Abstract
The activation of ATR-ATRIP in response to double-stranded DNA breaks (DSBs) depends upon ATM in human cells and Xenopus egg extracts. One important aspect of this dependency involves regulation of TopBP1 by ATM. In Xenopus egg extracts, ATM associates with TopBP1 and thereupon phosphorylates it on S1131. This phosphorylation enhances the capacity of TopBP1 to activate the ATR-ATRIP complex. We show that TopBP1 also interacts with the Mre11-Rad50-Nbs1 (MRN) complex in egg extracts in a checkpoint-regulated manner. This interaction involves the Nbs1 subunit of the complex. ATM can no longer interact with TopBP1 in Nbs1-depleted egg extracts, which suggests that the MRN complex helps to bridge ATM and TopBP1 together. The association between TopBP1 and Nbs1 involves the first pair of BRCT repeats in TopBP1. In addition, the two tandem BRCT repeats of Nbs1 are required for this binding. Functional studies with mutated forms of TopBP1 and Nbs1 suggested that the BRCT-dependent association of these proteins is critical for a normal checkpoint response to DSBs. These findings suggest that the MRN complex is a crucial mediator in the process whereby ATM promotes the TopBP1-dependent activation of ATR-ATRIP in response to DSBs.
INTRODUCTION
In eukaryotic cells, various checkpoint mechanisms operate to prevent the dissemination of genetic errors by cells that have suffered DNA lesions (Nyberg et al., 2002; Sancar et al., 2004). Two phosphoinositide kinase-related kinases (PIKKs) called ATM and ATR function near the apex of key checkpoint pathways. ATM is involved in the initial detection of double-stranded DNA breaks (DSBs) that occur, for example, after treatment of cells with ionizing radiation (IR) or various genotoxic chemicals (Shiloh, 2006). On the other hand, ATR responds principally to problems that arise at replication forks (Abraham, 2001; Kumagai and Dunphy, 2006; Cimprich and Cortez, 2008). Interestingly, ATR also has a role in response to DSBs that involves cooperation with ATM (Cuadrado et al., 2006; Jazayeri et al., 2006; Myers and Cortez, 2006; Yoo et al., 2007). Despite their structural and functional similarities, ATR is essential for cell viability, whereas ATM is expendable for normal cell proliferation.
One critical function of ATR involves activation of the downstream checkpoint effector kinase called Chk1 (Guo et al., 2000; Liu et al., 2000; Zhao and Piwnica-Worms, 2001). Once activated by ATR, Chk1 phosphorylates and modulates the activity of various proteins, including the cell cycle regulators Cdc25 and Wee1 (Perry and Kornbluth, 2007). The collective effect of these phosphorylations is the imposition of a cell cycle arrest until the cell has alleviated the DNA lesion that initially triggered the ATR-mediated response.
In order for ATR to undergo activation and recognize substrates, it must collaborate with various other proteins. For example, ATR possesses a stably bound partner protein called ATRIP (Cortez et al., 2001). Significantly, ATRIP can interact directly with RPA, a single-stranded DNA-binding protein (Zou and Elledge, 2003). RPA accumulates in significant amounts at stalled replication forks, which typically continue DNA unwinding in the absence of DNA polymerase activity (Walter and Newport, 2000; Byun et al., 2005). ATR-ATRIP also detects the occurrence of DSBs by associating with the RPA on single-stranded DNA that arises as a result of resection at the exposed DNA ends (Cimprich and Cortez, 2008). Thus, ATR-ATRIP recognizes an array of DNA lesions that share single-stranded DNA as one structural feature.
The docking of ATR-ATRIP onto RPA does not affect its catalytic activity. Instead, an activating protein known as TopBP1 subsequently associates with ATR-ATRIP and thereupon stimulates a large increase in its intrinsic kinase activity toward a range of different substrates (Kumagai et al., 2006). This process involves the interaction of ATR-ATRIP with a discrete ATR-activating domain (AAD) within TopBP1 (Kumagai et al., 2006; Mordes et al., 2008). Another important step in the checkpoint response to stalled replication forks entails the interaction between TopBP1 and the checkpoint clamp comprised of Rad9, Hus1, and Rad1 (the 9-1-1 complex) (Garcia et al., 2005; Delacroix et al., 2007; Lee et al., 2007). The 9-1-1 complex is deposited onto recessed DNA ends by a checkpoint clamp-loader complex that contains Rad17 and the four small subunits of replication factor C (RFC) (Parrilla-Castellar et al., 2004; Sancar et al., 2004). As is the case with single-stranded DNA, recessed DNA ends also accumulate at stalled replication forks. The independent interactions of ATR-ATRIP and TopBP1 with different structural features of stalled replication forks may contribute to the fidelity and efficiency of checkpoint signaling.
ATR-ATRIP also plays a role in checkpoint responses to DSBs that involves upstream regulation by ATM (Cuadrado et al., 2006; Jazayeri et al., 2006; Myers and Cortez, 2006). In particular, ATM appears to control the resection of DNA ends in a process that depends on the Xmre11-Xrad50-Xnbs1 (MRN) complex. Resection generates single-stranded DNA that can be recognized sequentially by RPA and ATR-ATRIP. In addition, our laboratory recently identified ATM as a TopBP1-interacting protein in Xenopus egg extracts undergoing a checkpoint response to DSBs (Yoo et al., 2007). We pursued this observation by showing that ATM phosphorylates TopBP1 on S1131 and thereby strongly stimulates the ATR-activating capacity of TopBP1. Significantly, phosphorylation of S1131 is required for a proper checkpoint response to DSBs but apparently not to stalled replication forks.
In this report, we have identified additional TopBP1-interacting proteins in egg extracts as the three components of the MRN complex. We have pursued this observation by showing that the MRN complex is necessary for the recruitment of ATM to TopBP1 in response to DSBs. This step is a key event in the process whereby ATM promotes the activation of ATR-ATRIP.
MATERIALS AND METHODS
Xenopus Egg Extracts
Xenopus egg extracts were prepared as described (Yoo et al., 2004a). Extracts were treated with 50 μg/ml dA70-dT70 (pA-pT) to elicit checkpoint responses as described (Kumagai and Dunphy, 2000). To prepare egg extracts containing chromatin with DSBs or stalled DNA replication forks, demembranated sperm nuclei (1000–3000/μl) were incubated in extracts containing 0.05 U/μl EcoRI or 50 μg/ml aphidicolin, respectively. For preparation of nuclear fractions, egg extracts (50 μl) containing 3000 sperm nuclei/μl were incubated under the indicated conditions. Nuclear fractions were isolated from the extracts as described previously (Yoo et al., 2006).
Antibodies
A DNA fragment encoding amino acids 532–762 of Xenopus Nbs1 (Xnbs1) was generated by PCR and cloned into a pET-His6 expression vector. The His6-Xnbs1(532–762) protein was expressed in Escherichia coli, isolated with nickel agarose, and used for production of rabbit antibodies at a commercial facility. Antiserum against Xmre11 was the generous gift of Dr. V. Costanzo (Clare Hall Laboratories, United Kingdom). An anti-human Nbs1 antibody was purchased from Calbiochem (La Jolla, CA). Affinity-purified antibodies against Xenopus versions of ATM (Xatm) and TopBP1 (XtopBP1) were described previously (Yoo et al., 2004b; Kumagai et al., 2006). Anti-human Chk1 phospho-Ser317 and phospho-Ser345 antibodies were obtained from Cell Signaling Technology (Beverly, MA). Anti-GST, anti-FLAG, anti-human α-tubulin, and control rabbit antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA), Sigma (St. Louis, MO), Oncogene Research Products (Boston, MA), and Zymed (South San Francisco, CA), respectively.
Production of Xenopus MRN Components
cDNA clones encoding full-length versions of Xenopus Mre11 (Xmre11) and Xnbs1 (Costanzo et al., 2001; You et al., 2005) were isolated from a Xenopus oocyte library by PCR. The sequence encoding Xenopus Rad50 (Xrad50) was cloned by PCR based on two expressed sequence tags with significant homology to the 5′ and 3′ regions of human Rad50, respectively. The full-length cDNA clone defined a complete open reading frame of 1312 amino acids (GenBank accession no. FJ436027). The human Nbs1 cDNA clone was generously supplied by Dr. T. Paull (University of Texas at Austin).
Immunoprecipitation and Immunodepletion
For immunoprecipitations, egg extracts (100 μl) were incubated under constant agitation for 45 min at 4°C with Affiprep-protein A beads (Bio-Rad, Richmond, CA) coated with anti-XtopBP1 (3 μg) or anti-Xnbs1 antibodies (3 μg). The beads were washed three times with buffer A (10 mM HEPES-KOH, pH 7.5, 150 mM NaCl, 0.5% NP-40, 2.5 mM EGTA, and 20 mM β-glycerolphosphate), twice with HEPES-buffered saline (HBS; 10 mM HEPES-KOH, pH 7.5, and 150 mM NaCl), and subjected to SDS-PAGE and immunoblotting. For immunodepletion of Xnbs1, interphase extracts (100 μl) were incubated at 4°C for 45 min with 20 μg of anti-Xnbs1 antibodies bound to 20 μl of Affiprep protein A beads. The same amount of control rabbit IgG was used for mock depletion. After the incubation, the beads were removed by centrifugation and the supernatants were treated again for a second round of depletion. XtopBP1 was immunodepleted as described previously (Yoo et al., 2007).
Production of Recombinant Proteins
PCR-generated DNA fragments encoding Xmre11, Xrad50, and Xnbs1 were cloned into a pFastBac-FLAG vector to express proteins with a FLAG tag at the C-terminal end. Untagged versions of Xmre11 and Xnbs1 were prepared by using a pFastBac that does not encode any tag. Fragments comprising residues 1-410 and 338-762 of Xnbs1 were prepared by using the pFastBac-GH vector, which encodes glutathione S-transferase (GST) and His6 tags at the C-terminal end (Kumagai and Dunphy, 2000). Recombinant baculoviruses were generated with the Bac-to-Bac system (Invitrogen, Carlsbad, CA). Recombinant baculovirus-expressed proteins were produced in Sf9 insect cells and purified using anti-FLAG agarose beads (Yoo et al., 2007). To produce the Xenopus MRN complex, Sf9 insect cell were coinfected with three recombinant baculoviruses (encoding Xmre11, Xrad50-FLAG, and Xnbs1). The complex was purified from the infected cells with anti-FLAG antibody beads. Recombinant, full-length HF-XtopBP1 with both hemagglutinin (HA) and His6 tags at the N-terminal end and a FLAG tag at the C-terminal end was produced in baculovirus-infected Sf9 cells by previously described methods (Kumagai and Dunphy, 2000). The ΔI-II and BRCT I-II versions of XtopBP1 lack residues 96-282 and 360-1485, respectively. These constructs were designed to include the last 28 amino acids of XtopBP1 (residues 1486-1513), which contain the nuclear localization sequence (NLS) of the protein. 35S-labeled proteins were synthesized in vitro with the TnT system (Promega, Madison, WI). The vector pcDNA3.1 was used for expression of FLAG-tagged human Nbs1 in tissue culture cells. Point mutants of XtopBP1, Xnbs1, and human Nbs1 were produced using the QuikChange kit (Stratagene, La Jolla, CA).
Pulldowns of Recombinant XtopBP1 from Xenopus Egg Extracts
Recombinant HF-XtopBP1 was incubated in egg extracts containing pA-pT and reisolated as described in a previous publication (Yoo et al., 2007). XtopBP1-interacting proteins were separated by SDS-PAGE and stained with Coomassie brilliant blue. Excised bands were cut into ∼1-mm3 pieces, in-gel digested with trypsin, and extracted from the gel matrix with acetonitrile and 5% formic acid (Shevchenko et al., 2006). Recovered peptides were subjected to LC-MS/MS analysis on an LTQ ion trap mass spectrometer (Thermo Fisher Scientific, San Jose, CA) under the conditions described in Waridel et al. (2007). The presence of components of the Xenopus MRN complex in the pulldowns was established by the following criteria. Xmre11 (accession no. AAD31866) was identified with 38 individual peptides matching to 152 MS/MS spectra and covering 52% of the sequence. Xrad50 (LOC495064 protein, accession no. AAH84223) was identified with 50 individual peptides matching to 168 MS/MS spectra and covering 53% of the available sequence. Finally, Xnbs1 (accession no. AAQ82670) was identified with 37 individual peptides matching to 59 MS/MS spectra and covering 43% of the sequence.
Assay for Binding of Xnbs1 to XtopBP1 in Egg Extracts
Recombinant XtopBP1 (1 μg) bound to anti-FLAG antibody beads was incubated for 100 min in egg extracts (50 μl) containing 100 μg/ml cycloheximide in the absence or presence of 50 μg/ml pA-pT. The beads were isolated by centrifugation and washed three times with buffer A and twice with HBS. Bound proteins were subjected to SDS-PAGE and immunoblotting.
Cell Culture and Small Interfering RNA Transfection
HEK293T cells and HeLa cells were maintained in DMEM containing 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin. A small interfering RNA (siRNA) duplex specific for Nbs1 with the sense strand sequence GGAGGAAGAUGUCAAUGUUUU was purchased from Dharmacon Research (Boulder, CO). siRNA was transfected into HeLa cells with Lipofectamine 2000 (Invitrogen). Constructs encoding siRNA-resistant versions of wild-type and 2A (T158A and K160A) human Nbs1 were generated by changing three nucleotides in the siRNA-targeting region.
Lentivirus Generation and Infection
Sequences encoding siRNA-resistant versions of wild-type and 2A-mutant human Nbs1 were cloned downstream of the human ubiquitin-C promoter in the lentivector plasmid FUW. Lentivirus production was performed as described (Qin et al., 2003). Briefly, HEK293T cells were transfected by using the calcium phosphate transfection method. HEK293T cells in 6-cm culture dishes were transfected with the appropriate lentiviral vector plasmid (5 μg) along with 2.5 μg each of the HIV-1 lentiviral packaging constructs pRSV-Rev and pMDLg/pRRE as well as the vesicular stomatitis virus glycoprotein (VSVG) expression plasmid pVSV. The viral supernatants were harvested 48 h after transfection and filtered through a 0.45-μm pore size filter. HeLa cells were infected with virus in presence of 4 μg/ml Polybrene (Sigma). Supernatant was removed after 8 h and replaced with growth medium. Cells were incubated for 1 d after infection and then transfected with control or Nbs1 siRNA. After an additional 2 d, cells were exposed to IR (5 Gy) or were mock-treated in parallel. Whole cell extracts were prepared 2 h later and processed for SDS-PAGE and immunoblotting.
Kinase Assays
Kinase assays of endogenous Xatm were performed as described previously with PHAS-I as the substrate (Yoo et al., 2004b).
RESULTS
XtopBP1 Interacts with the MRN Complex in Egg Extracts
As described previously, we searched for XtopBP1-interacting proteins by performing pulldown experiments with a recombinant version of XtopBP1 containing the FLAG epitope (HF-XtopBP1) (Yoo et al., 2007). For this purpose, we incubated HF-XtopBP1 in Xenopus egg extracts containing annealed oligomers comprised of dA70 and dT70 (henceforth referred to as pA-pT). This template elicits the robust activation of Xchk1 in a process that depends on key upstream regulators, including XtopBP1, Xatr-Xatrip, and Claspin (Kumagai and Dunphy, 2000; Kumagai et al., 2004; Hashimoto et al., 2006; Kumagai et al., 2006). This pathway also requires the activation of XtopBP1 by Xatm (Yoo et al., 2007). In our earlier studies, we identified two XtopBP1-interacting proteins as Xatm and Xatr. In further analyses, we noted that these pulldowns also contained all three subunits of the MRN complex (Xmre11, Xrad50, and Xnbs1) (see Materials and Methods).
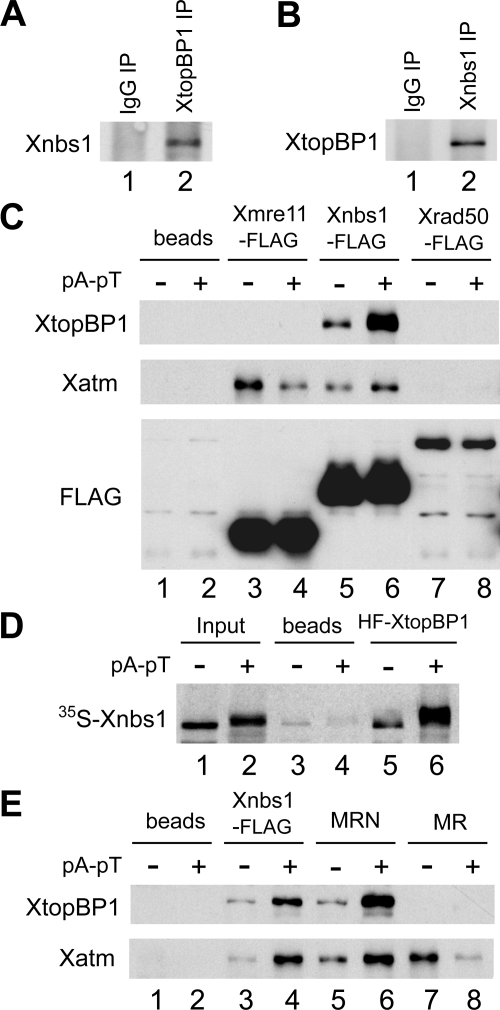
To assess the specificity of the observed interaction with the MRN components, we performed reciprocal immunoprecipitation experiments. For these studies, we prepared anti-Xnbs1 antibodies to detect the endogenous MRN complex in egg extracts. We subjected egg extracts containing pA-pT to immunoprecipitation with either anti-XtopBP1 or anti-Xnbs1 antibodies and then probed the immunoprecipitates for Xnbs1 or XtopBP1, respectively. We could readily detect Xnbs1 in anti-XtopBP1 immunoprecipitates and vice versa (Figure 1, A and B).
Figure 1.
XtopBP1 interacts in a regulated manner with the Xnbs1 subunit of the MRN complex in Xenopus egg extracts. (A) Control and anti-XtopBP1 immunoprecipitates (IP) from egg extracts containing pA-pT were immunoblotted for Xnbs1. (B) Control and anti-Xnbs1 immunoprecipitates from egg extracts containing pA-pT were immunoblotted for XtopBP1. (C) Anti-FLAG antibody beads containing no recombinant protein (lanes 1 and 2), Xmre11-FLAG (lanes 3 and 4), Xnbs1-FLAG (lanes 5 and 6), and Xrad50-FLAG (lanes 7 and 8) were incubated in egg extracts lacking or containing pA-pT. Beads were retrieved and immunoblotted with anti-XtopBP1, anti-Xatm, and anti-FLAG antibodies. (D) Anti-FLAG antibody beads containing no recombinant protein (lanes 3 and 4) or HF-XtopBP1 (lanes 5 and 6) were incubated in egg extracts containing 35S-labeled Xnbs1 with or without pA-pT. The beads were reisolated, and bound 35S-labeled Xnbs1 was detected by SDS-PAGE and phosphorimaging. Lanes 1 and 2, aliquots of initial extracts lacking or containing pA-pT. (E) Anti-FLAG antibody beads containing no recombinant protein (lanes 1 and 2), Xnbs1-FLAG (lanes 3 and 4), MRN complex (Xmre11, Xrad50-FLAG, and Xnbs1) (lanes 5 and 6), or MR complex (Xmre11 and Xrad50-FLAG) (lanes 7 and 8) were incubated in egg extracts lacking or containing pA-pT. Beads were retrieved and immunoblotted with antibodies against XtopBP1 and Xatm.
Next, we asked which component(s) of the MRN complex mediates the interaction with XtopBP1. For this purpose, we prepared C-terminally FLAG-tagged versions of Xmre11, Xrad50, and Xnbs1 in baculovirus-infected Sf9 insect cells. Full-length cDNA clones encoding Xmre11 and Xnbs1 had been described earlier (Costanzo et al., 2001; You et al., 2005). However, a Xenopus homologue of Rad50 (Xrad50) had not been characterized previously. Accordingly, we used DNA primers based on expressed sequence tags to isolate a full-length cDNA clone encoding Xrad50 from an oocyte cDNA library. The predicted sequence of Xrad50 encodes a polypeptide containing 1312 amino acids with 74.6% overall homology to human Rad50.
We incubated the FLAG-tagged versions of Xmre11, Xrad50, and Xnbs1 in egg extracts in the absence or presence of pA-pT. Next, we retrieved the tagged proteins from the extracts and probed for XtopBP1. We found that XtopBP1 associated only with the Xnbs1 subunit (Figure 1C). Furthermore, this binding was greatly increased in the presence versus absence of pA-pT. As expected, we could also observe binding of Xatm to both Xnbs1 and Xmre11 (Lee and Paull, 2007). These interactions did not depend on the presence of pA-pT. As another means to confirm the specificity of this interaction, we incubated a 35S-labeled form of Xnbs1 in egg extracts in the absence or presence of pA-pT and performed pulldowns with anti-FLAG antibody beads lacking or containing HF-XtopBP1. We also observed a specific, checkpoint-regulated interaction between Xnbs1 and XtopBP1 by this method (Figure 1D).
To pursue these observations further, we prepared the complete trimeric Xenopus MRN complex and the dimeric Xmre11-Xrad50 complex (MR) in Sf9 insect cells. It is known that Mre11 and Rad50 can form a functional complex with a subset of the activities of the whole MRN complex (Lee and Paull, 2007). We observed that XtopBP1 could associate with the Xenopus MRN but not MR complex (Figure 1E). Taken together, these experiments indicate that XtopBP1 engages in a checkpoint-regulated interaction with the Xenopus MRN complex through its Xnbs1 subunit and does not bind detectably to the Xmre11 or Xrad50 proteins.
Xnbs1 Associates with the BRCT I-II Region of XtopBP1
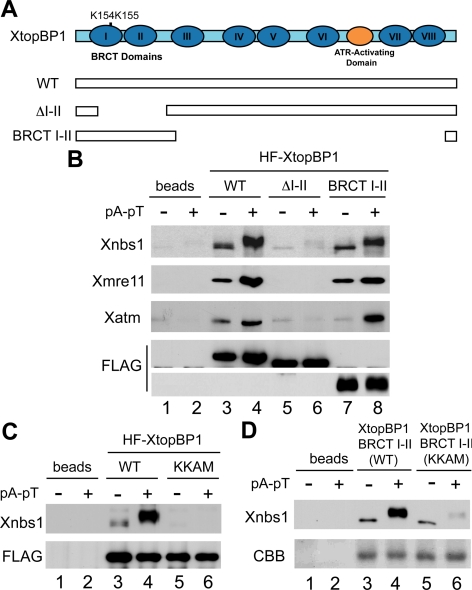
Next, we investigated which region of XtopBP1 is responsible for association with the MRN complex. XtopBP1 contains a total of eight BRCT repeats (see Figure 2A). We previously described various deletion mutants of XtopBP1 that lack one or more BRCT repeats or the ATR-activating domain (Lee et al., 2007). Using these mutants, we found that the version of XtopBP1 lacking the first two BRCT repeats (ΔI-II) was defective in the interaction with Xnbs1 (Figure 2B). Consistent with this observation, we found that there was also no binding of Xmre11 or Xatm to the ΔI-II mutant of XtopBP1. Furthermore, a FLAG-tagged fragment of XtopBP1 containing only the first two of the eight BRCT repeats could also bind to Xnbs1 in a checkpoint-regulated manner. Taken together, these experiments indicate that the BRCT I-II region of XtopBP1 is both necessary and sufficient for binding of the MRN complex. Interestingly, this same region of XtopBP1 is involved in the binding of the Xrad9 subunit of the Xenopus 9-1-1 complex (Lee et al., 2007).
Figure 2.
The BRCT I-II region of XtopBP1 is necessary and sufficient for association with Xnbs1. (A) Domains of XtopBP1. Roman numerals denote the BRCT domains of XtopBP1. Full-length HF-XtopBP1 (WT) and versions of XtopBP1 lacking residues 96-282 (ΔI-II) or 360-1485 (BRCT I-II) were produced in Sf9 cells. (B) Anti-FLAG antibody beads containing no recombinant protein (lanes 1 and 2) or the indicated versions of HF-XtopBP1 (lanes 3–8) were incubated in egg extracts in the absence or presence of pA-pT. Beads were retrieved and immunoblotted with anti-Xnbs1, anti-Xmre11, anti-Xatm, and anti-FLAG antibodies. (C) Wild-type and KKAM versions of HF-XtopBP1 on anti-FLAG antibody beads were incubated in egg extracts lacking or containing pA-pT. Beads were reisolated and immunoblotted with anti-Xnbs1 (top) and anti-FLAG antibodies (bottom). (D) Anti-FLAG antibody beads containing no recombinant protein (lanes 1 and 2) or either the wild-type or KKAM versions of the XtopBP1 BRCT I-II construct (lanes 3–6) were incubated in egg extracts in the absence or presence of pA-pT. Beads were retrieved and immunoblotted with the antibodies against Xnbs1 (top). The wild-type and KKAM versions of the BRCT I-II fragment on immunoblots were stained with Coomassie brilliant blue (CBB) (bottom).
Various studies have indicated that tandem BRCT repeats can form a phosphopeptide-binding domain (Caldecott, 2003; Glover et al., 2004). Structure-function analyses have identified critical residues that form the core of the phosphate-binding pocket. Through sequence alignments, we identified K155 in the first BRCT domain of XtopBP1 as a likely key residue in such a pocket. However, there is also a lysine at position 154. Accordingly, we decided to change both K154 and K155 to alanine and methionine, respectively, to create the XtopBP1-KKAM mutant. We incubated baculovirus-expressed, full-length versions of wild-type and KKAM-mutant HF-XtopBP1 in egg extracts in the absence or presence of pA-pT and then examined binding of Xnbs1. We observed that there was virtually no binding of Xnbs1 to the XtopBP1-KKAM protein (Figure 2C). We obtained similar results if we examined the binding of Xnbs1 to a KKAM mutant form of the XtopBP1 fragment containing BRCT repeats I-II (Figure 2D). Therefore, the putative phosphate-binding pocket in the BRCT I-II region of XtopBP1 appears to be critical for regulated binding to the MRN complex.
XtopBP1 Associates with the Tandem BRCT Repeats of Xnbs1
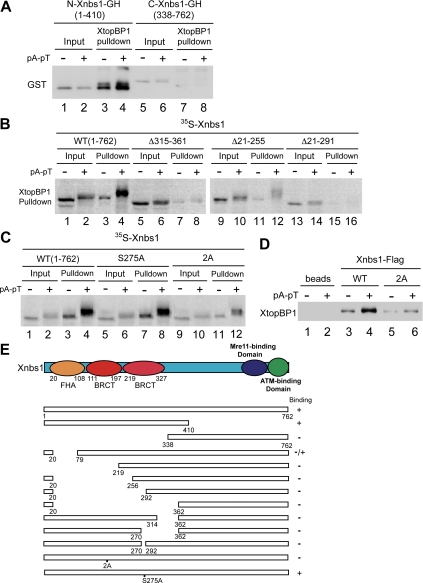
Having determined what region of XtopBP1 associates with Xnbs1, we also performed structure-function studies to elucidate what area of Xnbs1 participates in this interaction. Vertebrate forms of Nbs1 contain a number of identifiable functional domains (Lee and Paull, 2007; Williams et al., 2007; Rupnik et al., 2008). The N-terminal region of the protein contains one forkhead-associated (FHA) domain and two BRCT domains. The C-terminal area of the protein possesses two regions that interact with Mre11 and ATM, respectively. Accordingly, we prepared tagged fragments of Xnbs1 containing roughly the N-terminal (residues 1-410) and C-terminal (residues 338-762) halves of the protein. We incubated these fragments in egg extracts that also contained recombinant HF-XtopBP1. On retrieving the tagged XtopBP1 protein with anti-FLAG antibody beads, we observed that there was binding of the N-terminal but not C-terminal fragment of Xnbs1 (Figure 3A).
Figure 3.
The tandem BRCT domains of Xnbs1 are required for interaction with XtopBP1. (A) Recombinant HF-XtopBP1 on anti-FLAG antibody beads was incubated in egg extracts containing N-terminal (lanes 3 and 4) or C-terminal fragments of Xnbs1 (lanes 7 and 8) in the absence or presence of pA-pT. Beads were reisolated and immunoblotted for GST. Lanes 1, 2, 5, and 6, initial extract aliquots. (B) 35S-Labeled versions of full-length Xnbs1 (WT) (lanes 1–4) and the Δ315–361, Δ21–255, and Δ21–291 deletion mutants of Xnbs1 (lanes 5–16) were incubated in egg extracts containing HF-XtopBP1 bound to anti-FLAG antibody beads in the absence or presence of pA-pT. The beads were isolated, and bound 35S-labeled proteins were detected by SDS-PAGE and phosphorimaging (lanes 3, 4, 7, 8, 11, 12, 15, and 16). Lanes 1, 2, 5, 6, 9, 10, 13, and 14, initial extract aliquots. (C) 35S-Labeled versions of wild-type, S257A, and 2A Xnbs1 were analyzed for binding to XtopBP1 as described in B. (D) Wild-type and 2A mutant versions of Xnbs1-FLAG on anti-FLAG antibody beads were incubated in egg extracts lacking or containing pA-pT. Beads were reisolated and immunoblotted for XtopBP1. (E) Summary of the abilities of the indicated forms of Xnbs1 to interact with XtopBP1.
We proceeded to prepare various deletions of the N-terminal domain of Xnbs1 in order to define more precisely the requirements for binding to XtopBP1 (Figure 3, B and E). We observed that deletion of residues 21–78 of Xnbs1, which excises most of the FHA domain, significantly reduced but did not eliminate the binding of XtopBP1. However, any deletion that compromised either of the BRCT domains (residues 111-197 and 219-327, respectively) dramatically reduced interaction with XtopBP1 (Figure 3B). The results of these binding experiments are summarized in Figure 3E.
Next, we introduced point mutations into the BRCT-containing region of Xnbs1 in order to disable the phosphate-binding pocket that would be formed by the tandem BRCT domains. In particular, we changed both T156 and K158 of Xnbs1 to alanine to create the Xnbs1-2A mutant. Using two different assay methods, we observed that the Xnbs1-2A mutant displayed compromised binding to XtopBP1 (Figure 3, C and D). This mutant showed basal binding to XtopBP1 in the absence of the checkpoint-inducing DNA template pA-pT. However, there was little or no increase in the binding of Xnbs1-2A to XtopBP1 in the presence of pA-pT, in contrast to the strong increase in the binding of wild-type Xnbs1 that occurred under the same conditions. Finally, we mutated a potential ATM/ATR phosphorylation site at S275 in the second BRCT domain of Xnbs1 to a nonphosphorylatable form. This site is conserved in human Nbs1 (Lee and Paull, 2007). However, the resulting Xnbs1-S275A mutant displayed normal binding to XtopBP1. Taken together, these results indicate that Xnbs1 must contain intact tandem BRCT domains in order to undergo checkpoint-stimulated binding to XtopBP1.
The Xnbs1-associating Region of XtopBP1 Is Critical for a Checkpoint Response to DSBs in Egg Extracts
We next investigated whether the interaction between XtopBP1 and Xnbs1 is important for the activation of Xatr-Xatrip in response to DSBs in egg extracts. In one set of experiments, we focused on the KKAM mutant of XtopBP1 that cannot interact with Xnbs1 in a regulated manner. For these experiments, we immunodepleted the endogenous XtopBP1 from egg extracts and replaced it with either wild-type or KKAM versions of HF-XtopBP1 (Figure 4A). Subsequently, we examined checkpoint responses to DNA templates containing exposed double-stranded DNA ends. More specifically, we added pA-pT to egg extracts or treated egg extracts containing reconstituted sperm nuclei with EcoRI (to create DSBs in the chromatin) (Kobayashi et al., 2002; Yoo et al., 2004b). We observed that the phosphorylation of Xchk1 in response to either pA-pT or EcoRI was greatly compromised in egg extracts containing the XtopBP1-KKAM mutant (Figure 4, B and C). Thus, a normal checkpoint response to DSBs in egg extracts depends upon the ability of XtopBP1 to interact with the MRN complex in a DNA damage–dependent manner.
Figure 4.
The KKAM mutant of XtopBP1 is defective in the checkpoint response to DSBs. (A) Egg extracts were mock-depleted with control antibodies (lane 1) or immunodepleted with anti-XtopBP1 antibodies (lanes 2–4). Extracts were supplemented with no recombinant protein (lane 2), wild-type HF-XtopBP1 (lane 3), or KKAM HF-XtopBP1 (lane 4). (B) The extracts from A were incubated with 35S-Xchk1 in the absence or presence of pA-pT as indicated. Extracts were subjected to SDS-PAGE and phosphorimaging. (C) Egg extracts were mock-depleted with control antibodies (lanes 1 and 2) or immunodepleted with anti-XtopBP1 antibodies (lanes 3–5). Extracts were later supplemented with sperm chromatin and no recombinant protein (lanes 1–3), wild-type HF-XtopBP1 (lane 4), or KKAM HF-XtopBP1 (lane 5). Extracts were incubated with 35S-Xchk1 in the absence (lane 1) or presence of EcoRI (lanes 2–5). Nuclear fractions from the extracts were subjected to SDS-PAGE and processed for phosphorimaging (top) or immunoblotting with the indicated antibodies (middle and bottom).
Role of Xnbs1 in the Regulation of XtopBP1 by Xatm in Egg Extracts
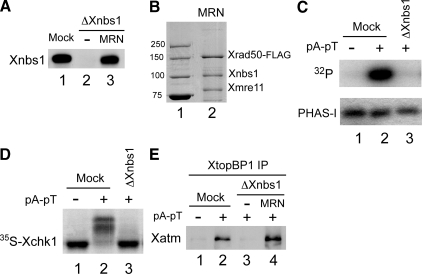
We then turned to the functional role of Xnbs1 in the Xatm-dependent activation of Xatr-Xatrip in egg extracts. For these experiments, we removed the endogenous Xnbs1 from egg extracts with anti-Xnbs1 antibodies. As shown in Figure 5A, we were able to achieve an essentially complete depletion of Xnbs1 from the extracts. Previous studies have indicated that the MRN complex is necessary for the activation of Xatm in response to DNA templates with exposed double-stranded DNA ends (Costanzo et al., 2004; You et al., 2005; You et al., 2007). Consistent with these findings, we found that there was no activation of Xatm in Xnbs1-depleted extracts containing the double-stranded oligonucleotide template pA-pT (Figure 5C). We also examined whether Xnbs1 is necessary for the activation of Xchk1 in response to DSBs. This issue had not been examined explicitly before. We observed that there was no phosphorylation of Xchk1 in Xnbs1-depleted extracts in response to pA-pT (Figure 5D).
Figure 5.
The MRN complex mediates association of Xatm with XtopBP1 in egg extracts containing DSBs. (A) Immunodepletion of Xnbs1. Egg extracts were mock-depleted with control antibodies (lane 1) or immunodepleted with anti-Xnbs1 antibodies (lanes 2 and 3). Subsequently, no recombinant protein (lane 2) or Xenopus MRN complex (Xmre11, Xrad50-FLAG, and Xnbs1) was added back to the depleted extracts (lane 3). Extracts were immunoblotted for Xnbs1. (B) A preparation of recombinant MRN complex (Xmre11, Xrad50-FLAG, and Xnbs1) (lane 2) that had been isolated from in Sf9 insect cells was stained with Coomassie brilliant blue. Lane 1 contains marker proteins (molecular mass indicated in kilodaltons). (C) Xatm was immunoprecipitated from mock-depleted (lanes 1 and 2) or Xnbs1-depleted extracts (lane 3) lacking or containing pA-pT. The anti-Xatm immunoprecipitates were incubated in the presence of [γ-32P]ATP with the substrate protein PHAS-I. Samples were processed for phosphorimaging (top) and staining with Coomassie brilliant blue (bottom). (D) Egg extracts were mock-depleted with control antibodies (lanes 1 and 2) or immunodepleted with anti-Xnbs1 antibodies (lane 3). The depleted extracts were incubated with 35S-Xchk1 in the absence or presence of pA-pT. Extracts were subjected to SDS-PAGE and phosphorimaging. (E) The indicated extracts from A were incubated in the absence (lane 1) or presence of pA-pT (lanes 2–4). XtopBP1 was immunoprecipitated from the extracts. Immunoprecipitates were subjected to SDS-PAGE and immunoblotted for Xatm.
Next, we asked whether binding of Xatm to XtopBP1 in egg extracts was dependent on Xnbs1. For this purpose, we immunoprecipitated XtopBP1 from mock-depleted and Xnbs1-depleted extracts that had been incubated in the absence or presence of pA-pT. We observed that there was no binding of Xatm to XtopBP1 in Xnbs1-depleted extracts (Figure 5E). Furthermore, we were able to rescue this binding by addition of a recombinant version of the Xenopus MRN complex composed of FLAG-tagged Xrad50 and untagged forms of Xmre11 and Xnbs1 (Figure 5, A, B, and E). This finding suggests that the MRN complex directly or indirectly bridges together Xatm and XtopBP1 in response to the occurrence of DSBs. Unfortunately, however, we were not able to rescue the activation of Xatm or Xchk1 in Xnbs1-depleted extracts containing pA-pT with the recombinant Xenopus MRN complex. Thus, this recombinant complex appears to be partly but not fully functional. Therefore, we were not able to test whether an MRN complex containing the Xnbs1-2A mutant is defective in checkpoint signaling.
Human Cells Harboring the Nbs1-2A Mutant Are Defective in the IR-induced Activation of Chk1
As an alternative means to assess the functional importance of the TopBP1-interacting region on Nbs1 for checkpoint signaling, we turned to an siRNA-based strategy in human cells. For this purpose, we treated HeLa cells with siRNA directed against Nbs1. Before this treatment, we infected the cells with recombinant lentiviruses encoding various forms of human Nbs1 that were resistant to the siRNA. For these experiments, we prepared a mutant form of human Nbs1 in which T158 and K160 were changed to alanine. This mutant, which we called Nbs1-2A, is equivalent to the Xnbs1-2A mutant from the Xenopus system (Figure 6A).
Figure 6.
Human cells harboring the Nbs1-2A mutant are defective in the checkpoint response to IR. (A) Sequence alignment of Xenopus and human Nbs1 around critical residues in the first BRCT domain. (B) HeLa cells were infected with lentiviruses expressing GFP (lanes 1–4) or either wild-type or 2A versions of human Nbs1 (lanes 5 and 6). Control or Nbs1 siRNA was transfected into these infected cells as indicated. Cells were mock-treated or exposed to IR. Cell extracts were prepared 2 h later and processed for immunoblotting with the indicated antibodies. (C) HeLa cells were transfected control vector (lanes 1 and 2) or a vector expressing either wild-type (lanes 3 and 4) or 2A versions (lanes 5 and 6) of FLAG-tagged human Nbs1. At 48 h after transfection, cells were treated as described in B. Anti-FLAG immunoprecipitates from cell lysates were immunoblotted with anti-TopBP1 and anti-FLAG antibodies.
As shown in Figure 6B, we used an siRNA that caused a pronounced depletion of Nbs1. Under the same conditions, a control siRNA had no effect on Nbs1. In these control cells, there was a significant increase in the phosphorylation of Chk1 on S317 in response to IR. This increase was abolished in cells treated the Nbs1 siRNA. Furthermore, the IR-induced phosphorylation of Chk1 on S317 was restored by expression of wild-type, siRNA-resistant Nbs1. However, there was no rescue of this phosphorylation in cells infected with a lentivirus that expressed the Nbs1-2A mutant. We verified that this human mutant protein was also defective for binding to TopBP1 (Figure 6C). Taken together, these experiments indicate that Nbs1 must possess an intact BRCT-containing region in order for cells to mount a normal checkpoint response to DSBs.
DISCUSSION
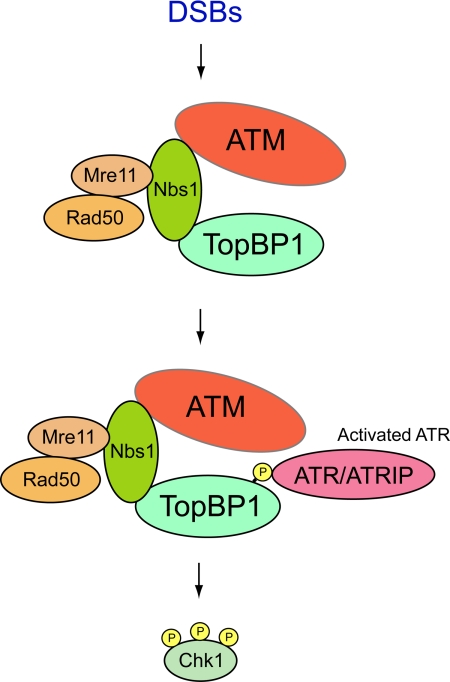
In this report, we have probed further into the regulation of XtopBP1 in response to DSBs. Toward this end, we have analyzed the functional significance of the interaction of XtopBP1 with additional proteins that we have identified in pulldown experiments with Xenopus egg extracts. Previously, we described a specific association between XtopBP1 and Xatm in egg extracts containing a DNA template that mimics DSBs (Yoo et al., 2007). We pursued this observation by showing that Xatm phosphorylates XtopBP1 on S1131 and thereby enhances its capacity to activate the Xatr-Xatrip complex. However, it was not clear whether Xatm bound directly to XtopBP1 or through an intermediary protein(s). In the present study, we have identified additional XtopBP1-associating proteins as the three subunits of the Xenopus MRN complex (Xmre11, Xrad50, and Xnbs1). Furthermore, we have established that this binding appears to occur largely if not exclusively through the Xnbs1 component of the complex. Recent studies have indicated that TopBP1 also associates with Nbs1 in human cells (Morishima et al., 2007). Our results indicate that this interaction is critical for the recruitment of Xatm to XtopBP1. In particular, there is no binding of Xatm to XtopBP1 in egg extracts that have been depleted of the Xnbs1 protein. Therefore, the MRN complex appears to be acting as a mediator between Xatm and XtopBP1 (see Figure 7).
Figure 7.
Model for functional interactions between ATM, the MRN complex, and TopBP1 in response to DSBs. See text for details. The indicated associations are not necessarily direct.
It is well established that the MRN complex plays crucial roles in the initial stages of cellular responses to DSBs (Lee and Paull, 2007; Williams et al., 2007; Rupnik et al., 2008). This complex can associate with broken DNA ends directly. Moreover, it accumulates in more substantial amounts at regions of damage in a manner that depends on phosphorylated H2AX. On arrival at DNA lesions, the MRN complex carries out multiple functions that include processing DNA structures and regulation of checkpoint signaling. For example, the MRN complex promotes the resection of broken DNA ends. This nucleolytic processing helps to amplify checkpoint signaling by creating significant amounts of single-stranded DNA in the proximity of DNA breaks. The MRN complex also appears to play a critical role in the process whereby ATM becomes converted from inactive dimers to active monomers. In the human and Xenopus systems, the activated form of ATM undergoes autophosphorylation on S1981, which serves as a useful marker for the presence of DSBs (Bakkenist and Kastan, 2003; You et al., 2005).
The MRN complex also promotes the activation of ATR-ATRIP in response to DSBs (Cuadrado et al., 2006; Jazayeri et al., 2006; Myers and Cortez, 2006). These initial studies established that this regulatory connection involved the ability of the MRN complex to mediate the resection of DNA at DSBs. The resulting stretches of single-stranded DNA would be able to recruit successively RPA and ATR-ATRIP. According to this scenario, ATM would promote the activation of ATR-ATRIP by facilitating the creation of a structure (i.e., RPA-coated, single-stranded DNA) that is important for the activation of ATR-ATRIP under usual circumstances. The MRN complex serves as an integral component in this train of events through its roles in both the activation of ATM and the resection of DNA.
However, our recent studies with Xenopus egg extracts have revealed additional aspects of this regulatory scheme. Consistent with the studies in human cells, the activation of Xatr-Xatrip in egg extracts containing DSBs is likewise dependent on Xatm. However, we recently discovered that XtopBP1 also plays a pivotal role this process (Yoo et al., 2007). In particular, Xatm associates with XtopBP1 and strongly stimulates its ATR-activating capacity; this step involves the Xatm-catalyzed phosphorylation of XtopBP1 on a well-conserved serine residue at position 1131. In the present study, we have shown that the Xenopus version of the MRN complex plays a role in the Xatm-dependent activation of Xatr-Xatrip in egg extracts apart from its presumed functions in resection of DNA and activation of Xatm. We have found that Xatm can no longer associate with XtopBP1 in DSB-containing egg extracts that lack a functional MRN complex (owing to depletion of the Xnbs1 subunit). Thus, the MRN complex appears to act as a mediator between Xatm and XtopBP1 in this context.
We have demonstrated that Xnbs1 interacts with a critical region of XtopBP1. More specifically, it associates with the N-terminal region of the protein that contains its first two BRCT repeats. The docking of Xnbs1 onto the BRCT I-II region of XtopBP1 appears to be functionally significant. In particular, a version of XtopBP1 containing point mutations in critical residues in the BRCT I-II region (the KKAM mutant) is defective in mediating a checkpoint response to DSBs in egg extracts. Intriguingly, this same region of TopBP1 interacts with the 9-1-1 complex at stalled replication forks in vertebrates (Delacroix et al., 2007; Lee et al., 2007). Therefore, it appears that the BRCT I-II region of XtopBP1 may be a conduit for the activation of Xatr-Xatrip in response to a variety of distinct DNA lesions. The fact that this region interacts with a number of different components (e.g., MRN, the 9-1-1 complex, and possibly other factors) does complicate somewhat the interpretation of our results. However, the MRN complex is a well-established regulatory binding partner of ATM. Therefore, we believe that it is reasonable to propose that the failure of the XtopBP1-KKAM mutant to interact with the MRN complex (and thus Xatm) would plausibly explain why this mutant cannot mediate the Xatm-dependent activation of Xatr in response to DSBs.
Furthermore, we have established that Xnbs1 must contain intact tandem BRCT domains in order to interact with XtopBP1 in a regulated manner. To evaluate the functional significance of this interaction with regard to the TopBP1-interacting site on Nbs1, we adopted an si-RNA–based approach in human cells. By this means, we were able to establish that Nbs1 must contain intact BRCT domains in order for human cells to carry out the activation of Chk1 in response to IR-induced DSBs.
In principle, the XtopBP1 and Xnbs1 proteins in egg extracts could associate directly by means of their BRCT-containing regions. However, the findings that this interaction is highly regulated and depends on intact phosphopeptide-binding pockets in the relevant BRCT domains of both proteins tend to argue against this model. Nonetheless, we cannot rule out the possibility that these mutations in XtopBP1 and Xnbs1 would compromise phosphorylation-independent interactions.
Another possibility is that an additional protein(s) bridges together XtopBP1 and the MRN complex in egg extracts. One potential candidate would be the putative Xenopus homologue of Mdc1 (Xmdc1). In human cells, localization of the MRN complex to IR-induced foci depends on Mdc1 (Goldberg et al., 2003; Stewart et al., 2003). Furthermore, Mdc1 interacts with Nbs1 in a highly specific manner (Chapman and Jackson, 2008; Melander et al., 2008; Spycher et al., 2008; Wu et al., 2008). Interestingly, Mdc1 and the MRN complex interact in a constitutive manner in human cells, but the binding of XtopBP1 and the MRN complex is highly regulated in egg extracts. Further studies will be required to elucidate the exact mechanism of the association between XtopBP1 and Xnbs1.
In closing, we have obtained further insight into how cells activate ATR-ATRIP in response to DSBs. In the future, it will be interesting to compare and contrast how responses to different checkpoint-inducing structures manifest themselves through distinct interactions of TopBP1 with other checkpoint proteins.
ACKNOWLEDGMENTS
We are grateful to J. Ramirez-Lugo and J. DeHart for comments on the manuscript. This work was supported by National Institutes of Health Grants GM043974 and GM070891 to W.G.D.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-12-1190) on March 11, 2009.
REFERENCES
- Abraham R. T. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- Bakkenist C. J., Kastan M. B. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- Byun T. S., Pacek M., Yee M. C., Walter J. C., Cimprich K. A. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldecott K. W. Cell signaling. The BRCT domain: signaling with friends? Science. 2003;302:579–580. doi: 10.1126/science.1091463. [DOI] [PubMed] [Google Scholar]
- Chapman J. R., Jackson S. P. Phospho-dependent interactions between NBS1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO Rep. 2008;9:795–801. doi: 10.1038/embor.2008.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich K. A., Cortez D. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D., Guntuku S., Qin J., Elledge S. J. ATR and ATRIP: partners in checkpoint signaling. Science. 2001;294:1713–1716. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- Costanzo V., Paull T., Gottesman M., Gautier J. Mre11 assembles linear DNA fragments into DNA damage signaling complexes. PLoS Biol. 2004;2:E110. doi: 10.1371/journal.pbio.0020110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo V., Robertson K., Bibikova M., Kim E., Grieco D., Gottesman M., Carroll D., Gautier J. Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Mol. Cell. 2001;8:137–147. doi: 10.1016/s1097-2765(01)00294-5. [DOI] [PubMed] [Google Scholar]
- Cuadrado M., Martinez-Pastor B., Murga M., Toledo L. I., Gutierrez-Martinez P., Lopez E., Fernandez-Capetillo O. ATM regulates ATR chromatin loading in response to DNA double-strand breaks. J. Exp. Med. 2006;203:297–303. doi: 10.1084/jem.20051923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delacroix S., Wagner J. M., Kobayashi M., Yamamoto K., Karnitz L. M. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 2007;21:1472–1477. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V., Furuya K., Carr A. M. Identification and functional analysis of TopBP1 and its homologs. DNA Repair. 2005;4:1227–1239. doi: 10.1016/j.dnarep.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Glover J. N., Williams R. S., Lee M. S. Interactions between BRCT repeats and phosphoproteins: tangled up in two. Trends Biochem. Sci. 2004;29:579–585. doi: 10.1016/j.tibs.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Goldberg M., Stucki M., Falck J., D’Amours D., Rahman D., Pappin D., Bartek J., Jackson S. P. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature. 2003;421:952–956. doi: 10.1038/nature01445. [DOI] [PubMed] [Google Scholar]
- Guo Z., Kumagai A., Wang S. X., Dunphy W. G. Requirement for Atr in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes Dev. 2000;14:2745–2756. doi: 10.1101/gad.842500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y., Tsujimura T., Sugino A., Takisawa H. The phosphorylated C-terminal domain of Xenopus Cut5 directly mediates ATR-dependent activation of Chk1. Genes Cells. 2006;11:993–1007. doi: 10.1111/j.1365-2443.2006.00998.x. [DOI] [PubMed] [Google Scholar]
- Jazayeri A., Falck J., Lukas C., Bartek J., Smith G. C., Lukas J., Jackson S. P. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Tada S., Tsuyama T., Murofushi H., Seki M., Enomoto T. Focus-formation of replication protein A, activation of checkpoint system and DNA repair synthesis induced by DNA double-strand breaks in Xenopus egg extract. J. Cell Sci. 2002;115:3159–3169. doi: 10.1242/jcs.115.15.3159. [DOI] [PubMed] [Google Scholar]
- Kumagai A., Dunphy W. G. Claspin, a novel protein required for the activation of Chk1 during a DNA replication checkpoint response in Xenopus egg extracts. Mol. Cell. 2000;6:839–849. doi: 10.1016/s1097-2765(05)00092-4. [DOI] [PubMed] [Google Scholar]
- Kumagai A., Dunphy W. G. How cells activate ATR. Cell Cycle. 2006;5:1265–1268. doi: 10.4161/cc.5.12.2834. [DOI] [PubMed] [Google Scholar]
- Kumagai A., Kim S. -M., Dunphy W. G. Claspin and the activated form of ATR-ATRIP collaborate in the activation of Chk1. J. Biol. Chem. 2004;279:49599–49608. doi: 10.1074/jbc.M408353200. [DOI] [PubMed] [Google Scholar]
- Kumagai A., Lee J., Yoo H. Y., Dunphy W. G. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- Lee J., Kumagai A., Dunphy W. G. The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. J. Biol. Chem. 2007;282:28036–28044. doi: 10.1074/jbc.M704635200. [DOI] [PubMed] [Google Scholar]
- Lee J. H., Paull T. T. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene. 2007;26:7741–7748. doi: 10.1038/sj.onc.1210872. [DOI] [PubMed] [Google Scholar]
- Liu Q., et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 2000;14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- Melander F., Bekker-Jensen S., Falck J., Bartek J., Mailand N., Lukas J. Phosphorylation of SDT repeats in the MDC1 N terminus triggers retention of NBS1 at the DNA damage-modified chromatin. J. Cell Biol. 2008;181:213–226. doi: 10.1083/jcb.200708210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordes D. A., Glick G. G., Zhao R., Cortez D. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev. 2008;22:1478–1489. doi: 10.1101/gad.1666208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishima K., Sakamoto S., Kobayashi J., Izumi H., Suda T., Matsumoto Y., Tauchi H., Ide H., Komatsu K., Matsuura S. TopBP1 associates with NBS1 and is involved in homologous recombination repair. Biochem. Biophys. Res. Commun. 2007;362:872–879. doi: 10.1016/j.bbrc.2007.08.086. [DOI] [PubMed] [Google Scholar]
- Myers J. S., Cortez D. Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J. Biol. Chem. 2006;281:9346–9350. doi: 10.1074/jbc.M513265200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyberg K. A., Michelson R. J., Putnam C. W., Weinert T. A. Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 2002;36:617–656. doi: 10.1146/annurev.genet.36.060402.113540. [DOI] [PubMed] [Google Scholar]
- Parrilla-Castellar E. R., Arlander S. J., Karnitz L. Dial 9-1-1 for DNA damage: the Rad9-Hus1-Rad1 (9-1-1) clamp complex. DNA Repair. 2004;3:1009–1014. doi: 10.1016/j.dnarep.2004.03.032. [DOI] [PubMed] [Google Scholar]
- Perry J. A., Kornbluth S. Cdc25 and Wee1: analogous opposites? Cell Div. 2007;2:12. doi: 10.1186/1747-1028-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X. F., An D. S., Chen I. S., Baltimore D. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc. Nat. Acad. Sci. USA. 2003;100:183–188. doi: 10.1073/pnas.232688199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupnik A., Grenon M., Lowndes N. The MRN complex. Curr. Biol. 2008;18:R455–457. doi: 10.1016/j.cub.2008.03.040. [DOI] [PubMed] [Google Scholar]
- Sancar A., Lindsey-Boltz L. A., Ünsal-Kaçmaz K., Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- Shevchenko A., Tomas H., Havlis J., Olsen J. V., Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protocol. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- Shiloh Y. The ATM-mediated DNA-damage response: taking shape. Trends Biochem. Sci. 2006;31:402–410. doi: 10.1016/j.tibs.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Spycher C., Miller E. S., Townsend K., Pavic L., Morrice N. A., Janscak P., Stewart G. S., Stucki M. Constitutive phosphorylation of MDC1 physically links the MRE11-RAD50-NBS1 complex to damaged chromatin. J. Cell Biol. 2008;181:227–240. doi: 10.1083/jcb.200709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart G. S., Wang B., Bignell C. R., Taylor A. M., Elledge S. J. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421:961–966. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- Walter J., Newport J. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol. Cell. 2000;5:617–627. doi: 10.1016/s1097-2765(00)80241-5. [DOI] [PubMed] [Google Scholar]
- Waridel P., Frank A., Thomas H., Surendranath V., Sunyaev S., Pevzner P., Shevchenko A. Sequence similarity-driven proteomics in organisms with unknown genomes by LC-MS/MS and automated de novo sequencing. Proteomics. 2007;7:2318–2329. doi: 10.1002/pmic.200700003. [DOI] [PubMed] [Google Scholar]
- Williams R. S., Williams J. S., Tainer J. A. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem. Cell Biol. 2007;85:509–520. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- Wu L., Luo K., Lou Z., Chen J. MDC1 regulates intra-S-phase checkpoint by targeting NBS1 to DNA double-strand breaks. Proc. Natl. Acad. Sci. USA. 2008;105:11200–11205. doi: 10.1073/pnas.0802885105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H. Y., Jeong S. -Y., Dunphy W. G. Site-specific phosphorylation of a checkpoint mediator protein controls its responses to different DNA structures. Genes Dev. 2006;20:772–783. doi: 10.1101/gad.1398806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H. Y., Kumagai A., Shevchenko A., Shevchenko A., Dunphy W. G. Adaptation of a DNA replication checkpoint response depends upon inactivation of Claspin by the Polo-like kinase. Cell. 2004a;117:575–588. doi: 10.1016/s0092-8674(04)00417-9. [DOI] [PubMed] [Google Scholar]
- Yoo H. Y., Kumagai A., Shevchenko A., Shevchenko A., Dunphy W. G. Ataxia-telangiectasia mutated (ATM)-dependent activation of ATR occurs through phosphorylation of TopBP1 by ATM. J. Biol. Chem. 2007;282:17501–17506. doi: 10.1074/jbc.M701770200. [DOI] [PubMed] [Google Scholar]
- Yoo H. Y., Shevchenko A., Shevchenko A., Dunphy W. G. Mcm2 is a direct substrate of ATM and ATR during DNA damage and DNA replication checkpoint responses. J. Biol. Chem. 2004b;279:53353–53364. doi: 10.1074/jbc.M408026200. [DOI] [PubMed] [Google Scholar]
- You Z., Bailis J. M., Johnson S. A., Dilworth S. M., Hunter T. Rapid activation of ATM on DNA flanking double-strand breaks. Nat. Cell Biol. 2007;9:1311–1318. doi: 10.1038/ncb1651. [DOI] [PubMed] [Google Scholar]
- You Z., Chahwan C., Bailis J., Hunter T., Russell P. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol. Cell. Biol. 2005;25:5363–5379. doi: 10.1128/MCB.25.13.5363-5379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Piwnica-Worms H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol. Cell. Biol. 2001;21:4129–4139. doi: 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Elledge S. J. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]