Abstract
The Chondrichthyes (cartilaginous fishes) are commonly accepted as being sister group to the other extant Gnathostomata (jawed vertebrates). To clarify gnathostome relationships and to aid in resolving and dating the major piscine divergences, we have sequenced the complete mtDNA of the starry skate and have included it in phylogenetic analysis along with three squalomorph chondrichthyans—the common dogfish, the spiny dogfish, and the star spotted dogfish—and a number of bony fishes and amniotes. The direction of evolution within the gnathostome tree was established by rooting it with the most closely related non-gnathostome outgroup, the sea lamprey, as well as with some more distantly related taxa. The analyses placed the chondrichthyans in a terminal position in the piscine tree. These findings, which also suggest that the origin of the amniote lineage is older than the age of the oldest extant bony fishes (the lungfishes), challenge the evolutionary direction of several morphological characters that have been used in reconstructing gnathostome relationships. Applying as a calibration point the age of the oldest lungfish fossils, 400 million years, the molecular estimate placed the squalomorph/batomorph divergence at ≈190 million years before present. This dating is consistent with the occurrence of the earliest batomorph (skates and rays) fossils in the paleontological record. The split between gnathostome fishes and the amniote lineage was dated at ≈420 million years before present.
Keywords: vertebrate evolution, Amniota, Chondrichthyes, starry skate, molecular dating
The relationship between gnathostomous fishes and their terrestrial relatives is of fundamental importance for the understanding of vertebrate evolution. Molecular analyses of this relationship have addressed in particular the question of whether, among extant fishes, the lungfishes or the coelacanth are the sister group to terrestrial vertebrates. However, although these analyses have differed with respect to the taxa included, a teleostean (1–4) or chondrichthyan (5) rooting of the gnathostome tree has been a common characteristic, and these studies have, in general, supported a sister group relationship between lungfishes and amniotes (or tetrapods). Because the application of rooting automatically gives evolutionary direction to a tree, it is essential that rooting is performed by using an outgroup that is unambiguously positioned without the ingroup taxa. The commonly applied teleostean rooting of the vertebrate tree is incompatible with piscine paleontology (6, 7) whereas the chondrichthyan rooting is subjective in the sense that it assumes a priori that chondrichthyans are the sister group of all other extant gnathostomes. Therefore, the application of either the teleostean or chondrichthyan rooting is inconsistent with the criterion that unequivocal outgroups should be used to establish the polarity of phylogenetic trees.
The conclusions based on the teleostean and chondrichthyan rooting have been challenged in two recent molecular studies (8, 9) in which the gnathostome tree was rooted by using non-gnathostome taxa. The first study indicated that the lungfishes have a basal position in the piscine tree and that the separation between extant bony fishes and amniotes preceded the divergence of the extant bony fishes. The second study refuted the commonly held belief that the Chondrichthyes are basal to other gnathostomes. This analysis, with only the spiny dogfish, Squalus acanthias, representing squalomorph chondrichthyans, did not, however, resolve the relationship between the coelacanth, the chondrichthyans, and the teleosts. To examine this relationship in greater detail, we have, in the present study, broken up the chondrichthyan branch by including mitochondrial genes of three other chondrichthyans, the common dogfish, Scyliorhinus canicula, (10), the star spotted dogfish, Mustelus manazo, (11), and the starry skate, Raja radiata, (present study). Thus, the chondrichthyans are represented by a total of three squalomorphs and one batomorph.
The divergence between squalomorphs (sharks) and batomorphs (skates and rays) is paleontologically dated to the early Jurassic (6, 7, 12). Even though this is the minimum age for the squalomorph/batoid divergence, the inclusion of the skate, in addition to strengthening the phylogenetic analysis, makes it possible to test a molecular estimate of the divergence time between the squalomorphs and the skate against the paleontological record of the Batomorphii.
MATERIALS AND METHODS
Enriched mtDNA was isolated from frozen liver of the starry skate, Raja radiata, following described procedures (13). The specimen was collected in Faxafloi, Iceland, by Oskar Gudmundsson. The mtDNA was digested separately with BlnI and BclI. Digested DNA fragments were separated on an agarose gel and were excised, electroeluted, and ligated. With the exception of parts of the NADH2 and NADH5 genes, which were PCR-amplified and direct-sequenced, natural clones covered the whole molecule. The mtDNA of the starry skate has been deposited in the GenBank database with accession number AF106038. Users of the sequence are kindly requested to refer to the present paper and not only to the accession number of the sequence.
The phylogenetic analyses included all published piscine mtDNAs together with a comprehensive selection of taxa represented by complete mtDNAs, namely sea lamprey, Petromyzon marinus (14); African lungfish, Protopterus dolloi (3); bichir, Polypterus ornatipinnis (2); coelacanth, Latimeria chalumnae (4); starry skate, Raja radiata, (present study); spiny dogfish, Squalus acanthias (9); common dogfish, Scyliorhinus canicula (10); star spotted dogfish, Mustelus manazo (11); loach, Crossostoma lacustre (15); rainbow trout, Onchorhynchus mykiss (16); carp, Cyprinus carpio; Atlantic cod, Gadus morhua (17); alligator, Alligator mississippiensis (18); ostrich, Struthio camelus (19); chicken, Gallus gallus (20); wallaroo, Macropus robustus (21); and cow, Bos taurus (22). Analyses also were carried out by using the same taxa with an addition of different outgroup sequences: the hagfish, Myxine glutinosa (8), the lancelet, Branchiostoma floridae (23), and three echinoderm sequences, Arbacia lixula (24), Strongylocentrotus purpuratus (25), and Asterina pectinifera (26).
The phylogenetic analyses [maximum likelihood (ML) (27), neighbor joining (NJ) (28), and maximum parsimony (MP) (29)] were performed on amino acid as well as nucleotide alignments of the concatenated sequences of 12 mitochondrial protein-coding genes. The nucleotide analyses were based on the combined data sets of both first (excluding synonymous leucine transitions) and second codon positions.
RESULTS AND DISCUSSION
General Features of the Mitochondrial Genome of the Starry Skate.
The size of the mitochondrial genome of the starry skate is 16,785 nt. The organization of the molecule is the same as generally found in mammals and gnathostomous fishes. With the exception of cytochrome oxidase subunit I, all protein-coding genes of the starry skate have a canonical methionine start codon, ATG or ATA. The start codon of cytochrome oxidase subunit I is GTG (valine). GTG is not a unique start codon among vertebrates and has, for example, been reported in the NADH4L gene of the blue whale and the NADH6 gene of Indian rhinoceros. The cytochrome oxidase subunit I and NADH4 genes have an incomplete stop codon, T, rather than a complete stop codon, TAA or TAG. Incomplete stop codons, TA or T, have been shown to occur in many mitochondrial protein-coding genes among the vertebrates. The tRNA genes of the starry skate conform with those characterizing other vertebrates, and all tRNAs can be folded into the common clover leaf structures. G-U base pairings occur in some stem structures, but base pairings of this kind were uncommon compared with standard Watson-Crick base pairings.
Phylogeny.
The phylogenetic analyses were carried out on an alignment of the 12 protein-coding genes encoded by the mitochondrial H-strand, excluding the L-strand encoded gene NADH6 (the composition of which deviates from that of the H-strand encoded genes). The different genes of each taxon were combined into one supergene because, as has been demonstrated by statistical analysis (30), in this manner, the stochastic effects of limited sequence data are reduced. The analyses were performed primarily on a data set that, in addition to the gnathostomes, included the sea lamprey Petromyzon marinus (14), but rooting of the gnathostome tree also was tested separately with the hagfish Myxine glutinosa (8), the lancelet Branchiostoma floridae (23), and three echinoderms—one starfish and two sea urchins—Arbacia lixula (24), Strongylocentrotus purpuratus (25), and Asterina pectinifera, (26).
After exclusion of gaps and ambiguous sites adjacent to gaps, the length of the lamprey alignment was 7,959 nt or 2,653 aa. The distance values of the amino acid data set are given in Table 1. Relative rate test between the gnathostomous fishes and the sea lamprey and between the gnathostomous fishes and the amniotes showed that the evolutionary rate of the lungfish is 10–15% higher than that of other gnathostomous fishes. This difference in evolutionary rate was compensated for in the subsequent molecular estimates of evolutionary divergence times by using the age of lungfish fossils as a calibration point. The distance values in Table 1 are consistent with all gnathostomous fishes being on a common branch separated from the amniotes. Thus, these values provide no support for the hypothesis that the lungfishes, as represented by the African lungfish, Protopterus dolloi, are the piscine sister group of the amniotes, as maintained in several other mtDNA studies (1, 3, 31, 32).
Table 1.
Pairwise distances among vertebrate taxa
| Sea lamprey | Chicken | Ostrich | Alligator | Cow | Wallaroo | Lungfish | Bichir | Coelacanth | Loach | Carp | Atlantic cod | Rainbow trout | Starry skate | Spiny dogfish | Star spotted dogfish | Common dogfish | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sea lamprey | – | 0.3809 | 0.3826 | 0.4585 | 0.3795 | 0.3739 | 0.3450 | 0.3329 | 0.3279 | 0.3312 | 0.3240 | 0.3371 | 0.3276 | 0.3305 | 0.3249 | 0.3582 | 0.3359 |
| Chicken | 760 | – | 0.0894 | 0.3400 | 0.3100 | 0.2993 | 0.3001 | 0.2878 | 0.2817 | 0.2625 | 0.2627 | 0.2773 | 0.2690 | 0.2898 | 0.2661 | 0.2849 | 0.2820 |
| Ostrich | 755 | 219 | – | 0.3540 | 0.3046 | 0.2961 | 0.3045 | 0.2947 | 0.2820 | 0.2682 | 0.2712 | 0.2828 | 0.2690 | 0.2861 | 0.2637 | 0.2868 | 0.2827 |
| Alligator | 858 | 681 | 699 | – | 0.3819 | 0.3832 | 0.3857 | 0.3808 | 0.3680 | 0.3615 | 0.3602 | 0.3712 | 0.3621 | 0.2861 | 0.3639 | 0.3774 | 0.3698 |
| Cow | 763 | 649 | 638 | 751 | – | 0.1764 | 0.3071 | 0.2951 | 0.2768 | 0.2784 | 0.2760 | 0.2798 | 0.2787 | 0.2802 | 0.2682 | 0.2845 | 0.2768 |
| Wallaroo | 752 | 632 | 624 | 760 | 415 | – | 0.3011 | 0.2920 | 0.2850 | 0.2764 | 0.2722 | 0.2794 | 0.2801 | 0.2814 | 0.2637 | 0.2893 | 0.2776 |
| Lungfish | 696 | 632 | 634 | 757 | 643 | 633 | – | 0.2450 | 0.2273 | 0.2217 | 0.2175 | 0.2282 | 0.2140 | 0.2332 | 0.2205 | 0.2437 | 0.2205 |
| Bichir | 682 | 615 | 620 | 748 | 625 | 619 | 536 | – | 0.2117 | 0.1923 | 0.1890 | 0.2041 | 0.1869 | 0.2185 | 0.1985 | 0.2195 | 0.2106 |
| Coelacanth | 686 | 606 | 600 | 736 | 599 | 611 | 502 | 478 | – | 0.1704 | 0.1622 | 0.1921 | 0.1616 | 0.1970 | 0.1666 | 0.1856 | 0.1805 |
| Loach | 682 | 567 | 574 | 715 | 595 | 594 | 487 | 434 | 395 | – | 0.0738 | 0.1266 | 0.1036 | 0.1743 | 0.1435 | 0.1682 | 0.1637 |
| Carp | 673 | 569 | 580 | 718 | 593 | 587 | 479 | 432 | 380 | 184 | – | 0.1245 | 0.0982 | 0.1649 | 0.1377 | 0.1634 | 0.1616 |
| Atlantic cod | 699 | 595 | 596 | 730 | 595 | 597 | 503 | 457 | 439 | 303 | 299 | – | 0.1053 | 0.1879 | 0.1690 | 0.1921 | 0.1914 |
| Rainbow trout | 677 | 576 | 576 | 721 | 594 | 600 | 472 | 425 | 379 | 253 | 241 | 255 | – | 0.1658 | 0.1444 | 0.1634 | 0.1583 |
| Starry skate | 685 | 613 | 600 | 714 | 597 | 604 | 510 | 486 | 447 | 402 | 383 | 426 | 383 | – | 0.1034 | O.1210 | 0.1143 |
| Spiny dogfish | 675 | 573 | 567 | 723 | 578 | 594 | 488 | 449 | 388 | 339 | 326 | 390 | 340 | 252 | – | 0.0827 | 0.0774 |
| Star spotted dogfish | 726 | 604 | 603 | 742 | 603 | 595 | 529 | 487 | 424 | 388 | 378 | 434 | 377 | 287 | 203 | – | 0.0692 |
| Common dogfish | 693 | 602 | 599 | 734 | 595 | 572 | 513 | 472 | 418 | 382 | 377 | 435 | 369 | 276 | 194 | 171 | – |
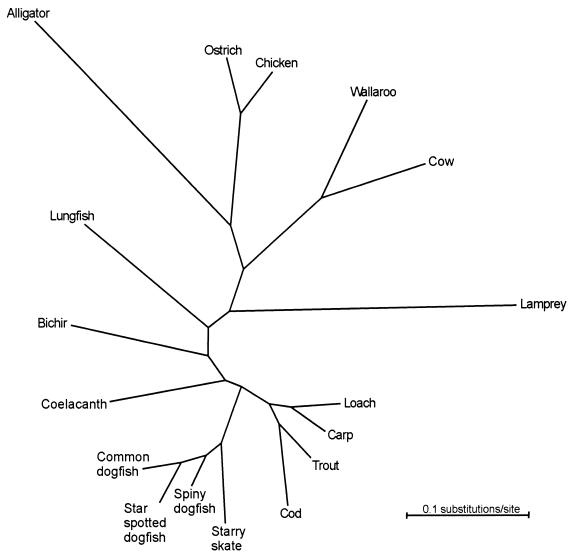
The present analyses concentrated on the relationship between gnathostomous fishes and amniotes rather than that between gnathostomous fishes and tetrapods. This is because the amniotes constitute a monophyletic group, inter alia, characterized by the synapomorph amnion whereas the monophyly of amphibians, and hence the tetrapods as a whole, has been questioned (33). The amphibians currently are represented by only one complete mtDNA, that of Xenopus laevis (34), and the present study did not, therefore, allow analysis of basal amphibian or tetrapod relationships. Furthermore, previous analyses of the mtDNA of Xenopus have shown that the molecule is unstable in the vertebrate tree (3, 4, 8, 35, 36), an observation that was confirmed in the present study because Xenopus was the only species in the study that did not maintain a constant position in different analyses. After the exclusion of Xenopus, the different data sets (amino acids or nucleotides) and the different methods of phylogenetic reconstruction (ML, NJ, and MP) yielded one consistent tree topology. Fig. 1 shows the unrooted tree reconstructed by ML analysis of the amino acid alignment, assuming rate homogeneity. As is evident, the teleosts and the chondrichthyans constitute sister groups. This relationship, along with the other phylogenetic relationships in Fig. 1, is consistent with the distance values shown in Table 1. Fig. 2 shows the NJ tree of the same data set, but rooted by using the position of the sea lamprey. Again, consistent with the distance values in Table 1, all gnathostomous fishes are on a common branch, with the lungfish in a basal position on that branch. Rooting of the gnathostome data set with either the hagfish, the lancelet, or the echinoderms reconstructed the same split between amniotes and fishes as in the lamprey data set, but the position of the lungfish was not resolved in all MP analyses. The same results were obtained in analyses taking into account different evolutionary rates by use of discrete Γ distribution with five rate categories (37).
Figure 1.
An unrooted ML tree including gnathostomes and the sea lamprey. The tree was reconstructed from analyses of the concatenated amino acid sequences of 12 mitochondrial protein-coding genes as described in the text. For scientific names, see Materials and Methods. The sea lamprey was replaced by the hagfish, the lancelet, or three echinoderms in three parallel analyses. Like the lamprey, all of these taxa received a position between the lungfish and the amniotes.
Figure 2.
A NJ tree showing the evolutionary polarity of the gnathostome tree as established by rooting with the sea lamprey. The primary gnathostome split is, as shown here, between fishes and amniotes. The lungfish has a basal position on the piscine branch. The four chondrichthyans, the common dogfish, the spiny dogfish, the star spotted dogfish, and the starry skate have a terminal position in the piscine tree. Maximal support values are indicated by an asterisk on individual branches whereas other values are given in Table 2. The relationship among the piscine taxa was maintained whether or not the amniotes were included in the analyses.
Table 2 gives the support values for different branches (labeled a–k) in Fig. 2. The support values for the sister group relationship between chondrichthyans and teleosts (branch d) are 78% (ML), 79% (NJ), and 70% (MP), respectively. The terminal position of the Chondrichthyes in the piscine tree is consistent with a previous Kishino-Hasegawa (ML) test (38), which refuted with statistical significance a basal position of the spiny dogfish among the Gnathostomata (9). It should be observed that the chondrichthyan/teleostean split does not automatically imply teleostean origin because the present analysis does not exclude the possibility that the branch leading to the teleosts may be split by some neopterygians (e.g., bowfin and gars) or acipenseriforms (sturgeons and paddlefish).
Table 2.
Support values in percent for the branches of the tree in Fig. 2
| Method | a | b | c | d | e | f | g | h | i | j | k |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NJ | 98 | 92 | 95 | 79 | 100 | 98 | 100 | 100 | 100 | 99 | 100 |
| MP | 37 | 39 | 74 | 70 | 99 | 96 | 100 | 90 | 100 | 98 | 99 |
| ML | 100 | 99 | 97 | 78 | 100 | 100 | 96 | 97 | 98 | 100 | 100 |
NJ and MP values were calculated by using 100 bootstrap replicates whereas ML values were calculated by using 1,000 Quartet puzzling steps (44).
The terminal position of chondrichthyans and teleosts in the piscine tree is evident in Fig. 2. Therefore, if the piscine tree is rooted with either chondrichthyans or teleosts, the tree will be inverted, placing the lungfish at the top of that tree. As a consequence of such a rooting, and in the absence of the sea lamprey, the lungfish automatically would become the sistergroup of the amniotes whereas, in the presence of the lamprey, the teleostean or chondrichthyan rootings would place the lamprey as the sistergroup of the amniotes. The artificial reconstruction of a topology with the lungfish as the sistergroup of the amniotes is consistent with the conclusions of a series of molecular studies applying either teleostean or chondrichthyan rooting (1–4, 11, 31). These rootings are incompatible, however, with the use of an unequivocal outgroup for establishing the polarity of phylogenetic trees.
Compared with the previous study (9), the present analysis improved the resolution of the relationship between the coelacanth, the chondrichthyans, and the teleosts. In other respects, the phylogenetic findings of this study were consistent with recent findings applying unambiguous rooting of the gnathostome tree (8, 9), showing, inter alia, that the divergence between amniotes and extant gnathostomous fishes took place before the diversification of extant gnathostome fishes.
The present findings challenge the commonly accepted understanding of basal gnathostome divergences and relationships. It might be argued that protein-coding mtDNA genes do not have the capacity to correctly resolve early relationships such as deep piscine divergences or the divergence between fishes and amniotes. It has been claimed that MP analysis of mitochondrial protein-coding genes may provide strong support for incorrect phylogenies (39). This conclusion was based on MP analysis of an alignment, >12,000 nt long, of concatenated mitochondrial protein-coding sequences, extending from nematodes to mammals. We have collected and reanalyzed the same set of taxa. This reanalysis was performed according to the conservative approach used in the present study, i.e., by removing gaps and all ambiguous sites adjacent to gaps. After these precautionary steps, which aim at ensuring comparison of truly homologous sites, the length of the alignment was 5,127 nt, i.e., <40% of that used in the previous nematode/vertebrate study. Consistent with several other mtDNA analyses, the position of Xenopus remained unresolved. Apart from this, our analysis of the more conservative data set (first and second codon positions) yielded a MP tree consistent with that championed as being the correct one (39). Thus, contrary to the conclusions based on the relaxed (>12,000 nt) nematode/vertebrate alignment, our analysis of the same data set suggests that conservative alignments of concatenated mtDNA sequences have the capacity to reconstruct correct topologies for even deep evolutionary divergences. In the nematode/vertebrate study (39), the piscine presence was limited to two teleosts. The analysis did not, therefore, address the relationship between lungfishes and tetrapods or have the potential to determine the polarity of the piscine tree.
In a recent molecular study (10) including the common dogfish, but not the lungfish or the bichir, the vertebrate tree was rooted with the lancelet. Like in the nematode/vertebrate study (39), the length of the alignment exceeded 12,000 nt. In this instance, the requirement of unequivocal rooting was fulfilled, but the phylogenetic analysis, which did not conclusively resolve the position of the common dogfish, included gaps and third codon position, thereby introducing considerable amount of noise into the data set. Exclusion of gaps and third codon positions from the data set reconstructed, with maximal support, a phylogenetic tree with a basal gnathostome split between the single tetrapod included (Gallus gallus) and all gnathostome fishes. It is also noteworthy that NJ reanalysis of the distance values given in Table 1 of that paper (10) place the common dogfish at a terminal position of the tree rather than at the position shown in the phylogenetic tree depicted in the paper.
Estimates of Divergence Times.
We have estimated the time of the divergence between the dogfishes and the starry skate, along with that of other piscine divergences, by using two independent molecular/paleontological calibration points. The first is the age of the oldest undisputed lungfish fossils (400 million years) whereas the second is the divergence of the Diapsida/Synapsida, 310 million years before present (MYBP) (40). Use of the lungfish fossil reference gives a squalomorph/batomorph divergence time of ≈190 MYBP. By using the same reference, the divergence between the chondrichthyan and teleostean branches was estimated at ≈290 MYBP, the divergence between the coelacanth and the branch leading to teleosts and chondrichthyans was estimated at ≈310 MYBP, and the divergence between the cladistians (bichir) and coelacanth/teleosts/chondrichthyans was estimated at ≈380 MYBP. The divergence between amniotes and gnathostomous fishes was estimated at ≈420 MYBP. By rooting the tree with echinoderm sequences, the divergence between agnathans (Petromyzon) and gnathostomes was estimated at ≈550 MYBP. The calculations based on the Diapsida/Synapsida calibration point (310 MYBP) yielded datings that were generally ≈10% more recent than those based on the lungfish reference. This discrepancy may be caused by insufficient correction for the faster evolutionary rate among amniotes than among fishes, a too-recent dating of the Diapsida/Synapsida reference, or a combination of these factors.
The oldest batoid fossils are of Jurassic age, ≈190 million years old (6, 7, 12). Thus, the lungfish dating of the divergence between the squalomorphs and the skate, given above, is consistent with the paleontological record whereas the corresponding dating of the origin of the Chondrichthyes, ≈290 MYBP, is much more recent than the age (Devonian) of Cladoselache and Leonodus fossils (7, 40), which commonly are accepted as ancestral chondrichthyans. However, Leonodus is only represented by teeth of somewhat uncertain identity whereas the relationship between Cladoselache and other gnathostomous fishes is unsettled (7). Even though cladoselachids were recognizably sharklike, their position as the ancestors of recent chondrichthyans has been questioned (7, 12, 40). The present findings suggest that the origin of the extant chondrichthyans included in the present study is unrelated to the cladoselachian lineage.
Use of the lungfish calibration point places the intrateleostean divergence between the clades containing the rainbow trout and the loach at ≈190 MYBP. It is probable that more extensive teleostean sampling will reveal other teleostean divergences that are even deeper than the rainbow trout/loach split. In a recent study (42), the split between Chondrichthyes and the other gnathostomes was dated molecularly at 530 MYBP. The present findings are inconsistent with that dating, and the phylogeny underlying that dating, which placed the Chondrichthyes basal to other extant gnathostomes.
Implications and Conclusions.
Providing that the phylogeny and the estimated datings of various divergences given here are indeed correct, these findings have major implications for both the monophyly of Chondrichthyes and the evolutionary direction of several morphological characters commonly used for reconstructing vertebrate relationships. Chondrichthyan relationships, based on morphological comparisons, have been reviewed thoroughly (41) and will not be detailed here. The author (41) concluded that all sharklike chondrichthyans are not necessarily elasmobranchs and defined the latter as extant sharks, skates, and rays plus several fossil taxa, such as Paleospinax, Synechodus, Hybodus, Xenacanthus, and Ctenacanthus. The phylogeny plus the molecular datings presented here, however, indicate that neither Xenacanthus nor Ctenacanthus are true elasmobranchs because the fossils representing these taxa are considerably older than the molecularly estimated elasmobranch origin, ≈300 MYBP. This suggests that reevaluation of the morphological characters uniting the Chondrichthyes will be necessary.
Considering the currently reconstructed phylogenetic tree in its entirety, our findings suggest the following interpretations of the polarity of some main anatomical/morphological gnathostome characters: (i) that the cartilaginous skeleton of modern Chondrichthyes is not ancestral to the osteichthyan skeleton; (ii) that the exoskeleton of chondrichthyans (almost entirely composed of minute, simple scales and independent teeth) is a derived condition that has arisen from the larger dermal plates of osteichthyans; (iii) that the lungs of both fishes and terrestrial vertebrates are ancestral to the swim bladder or the absence of that organ in the Chondrichthyes; (iv) that the separate gill slits of elasmobranchs, which have been regarded as a primitive condition because they are found widely among jawless vertebrates, are, in fact, secondary and are derived from a single operculate branchial opening as found in osteichthyans and holocephalans; and (v) that the polybasal fins (with several rays attached separately on the girdles) of cladistians, actinopterygians, and elasmobranchs are derived relative to the monobasal, lobed fins. Even though these interpretations differ radically from current views on gnathostome evolution, it is noteworthy that the presently proposed polarities of the lung/swim bladder, the bone/cartilage, and the exoskeletal characters have been advocated in basic texts (43), although, at that time, they were not put into the context of the phylogenetic relationships of early vertebrates.
The presently proposed polarity of the lung/swim bladder character may suggest that the early evolution of extant gnathostomes took place in shallow waters from which both deeper waters and terrestrial environments subsequently were colonized. Colonization of deep waters thus would have led to the rudimentation of the lungs as a breathing organ whereas the colonization of land would have been associated with extended development of the lungs. Given the present results, it also follows that the origin of the lineage leading to the amniotes is considerably older than 400 million years: i.e., the age of the oldest lungfishes. This suggests that piscine fossils younger than 400 million years, which have been linked to amniote origin, are either on the piscine branch and therefore unrelated to amniote origin or, in the phylogenetic sense, are ancestral amniotes with pronounced piscine characteristics.
Acknowledgments
We express our gratitude to Mr. Oskar Gudmundsson, Reykjavik, Iceland, for samples, to Dr. Philippe Janvier for valuable comments and suggestions, and to Ms. Kerryn Slack for help with the manuscript. The work has been supported by the Swedish Natural Sciences Research Council, the Jörgen Lindström Foundation, and the Nilsson-Ehle Foundation.
ABBREVIATIONS
- MYBP
million years before present
- ML
maximum likelihood
- MP
maximum parsimony
- NJ
neighbor joining
- NADH1-6
subunits 1–6 of nicotinamide adenine dinucleotide dehydrogenase
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF106038).
References
- 1.Meyer A, Wilson A C. J Mol Evol. 1990;31:359–364. doi: 10.1007/BF02106050. [DOI] [PubMed] [Google Scholar]
- 2.Noack K, Zardoya R, Meyer A. Genetics. 1996;144:1165–1180. doi: 10.1093/genetics/144.3.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zardoya R, Meyer A. Genetics. 1996;142:1249–1263. doi: 10.1093/genetics/142.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zardoya R, Meyer A. Genetics. 1997;146:995–1010. doi: 10.1093/genetics/146.3.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hedges S B, Moberg K D, Maxson L R. Mol Biol Evol. 1990;7:607–633. doi: 10.1093/oxfordjournals.molbev.a040628. [DOI] [PubMed] [Google Scholar]
- 6.Colbert E H, Morales M. Evolution of the Vertebrates. New York: Wiley–Liss; 1994. [Google Scholar]
- 7.Janvier P. Early Vertebrates. Oxford: Oxford Univ. Press; 1996. [Google Scholar]
- 8.Rasmussen A-S, Janke A, Arnason U. J Mol Evol. 1998;46:382–387. doi: 10.1007/pl00006317. [DOI] [PubMed] [Google Scholar]
- 9.Rasmussen A-S, Arnason U. J Mol Evol. 1999;48:118–123. doi: 10.1007/pl00006439. [DOI] [PubMed] [Google Scholar]
- 10.Delabre C, Spruyt N, Delmarre C, Gallut C, Barriel V, Janvier P, Laudet V, Gachelin G. Genetics. 1998;150:331–344. doi: 10.1093/genetics/150.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao Y, Waddell P J, Okada N, Hasegawa M. Mol Biol Evol. 1998;15:1637–1646. doi: 10.1093/oxfordjournals.molbev.a025891. [DOI] [PubMed] [Google Scholar]
- 12.Helfman G S, Colette B B, Facey D E. The Diversity of Fishes. Oxford: Blackwell Scientific; 1997. [Google Scholar]
- 13.Arnason U, Gullberg A, Widegren B. J Mol Evol. 1991;33:556–568. doi: 10.1007/BF02102808. [DOI] [PubMed] [Google Scholar]
- 14.Lee W-J, Kocher T D. Genetics. 1995;139:873–887. doi: 10.1093/genetics/139.2.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tzeng C S, Hui C F, Shen S C, Huang P C. Nucleic Acids Res. 1992;20:4853–4858. doi: 10.1093/nar/20.18.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zardoya R, Garrido-Pertierra A, Bautista J M. J Mol Evol. 1995;41:942–951. doi: 10.1007/BF00173174. [DOI] [PubMed] [Google Scholar]
- 17.Johansen S, Bakke I. Mol Marine Biol Biotech. 1996;5:203–214. [PubMed] [Google Scholar]
- 18.Janke A, Arnason U. Mol Biol Evol. 1997;14:1266–1272. doi: 10.1093/oxfordjournals.molbev.a025736. [DOI] [PubMed] [Google Scholar]
- 19.Härlid A, Janke A, Arnason U. Mol Biol Evol. 1997;14:754–761. doi: 10.1093/oxfordjournals.molbev.a025815. [DOI] [PubMed] [Google Scholar]
- 20.Desjardins P, Morais R. J Mol Biol. 1990;212:599–634. doi: 10.1016/0022-2836(90)90225-B. [DOI] [PubMed] [Google Scholar]
- 21.Janke A, Xu X, Arnason U. Proc Natl Acad Sci USA. 1997;94:1276–1281. doi: 10.1073/pnas.94.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson S, de Brujin M L H, Coulson A R, Eperon I C, Sanger F, Young G. J Mol Biol. 1982;156:683–717. doi: 10.1016/0022-2836(82)90137-1. [DOI] [PubMed] [Google Scholar]
- 23.Naylor G J P, Brown W M. Syst Biol. 1998;47:61–76. doi: 10.1080/106351598261030. [DOI] [PubMed] [Google Scholar]
- 24.De Giorgi C, Martiradonna A, Lanave C, Saccone C. Mol Phylogenet Evol. 1996;5:323–332. doi: 10.1006/mpev.1996.0027. [DOI] [PubMed] [Google Scholar]
- 25.Jacobs H T, Elliott D J, Math V B, Farquharson A. J Mol Biol. 1988;202:185–217. doi: 10.1016/0022-2836(88)90452-4. [DOI] [PubMed] [Google Scholar]
- 26.Asakawa S, Himeno H, Miura K, Watanabe K. Genetics. 1995;140:1047–1060. doi: 10.1093/genetics/140.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Felsenstein J. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 28.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 29.Fitch W M. Syst Zool. 1971;20:406–415. [Google Scholar]
- 30.Cao Y, Adachi J, Janke A, Pääbo S, Hasegawa M. J Mol Evol. 1994;39:519–527. doi: 10.1007/BF00173421. [DOI] [PubMed] [Google Scholar]
- 31.Hedges S B, Hass C A, Maxson L R. Nature (London) 1993;363:501–502. doi: 10.1038/363501b0. [DOI] [PubMed] [Google Scholar]
- 32.Zardoya R, Cao Y, Hasegawa M, Meyer A. Mol Biol Evol. 1998;15:506–517. doi: 10.1093/oxfordjournals.molbev.a025950. [DOI] [PubMed] [Google Scholar]
- 33.Jarvik E. In: Studies in Herpetology. Rocek Z, editor. Prague: Univerzita Karlova; 1986. pp. 1–24. [Google Scholar]
- 34.Roe R A, Ma D-P, Wilson R K, Wong J F-H. J Biol Chem. 1985;260:9759–9774. [PubMed] [Google Scholar]
- 35.Adachi J. Ph.D. thesis. Tokyo: School of Mathematical and Physical Science; 1995. [Google Scholar]
- 36.Lê H L V, Lecointre G, Perasso R. Mol Phylogenet Evol. 1993;2:31–51. doi: 10.1006/mpev.1993.1005. [DOI] [PubMed] [Google Scholar]
- 37.Yang Z. J Mol Evol. 1994;39:306–314. doi: 10.1007/BF00160154. [DOI] [PubMed] [Google Scholar]
- 38.Kishino H, Hasegawa M. J Mol Evol. 1989;29:170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- 39.Naylor G J, Brown W M. Nature (London) 1997;388:527–528. doi: 10.1038/41460. [DOI] [PubMed] [Google Scholar]
- 40.Benton M J. J Mol Evol. 1990;30:409–424. doi: 10.1007/BF02101113. [DOI] [PubMed] [Google Scholar]
- 41.Maisey J G. J Vertebrate Paleontol. 1984;4:359–371. [Google Scholar]
- 42.Kumar S, Hedges S B. Nature (London) 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- 43.Romer A S. The Vertebrate Body. Philadelphia: Saunders; 1955. [Google Scholar]
- 44.Strimmer K, von Heaseler A. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- 45.Adachi J, Hasegawa M. J Mol Evol. 1996;42:459–468. doi: 10.1007/BF02498640. [DOI] [PubMed] [Google Scholar]