A protease-resistant mutant form of human galectin-8 has been crystallized and diffraction data have been collected to 3.4 Å resolution.
Keywords: galectin-8, carbohydrate-recognition domains, tandem-repeat-type galectins
Abstract
A crystal of a protease-resistant mutant form of human galectin-8, a tandem-repeat-type galectin with two carbohydrate-recognition domains, was obtained using the hanging-drop method and was found to belong to the tetragonal space group P43212, with unit-cell parameters a = 78.93, b = 78.93, c = 132.05 Å. Diffraction data were collected to a resolution of 3.4 Å.
1. Introduction
The galectins are a family of animal lectins which are defined based on their affinity for β-galactosides and their conserved sequence elements for carbohydrate recognition. To date, ten members of the human galectin family have been discovered and the family members can be classified into three subtypes on the basis of their structure. The prototype galectins (galectin-1, galectin-2, galectin-7, galectin-10 and galectin-13) and chimera-type galectins (galectin-3) have a carbohydrate-recognition domain (CRD) that usually forms a noncovalent homodimer with two identical sugar-binding sites. The tandem-repeat-type galectins (galectin-4, galectin-8, galectin-9 and galectin-12) have two CRDs and these domains are joined by an ∼30-amino-acid linker peptide. Since different CRDs usually show different sugar-binding specificities, tandem-repeat-type galectins can cross-link a variety of glycoconjugates owing to their heterobifunctional properties.
Galectin-8 was first identified in a rat liver cDNA library (Hadari et al., 1995 ▶) and human galectin-8 cDNA was identified by two independent studies concerning tumour-associated antigens (Su et al., 1996 ▶; Bassen et al., 1999 ▶). In contrast to other tandem-repeat-type galectins, galectin-8 is widely expressed in normal tissues, including the liver, heart, lungs, muscle and brain, in addition to tumour cells. Galectin-8 is expected to be involved in malignant transformation and cell–matrix interaction (Danguy et al., 2001 ▶; Hadari et al., 2000 ▶; Levy et al., 2001 ▶) and has also been reported to regulate neutrophil adhesion by interacting with target molecules (Nishi et al., 2003 ▶; Yamamoto et al., 2008 ▶).
The tandem-repeat-type galectins are susceptible to proteolysis owing to the presence of the linker peptides, making it difficult to carry out in vivo experiments using recombinant proteins. Therefore, protease-resistant tandem-repeat-type galectin-8 and galectin-9 were developed by modification of their linker peptide (Nishi et al., 2005 ▶). A mutant form of human galectin-8 that lacks the entire linker region (G8Null; 291 amino-acid residues; 33 065 Da) has a high protease resistance compared with wild-type galectin-8; interestingly, it can induce neutrophil adhesion in the same manner as wild-type galectin-8. This suggests that the linker peptide is not essential for interaction with target molecules in the regulation of cell adhesion.
To elucidate the relationship between the structure and biological activity of galectin-8, the structure of galectin-8 with both N- and C-terminal CRDs is very important. In this study, we succeeded in obtaining a crystal of G8Null and carried out preliminary X-ray diffraction analysis. To date, the crystal structures of single CRDs of tandem-repeat-type galectins (galectin-4, galectin-8 and galectin-9) have been determined. This is the first report of an X-ray diffraction study of a tandem-repeat-type galectin that includes both CRDs.
2. Materials and methods
2.1. Purification and crystallization
The expression and purification of G8Null have previously been reported (Nishi et al., 2005 ▶). Briefly, G8Null was expressed in Escherichia coli BL21 (DE3) cells and purified by affinity chromatography on a lactose–agarose column (Seikagaku Corp., Tokyo, Japan). After dialysis against buffer solution (10 mM Tris–HCl pH 7.5, 50 mM NaCl) overnight, the purified protein solution was concentrated to 4.26 mg ml−1 using an Amicon Ultra-4 10 kDa Ultracel (Millipore, Billerica, Massachusetts, USA).
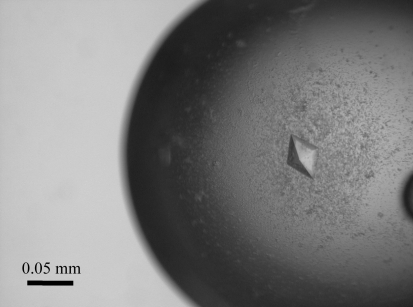
Initial crystallization screening for G8Null was performed using Crystal Screens I and II, PEG/Ion Screen, SaltRX Screen, Index Screen (Hampton Research Corp., California, USA) and Wizard I, II and III (Emerald BioSystems, Washington, USA) by the sitting-drop method with 96-well plates (Corning Inc., New York, USA) at 293 K. Small crystals appeared in two weeks during the equilibration of a droplet containing the same volumes (1 µl) of protein solution (4.26 mg ml−1 in 10 mM Tris–HCl pH 7.5, 50 mM NaCl) and reservoir solution [2%(v/v) 1,4-dioxane, 10%(w/v) PEG 20 000 and 0.1 M bicine pH 9.0; Crystal Screen II condition No. 48) against 100 µl reservoir solution. To obtain a crystal that was suitable for an X-ray diffraction experiment, the hanging-drop method was used with a 24-well plate (TPP, Switzerland). As shown in Fig. 1 ▶, a crystal with dimensions of 0.05 × 0.05 × 0.05 mm grew after 6 d in a droplet containing 2 µl each of protein solution and the above reservoir solution (Crystal Screen II condition No. 48) equilibrated against 500 µl reservoir solution.
Figure 1.
A crystal of G8Null with dimensions of 0.05 × 0.05 × 0.05 mm.
2.2. X-ray diffraction data collection
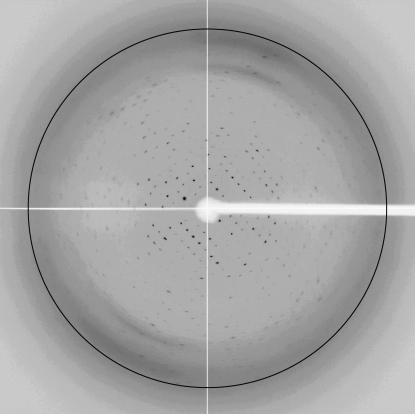
Data collection was carried out at KEK PF-AR NW12A (Tsukuba, Japan) using an ADSC Quantum 210 CCD detector at a wavelength of 1.0 Å. A crystal mounted in a loop was soaked in crystallization solution additionally containing 30%(v/v) glycerol and was flash-cooled in a stream of evaporating nitrogen at 100 K. The crystal of G8Null diffracted to a resolution of 3.4 Å as shown in Fig. 2 ▶. All data were processed using the HKL-2000 system (Otwinowski & Minor, 1997 ▶).
Figure 2.
Diffraction image of human G8Null with 3.4 Å resolution marked.
3. Results and discussion
A complete data set was successfully collected to 3.4 Å resolution as summarized in Table 1 ▶. The crystal belonged to the tetragonal space group P41212 or P43212, with unit-cell parameters a = 78.93, b = 78.93, c = 132.05 Å. The asymmetric unit is expected to contain one molecule, with a crystal volume per unit molecular weight (V M) of 3.1 Å3 Da−1 (Matthews, 1968 ▶). The solvent content of 60.5% in the crystal of G8Null is relatively high compared with the 46.2% solvent content found for the human galectin-8 N-terminal CRD (N-CRD; PDB code 2yv8; S. Kishishita, A. Nishino, K. Murayama, T. Terada, M. Shirouzu & S. Yokoyama, unpublished work), suggesting that the molecular surface of G8Null is largely exposed to solvent molecules in the crystal.
Table 1. Data-collection statistics.
Values in parentheses are for the highest resolution bin (3.52–3.40 Å).
| Beamline | PF-AR NW12A |
| Temperature (K) | 100 |
| Wavelength (Å) | 1.0 |
| Resolution range (Å) | 50–3.4 |
| No. of measured reflections | 70218 |
| No. of unique reflections | 6233 |
| Redundancy | 11.8 (9.2) |
| Completeness (%) | 95.8 (72.4) |
| Mean Io/σ(Io) | 9.2 (3.5) |
| Rmerge† (%) | 10.5 (38.4) |
| Space group | P43212 |
| Unit-cell parameters | |
| a (Å) | 78.93 |
| b (Å) | 78.93 |
| c (Å) | 132.1 |
| VM (Å3 Da−1) | 3.1 |
| Solvent content (%) | 60.5 |
R
merge = 
 , where I
i(hkl) is the ith measurement and 〈I(hkl)〉 is the weighted mean of all measurements of I(hkl).
, where I
i(hkl) is the ith measurement and 〈I(hkl)〉 is the weighted mean of all measurements of I(hkl).
Using the structure of human galectin-8 N-CRD, which has a β-sandwich structure, a molecular-replacement method was applied using the MOLREP program from the CCP4 program suite (Collaborative Computational Project, Number 4, 1994 ▶). Human galectin-8 N-CRD and C-terminal CRD (C-CRD; PDB code 2yro; T. Tomizawa, S. Koshiba, M. Inoue, T. Kigawa & S. Yokoyama, unpublished work) have 37% amino-acid sequence identity with a similar three-dimensional structure. A clear solution of the rotation and translation parameters for two CRDs in space group P43212 was obtained (the R factor and score of the best solution were 0.528 and 0.514, respectively; those of the second best solution were 0.591 and 0.407), while no plausible solutions were obtained in space group P41212 (the R factor and score of the best solution were 0.587 and 0.390, respectively; those of the second solution were 0.621 and 0.355). Because the calculated electron densities based on the best solution in space group P43212 appeared to be suitable for model building, the space group of G8Null was determined to be P43212.
We are currently refining the crystallization conditions in order to obtain crystals that diffract to higher resolution.
Acknowledgments
This research was supported in part by Kagawa University Characteristic Prior Research fund 2009 and was performed with the approval of the Photon Factory Advisory Committee and the National Laboratory for High Energy Physics, Japan.
References
- Bassen, R., Brichory, F., Caulet-Maugendre, S., Bidon, N., Delaval, P., Desrues, B. & Dazord, L. (1999). Anticancer Res.19, 5429–5433. [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Danguy, A., Rorive, S., Decaestecker, C., Bronckart, Y., Kaltner, H., Hadari, Y. R., Goren, R., Zich, Y., Petein, M., Salmon, I., Gabius, H. J. & Kiss, R. (2001). Histol. Histopathol.16, 861–868. [DOI] [PubMed]
- Hadari, Y. R., Arbel-Goren, R., Levy, Y., Amsterdam, A., Alon, R., Zakut, R. & Zick, Y. (2000). J. Cell Sci.113, 2385–2397. [DOI] [PubMed]
- Hadari, Y. R., Paz, K., Dekel, R., Mestrovic, T., Accili, D. & Zick, Y. (1995). J. Biol. Chem.270, 3447–3453. [DOI] [PubMed]
- Levy, Y., Arbel-Goren, R., Hadari, Y. R., Eshhar, S., Ronen, D., Elhanany, E., Geiger, B. & Zick, Y. (2001). J. Biol. Chem.276, 31285–31295. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Nishi, N., Itoh, A., Fujiyama, A., Yoshida, N., Araya, S., Hirashima, M., Shoji, H. & Nakamura, T. (2005). FEBS Lett.579, 2058–2064. [DOI] [PubMed]
- Nishi, N., Shoji, H., Seki, M., Itoh, A., Miyanaka, H., Yuube, K., Hirashima, M. & Nakamura, T. (2003). Glycobiology, 13, 755–763. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Su, Z. Z., Lin, J., Shen, R., Fisher, P. E., Goldstein, N. I. & Fisher, P. I. (1996). Proc. Natl Acad. Sci. USA, 93, 7252–7257. [DOI] [PMC free article] [PubMed]
- Yamamoto, H., Nishi, N., Shoji, H., Itoh, A., Lu, L.-H., Hirashima, M. & Nakamura, T. (2008). J. Biochem.143, 311–324. [DOI] [PubMed]