir solution. Each well contained 1 ml reservoir solution. The best-diffracting crystals grew in 13–18%(w/v) PEG 6000, 0.2 M CaCl2 and 100 mM MES buffer pH 6.5. Crystals grew to maximal dimensions of 0.15 × 0.1 × 0.8 mm (Fig. 1 ▶ b) within three weeks.
4. Data collection and preliminary X-ray analysis
4.1. TeGST
Crystals of TeGST were taken from the crystallization drop and flash-cooled at 100 K in a stream of liquid nitrogen. The crystals diffracted to 1.4 Å resolution and a data set was collected on beamline 14-BM-C of the Advanced Photon Source synchrotron, USA. The diffraction images were recorded on an ADSC Quantum-4 CCD image-plate detector. The crystals belonged to space group P6522, with unit-cell parameters a = b = 64.2, c = 196.9 Å. The data were indexed and scaled with the HKL-2000 suite (Otwinowski & Minor, 1997 ▶) and converted to CCP4 format (Collaborative Computational Project, Number 4, 1994 ▶). Diffraction data statistics are summarized in Table 1 ▶. Molecular replacement was performed with AMoRe (Navaza, 2001 ▶) using a polyalanine model of P. mirabilis GST (PDB code 1pmt; Rossjohn et al., 1998 ▶). One GST monomer was found in the asymmetric unit, corresponding to a Matthews coefficient (Matthews, 1968 ▶) of 2.85 Å3 Da−1 with an estimated solvent content of 58%. Refinement of the model is in progress.
Table 1. Crystallographic data-processing statistics.
Values in parentheses are for the highest resolution bin (approximate interval of 0.1 Å).
| TeGST | SeGST | |
|---|---|---|
| Space group | P6522 | P212121 |
| Unit-cell parameters (Å) | a = 64.2, b = 64.2, c = 196.9 | a = 76.7, b = 94.3, c = 101.4 |
| Resolution (Å) | 1.45 | 2.0 |
| No. of observations | 391235 | 612041 |
| No. of unique reflections | 43959 | 47422 |
| Redundancy | 8.9 (9.0) | 12.9 (7.0) |
| Data completeness (%) | 99.9 (100) | 94.3 (72.3) |
| I/σ(I) | 28.9 (3.3) | 28.5 (3.0) |
| Rmerge† (%) | 7.2 (68.1) | 8.1 (42.2) |
R
merge = 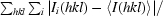
 , where I
i(hkl) is the intensity of the ith measurement of an equivalent reflection with indices hkl.
, where I
i(hkl) is the intensity of the ith measurement of an equivalent reflection with indices hkl.
4.2. SeGST
Prior to flash-freezing, the crystals were quickly dipped in mother liquor containing 15%(v/v) glycerol. Diffraction data were collected at 100 K and recorded on an ADSC Quantum-4 CCD image-plate detector on beamline GM/CA-CAT of the Advanced Photon Source synchrotron (Chicago, USA). The crystals diffracted to 2.0 Å resolution and belonged to the orthorhombic space group P212121, with unit-cell parameters a = 76.7, b = 94.3, c = 101.4 Å. The data were processed with the HKL-2000 suite (Otwinowski & Minor, 1997 ▶) and converted to CCP4 format. Data-collection and processing statistics are shown in Table 1 ▶. The sequence identity between SeGST and TeGST is 60%, but that between SeGST and P. mirabilis GST is only 28%. Owing to the higher sequence identity between SeGST and TeGST, a preliminary model of TeGST was chosen as a search model for molecular replacement. Molecular replacement was carried out using the program Phaser (Storoni et al., 2004 ▶). Four GST monomers were found in the asymmetric unit, corresponding to a Matthews coefficient (Matthews, 1968 ▶) of 2.29 Å3 Da−1 with an estimated solvent content of 46%. Refinement of the model is in progress.
Acknowledgments
This work was supported by the Australian Synchrotron Research Program, which is funded by the Commonwealth of Australia under the Major National Research Facilities Program. Use of the Advanced Photon Source was supported by the US DOE, Basic Energy Sciences, Office of Energy Research. GM/CA CAT has been funded in whole or in part with US Federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Science (Y1-GM-1104). This work was also supported by a grant from the Australian Research Council. SCF is supported by a National Health and Medical Research Council of Australia (NHMRC) Industry Fellowship. MWP is an Australian Research Council Federation Fellow and an NHMRC Honorary Fellow.
References
- Allocati, N., Federici, L., Masulli, M. & Di Ilio, C. (2009). FEBS J.276, 58–75. [DOI] [PubMed]
- Armstrong, R. (1997). Chem. Res. Toxicol.10, 2–18. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Federici, L., Masulli, M., Bonivento, D., Di Matteo, A., Gianni, S., Favaloro, B., Di Ilio, C. & Allocati, N. (2007). Biochem. J.403, 267–274. [DOI] [PMC free article] [PubMed]
- Hayes, J. D., Flanagan, J. U. & Jowsey, I. R. (2005). Annu. Rev. Pharmacol. Toxicol.45, 51–88. [DOI] [PubMed]
- Martin, J. L. (1995). Structure, 3, 245–250. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Navaza, J. (2001). Acta Cryst. D57, 1367–1372. [DOI] [PubMed]
- Nishida, M., Harada, S., Noguchi, S., Satow, Y., Inoue, H. & Takahashi, K. (1998). J. Mol. Biol.281, 135–147. [DOI] [PubMed]
- Nishida, M., Kong, K.-H., Inoue, H. & Takahashi, K. (1994). J. Biol. Chem.269, 32536–32541. [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Perito, B., Allocati, N., Casalone, E., Masulli, M., Dragani, B., Polsinelli, M., Aceto, A. & Di Ilio, C. (1996). Biochem. J.318, 157–162. [DOI] [PMC free article] [PubMed]
- Remmerie, B., Vandenbroucke, K., De Smet, L., Carpentier, W., De Vos, D., Stout, J., Van Beeumen, J. & Savvides, S. N. (2008). Acta Cryst. F64, 548–553. [DOI] [PMC free article] [PubMed]
- Rossjohn, J., Polekhina, G., Feil, S. C., Allocati, N., Masulli, M., Di Ilio, C. & Parker, M. W. (1998). Structure, 6, 721–734. [DOI] [PubMed]
- Sheehan, D., Meade, G., Foley, V. M. & Dowd, C. A. (2001). Biochem. J.360, 1–16. [DOI] [PMC free article] [PubMed]
- Storoni, L. C., McCoy, A. J. & Read, R. J. (2004). Acta Cryst. D60, 432–438. [DOI] [PubMed]
- Tocheva, E. I., Fortin, P. D., Eltis, L. D. & Murphy, M. E. P. (2006). J. Biol. Chem.281, 30933–30940. [DOI] [PubMed]
- Wiktelius, E. & Stenberg, G. (2007). Biochem. J.406, 115–123. [DOI] [PMC free article] [PubMed]
- Wilce, M. C. J. & Parker, M. W. (1994). Biochim. Biophys. Acta, 1205, 1–18. [DOI] [PubMed]