Abstract
Myosin II is an essential component of the actomyosin contractile ring and plays a crucial role in cytokinesis by generating the forces necessary for contraction of the actomyosin ring. Cdc4 is an essential myosin II light chain in fission yeast and is required for cytokinesis. In various eukaryotes, the phosphorylation of myosin is well documented as a primary means of activating myosin II, but little is known about the regulatory mechanisms of Cdc4. Here, we isolated Nrd1, an RNA-binding protein with RNA-recognition motifs, as a multicopy suppressor of cdc4 mutants. Notably, we demonstrated that Nrd1 binds and stabilizes Cdc4 mRNA, thereby suppressing the cytokinesis defects of the cdc4 mutants. Importantly, Pmk1 mitogen-activated protein kinase (MAPK) directly phosphorylates Nrd1, thereby negatively regulating the binding activity of Nrd1 to Cdc4 mRNA. Consistently, the inactivation of Pmk1 MAPK signaling, as well as Nrd1 overexpression, stabilized the Cdc4 mRNA level, thereby suppressing the cytokinesis defects associated with the cdc4 mutants. In addition, we demonstrated the cell cycle–dependent regulation of Pmk1/Nrd1 signaling. Together, our results indicate that Nrd1 plays a role in the regulation of Cdc4 mRNA stability; moreover, our study is the first to demonstrate the posttranscriptional regulation of myosin expression by MAPK signaling.
INTRODUCTION
Cytokinesis, the physical separation of a mother cell into two daughter cells after mitosis, is achieved through the constriction of a medially placed contractile ring composed of actin, myosin, and numerous other proteins (Satterwhite and Pollard, 1992; Guertin et al., 2002; Glotzer, 2005). Myosin II is an essential component of the actomyosin contractile ring and plays a crucial role in cytokinesis by generating the forces necessary for contraction of the actomyosin ring (Satterwhite and Pollard, 1992). Type II myosin consists of a dimer comprising two heavy chains, each having two associated proteins: the essential light chain and regulatory light chain (Guertin et al., 2002). In various eukaryotes, the phosphorylation of both the heavy and light chains of myosin has been well documented as a primary means of activating myosin II, which is known to be crucial for cytokinesis (Satterwhite et al., 1992; Brzeska and Korn, 1996; Kamm and Stull, 2001).
The genetically tractable fission yeast Schizosaccharomyces pombe is an excellent organism for studying cytokinesis because it undergoes cell division by medial fission after assembly and contraction of the actomyosin contractile ring, which may be considered functionally analogous to the cleavage furrow in mammalian cells (Bezanilla et al., 1997; Balasubramanian et al., 2004; Wolfe and Gould, 2005). Moreover, numerous mutations that result in defects in each step of the cell division process are available; these are highly useful for clarifying the molecular pathways and regulatory mechanisms of the cell cycle including cytokinesis (Nasmyth and Nurse, 1981; Balasubramanian et al., 1998).
Analysis of mutants containing defects in medial ring formation led to the identification of several components of myosin II in fission yeast. These include an essential myosin light chain encoded by the cdc4+ gene (McCollum et al., 1995) and a type II myosin heavy chain encoded by the myo2+ gene (Kitayama et al., 1997; Balasubramanian et al., 1998). It has been suggested that Myo2 an essential component of the cytokinetic actomyosin ring provides the contractile force required for cytokinesis (Kitayama et al., 1997; Balasubramanian et al., 1998). The second type II myosin in S. pombe, Myp2, which localizes in the ring at a much later stage of cytokinesis, has been proposed to increase the efficiency of cytokinesis (Bezanilla et al., 1997; May et al., 1997; Motegi et al., 2000). Two myosin light chains, Cdc4 and Rlc1 have been characterized in S. pombe. Cdc4, an essential light chain of the myosin superfamily, is a component of the contractile ring and plays a crucial role in cytokinesis (McCollum et al., 1995; Hou and McCollum, 2002; Guertin et al., 2002), and it has been shown that Rlc1 is required for cytokinesis at lower temperatures (Le et al., 2000; Naqvi et al., 2000). A study by D'souza et al. (2001) reported that in addition to its association with Myo2 and Myp2, Cdc4 interacts with the IQ domain containing actomyosin ring components Myp51 and Rng2. Notably, the Cdc4 protein has been shown to be phosphorylated, but this modification is not required for cytokinesis (McCollum et al., 1999). Hence, the mechanism underlying the regulatory activity of Cdc4 in cytokinesis remains unclear.
To identify novel genes that are involved in Cdc4 function, we searched for multicopy suppressors of the temperature sensitivity of the cdc4-8 mutants and identified Nrd1, an RNA-binding protein with RNA-recognition motifs (RRMs; Tsukahara et al., 1998; Jeong et al., 2004). The nrd1+/msa2+ gene was previously isolated as a high-dosage suppressor of pat1-114 or sam1 mutants, both of which exhibit the hyperconjugation phenotype (Yamamoto et al., 1999;Jeong et al., 2004b). Nrd1 and its mammalian counterpart Rod1 have been reported to negatively regulate sexual differentiation (Tsukahara et al., 1998; Yamamoto et al., 1999). The biological role of Nrd1 is to block the onset of sexual differentiation by repressing the Ste11-regulated genes essential for conjugation and meiosis, but its cellular mRNA targets remain unknown.
Here, we show that Nrd1 directly binds to and regulates Cdc4 mRNA stability. Notably, Pmk1, the mitogen-activated protein kinase (MAPK), which regulates cell integrity (Toda et al., 1996; Sugiura et al., 1999; Sugiura et al., 2003), directly phosphorylates Nrd1, thereby negatively regulating the activity of Nrd1 to bind to and stabilize Cdc4 mRNA. We propose that the MAPK-dependent phosphorylation of the RNA-binding protein Nrd1 may serve as a novel mechanism for the regulation of myosin mRNA and cytokinesis in fission yeast.
MATERIALS AND METHODS
Strains, Media, and Genetic and Molecular Biology Methods
S. pombe strains used in this study are listed in Table 1. The complete medium YPD (yeast extract-peptone-dextrose) and the minimal medium EMM (Edinburgh minimal medium) have been described previously (Toda et al., 1996). Standard genetic and recombinant DNA methods (Moreno et al., 1991) were used except where noted.
Table 1.
Schizosaccharomyces pombe strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| HM123 | h− leu1-32 | Our stock |
| HM528 | h+ his2 | Our stock |
| KP928 | h+ his2 leu1-32 ura4-D18 | Our stock |
| KP208 | h− leu1-32 ura4-D18 pmk1∷ura4+ | Our stock |
| KP2118 | h− leu1-32 ura4-D18 pmk1∷KanMx6 | This study |
| KP2181 | h+leu1-32 his2 pmk1∷KanMx6 | This study |
| KP2178 | h− leu1-32 pmk1∷KanMx6 | This study |
| KP403 | h− leu1-32 cdc4-8 | Our stock |
| KP616 | h− leu1–32 ura4-D18 nrd1∷ura4+ | This study |
| KP622 | h− leu1-32 pat1-114 | Our stock |
| KP654 | h− leu1-32 ura4-D18 cdc4-8 nrd1∷ura4+ | This study |
| SP161 | h− leu1-32 ura4-D18 cdc4-8 pmk1∷ura4+ | This study |
| SP167 | h− leu1-32 ura4-D18 cdc4-8 spk1∷ura4+ | This study |
| SP310 | h− leu1-32 ura4-D18 cdc4-8 spc1∷ura4+ | This study |
| SP873 | h− leu1-32 ura4-D18 cdc4-8 pmk1∷KanMx6 nrd1∷ura4+ | This study |
| SP556 | h− leu1--32 ura4-D18 pmk1-GST∷KanMx6 | Takada et al. (2007) |
| SP628 | h− leu1-32 ura4-D18 cdc25-22 pmk1-GST∷KanMx6 | This study |
| FC129 | h+ cdc4-312 | Chang et al.(1996) |
| SP920 | h− leu1-32 ura4-D18 cdc4-312 nrd1∷ura4+ | This study |
| SP912 | h− leu1-32 cdc4-312 pmk1∷KanMx6 | This study |
| MBY142 | h− leu1-32 ade6-210 ura4-D18 rng2-D5 | Eng et al. (1998) |
| SP817 | h− leu1-32 ura4-D18 rng2-D5 nrd1∷ura4+ | This study |
| SP820 | h− leu1-32 rng2-D5 pmk1∷KanMx6 | This study |
| MBY517 | h+ leu1-32 ade6–21 × ura4-D18 rng3-GFP∷ura4+ myo2-E1 | Wong et al. (2000) |
| SP919 | h− leu1-32 ura4-D18 myo2-E1 nrd1∷ura4+ | This study |
| SP931 | h− leu1-32 myo2-E1 pmk1∷KanMx6 | This study |
| MBY691 | h+ leu1-32 ura4-D18 his3-D1 ade6-M216 rlc1∷ura4+ | Naqvi et al. (2000) |
| SP927 | h− leu1-32 ura4-D18 arg1 rlc1∷ura4+ nrd1∷arg1+ | This study |
| SP847 | h− leu1-32 ura4-D18 rlc1∷ura4+ pmk1∷ KanMx6 | This study |
| MBY113 | h+ leu1-32 ade6–210 rng3-65 | Wong et al. (2000) |
| SP842 | h− leu1–32 ura4-D18 rng3-65 nrd1∷ura4+ | This study |
| SP816 | h− leu1-32 rng3-65 pmk1∷KanMx6 | This study |
| MBY142 | h− rng2-D5 ade6-210 leu1-32 ura4-D18 | Eng et al. (1998) |
| KP413 | h− cdc12-112 leu1-32 | Chang et al. (1996) |
| SP781 | h− cdc12-112 leu1-32 ura4-D18 nrd1∷ura4+ | This study |
| SP827 | h− leu1-32 ura4-D18 cdc12–112 pmk1∷ura4+ | This study |
| KP1304 | h− leu1-32 ura4-D18 myo3/cis2∷ ura4+ | Fujita et al. (2002) |
| SP845 | h− leu1-32 ura4-D18 cis2∷ ura4+ pmk1∷ KanMx6 | This study |
| SP886 | h− leu1-32 ura4-D18 arg1 cis2∷ ura4+ nrd1∷arg1+ | This study |
Gene Disruption of nrd1+
A 3.4-kb HindIII fragment containing nrd1+ was subcloned into pGEM-13Zf to create pGEM-nrd1. pGEM-nrd1 was cleaved at the single BamHI site in nrd1+ and ligated to S. pombe ura4+ (1.8-kb). This construct was used to transform haploid cells (Rothstein, 1983).
Northern Blot Analyses
Total RNA was isolated using the method of Kohrer and Domdey (1991). A 20-μg sample of total RNA per lane was subjected to electrophoresis on denaturing formaldehyde 1% agarose gels and transferred to nylon membranes. Hybridization was performed using digoxigenin (DIG)-labeled antisense cRNA probes coding for cdc4 and leu1 as described previously (Hirayama et al., 2003). The DIG-labeled hybrids were detected by an enzyme-linked immunoassay by using an anti-DIG–alkaline phosphatase antibody conjugate. The hybrids were visualized by chemiluminescence detection on a light-sensitive film according to the manufacturer's instructions (Roche Applied Science, Indianapolis, IN).
mRNA Protein-binding Assay
The RNA protein-binding assay was carried out as described previously (Irie et al., 2002). Briefly, exponentially growing cells (2 × 108) expressing glutathione S-transferase (GST)-Nrd1 were disrupted with glass beads in 200 μl extraction buffer (25 mM HEPES-KOH, pH 7.5, 150 mM KCl, and 2 mM MgCl2 containing 20 mM vanadyl ribonucleoside complexes, 200 U/ml RNasin, 0.1% NP-40, 1 mM DTT, and a mixture of protease inhibitors [1 mM phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, 10 μg/ml pepstatin A, and 10 μg/ml leupeptin]). Extracts were cleared by centrifugation (10 min at 4000 × g). Glutathione beads were added to the cleared extracts, which were incubated for 1 h at 4°C. Beads were washed four times in wash buffer (25 mM HEPES-KOH, pH 7.5, 150 mM KCl, and 2 mM MgCl2) and were eluted in 100 mM Tris-HCl pH 7.5, 150 mM NaCl, 12.5 mM EDTA, 0.1% SDS for 10 min at 65°C. Eluted samples were extracted with phenol-chloroform, ethanol-precipitated, resuspended in DNase buffer, and treated with RNase-free DNase. The remaining RNA was extracted, precipitated, and resuspended in diethylpyrocarbonate water. RT-PCR was performed using 1 μl RNA as template using Cdc4-specific primers. The number of amplification cycles was adjusted to avoid reaching a plateau phase during PCR.
In Vitro RNA-binding Assay
The affinity capillary electrophoresis mobility shift assay was carried out as described previously (Mucha et al., 2002; Taga et al., 2004) with some modifications. Capillary electrophoresis was performed using a CAPI-3100 capillary electrophoresis system of Photal, equipped with an autosampler, a photodiode array multiwavelength UV detector and a capillary oven thermostated by circulating air. The FunCap-CE/Type C (total length, 72 cm; effective length, 60 cm; internal diameter, 50 μm) from GL Sciences (Tokyo, Japan) for use as a carboxylated capillary was installed in the apparatus. A stock solution of phosphate buffer was prepared by adding 50 mM disodium hydrogen phosphate aqueous solution to a solution of 50 mM sodium dihydrogen phosphate until a solution with pH 6.8 was reached using a pH meter. An RNA solution was prepared by dissolving Cdc4 mRNA to a phosphate buffer at a concentration of 100 μg/ml. The RNA solution was diluted with the same buffer to appropriate concentrations to use as the RNA containing electrophoretic solutions for affinity capillary electrophoresis mobility shift assay. GST-fusion proteins were analyzed in the electrophoretic solutions containing mRNA at various concentrations. The electrophoretic solutions were filtered through a 0.45-μm membrane filter, followed by degassing by sonication for 10 min before use. Sample solutions were introduced from the anodic end of the capillary for 30 s by the hydrostatic introduction system. Potential of 20 kV was applied between both ends of the capillary. The capillary tube was conditioned by rinsing with an aqueous solution of 1 M sodium chloride for 1 min followed by an electrophoretic solution for 4 min using the flush mode of the introduction system before each sample introduction. Detection was carried out using a UV detector at a 225-nm wavelength, which is the λmax of GST-fusion proteins. The capillary electrophoretic analyses were carried out at 25 ± 0.1°C. In this method migration times were reflected to molar ratios of complex types. Therefore, migration time shifts indicate the strength of interaction between a protein sample and a ligand in an electrophoretic solution.
Protein Expression, Site-directed Mutagenesis, and Phosphorylation Assay
For protein expression in yeast, the thiamine-repressible nmt1 promoter was used (Maundrell, 1990). Expression was repressed by the addition of 4 μg/ml thiamine to EMM and was induced by washing and incubating the cells in EMM lacking thiamine. The GST- or the green fluorescent protein (GFP)-fused gene was subcloned into the pREP1, or pREP41 vectors, obtaining a maximum expression of the fused gene using pREP1, whereas pREP41 contained an attenuated version of the nmt1 promoter. For protein expression in bacteria, the full-length Nrd1 cDNA was amplified by RT-PCR from the wild-type fission yeast total RNA. GST-fusion proteins encoding Nrd1 was constructed using pGEX-6P (GE Healthcare, Waukesha, WI), expressed in Escherichia coli XL1-blue, and purified using glutathione-Sepharose beads as previously described (Sugiura et al., 1998), and the purified GST-fusion proteins were used for in vitro kinase reactions. The site-directed mutagenesis was performed using the Quick Change Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). The cdc4M1M2 mutant cells were obtained after transformation with the integrative plasmid pJK-cdc4C-leu (containing the latter half of the Cdc4 mRNA) previously digested with BglII at the unique site within the Cdc4 mRNA. The linear fragment was transformed into haploid strains so as to target integration at the cdc4+ locus. Leucine prototrophs were selected, and the identification of strain containing one copy of cdc4M1M2 expressed from the genomic cdc4+ promoter was verified by PCR and immunoblot analysis. In vitro kinase reactions were performed as previously described (Takada et al., 2007) with some modifications. Bacterially expressed and purified GST-Nrd1 in place of Atf1-FLAG was used as a substrate. Anti-phospho-Nrd1 antibodies were produced by immunization of rabbits with a synthetic phospho-peptide (keyhole limpet hemocyanin—coupled) corresponding to residues surrounding Thr40 or Thr126 of Nrd1.
mRNA Stability Assay
To analyze Cdc4 mRNA stability, cells were transformed with a reporter plasmid pREP41-GFP-Cdc4 3′-untranslated region (UTR) fusion gene (which included the entire cdc4 coding region plus the 3′-UTR) and then cultured in EMM. The mRNA stability was determined as described in Sugiura et al. (2003). The fractions of mRNA remaining were normalized to the level of Leu1 mRNA to control for loading error. The graphs show the averages of three different experiments.
Synchronization
For synchronization, cdc25-22 strains were grown at 25°C to an optical density at 595-nm wavelength (OD 595) of 0.3 and shifted to 37°C for 4 h. Then, cells were released into the cell cycle by quick transfer to 25°C. Septation index was determined by calculating the percentage of cells with a septum.
RESULTS
Nrd1 Is a Multicopy Suppressor of Cdc4
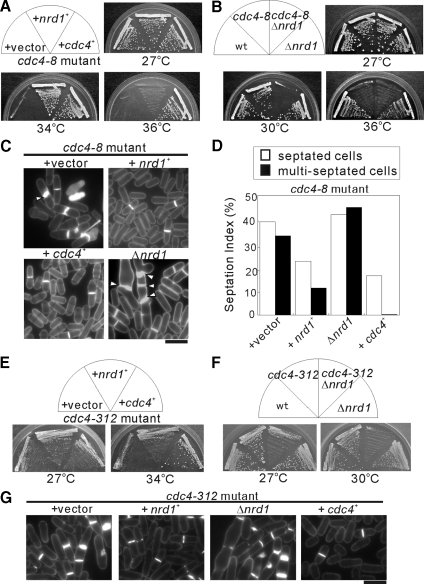
The fission yeast cdc4-8 mutant cells grew at the permissive temperature of 27°C, but the cells failed to grow at the restrictive temperature of 34°C (Figure 1A, +vector). To identify novel genes that are involved in Cdc4 function, we screened a fission yeast genomic library to isolate genes that when overexpressed could suppress the temperature sensitivity of cdc4-8 mutants. One of the genes that was previously identified is the nrd1+ gene that encodes a highly conserved RNA-binding protein with four repeats of the typical RRM (Tsukahara et al., 1998; Jeong et al., 2004b). As shown in Figure 1A, overexpression of the nrd1+ gene suppressed the temperature sensitivity of the cdc4-8 mutants at 34°C (Figure 1A, +nrd1+). However, cdc4-8 mutants overexpressing the nrd1+ gene failed to grow at 36°C, whereas cdc4-8 mutants expressing the cdc4+ gene grew well (Figure 1A, +cdc4+), indicating that Nrd1 partially suppresses the temperature sensitivity of cdc4-8 mutants.
Figure 1.
Nrd1 was isolated as a multicopy suppressor of the cdc4 mutant cells. (A) Overexpression of nrd1+ suppresses the temperature sensitivity of the cdc4-8 mutants. Cells were transformed with a multicopy plasmid containing the indicated genes, streaked onto YPD plates and then incubated at the temperatures as indicated. (B) Nrd1 deletion exacerbated the temperature sensitivity of the cdc4-8 mutants. Cells as indicated were streaked onto YPD plates and incubated at the temperature as indicated. (C) Nrd1 affects the cytokinesis defects of the cdc4-8 mutants. Cells as indicated were incubated at 27°C and then stained with calcofluor to visualize the cell wall and septum. Arrows indicate cells with abnormally thickened septa (+vector) or multiseptated cells (Δnrd1), respectively. Bar, 10 μm. (D) Cells as indicated were incubated at 36°C for 8 h, and the septation index was determined microscopically with calcofluor staining. (E) Overexpression of nrd1+ suppresses the temperature sensitivity of the cdc4-312 mutants. Cells were transformed with a multicopy plasmid containing the indicated genes and analyzed as described in A. (F) Nrd1 deletion exacerbated the temperature sensitivity of the cdc4-312 mutants. Cells as indicated were analyzed as described in B. (G) Nrd1 affects the cytokinesis defects of the cdc4-312 mutants. Cells as indicated were incubated at 27°C and then stained with calcofluor to visualize the cell wall and septum. Bar, 10 μm.
We also investigated the effect of nrd1+ deletion on the temperature sensitivity of cdc4-8 mutants. The Nrd1-deletion cells were viable and did not exhibit temperature-sensitive growth defect (Figure 1B, Δnrd1); the cdc4-8 mutants grew well at 30°C; however, the cdc4-8Δnrd1 double mutants failed to grow at 30°C, indicating that the Nrd1 deletion exacerbated the temperature sensitivity of the cdc4-8 mutants (Figure 1B).
To explore the functional relationship between Nrd1 and cytokinesis, we investigated the effects of both the overexpression and deletion of nrd1+ on the cytokinesis phenotypes of cdc4-8 mutants. Microscopic observation revealed that the cdc4-8 mutants, even at the permissive temperature, exhibited an increase in septation index as well as an increase in cells with irregular thickened septum that was brightly stained with calcofluor (Figure 1C, +vector, white arrowhead) and that was negligible in cdc4-8 mutants expressing the cdc4+ gene (Figure 1C, +cdc4+). In cdc4-8 mutant cells harboring the nrd1+ gene, however, the number of septated cells significantly decreased, and the cytokinesis defects associated with the cdc4-8 mutants were not observed (Figure 1C, +nrd1+). Moreover, compared with the cdc4-8 single mutants (Figure 1C, +vector), in the cdc4-8Δnrd1 double mutants, abnormal multiseptated cells were frequently observed (Figure 1C, Δnrd1, white arrowheads), indicating that Nrd1 deletion enhanced the abnormalities in cytokinesis.
As shown in Figure 1D, upon temperature upshift to 36°C, in the cdc4-8 mutants bearing the control vector alone (Figure 1D, +vector), the frequency of septated cells markedly increased (39%), and multiseptated cells were frequent (33%). In cdc4-8 mutant cells harboring the nrd1+ gene (Figure 1D, +nrd1+), the frequencies of septated cells (23%) and multiseptated cells (10%) were lower. Notably, in the cdc4-8Δnrd1 double mutants (Figure 1D, Δnrd1) the frequency of septated cells (42%) and multiseptated cells (45%) further increased compared with the single cdc4-8 mutants. Thus, both the overexpression and deletion of Nrd1 had an effect on the cytokinesis phenotypes of the cdc4-8 mutant cells.
We next examined whether Nrd1 overproduction can rescue other cdc4 mutations. The results showed that Nrd1 overproduction partly rescued the temperature sensitivity and the cytokinesis defect of the cdc4-312 mutation (Figure 1, E and G). In addition, the cdc4-312Δnrd1 cells failed to grow at 30°C, whereas all the single mutants grew well (Figure 1F). Even at the permissive temperature of 27°C, the cdc4-312Δnrd1 cells exhibited poor growth (Figure 1F), and Nrd1 deletion enhanced the cytokinesis defects observed in the cdc4-312 mutants (Figure 1G, Δnrd1), indicating that Nrd1 genetically interacts with the cdc4-312 mutation as well as with the cdc4-8 mutation. Thus, Nrd1 globally affects cdc4 function and is not an allele-specific suppressor of cdc4 mutants.
Genetic Interactions between Nrd1 and Other Cytokinesis Mutants
To examine the effect of Nrd1 on Cdc4, we investigated possible genetic interactions between nrd1+ and other genes encoding proteins that form the actomyosin ring. To this end, we carried out genetic crosses between Δnrd1 and actomyosin ring assembly/function mutants, namely, cdc12-112, rng2-D5, rng3-65, myo2-E1, Δrlc1, and Δmyo3/Δmyp2/Δcis2 (Supplementary Table S1). Except for Δrlc1, these ring mutants show temperature-sensitive phenotypes. Because Δrlc1 cells are cold sensitive (Le et al., 2000), they grew well at 30°C (the permissive temperature for Δrlc1) but failed to grow at 23°C (the nonpermissive temperature for Δrlc1). We therefore chose 33°C as the nonpermissive temperature for temperature-sensitive ring mutants and 23°C as the nonpermissive temperature for Δrlc1. It was noted that Δnrd1 cells display synthetic growth defects under restrictive conditions when combined with myosin II heavy chain myo2-E1, myosin regulatory light chain rlc1Δ, and the type II myosin heavy chain Δmyo3/Δmyp2/Δcis2 (Supplementary Table S1, Supplementary Figure S1, A and B). In addition, the double mutants displayed more severe cell separation phenotypes compared with that of the each single mutant (Supplementary Table S1 and Supplementary Figure S1, C and D). Thus, these genetic interactions with other ring mutants suggest that Cdc4 function is compromised in nrd1-deletion cells.
Nrd1 Regulates Cdc4 mRNA
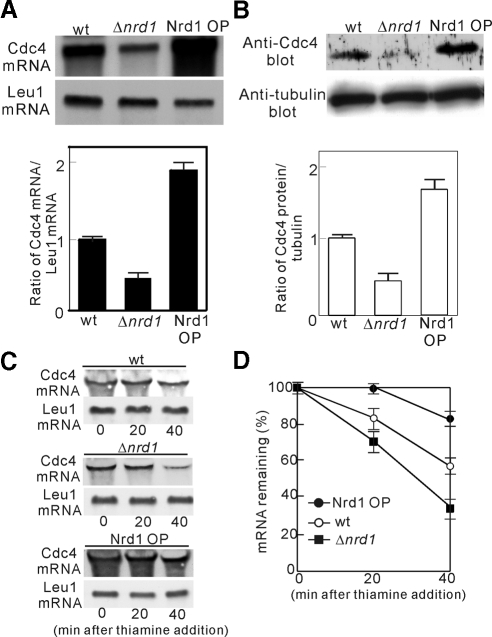
How does Nrd1 overexpression and deletion affect the phenotypes of cdc4 mutant cells? Because the nrd1+ gene encodes an RNA-binding protein, Nrd1 might control the metabolism of Cdc4 mRNA, thereby affecting the temperature sensitivity and cytokinesis phenotypes of the cdc4 mutants. To investigate the effects of the overexpression and deletion of Nrd1 on the amount of Cdc4 mRNA, further experiments were conducted. In nrd1-knockout cells (Figure 2A, Δnrd1), the level of Cdc4 mRNA was significantly low compared with that of wild-type cells, whereas in Nrd1-overproducing cells (Figure 2A, Nrd1 OP), the level of Cdc4 mRNA was markedly increased (Figure 2A), thus indicating that Nrd1 regulates the levels of Cdc4 mRNA. We also examined the protein levels of Cdc4 in these cells by immunoblotting with anti-Cdc4 antibodies (Figure 2B). The immunoblot analysis revealed that Nrd1 deletion decreased and Nrd1 overexpression increased the Cdc4 protein level (Figure 2B), thus indicating the correlation with those of the Cdc4 mRNA abundance (Figure 2B).
Figure 2.
Nrd1 regulates the stability of Cdc4 mRNA. (A) Top panel, Northern blot analysis of the Cdc4 mRNA levels in wild-type cells (wt), Nrd1-deletion cells (Δnrd1), and cells overproducing Nrd1 (Nrd1 OP). Bottom panel, quantification of Cdc4 mRNA levels by densitometry of the bands was expressed relative to that of the control mRNA (leucine) . (B) Top panel. immunoblot analysis of the Cdc4 protein in cells as indicated. The whole-cell lysates were analyzed by immunoblotting with anti-Cdc4 antibodies. Bottom panel, the Cdc4 protein levels (relative to tubulin) were obtained by densitometry. (C) The effect of Nrd1 on the stability of Cdc4 mRNA. Cdc4 mRNA levels after expression from the nmt1/cdc4 construct that included the entire coding region plus the 3′-UTR and transcription repression with thiamine. (D) Quantification of Cdc4 mRNA levels from C. Cdc4 mRNA levels at each time point were calculated as a percentage of time (0 min = 100%) after normalization to leu1 levels. Shown are the mean values (± SD) from three independent experiments, identical to those shown in C.
To examine if the above changes in the Cdc4 mRNA levels are due to alterations in Cdc4 mRNA stability, we examined the mRNA level of GFP-Cdc4 transcribed from the thiamine-repressible nmt1 promoter (Maundrell, 1990). For this, we created a reporter construct that included the cdc4-coding region plus the 3′-UTR (see Materials and Methods). The levels of GFP-Cdc4 mRNA were significantly low in Δnrd1 cells and markedly high in Nrd1-overproducing cells as compared the wild-type cells (Figure 2C). Furthermore, the GFP-Cdc4 mRNA level in Δnrd1 cells decreased more rapidly, whereas that in Nrd1-overproducing cells decreased more slowly than that in the wild-type cells after transcription was shut off (Figure 2D). Thus, Cdc4 mRNA is destabilized by Δnrd1 mutation and stabilized by Nrd1 overproduction. Together, these results suggest that Nrd1 affects the phenotypes of the cdc4-8 mutants by regulating Cdc4 mRNA stability.
Nrd1 Binds to Cdc4 mRNA In Vivo and In Vitro
The above findings raise the possibility that the Nrd1 protein associates with Cdc4 mRNA. To test this, we expressed GST with a control vector or GST-Nrd1 with either a control vector or the Cdc4 open reading frame (ORF) with or without the 3′-UTR. We then purified GST-Nrd1 or GST, and the mRNAs bound to the precipitates were detected by RT-PCR analysis. As shown in Figure 3A, Cdc4 mRNA was detectable in the GST-Nrd1 pulldowns but not in the GST pulldowns, indicating that Nrd1 associates with Cdc4 mRNA in the presence (+Cdc4 ORF with the 3′-UTR) or absence (+Cdc4 ORF) of its 3′-UTR. The PCR product was not obtained when reverse transcriptase was omitted, indicating that formation of this band is dependent on RNA (data not shown). Thus, the Nrd1 protein interacts with Cdc4 mRNA in vivo.
Figure 3.
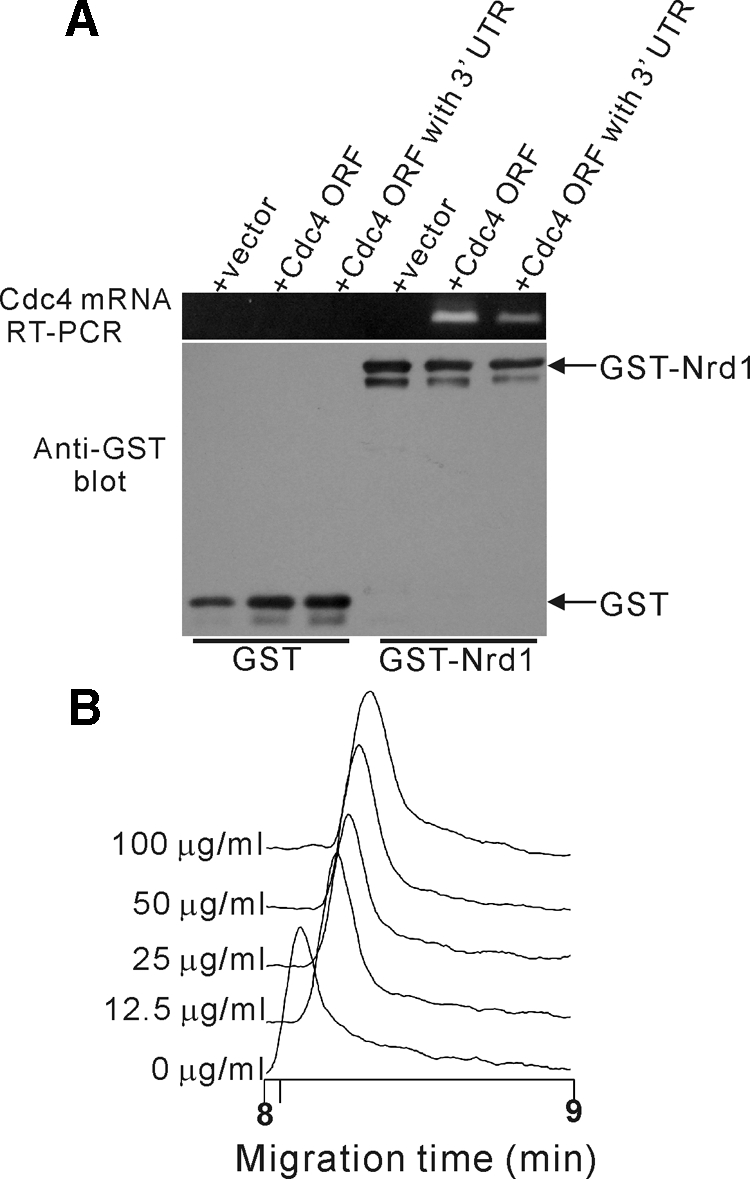
Nrd1 binds to Cdc4 mRNA in vivo and in vitro. (A) GST-Nrd1 associates with Cdc4 mRNA in vivo. Wild-type cells expressing GST with either a control vector or Cdc4 with or without its 3′-UTR or GST-Nrd1 with either a control vector or Cdc4 with or without its 3′-UTR were analyzed for their mRNA binding as described in Materials and Methods. (B) GST-Nrd1 associates with Cdc4 mRNA in vitro. Affinity capillary electrophoresis mobility shift assay of the recombinant Nrd1 protein concentrations of Cdc4 mRNA as indicated were analyzed as described in Materials and Methods.
To investigate the Nrd1 protein-Cdc4 mRNA interaction in vitro, we used the affinity capillary electrophoresis mobility shift assay (Taga et al., 2004) and developed a method to detect protein–RNA interaction (see Materials and Methods). We expressed and purified the recombinant Nrd1 protein from E. coli and examined the effects of various concentrations of in vitro–synthesized Cdc4 mRNA on the migration of the Nrd1 protein. As shown in Figure 3B, mobility shift (migration time; ∼0.2 min) was observed in the presence of Cdc4 mRNA (12.5 μg/ml) compared with that of the control (0 μg/ml). This migration time delay of 0.2 min is equivalent to ∼2.4% of migration time in the absence of mRNA. It was caused by the addition of Cdc4 mRNA to the electrophoretic solution, because this method has high repeatability with a relative SD of a migration time of <0.40% (n = 6). Moreover, the mobility shift was observed in an RNA concentration–dependent manner, suggesting the formation of an RNA–protein complex. No mobility shift was observed in the presence of 100 μg/ml anti-sense Cdc4 mRNA (data not shown). Thus, Nrd1 directly binds with Cdc4 mRNA in vivo and in vitro.
Mutation in the UCUU Sequences in Cdc4 mRNA Affects Nrd1 Binding and mRNA Stability
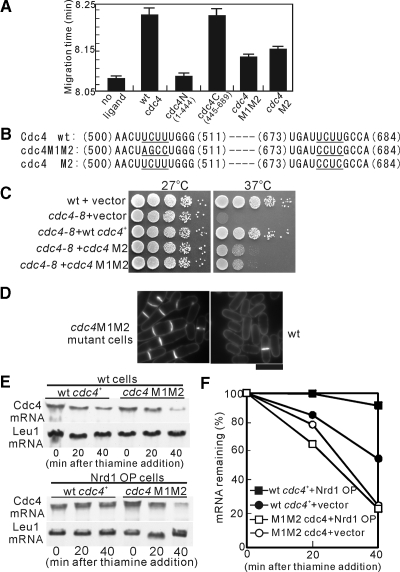
As a first step toward locating the Nrd1-binding site in the Cdc4 mRNA, we divided the Cdc4-coding region into two fragments, N (nucleotides 1–444; numbering starts at the first nucleotide of the start codon) and C (nucleotides 445–689), and performed in vitro binding experiments using these truncated forms of in vitro–synthesized Cdc4 mRNAs as ligands. As shown in Figure 4A, the mobility shift in the presence of the C fragment of the Cdc4 mRNA (cdc4C, migration time; ∼0.20 min) was comparable to that of the full-length Cdc4-coding region (wt cdc4), indicating that fragment C of the cdc4-coding region maintains strong binding activity with Nrd1. In contrast, the mobility shift in the presence of fragment N (cdc4N) was almost equivalent to that in the absence of a ligand (no ligand), suggesting that fragment C is responsible for the binding with Nrd1 (Figure 4A).
Figure 4.
Mutation in the UCUU sequences in Cdc4 mRNA affects Nrd1 binding and mRNA stability. (A) Affinity capillary electrophoresis mobility shift assay, as described in Figure 3B, was performed with the wild-type Cdc4 mRNA and its mutant versions. Shown are the mean values (±SD) of migration time in each sample from three independent experiments. (B) The UCUU sequences in the Cdc4 mRNA. The UCUU elements and their mutated sequences are underlined. The nucleotide number from the start codon is indicated. (C) Cells transformed with the pREP41 vector containing the indicated genes were spotted onto EMM plates containing thiamine (repressed condition) and then incubated at 27 or 37°C. (D) Cytokinesis defects of the cdc4 M1M2 mutant cells. Cells as indicated were analyzed as described in Figure 1C. Bar, 10 μm. (E) Mutation in UCUU sequences results in the destabilization of Cdc4 mRNA. Wild-type cells or wild-type cells overproducing Nrd1 (Nrd1 OP cells) and expressing either pREP41-GFP-Cdc4 (wt cdc4+) or pREP41-GFP-Cdc4M1M2 (cdc4M1M2) were analyzed as described in Figure 2C and Materials and Methods. (F) Quantification of Cdc4 mRNA levels from E. The mRNA stability of Cdc4 was examined as shown in Figure 2C. Results are expressed as the mean value of the results of three independent experiments.
We noted the presence of two UCUU sequences within the C fragment of the Cdc4 mRNA (Figure 4B; Cdc4 wt, underlined). Notably, Nrd1 shows significant sequence similarity with TIA-1 and the TIA-1–related (TIAR) proteins in higher eukaryotes (Dember et al., 1996; Kedersha et al., 1999). It has been reported that both TIA-1 and TIAR bind to RNA, with the preferred binding sequence being U-rich, including the UCUU motifs present in the mRNAs (Forch et al., 2000; Gatto-Konczak et al., 2000). To test whether a mutation in the UCUU sequences affects the binding between Cdc4 mRNA and the Nrd1 protein, we generated mutant versions of Cdc4 mRNA without alteration to the reading frame (Figure 4B; cdc4M1M2 and cdc4M2, underlined). We then performed in vitro–binding experiments using the mutant versions of in vitro–synthesized Cdc4 mRNAs as ligands. Notably, the mutation in the two UCUU sequences (cdc4M1M2) abrogated the binding between Nrd1 and Cdc4 mRNA, as shown by the migration time of the M1M2 mutant, which was almost half that of the full-length Cdc4 (Figure 4A, cdc4M1M2). The migration time of the M2 mutant (Figure 4A, cdc4M2) increased only slightly compared with that of the M1M2 mutant but decreased significantly compared with that of wild-type Cdc4.
Consistently, the cdc4M1M2 and cdc4M2 mutants only weakly suppressed the temperature sensitivity of the cdc4-8 mutant, whereas the wild-type cdc4 fully suppressed these phenotypes, indicating a good correlation between the Nrd1 binding and the suppression ability of Cdc4 (Figure 4C). We then created the cdc4 M1M2 mutant cells by altering these sites in the Cdc4 mRNA and examined the impact of the mutation on the morphology. For this, the cdc4 mutant strain with one copy of cdc4M1M2 expressed from the genomic cdc4+ promoter was obtained. Notably, the cdc4M1M2 mutant cells exhibited cell separation defects (Figure 4D), suggesting that the loss of Nrd1 binding affects cytokinesis.
We further examined the mRNA stability of the cdc4M1M2 mutant, and the results revealed that the cdc4M1M2 mutant mRNA exhibited significantly reduced stability compared with that of the wild-type Cdc4 (Figure 4, E and F). In addition, the overproduction of Nrd1 (Nrd1 OP) stabilized the wild-type Cdc4 mRNA levels, whereas the M1M2 mutant mRNA exhibited minimal response to Nrd1 overexpression (Figure 4, E and F). Thus, mutation in these UCUU sequences affects Nrd1 binding, thereby resulting in the destabilization of the Cdc4 mRNA.
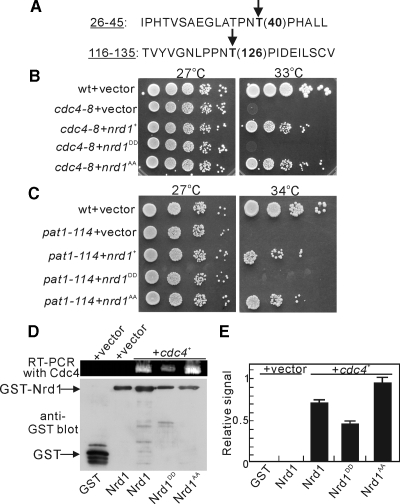
The RNA-binding Activity of Nrd1 is Phosphorylation Dependent
In the Nrd1 amino acid sequence, we noted the presence of two potential proline-directed MAPK phosphorylation sites Thr40 (T40) and Thr126 (T126; Figure 5A, arrows). To assess the functional significance of the phosphorylation sites in Nrd1, we first constructed the substitution mutant Nrd1AA by replacing two threonine residues with alanine and expressed this in cdc4-8 mutants. The expression of Nrd1 and its phosphorylation mimic versions was induced by using the thiamine-repressible nmt1 promoter. The expression of the unphosphorylatable Nrd1AA mutant suppressed the temperature sensitivity of the cdc4-8 mutants to a slightly greater degree than that of the wild-type Nrd1 (Figure 5B, cdc4-8+nrd1AA). Second, we also constructed the Nrd1DD mutant by replacing T40 and T126 with aspartic acid. In contrast, the expression of the Nrd1DD, a phosphorylation mimic version of Nrd1, failed to suppress the cdc4-8 mutants (Figure 5B, cdc4-8+nrd1DD). Because the nrd1+ gene was isolated as a multicopy suppressor of the pat1-114 mutants, we examined if the suppression of the temperature sensitivity of the pat1-114 mutants was also phosphorylation dependent. The unphosphorylated mutant version of Nrd1, Nrd1AA, suppressed the pat1-114 mutants, whereas the phosphorylation mimic version of Nrd1, Nrd1DD, failed to suppress the pat1-114 mutants (Figure 5C). Notably, the suppression ability of Nrd1 (or Nrd1AA) was observed even in the presence of thiamine (i.e., under the repressed condition), suggesting that only a modest increase in Nrd1 expression is sufficient for the suppression of these mutant cells.
Figure 5.
The RNA-binding ability of Nrd1 is phosphorylation dependent. (A) The amino acid sequence containing the two putative MAPK phosphorylation sites. The identified phosphorylation sites, T(40) and T(126), are denoted by arrows. (B) Phosphorylation of T(40) and T(126) abrogates the ability of Nrd1 to suppress the cdc4-8 mutant. Cells transformed with the pREP1 vector containing the indicated genes were spotted onto the EMM plates containing thiamine (repressed condition) and then incubated at 27 or 33°C. (C) Phosphorylation of T(40) and T(126) abrogates the ability of Nrd1 to suppress the temperature sensitivity of the pat1-114 mutants. Cells transformed with the pREP1 vector containing the indicated genes were spotted onto EMM plates containing thiamine (repressed condition) and then incubated at 27 or 34°C. (D) Wild-type cells expressing GST with a control vector, expressing GST-Nrd1 with a control vector, or expressing either GST-Nrd1, GST-Nrd1DD, or GST-Nrd1AA with Cdc4 were analyzed as shown in Figure 3A. (E) The amount of Cdc4 mRNA bound by either GST, GST-Nrd1, or GST-Nrd1 mutants was quantified by densitometry of the bands from D and was expressed relative to the control GST pulldown precipitates run in the same experiment.
Third, we performed in vivo RNA-binding experiments by utilizing the phosphorylation mutant versions of Nrd1 and comparing them with the wild-type Nrd1. As expected, the amount of Cdc4 mRNA bound to the Nrd1AA mutant was increased compared with that bound to wild-type Nrd1, and the amount of Cdc4 mRNA bound to the Nrd1DD mutant was lower (Figure 5, D and E). Collectively, these results suggest that the phosphorylation of T40 and T126 affects the ability of Nrd1 to bind to and stabilize the target RNA(s) and that the unphosphorylated version of Nrd1 strongly binds to Cdc4 mRNA, thereby suppressing the cdc4-8 mutant cells.
Nrd1 is a Direct Target of MAPK Pmk1
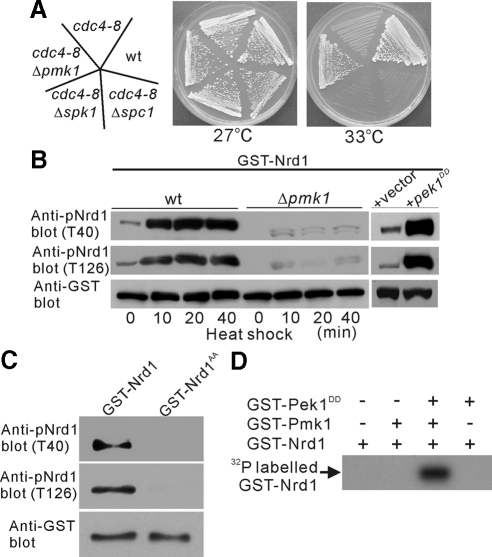
The above results lead to the hypothesis that the inactivation of a MAPK pathway might positively control the function of Nrd1, thereby suppressing the phenotype of the cdc4-8 mutant cells. To test this possibility, we examined the effects of a series of MAPK-deletion strains on the temperature-sensitive phenotype of the cdc4-8 mutants. Notably, the deletion of Pmk1 MAPK, which regulates cell integrity (Toda et al., 1996; Sugiura et al., 1999, 2003), suppressed the temperature sensitivity of the cdc4-8 mutant cells (Figure 6A, cdc4-8Δpmk1). In contrast, the deletion of Spk1 MAPK, which regulates meiosis (Gotoh et al., 1993; Figure 6A, cdc4-8Δspk1), or the deletion of Spc1/Sty1 MAPK, a homologue of p38 in fission yeast (Millar et al., 1995; Degols et al., 1996), did not suppress the temperature-sensitive growth of cdc4-8 mutant cells (Figure 6A, cdc4-8Δspc1). Thus, the deletion of Pmk1 specifically suppressed the temperature-sensitive phenotype of the cdc4-8 mutants, suggesting that the Pmk1 MAPK may phosphorylate Nrd1, thus controlling the function of Nrd1.
Figure 6.
Nrd1 is a direct target of MAPK Pmk1. (A) Inactivation of Pmk1 specifically suppressed the temperature sensitivity of the cdc4-8 mutants. The strains were streaked onto the YPD plates and incubated as indicated. (B) Phosphorylation of Nrd1 is dependent on Pmk1 signaling in vivo. The cells as indicated were transformed with pREP1-GST-Nrd1, with either the control vector or the constitutively active pek1DD, and analyzed for T40 or T126 phosphorylation. (C) Reactivity of phospho-specific Nrd1 antibodies with wild-type Nrd1 and the mutant Nrd1AA proteins. Wild-type cells expressing pREP1-GST-Nrd1, or the pREP1-GST-Nrd1AA mutant protein were analyzed by immunoblotting with anti-pNrd1 (T40), anti-pNrd1 (T126), or anti-GST antibodies. (D) Pmk1 MAPK directly phosphorylates Nrd1 in vitro. GST-Nrd1 phosphorylation activity of the recombinant GST-Pmk1 was analyzed as described in Materials and Methods.
To investigate the effects of Pmk1 signaling on the phosphorylation levels of Nrd1, we first created anti-phospho Nrd1 T40 antibodies and anti-phospho Nrd1 T126 antibodies that recognize the phosphorylation state of T40 or T126, respectively. Results showed that T40 phosphorylation or T126 phosphorylation of GST-Nrd1 was detected in wild-type cells (wt) but not in Pmk1-deletion cells (Δpmk1; Figure 6B, 0 min). Moreover, heat-shock treatment induced the phosphorylation of Nrd1 on both T40 and T126 in wild-type cells, whereas the phosphorylation was detected only faintly in Δpmk1 cells (Figure 6B). Second, we examined whether the activation of Pmk1 by the overexpression of Pek1DD, a constitutively active form of MAPKK for Pmk1 induces the phosphorylation of Nrd1. As expected, Nrd1 is heavily phosphorylated on both T40 and T126 by the overexpression of constitutively active Pek1DD (Figure 6B, +pek1DD). It should be noted that anti-phospho Nrd1 T40 antibodies or anti-phospho Nrd1 T126 antibodies recognize the wild-type GST-Nrd1 protein but not the GST-Nrd1AA mutant protein (Figure 6C), indicating that these antibodies are highly specific and did not cross-react with nonphosphorylated Nrd1. Finally, we carried out the in vitro kinase assay to evaluate whether the Pmk1-dependent phosphorylation of Nrd1, as demonstrated above, might involve the direct phosphorylation of Nrd1 by Pmk1 kinase. The results shown in Figure 6D indicate that the addition of purified Pek1DD stimulated the kinase activity of GST-Pmk1, thereby resulting in the phosphorylation of GST-Nrd1. It should be noted that the addition of GST-Pek1DD alone did not result in the phosphorylation of GST-Nrd1 (Figure 6D). Altogether, these results indicate that Pmk1 directly phosphorylates Nrd1 in vivo and in vitro, and that Nrd1 is a novel target of Pmk1 MAPK.
Pmk1-dependent Phosphorylation Negatively Regulates the Ability of Nrd1 to Bind and Regulate Cdc4 mRNA
Next, we attempted to examine whether Pmk1-dependent phosphorylation affects the binding of Nrd1 to Cdc4 mRNA. First, we observed the effects of the overexpression of constitutively active Pek1DD and Pmk1 deletion on the amount of Cdc4 mRNA bound to Nrd1. The overexpression of Pek1DD reduced the amount of Cdc4 mRNA bound to Nrd1 (Figure 7A), which is consistent with the data obtained with the phosphorylation mimic version of Nrd1DD (Figure 5D). The Pmk1 deletion, in contrast, increased the amount of Cdc4 mRNA associated with Nrd1 because Cdc4 mRNA was detected even without the coexpression of Cdc4 (Figure 7A, Δpmk1). Second, we performed in vitro RNA-binding experiments utilizing bacterially expressed Nrd1 phosphorylated with the activated Pmk1 MAPK. We found that the phosphorylated Nrd1 lost its ability to bind to Cdc4 mRNA because almost no mobility shift was detected even in the presence of 100 μg/ml Cdc4 mRNA (Figure 7B). Again, we observed that the assay was highly reproducible with a relative SD of a migration time of <0.27% (n = 3). Thus, Pmk1 phosphorylation of Nrd1 negatively regulates the association of Nrd1 with Cdc4 mRNA.
Figure 7.
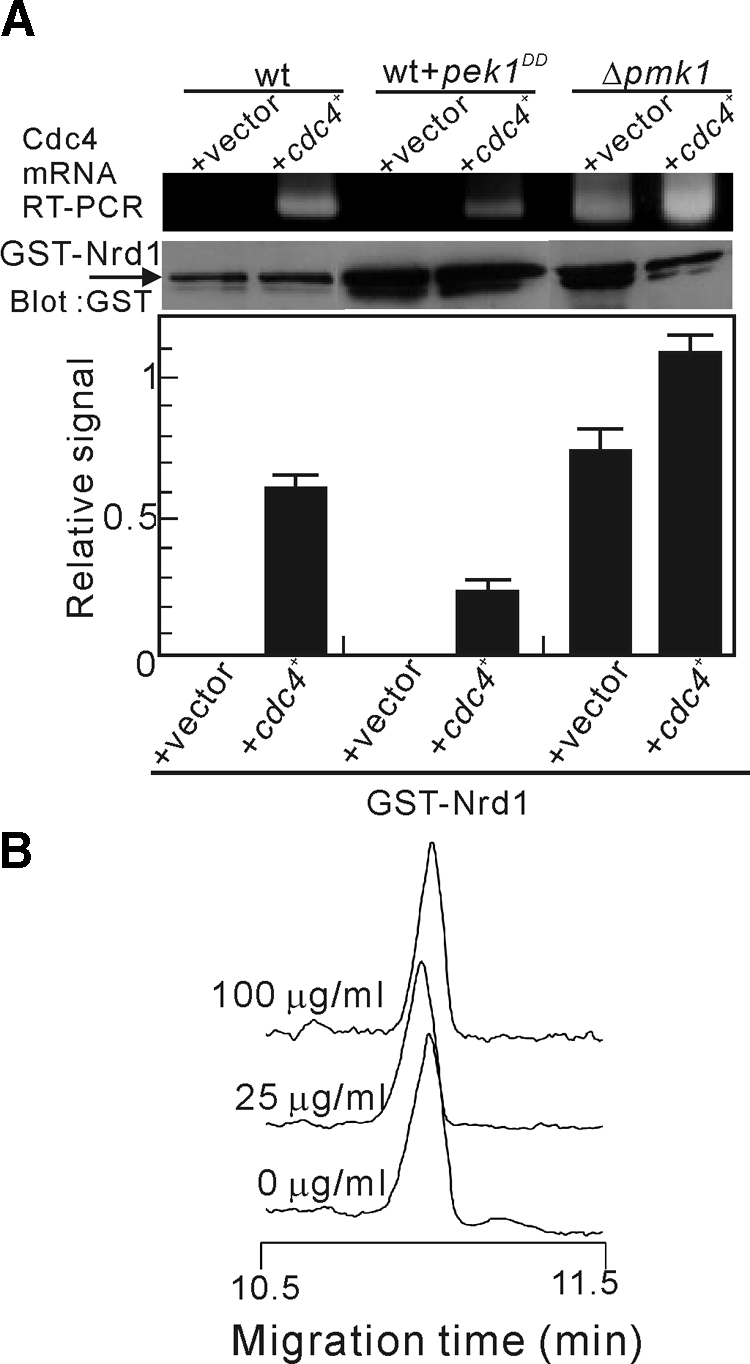
Pmk1-mediated phosphorylation negatively regulates the ability of Nrd1 to bind and regulate Cdc4 mRNA. (A) Pmk1 phosphorylation of Nrd1 impairs the binding activity of Nrd1 to Cdc4 mRNA. The cells indicated are transformed either with the control vector or cdc4+ and analyzed as shown in Figure 3A. Data were quantified and expressed as shown in Figure 5D. (B) Affinity capillary electrophoresis mobility shift assay as shown in Figure 3B was performed with the Nrd1 protein phosphorylated by the activated Pmk1 MAPK.
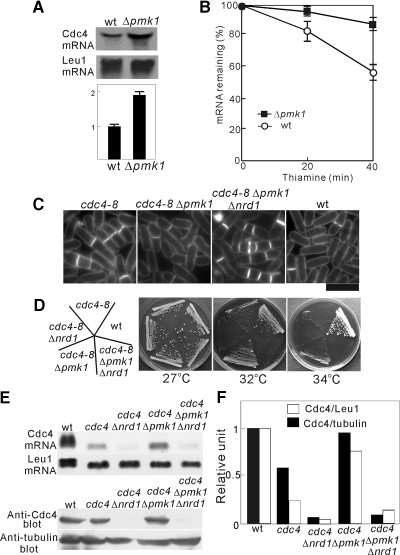
We then attempted to assess the effect of the MAPK-dependent regulation of Nrd1 function on the amount of Cdc4 mRNA and the cytokinesis phenotypes of the cdc4-8 mutants. As shown above, Pmk1 deletion suppressed the temperature-sensitive growth defect of the cdc4-8 mutant (Figure 6A). If Pmk1 MAPK negatively regulates the Nrd1 activity thereby affecting the temperature-sensitive phenotype of cdc4-8 mutants, it would be expected that Pmk1 deletion, like Nrd1 overexpression, could stabilize Cdc4 mRNA and suppress the cytokinesis defects observed in the cdc4-8 mutants. As expected, the amount of Cdc4 mRNA was significantly increased in the Pmk1-deletion cells compared with the wild-type cells (Figure 8A). Moreover, after transcription was shut off, the GFP-Cdc4 mRNA level decreased more slowly in the Δpmk1 cells than in the wild-type cells (Figure 8B). Consistently, Pmk1 deletion suppressed the cytokinesis defects of the cdc4-8 mutants, because the abnormal multiseptated phenotypes associated with the cdc4-8 mutants improved considerably in the cdc4-8Δpmk1 mutant cells (Figure 8C).
Figure 8.
The Pmk1 MAPK pathway modulates the level of Cdc4 mRNA. (A) Inactivation of Pmk1 stabilized Cdc4 mRNA. Northern blot analysis of the Cdc4 mRNA levels in wild-type cells (wt) and Pmk1-deletion cells (Δpmk1) (top panel). Quantification of Cdc4 mRNA levels (bottom panel). The above data were quantified by densitometry and were expressed as shown in Figure 2A. (B) The effect of Pmk1 on the stability of Cdc4 mRNA. The stability of Cdc4 mRNA was examined as shown in Figure 2, C and D. (C) Inactivation of Pmk1 ameliorated the cytokinesis defects of the cdc4-8 mutants. The indicated strains were incubated at 27°C and then stained with calcofluor as shown in Figure 1C. Bar, 10 μm. (D) The Nrd1 deletion reverted the phenotypes of the cdc4-8Δpmk1 cells. Cells as indicated were streaked onto YPD plates and incubated at the temperature as indicated. (E) Top panel, Northern blot analysis of the Cdc4 mRNA levels in the indicated cells, as described in Figure 2A. Bottom panel, immunoblot analysis of the Cdc4 protein in the indicated cells. The whole-cell lysates were analyzed by immunoblotting with anti-Cdc4 antibodies. (F) Cdc4 mRNA and protein levels were quantified as described in Figure 2, A and B.
To investigate if this rescue of the cdc4-8 mutant phenotypes induced by Pmk1 deletion occurs through a pathway involving Nrd1, we created cdc4-8Δpmk1Δnrd1 triple mutants. The cdc4-8Δpmk1Δnrd1 mutants failed to grow at 32°C, whereas the cdc4-8Δpmk1 mutants grew (Figure 8D), indicating that the Nrd1 deletion reverted the effect of Pmk1 deletion on the cdc4-8 phenotypes. Moreover, the cdc4-8Δpmk1Δnrd1 mutant displayed an abnormal morphology and cytokinesis defects (Figure 8C). Consistent with the aggravation of the phenotypes, the Cdc4 mRNA and protein levels were decreased by Nrd1 deletion in the cdc4-8Δpmk1Δnrd1 mutants (Figure 8, E and F). Thus, the RNA-binding protein Nrd1 transmits Pmk1 MAPK signaling to an important actin ring component, Cdc4.
Pmk-Deletion Mutants Genetically Interact with Other Cytokinesis Mutants
The findings that Pmk1 deletion, like Nrd1 overexpression, could stabilize Cdc4 mRNA and rescue the cdc4 mutant phenotypes prompted us to investigate genetic interactions between Δpmk1 and actomyosin ring mutants. For this, we used 33°C as the nonpermissive temperature for temperature-sensitive ring mutants and 23°C as the nonpermissive temperature for Δrlc1 cells. First, deletion of Pmk1, similar to Nrd1 overproduction, rescued the temperature sensitivity and cytokinesis defects of the cdc4-312 mutant (Supplementary Figure 2, A and B). Furthermore, Δpmk1 displayed genetic interactions with Δrlc1, myo2-E1, and Δmyo3 mutations. The Pmk1 deletion, rescued the growth defects under the restrictive conditions of the Δrlc1, myo2-E1, and Δmyo3 mutants (Supplementary Table S2 and Supplementary Figure S2, C and E). This is consistent with the data that Δnrd1 cells display synthetic growth defects when combined with myo2-E1, rlc1Δ, and Δmyo3/Δmyp2/Δcis2 (Supplementary Table S1, Supplementary Figure S1). In line with this view, cdc4-8 showed strong synthetic lethal interactions with the myo2-E1 (Naqvi et al., 1999) and rlc1-981 mutants (Le et al., 2000). Thus, these genetic interactions between Pmk1 and ring mutants are in good agreement with the observation that Nrd1 affects cdc4 mRNA stability; therefore, a reduction in cdc4 function would be expected to exacerbate the mutations in these ring components.
Cell Cycle-dependent Regulation of Pmk1-Nrd1 Signaling
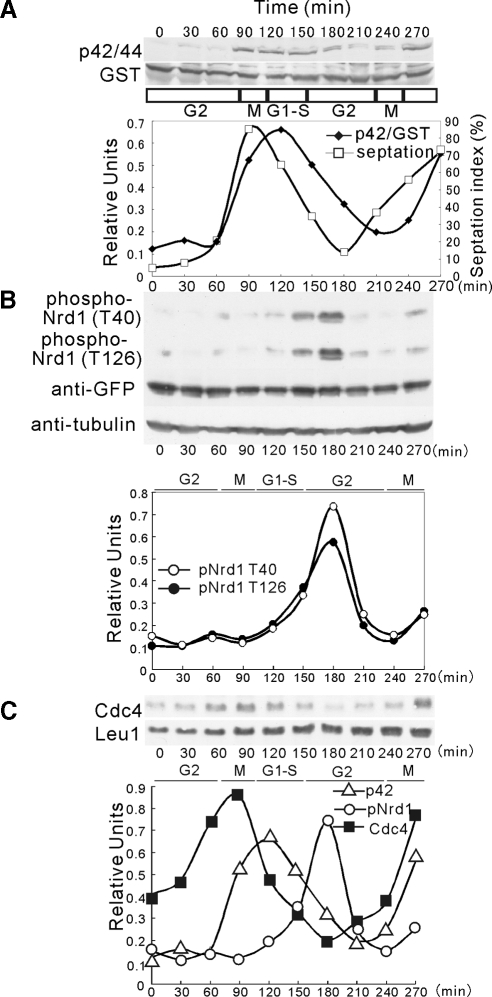
In a recent study, Madrid et al. (2007) reported that the phosphorylation state of Pmk1 oscillates as a function of the cell cycle. This prompted us to examine the hypothesis that Nrd1 RNA-binding activity and Cdc4 mRNA abundance are also regulated by the cell cycle. First, we examined the phosphorylation state of Pmk1 during the cell cycle by introducing the Pmk1-GST fusion into the cdc25--22 strain. Cells from this mutant were grown to the log phase at 25°C, shifted to 37°C for 4 h to synchronize the cells in the G2 phase, and then shifted back to 25°C. As shown in Figure 9A, changes in Pmk1 phosphorylation as detected by anti-phospho-p42/44 antibodies were noted during the cell cycle, reaching a maximum during cytokinesis, which is consistent with previous observations (Madrid et al., 2007).
Figure 9.
Cell cycle–dependent regulation of Pmk1-Nrd1 signaling. (A) Cell cycle–dependent activation of Pmk1 MAPK. Top, cells from SP628 strains (cdc25-22 pmk1-GST∷KanMx6) were grown at 25°C, shifted to 37°C for 4 h, and then released from growth arrest by transferring them back to 25°C. Aliquots were obtained at different time points, and activated or total Pmk1 was detected by immunoblotting with anti-phospho-p42/44 or anti-GST antibodies, respectively. Bottom, quantification of the Pmk1 activity during the cell cycle by normalization to the loading control value. The septation index (□) is also shown, which indicates good cell cycle synchrony in the culture. (B) Cell cycle–dependent phosphorylation of Nrd1. Top, cells from strain cdc25-22 were transformed with pREP1-GFP-Nrd1, grown in EMM containing thiamine, and analyzed as shown in A. Phosphorylated or total Nrd1 was detected by immunoblotting with anti-phospho-Nrd1 (T40), anti-phospho-Nrd1 (T126), or anti-GFP antibodies. Tubulin served as a loading control. Bottom, quantification of Nrd1 phosphorylation by normalization to the loading control value. (C) Cell cycle–dependent oscillation of Cdc4 mRNA. Top, cells from the SP628 strains were grown at 25°C, shifted to 37°C for 4 h, and then released from growth arrest by transferring them back to 25°C. Aliquots were obtained at different time points, and Cdc4 mRNA was detected by Northern blot analysis as described in Figure 2A. Bottom, quantification of Pmk1 activation (▵), T40 Nrd1 phosphorylation (○), and Cdc4 mRNA levels (■).
Next, we examined if the phosphorylation levels of Nrd1 are also cell cycle–dependent. As shown in Figure 9B, the Nrd1 phosphorylation, as measured by the phospho-T40 or phospho-T126 Nrd1 antibodies, oscillated during the cell cycle, increasing during the G1/S phases and reaching a maximum during the G2 phase (180 min). This pattern of oscillation in Nrd1 phosphorylation is consistent with our data that Pmk1 mediates signal transmission by phosphorylating Nrd1, further suggesting that Pmk1/Nrd1 signaling is regulated in a cell cycle–dependent manner.
Finally, we measured the Cdc4 mRNA levels during the cell cycle. As shown in Figure 9C, the Cdc4 mRNA levels also oscillate with a peak around the M phase to G1 phases, reaching a minimum around the G2 phase (180 min). These results are in agreement with those of genome-wide studies on cell cycle–dependent gene expression in fission yeast (Rustici et al., 2004; Peng et al., 2005). Notably, a decrease in the Cdc4 expression levels coincided with an increase in Nrd1 phosphorylation, further supporting our hypothesis that Nrd1 binds to and stabilizes Cdc4 mRNA and that Nrd1 RNA-binding activity is negatively regulated by its phosphorylation.
DISCUSSION
In various eukaryotes, phosphorylation or dephosphorylation of myosin has been well documented as a regulatory mechanism of myosin II (Satterwhite et al., 1992; Brzeska and Korn, 1996; Kamm and Stull, 2001; Loo and Balasubramanian, 2008). Notably, it has been reported that the phosphorylation of the Cdc4 protein is not required for cytokinesis (McCollum et al., 1999). This raises a question as to what is the mechanism that regulates the essential myosin light chain in fission yeast. Here in fission yeast, we demonstrate that Nrd1, as an RNA-binding protein, is implicated in the regulation of Cdc4 mRNA stability. The cdc4-8 mutation results in the substitution of serine for glycine at amino acid 107 (G107S), indicating that the cdc4-8 mutant mRNA contains a missense codon (McCollum et al., 1995). One possible explanation for the suppression mechanism mediated by Nrd1 is that the Cdc4 G107S mRNA (and/or protein) is more destabilized than the wild-type Cdc4 mRNA. In line with these expectations, we demonstrated that the cdc4-8 mRNA was indeed more destabilized than the wild-type Cdc4 and that Nrd1 deletion further decreased the amount of Cdc4 mRNA (Figure 8, E and F), which might explain the synthetic growth defect of the cdc4-8 mutants when combined with Δnrd1. Thus, the overexpression of nrd1+ may suppress the cdc4-8 mutant (G107S) by stabilizing its mRNA. Consistent with this hypothesis, the overexpression of the mutant cdc4 G107S partially suppressed the temperature-sensitive growth of the cdc4-8 mutants (data not shown).
We also demonstrated that Nrd1 exhibits genetic interactions with the components of type II myosin. First, Nrd1 overexpression suppressed the temperature-sensitive growth and cell separation defects of the cdc4-312 mutant as well as the cdc4-8 mutant. Second, Δnrd1 cells display synthetic growth defects when combined with cdc4-312, myosin II heavy chain myo2-E1, myosin regulatory light-chain rlc1Δ, and type II myosin heavy-chain Δmyo3 mutants, indicating that Δnrd1 cells are supersensitive to the mutations that affect myosin function. These genetic interactions among myosin mutants and nrd1-deletion cells, together with biochemical data, suggest that Nrd1 stabilization of Cdc4 mRNA is a bona fide mechanism for enhancing Cdc4 levels and that Nrd1 has a general function in cytokinesis. To the best of our knowledge, this is the first report on the involvement of an RNA-binding protein in the regulation of myosin function. Although previous reports have identified Nrd1 as a negative regulator of differentiation, no physiological target mRNA of Nrd1 has been identified. Here, by unraveling the functional interaction between Nrd1 and myosin mRNA, we have discovered a novel role of Nrd1 and a mechanism for the fine tuning of cytokinesis.
Relationship between Nrd1 and the Pmk1 MAPK Pathway
We have also demonstrated that Nrd1 is a novel target of Pmk1 MAPK and that the Pmk1 MAPK cell integrity signaling pathway negatively regulates the ability of Nrd1 to bind and stabilize Cdc4 mRNA. Consistently, Pmk1 deletion suppressed the phenotypes of the cdc4-8 mutants, including temperature sensitivity and cytokinesis defects, by stabilizing the Cdc4 mRNA. The finding that the Nrd1 deletion reverted the effects of Pmk1 deletion on the phenotypes of the cdc4-8 mutants further supports this model. Notably, Pmk1 deletion also rescued other myosin mutants, including cdc4-312 and Δrlc1 mutants. These data are in good agreement with the genetic interactions between Nrd1 deletion and myosin mutants and further strengthen the hypothesis that Pmk1 transmits signals via the phospho-regulation of Nrd1. It should be noted, however, that Pmk1 deletion rescued the growth defects under restrictive conditions but not the cell separation phenotypes of the myo2-E1 and Δmyo3 mutants (Supplementary Table S2,Supplementary Figure S2, E and F). This suggests that Pmk1 might influence cytokinesis through multiple targets other than Nrd1, such as targets involved in the regulation of cell wall integrity and/or stress responses.
The Pmk1 MAPK pathway plays a key role in cytokinesis, cell wall integrity, and ion homeostasis (Toda et al., 1996; Sugiura et al., 1999, 2003); however, no physiological Pmk1 substrate involved in cytokinesis has been identified. Nrd1, by serving as a MAPK target and as an RNA-binding factor of an essential myosin light chain, responds to extracellular signals mediated by MAPK signaling and fine tunes myosin expression by switching off its RNA-binding property. Furthermore, we demonstrated the cell cycle–dependent regulation of Pmk1/Nrd1/Cdc4 signaling. Pmk1 kinase activity is highest at the M phase and steadily declines through G2. In contrast Nrd1 phosphorylation is highest at G2, and Cdc4 mRNA abundance is the lowest at G2, thus showing the nice agreement of Nrd1 phosphorylation and Cdc4 mRNA abundance. It should be noted, however, that Pmk1 activity does not correlate with Nrd1 phosphorylation and Cdc4 mRNA abundance. One possible explanation for this discrepancy is the presence of a phosphatase that dephosphorylates Nrd1, thus counteracting Pmk1 kinase activity; the activity of this phosphatase is high around the M phase and declines through G2.
In higher eukaryotes, MAPKs have been shown to regulate several RNA-binding proteins including TTP (Mahtani et al., 2001), nucleolin (Yang et al., 2002), and hnRNP-K (Habelhah et al., 2001). Moreover, in our previous study, we identified Rnc1 a KH-type RNA-binding protein that acts as a negative regulator of Pmk1 signaling by binding the mRNA of Pmp1, the MAPK phosphatase for Pmk1 (Sugiura et al., 2003). Thus, RNA-binding proteins are important targets of MAPK signaling and play a crucial role in cell signaling by controlling posttranscriptional mRNA regulation. Further studies are, however, required to clarify how the MAPK phosphorylation of these RNA-binding proteins affects their RNA-binding activity.
Nrd1 as a Molecular Switch Regulating Cytokinesis and Differentiation
Our data suggest the possibility of the phosphorylation-dependent Nrd1 regulation of two physiological processes, namely, cytokinesis and differentiation. This implies that there exists a mechanism that switches Nrd1 function between cytokinesis and differentiation. Intriguingly, in glucose starvation conditions, under which meiosis is induced, Nrd1 is phosphorylated even in Pmk1-deletion cells (data not shown), indicating that other kinase(s) are responsible for the Nrd1 phosphorylation event during the phase of sexual differentiation. It is notable that the overexpression of human ROD1 (hROD1) and rat ROD1 (rROD1) failed to suppress the temperature sensitivity of the cdc4-8 mutants, whereas they rescued the pat1-114 mutants (data not shown). This finding suggests the possibility that mammalian ROD1 may act as a functional homologue of the nrd1+ gene with respect to sexual differentiation. Moreover, TIA-1 or TIAR, both of which have significant sequence similarity and share a common preferred RNA-binding sequence with Nrd1, might act as a mammalian homologue of Nrd1 in the process of cytokinesis. Future studies are required in order to unravel the regulatory mechanism that switches the Nrd1 function between cytokinesis and meiosis by isolating Nrd1 targets in both physiological processes.
In conclusion, this is the first study to demonstrate that myosin, a key player in cytokinesis, is regulated at the posttranscriptional level through MAPK signaling. Given the remarkable conservation of myosin and MAPK, a similar mechanism may regulate myosin and cytokinesis in other eukaryotes. Further functional and molecular characterization of Nrd1 function may help in gaining an understanding of how eukaryotic cells integrate signaling information to regulate the mitotic cycle and differentiation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. M. Yanagida (Kyoto University), T. Toda (Cancer Research, UK), K. Gould (Howard Hughes Medical Institute, Vanderbilt University School of Medicine), H. Okayama (University of Tokyo), M. K. Balasubramanian (National University of Singapore), D. McCollum (University of Massachusetts Medical School), P. G. Young (Queens University, Ontario, Canada), and Yeast Resource Centre (YGRC/NBRP; http://yeast.lab.nig.ac.jp/nig) for providing strains and plasmids; K. Irie and T. Inada for helpful discussions; and Susie O. Sio and Matthew Thornton for critical reading of the manuscript. We are grateful to the members of the Laboratory of Molecular Pharmacogenomics for their support. This work was supported by the Asahi Glass Foundation, the Uehara Memorial Foundation, and research grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (R.S.). This work was also supported in part by the “Academic Frontier” Project for Private Universities: matching fund subsidy from the Ministry of Education, Culture, Sports, Science and Technology, 2005–2007.
Abbreviations used:
- MAPK
mitogen-activated protein kinase
- EMM
Edinburgh minimal medium
- YPD
yeast extract-peptone-dextrose
- GFP
green fluorescent protein
- ORF
open reading frame
- GST
glutathione-S-transferase
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-09-0893) on March 11, 2009.
REFERENCES
- Balasubramanian M. K., Bi E., Glotzer M. Comparative analysis of cytokinesis in budding yeast, fission yeast and animal cells. Curr. Biol. 2004;14:R806–R818. doi: 10.1016/j.cub.2004.09.022. [DOI] [PubMed] [Google Scholar]
- Balasubramanian M. K., McCollum D., Chang L., Wong K. C., Naqvi N. I., He X., Sazer S., Gould K. L. Isolation and characterization of new fission yeast cytokinesis mutants. Genetics. 1998;149:1265–1275. doi: 10.1093/genetics/149.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla M., Forsburg S. L., Pollard T. D. Identification of a second myosin-II in Schizosaccharomyces pombe: Myp2p is conditionally required for cytokinesis. Mol. Biol. Cell. 1997;8:2693–2705. doi: 10.1091/mbc.8.12.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzeska H., Korn E. D. Regulation of class I and class II myosins by heavy chain phosphorylation. J. Biol. Chem. 1996;271:16983–16986. doi: 10.1074/jbc.271.29.16983. [DOI] [PubMed] [Google Scholar]
- Chang F., Woollard A., Nurse P. Isolation and characterization of fission yeast mutants defective in the assembly and placement of the contractile actin ring. J. Cell Sci. 1996;109:131–142. doi: 10.1242/jcs.109.1.131. [DOI] [PubMed] [Google Scholar]
- D'souza V. M., Naqvi N. I., Wang H., Balasubramanian M. K. Interactions of Cdc4p, a myosin light chain, with IQ-domain containing proteins in Schizosaccharomyces pombe. Cell Struct. Funct. 2001;26:555–565. doi: 10.1247/csf.26.555. [DOI] [PubMed] [Google Scholar]
- Degols G., Shiozaki K., Russell P. Activation and regulation of the Spc1 stress-activated protein kinase in Schizosaccharomyces pombe. Mol. Cell. Biol. 1996;16:2870–2877. doi: 10.1128/mcb.16.6.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dember L. M., Kim N. D., Liu K. Q., Anderson P. Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J. Biol. Chem. 1996;271:2783–2788. doi: 10.1074/jbc.271.5.2783. [DOI] [PubMed] [Google Scholar]
- Eng K., Naqvi N. I., Wong K. C., Balasubramanian M. K. Rng2p, a protein required for cytokinesis in fission yeast, is a component of the actomyosin ring and the spindle pole body. Curr. Biol. 1998;8:611–621. doi: 10.1016/s0960-9822(98)70248-9. [DOI] [PubMed] [Google Scholar]
- Forch P., Puig O., Kedersha N., Martinez C., Granneman S., Seraphin B., Anderson P., Valcarcel J. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol. Cell. 2000;6:1089–1098. doi: 10.1016/s1097-2765(00)00107-6. [DOI] [PubMed] [Google Scholar]
- Gatto-Konczak F., Bourgeois C. F., Le, Guiner C., Kister L., Gesnel M. C., Stevenin J., Breathnach R. The RNA-binding protein TIA-1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5' splice site. Mol. Cell. Biol. 2000;20:6287–6299. doi: 10.1128/mcb.20.17.6287-6299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M. The molecular requirements for cytokinesis. Science. 2005;307:1735–1739. doi: 10.1126/science.1096896. [DOI] [PubMed] [Google Scholar]
- Gotoh Y., Nishida E., Shimanuki M., Toda T., Imai Y., Yamamoto M. Schizosaccharomyces pombe Spk1 is a tyrosine-phosphorylated protein functionally related to Xenopus mitogen-activated protein kinase. Mol. Cell. Biol. 1993;13:6427–6434. doi: 10.1128/mcb.13.10.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin D. A., Trautmann S., McCollum D. Cytokinesis in eukaryotes. Microbiol. Mol. Biol. Rev. 2002;66:155–178. doi: 10.1128/MMBR.66.2.155-178.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habelhah H., Shah K., Huang L., Ostareck-Lederer A., Burlingame A. L., Shokat K. M., Hentze M. W., Ronai Z. ERK phosphorylation drives cytoplasmic accumulation of hnRNP-K and inhibition of mRNA translation. Nat. Cell Biol. 2001;3:325–330. doi: 10.1038/35060131. [DOI] [PubMed] [Google Scholar]
- Hirayama S., Sugiura R., Lu Y., Maeda T., Kawagishi K., Yokoyama M., Tohda H., Hama Y. G., Shuntoh H., Kuno T. Zinc finger protein Prz1 regulates Ca2+ but not Cl− homeostasis in fission yeast: Identification of distinct branches of calcineurin signaling pathway in fission yeast. J. Biol. Chem. 2003;278:18078–18084. doi: 10.1074/jbc.M212900200. [DOI] [PubMed] [Google Scholar]
- Hou M. C., McCollum D. Cytokinesis: myosin spots the ring. Curr. Biol. 2002;12:R334–RR336. doi: 10.1016/s0960-9822(02)00834-5. [DOI] [PubMed] [Google Scholar]
- Irie K., Tadauchi T., Takizawa P. A., Vale R. D., Matsumoto K., Herskowitz I. The Khd1 protein, which has three KH RNA-binding motifs, is required for proper localization of ASH1 mRNA in yeast. EMBO J. 2002;21:1158–1167. doi: 10.1093/emboj/21.5.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H. T., Oowatari Y., Abe M., Tanaka K., Matsuda H., Kawamukai M. Interaction between a negative regulator (Msa2/Nrd1) and a positive regulator (Cpc2) of sexual differentiation in Schizosaccharomyces pombe. Biosci. Biotechnol. Biochem. 2004;68:1621–1626. doi: 10.1271/bbb.68.1621. [DOI] [PubMed] [Google Scholar]
- Kamm K. E., Stull J. T. Dedicated myosin light chain kinases with diverse cellular functions. J. Biol. Chem. 2001;276:4527–4530. doi: 10.1074/jbc.R000028200. [DOI] [PubMed] [Google Scholar]
- Kedersha N. L., Gupta M, Li W., Miller I., Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitayama C., Sugimoto A., Yamamoto M. Type II myosin heavy chain encoded by the myo2 gene composes the contractile ring during cytokinesis in Schizosaccharomyces pombe. J. Cell Biol. 1997;137:1309–1319. doi: 10.1083/jcb.137.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohrer K., Domdey H. Preparation of high molecular weight RNA. Methods Enzymol. 1991;194:398–405. doi: 10.1016/0076-6879(91)94030-g. [DOI] [PubMed] [Google Scholar]
- Le G. X., Motegi F., Salimova E., Mabuchi I., Simanis V. The S.pombe rlc1 gene encodes a putative myosin regulatory light chain that binds the type II myosins myo3p and myo2p. J. Cell Sci. 2000;113:4157–4163. doi: 10.1242/jcs.113.23.4157. [DOI] [PubMed] [Google Scholar]
- Loo T. H., Balasubramanian M. Schizosaccharomyces pombe Pak-related protein, Pak1p/Orb2p, phosphorylates myosin regulatory light chain to inhibit cytokinesis. J. Cell Biol. 2008;183:785–793. doi: 10.1083/jcb.200806127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid M., Núñez A., Soto T., Vicente S. J., Gacto M., Cansado J. Stress-activated protein kinase-mediated down-regulation of the cell integrity pathway mitogen-activated protein kinase Pmk1p by protein phosphatases. Mol. Biol. Cell. 2007;18:4405–4419. doi: 10.1091/mbc.E07-05-0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahtani K. R., Brook M., Dean J. L., Sully G., Saklatvala J., Clark A. R. Mitogen-activated protein kinase p38 controls the expression and posttranslational modification of tristetraprolin, a regulator of tumor necrosis factor alpha mRNA stability. Mol. Cell. Biol. 2001;21:6461–6469. doi: 10.1128/MCB.21.9.6461-6469.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maundrell K. nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J. Biol. Chem. 1990;265:10857–10864. [PubMed] [Google Scholar]
- May K. M., Watts F. Z., Jones N., Hyams J .S. Type II myosin involved in cytokinesis in the fission yeast, Schizosaccharomyces pombe. Cell Motil. Cytoskelet. 1997;38:385–396. doi: 10.1002/(SICI)1097-0169(1997)38:4<385::AID-CM8>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- McCollum D., Balasubramanian M. K., Pelcher L. E., Hemmingsen S. M., Gould K. L. Schizosaccharomyces pombe cdc4+ gene encodes a novel EF-hand protein essential for cytokinesis. J. Cell Biol. 1995;130:651–660. doi: 10.1083/jcb.130.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCollum D., Feoktistova A., Gould K. L. Phosphorylation of the myosin-II light chain does not regulate the timing of cytokinesis in fission yeast. J. Biol. Chem. 1999;274:17691–17695. doi: 10.1074/jbc.274.25.17691. [DOI] [PubMed] [Google Scholar]
- Millar J. B., Buck V., Wilkinson M. G. Pyp1 and Pyp2 PTPases dephosphorylate an osmosensing MAP kinase controlling cell size at division in fission yeast. Genes Dev. 1995;9:2117–2130. doi: 10.1101/gad.9.17.2117. [DOI] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Motegi F., Nakano K., Mabuchi I. Molecular mechanism of myosin-II assembly at the division site in Schizosaccharomyces pombe. J. Cell Sci. 2000;113:1813–1825. doi: 10.1242/jcs.113.10.1813. [DOI] [PubMed] [Google Scholar]
- Mucha P., Szyk A., Rekowski P., Guenther R., Agris P. F. Interaction of RNA with phage display selected peptides analyzed by capillary electrophoresis mobility shift assay. RNA. 2002;8:698–704. doi: 10.1017/s1355838202020319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K., Nurse P. Cell division cycle mutants altered in DNA replication and mitosis in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 1981;182:119–124. doi: 10.1007/BF00422777. [DOI] [PubMed] [Google Scholar]
- Naqvi N. I., Eng K., Gould K. L., Balasubramanian M. K. Evidence for F-actin-dependent and -independent mechanisms involved in assembly and stability of the medial actomyosin ring in fission yeast. EMBO J. 1999;18:854–862. doi: 10.1093/emboj/18.4.854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi N. I., Wong K. C., Tang X., Balasubramanian M. K. Type II myosin regulatory light chain relieves auto-inhibition of myosin-heavy-chain function. Nat. Cell Biol. 2000;2:855–858. doi: 10.1038/35041107. [DOI] [PubMed] [Google Scholar]
- Peng X., et al. Identification of cell cycle-regulated genes in fission yeast. Mol. Biol. Cell. 2005;16:1026–1042. doi: 10.1091/mbc.E04-04-0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Rustici G., Mata J., Kivinen K., Lio P., Penkett C. J., Burns G., Hayles J., Brazma A., Nurse P., Bahler J. Periodic gene expression program of the fission yeast cell cycle. Nat. Genet. 2004;36:809–817. doi: 10.1038/ng1377. [DOI] [PubMed] [Google Scholar]
- Satterwhite L. L., Lohka M. J., Wilson K. L., Scherson T. Y., Cisek L. J., Corden J. L., Pollard T. D. Phosphorylation of myosin-II regulatory light chain by cyclin-p34cdc2, a mechanism for the timing of cytokinesis. J. Cell Biol. 1992;118:595–605. doi: 10.1083/jcb.118.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterwhite L. L., Pollard T. D. Cytokinesis. Curr. Opin. Cell Biol. 1992;4:43–52. doi: 10.1016/0955-0674(92)90057-j. [DOI] [PubMed] [Google Scholar]
- Sugiura R., Kita A., Shimizu Y., Shuntoh H., Sio S. O., Kuno T. Feedback regulation of MAPK signalling by an RNA-binding protein. Nature. 2003;424:961–965. doi: 10.1038/nature01907. [DOI] [PubMed] [Google Scholar]
- Sugiura R., Toda T., Dhut S., Shuntoh H., Kuno T. The MAPK kinase Pek1 acts as a phosphorylation-dependent molecular switch. Nature. 1999;399:479–483. doi: 10.1038/20951. [DOI] [PubMed] [Google Scholar]
- Sugiura R., Toda T., Shuntoh H., Yanagida M., Kuno T. pmp1+, a suppressor of calcineurin deficiency, encodes a novel MAP kinase phosphatase in fission yeast. EMBO J. 1998;17:140–148. doi: 10.1093/emboj/17.1.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taga A., Yamamoto Y., Maruyama R., Honda S. Search for serum protein-binding disaccharides and disaccharide-binding serum proteins by affinity capillary electrophoresis. Electrophoresis. 2004;25:876–881. doi: 10.1002/elps.200305765. [DOI] [PubMed] [Google Scholar]
- Takada H., et al. Atf1 is a target of the mitogen-activated protein kinase Pmk1 and regulates cell integrity in fission yeast. Mol. Biol. Cell. 2007;18:4794–4802. doi: 10.1091/mbc.E07-03-0282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T., Dhut S., Superti F. G., Gotoh Y., Nishida E., Sugiura R., Kuno T. The fission yeast pmk1+ gene encodes a novel mitogen-activated protein kinase homolog which regulates cell integrity and functions coordinately with the protein kinase C pathway. Mol. Cell. Biol. 1996;16:6752–6764. doi: 10.1128/mcb.16.12.6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukahara K., Yamamoto H., Okayama H. An RNA binding protein negatively controlling differentiation in fission yeast. Mol. Cell. Biol. 1998;18:4488–4498. doi: 10.1128/mcb.18.8.4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe B. A., Gould K. L. Split decisions: coordinating cytokinesis in yeast. Trends Cell Biol. 2005;15:10–18. doi: 10.1016/j.tcb.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Wong K. C., Naqvi N. I., Iino Y., Yamamoto M., Balasubramanian M. K. Fission yeast Rng3p: an UCS-domain protein that mediates myosin II assembly during cytokinesis. J. Cell Sci. 2000;113:2421–2432. doi: 10.1242/jcs.113.13.2421. [DOI] [PubMed] [Google Scholar]
- Yamamoto H., Tsukahara K., Kanaoka Y., Jinno S., Okayama H. Isolation of a mammalian homologue of a fission yeast differentiation regulator. Mol. Cell. Biol. 1999;19:3829–3841. doi: 10.1128/mcb.19.5.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Maiguel D. A., Carrier F. Identification of nucleolin and nucleophosmin as genotoxic stress-responsive RNA-binding proteins. Nucleic Acids Res. 2002;30:2251–2260. doi: 10.1093/nar/30.10.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.