Abstract
In both experimental animals and humans, aflatoxin B1 (AFB1) is a potent hepatic toxin and carcinogen against which a variety of antioxidants and experimental or therapeutic drugs (e.g., oltipraz, related dithiolethiones, and various triterpenoids) protect from both acute toxicity and carcinogenesis. These agents induce several hepatic glutathione S-transferases (GST) as well as aldo-keto reductases (AKR) which are thought to contribute to protection. Studies were undertaken in transgenic rats to examine the role of one inducible enzyme, AKR7A1, for protection against acute and chronic actions of AFB1 by enhancing detoxication of a reactive metabolite, AFB1 dialdehyde, by reduction to alcohols. The AFB1 dialdehyde forms adducts with protein amino groups by a Schiff base mechanism and these adducts have been theorized to be at least one cause of the acute toxicity of AFB1 and to enhance carcinogenesis. A liver-specific AKR7A1 transgenic rat was constructed in the Sprague-Dawley strain and two lines, AKR7A1Tg2 and AKR7A1Tg5, were found to overexpress AKR7A1 by 18- and 8-fold, respectively. Rates of formation of AFB1 alcohols, both in hepatic cytosols and as urinary excretion products, increased in the transgenic lines with AKR7A1Tg2 being the highest. Neither line offered protection against acute AFB1-induced bile duct proliferation, a functional assessment of acute hepatotoxicity by AFB1, nor did they protect against the formation of GST-P positive putative preneoplastic foci as a result of chronic exposure to AFB1. These results imply that the prevention of protein adducts mediated by AKR are not critical to protection against AFB1 tumorigenicity.
Keywords: aflatoxin; aldo-keto reductase; chemoprevention, hepatic carcinogenesis; hepatotoxicity; transgenic rat
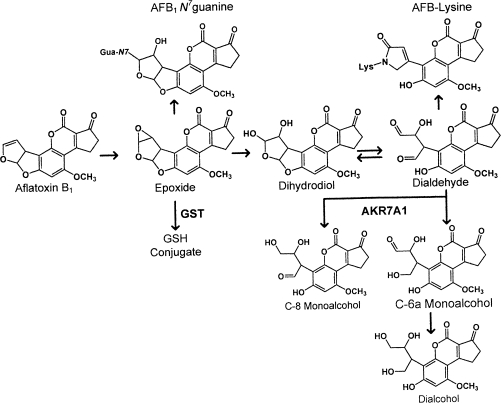
Aflatoxin B1 (AFB1), a secondary metabolic product of the mold Aspergillus flavus, is a potent hepatotoxin and carcinogen in both experimental animals and humans (Kensler et al., 1999). In the liver, the metabolic activation of AFB1 is mediated by microsomal cytochromes P450, CYP1A2, and CYP3A4, to form the AFB1-8,9-exo and -8,9-endo epoxides (see Fig. 1) (Gallagher et al., 1996; Johnson and Guengerich, 1997). Only the AFB1-8,9-exo isomer reacts with DNA, forming the N7-guanine adduct and the derivative AFB1 formamidopyrimidine adducts (Johnson and Guengerich, 1997). If not repaired, these DNA adducts or the abasic sites that remain from the spontaneous depurination of the N7-guanine adduct can lead to DNA mutations. Both of the AFB1 epoxides are substrates for glutathione S-transferases (GSTs), which catalyze the conjugation of the epoxide with reduced glutathione, thus mitigating the formation of DNA adducts (Eaton and Bammler, 1999). In addition to GSTs, other detoxication pathways may contribute to resistance to this mycotoxin. Both AFB1 epoxides undergo rapid hydrolysis in water forming AFB1 dihydrodiol. The AFB1 dihydrodiol undergoes a base-catalyzed rearrangement to, and is in equilibrium with, AFB1 dialdehyde (Johnson et al., 1996). AFB1 dialdehyde can form Schiff base derived adducts with protein amino groups, particularly lysine (Groopman et al., 1980; Guengerich et al., 2002; Sabbioni et al., 1987) and these protein adducts have been implicated in the acute toxicity of AFB1 (Ellis et al., 1993; Guengerich et al., 2002).
FIG. 1.
Schematic of enzymatic and chemical conversion of AFB1 to the reactive dialdehyde and its reduction by AKR7A1 to three possible aflatoxin alcohols.
Aflatoxin aldehyde reductase was first identified as an ethoxyquin-inducible member of the nicotinamide adenine dinucleotide phosphate (reduced) (NADPH)–dependent aldo-keto reductase (AKR) superfamily. This enzyme, now designated AKR7A1, was purified from rat liver and shown to catalyze the reduction of the protein reactive AFB1 dialdehyde to the dialcohol product (Ellis et al., 1993; Hayes et al., 1993). Large increases in the levels of AKR7A1 protein have been observed in livers of rats following administration of ethoxyquin, dithiolethiones and other classes of inducers (Guengerich et al., 2001; Ireland et al., 1998; Knight et al., 1999). Aflatoxin aldehyde reductases, specifically rat AKR7A1 (Ellis et al., 1993; Guengerich et al., 2001; Knight et al., 1999), human AKR7A3 (Guengerich et al., 2001; Ireland et al., 1998; Knight et al., 1999), and mouse AKR7A5 (Hinshelwood et al., 2003), are known to catalyze the reduction of the reactive AFB1-derived dialdehyde at similarly high rates of activity. In a study of rat AKR7A1 and human AKR7A3 enzyme kinetics, it was shown that the AFB1 dialdehyde was preferentially reduced to a C-8 monoalcohol, corresponding to reduction of the C-8 carbon. Production of a C-6a monoalcohol occurred at a lower rate, and the formation of the dialcohol was not rapid in these reactions (Guengerich et al., 2001).
Recently, we reported on the protection against the formation of AFB1 dialdehyde protein adducts and cytotoxicity in cell culture by recombinant expression of cloned rat AKR7A1 and human AKR7A3 (Bodreddigari et al., 2008). Because these results indicated a role of AKR in the protection against AFB1-induced cytotoxicity in vitro and because rat AKR7A1 is highly induced by compounds such as oltipraz that are known protectors against the toxicity and carcinogenicity of AFB1 in the rat (Liu et al., 1988), a better understanding of this pathway in vivo is warranted. Induction of AKR7A1 may limit AFB1-induced cytotoxicity in vivo by inhibiting the compensatory hyperplasia that occurs following cellular necrosis, thereby, attenuating several steps in AFB1 carcinogenesis including events involved in initiation, promotion and progression (Roebuck, 2004). Based on these observations, we report here the development of a liver-specific AKR7A1 transgenic rat and studies to reveal the impact of AKR7A1 in vivo in hepatic AFB1 metabolism, toxicity, and carcinogenicity.
MATERIALS AND METHODS
AKR7A1 transgenic rat.
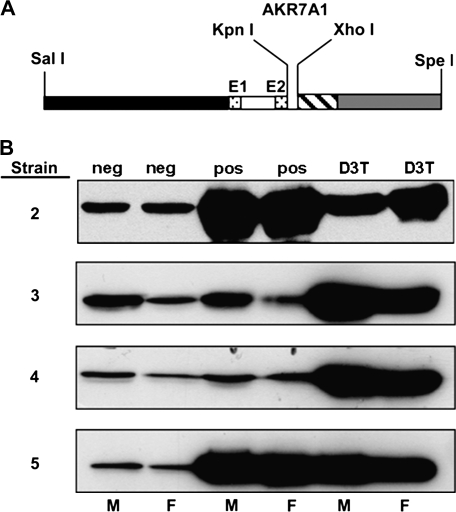
The DNA construct for expression of the rat AKR7A1 gene was produced using the cDNA clone of rat AKR7A1 (Knight et al., 1999) and the liver-specific expression vector pLiv-7, kindly provided by Dr John Taylor, UCSF (Fan et al., 1994; Yamanaka et al., 1995). This vector was designed for liver-specific expression and contains sequences from the human APOE gene (Fan et al., 1994). A schematic of the construct is shown in Figure 2A. The SpeI site in the AKR7A1 polylinker was opened by restriction endonuclease digestion. The linearized plasmid was filled in with T4 DNA polymerase and the plasmid was closed by blunt-end ligation. The AKR7A1 KpnI–XhoI containing insert, 5′ AUG sequence towards the KpnI site, was then purified from the resulting plasmid and ligated into the pLiv-7 vector that had been digested with KpnI and XhoI. The resulting plasmid was purified and the orientation of the AKR7A1 insert verified by DNA sequence analysis using rat AKR7A1 specific primers, which were used to sequence into the pLiv-7 vector. The primer, 5′-ACGCGGAGCTGGAGGTCA-3′, annealed to the 5′ end of AKR7A1 and was used to sequence into pLiv-7. To confirm the orientation, a second primer, 5′-GTGGATGCCTTTGACCAAG-3′, which anneals to the 3′ end of AKR7A1, was used to sequence into pLiv-7. The SalI–SpeI DNA fragment (pLivAKR7A1 transgene construct) was isolated by gel electrophoresis and further purified using a Qiaquick column (Qiagen Inc., Valencia, CA). Transgene specific PCR primers were synthesized: one primer to the AKR7A1 cDNA sequence, 5′-AATTTGAACCCCGGGAGAGGAAGA-3′, the other corresponding to the 5′ APOE sequence, 5′-ACGCGGAGCTGGAGGTCACATC-3′. These primers produced a single PCR product corresponding to the transgene when the transgene was spiked at varying concentration into rat genomic DNA at ratios ranging from 1 to 10 gene copies per cell equivalent of DNA. The concentration of the pLivAKR7A1 DNA fragment was confirmed by electrophoresis versus plasmid DNA of a standard concentration and sent frozen to Xenogen Biosciences (Cranbury, NJ).
FIG. 2.
Transgene expression of AKR7A1. (A) The DNA construct was designed for liver-specific expression using the pLiv-7 vector containing sequences from the human APOE gene: 3 kb of 5′-flanking region (black); the first exon (E1, dotted), first intron (open), and first six nucleotides of the second exon (E2, dotted); 0.25 kb of 3′-flanking region including the polyadenylation signal (dashed) and a 1.7-kb hepatic control region of the APOE/C-I gene locus (gray). The AKR7A1 cDNA was inserted into the polylinker region using the KpnI and XhoI restriction sites. (B) Transgene expression of AKR7A1. Protein levels of AKR7A1 in samples of liver cytosol (30 μg protein/lane) from one male (M) and one female (F) of four separate AKR7A1 transgenic rat lines were analyzed by immunoblot. The samples, identified at the top, are nontransgenic genetic control animals (neg), transgenic animals (pos), and nontransgenic rats pretreated with 3H-1,2-dithiole-3-thione (D3T) a known inducer of AKR7A1.
Xenogen Biosciences produced four founder rats containing the pLivAKR7A1 transgene. Briefly, zygotes from superovulated Sprague-Dawley rats were flushed from the oviduct and the transgene delivered by pronuclear microinjection. The injected zygotes were implanted into pseudopregnant foster mothers and the resulting founder pups were genotyped by PCR using the primers described above. Four transgenic rats (two males and two females) were produced and shipped to Dartmouth Medical School. These rats were bred to provide both transgenic positive and negative (genetic control) rats for subsequent experiments. Breeders were fed a commercial, natural ingredient diet. All rats used in experimental studies were fed the AIN-76A purified diet without the antioxidant ethoxyquin which is usually added to this diet. The latter diet was fed to maintain the hepatic drug metabolizing enzymes at relatively stable levels since the aflatoxins are activated and detoxified by a series of these enzymes. In all experiments, transgenic rats were matched with a genetic control, litter mate of the same sex. All experiments were approved by the Animal Care and Use Committee of Dartmouth College.
Estimation of integrated gene copy number and genotyping.
Genomic DNA was isolated from rat livers using DNeasy tissue kit (Qiagen, Inc., Valencia, CA). PCR was performed using the Bio-Rad iCycler (Hercules, CA) and a forward primer to the AKR7A1 (FP)-5′-AATTTGAACCCCGGGAGAGGAAGA-3′ sequence and a reverse primer (RP)-5′-ACGCGGAGCTGGAGGTCACATC-3′ to the APOE sequence (IDT, Coralville, IA). The PCR mix consisted of 2× iQ SYBR Green supermix (2× reaction buffer, iTaq DNA polymerase, dNTPs, 6mM MgCl2, SYBR Green I, and stabilizers) (Bio-Rad, Hercules, CA), 0.2μM FP, 0.2μM RP, and 300 ng of genomic DNA in a final volume of 50 μl. Amplification was carried out as follows: denaturation for 3 min at 95°C, 40 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s, with a final extension at 72°C for 2 min. The gene copy number was calculated using 300 ng of genomic DNA and a standard curve containing 1, 3, 10, and 30 copies of cDNA (AKR7A1) and 300 ng of genomic DNA from a nontransgenic rat. The genotype for transgenic positive and negative rats was determined using the primers listed above.
Preparation of hepatic cytosol, immunoblot, and enzyme assay.
Liver cytosolic proteins were prepared from untreated, D3T-treated (an oral gavage dose of 0.3 mmol/kg body weight on 5, 3, and 1 day prior to autopsy), or transgenic animals and analyzed by immunoblot as described previously (Egner et al., 2006; Knight et al., 1999). 9,10-Phenanthrene quinone reductase activity was determined by monitoring the oxidation of NADPH at 340 nm (ϵ = 6270/M/cm), as described previously (Knight et al., 1999) in a 1 ml of reaction containing 25 μg of cytosolic protein and 30μM 9,10-phenanthrene quinone. When using AFB1 dialdehyde as substrate, incubation mixtures consisted of 15 μl of hepatic cytosol, 50mM potassium N-(2-hydroxyethyl) piperazine-N-(2-ethanesulfonate) buffer pH 7.4, 5mM MgCl2, 0.2mM NADPH, and 7μM AFB1 dialdehyde in a total volume of 100 μl. Mixtures were incubated at 37°C, initiated by the addition of cytosol, and terminated after 3 min by addition of 10 μl of concentrated acetic acid. AFB1 dialdehyde was formed by the dilution of AFB1 diol into CHES buffer, pH 10. The postincubation mixtures were clarified by centrifugation (10,000 × g, 3 min) and 10 μl aliquots of the supernatants were diluted with 15 μl of high-performance liquid chromatography (HPLC) initial mobile phase prior to isotope dilution mass spectrometry analysis. Specific activity results are reported as pmol of AFB1 alcohol metabolites formed/min/mg cytosolic protein and represent the mean ± SE from three independent experiments. Hepatic GST specific activities were determined spectrophotometrically by monitoring metabolism of the 1-chloro-2,4-dinitrobenzene substrate at 340 nm over 2 min as previously described (Habig et al., 1974). Cytosolic protein concentrations were determined by the Bio-Rad assay (Bio-Rad Corporation, Hercules, CA).
Aflatoxin metabolism.
Urine was collected on dry ice over a 24-h period from male rats of transgenic positive and negative litter mates of approximately 5 weeks of age and 125 g body weight. This age and weight corresponded to those of rats in the carcinogenicity experiments (see below) when they received the first dose of AFB1. AFB1 was obtained from Sigma-Aldrich (St Louis, MO). Rats were gavaged with a single dose of AFB1 (25 μg/rat) and housed singly in glass metabolic cages with access to water and diet ad libitum. The 24-h urine samples were stored at -80°C prior to analysis. Urine samples from animals were thawed and a 5-ml aliquot adjusted to pH 3.5 with acetic acid prior to centrifugation at 500 × g for 10 min. Samples were then spiked with 2 ng of 13C17-AFB1 dialcohol internal standard and extracted using 6-cm3 Waters MCX Oasis SPE columns (Waters Corp., Milford, MA) equilibrated with 1 column volume of methanol followed by 1 column volume of water prior to loading the urine sample. AFB1 alcohols were eluted from the SPE columns with 10 ml of 100% methanol and reduced to a final volume of approximately 50 μl using high-purity nitrogen. Samples were then diluted to approximately 600 μl with water and loaded onto an aflatoxin-specific preparative monoclonal antibody immunoaffinity column, as previously described (Egner et al., 2006). The immunoaffinity column consisted of a 2:1 mix of aflatoxin-lysine and aflatoxin-2B11 antibodies. The affinity column was washed with phosphate-buffered saline and water to remove nonspecifically bound materials and the AFB1 alcohols were eluted from the immunoaffinity column with 6 ml of 70% dimethylsulfoxide/water (vol/vol) followed by another two volumes of water. The dimethylsulfoxide and water fractions were combined, diluted with water and then applied to a 3-cm3 Varian Bond-Elut LRC C18 SPE column (Varian, Inc., Walnut Creek, CA). AFB1 alcohols were eluted from the SPE with 5 ml of a 50/50 mixture of 1% acetic acid/methanol followed by 5 ml of 100% methanol and concentrated to approximately 40 μl under a nitrogen stream: 1 μl of final urinary extract was diluted with 24 μl of HPLC initial mobile phase prior to analysis. The AFB1 dialdehyde metabolites (C-6a monoalcohol, C-8 monoalcohol, and dialcohol) were then quantified by isotope dilution tandem mass spectrometry as recently described (Johnson et al., 2008). Urinary creatinine levels were determined by Eagle Diagnostics Creatinine Direct Reagent Sets (De Soto, TX). Subsequent to urine collection, the rats were autopsied and liver samples were snap frozen. DNA was isolated and analyzed for AFB1 N7-guanine adducts by isotope dilution mass spectrometry as previously described (Egner et al., 2006). Plasma samples were processed and analyzed for AFB1 lysine adducts by isotope dilution mass spectrometry as previously described (McCoy et al., 2005).
Acute aflatoxin toxicity.
Hepatotoxicity was assessed by evaluating the classic phenomenon of bile duct proliferation in response to acute toxic doses of AFB1 (Maxuitenko et al., 1994; Newberne, 1973; Roebuck and Maxuitenko, 1996). The quantification of AFB1-induced bile duct proliferation has been described and used as a screening method for selecting chemoprotective dithiolethiones (Maxuitenko et al., 1996). At 7–8 weeks of age, rats received two doses of AFB1 (500 μg/kg body weight, i.p.) on two successive days, followed on day three with two doses (100 mg/kg body weight, i.p.) of 5-bromo-2′deoxyuridine (BrdU) given 5 and 2 h prior to autopsy. Livers were fixed in formalin and processed by routine histological methods. Hepatic tissue sections (5 μm thick) were stained immunohistochemically for BrdU (BrdU In-Situ Detection Kit, BD Biosciences, San Jose, CA). Small bile ducts of less than 15 cells were evaluated by light microscopy to determine a BrdU labeling index. Approximately 500 bile duct cells were counted per rat liver and a bile duct labeling index was calculated.
Hepatic carcinogenicity.
Historical data predicted (Busby and Wogan, 1984) and preliminary data (not shown) confirmed that transgenic rats derived from of the Sprague-Dawley rat strain were less sensitive to AFB1 than the F344 rats for which extensive data exists regarding the induction of putative preneoplastic foci (Busby and Wogan, 1984; Roebuck and Maxuitenko, 1994; Roebuck et al., 2003; Yates et al., 2006). Therefore to have adequate numbers of putative preneoplastic foci (henceforth, termed foci) in livers for quantitative evaluation, the total dose of AFB1 was doubled by extending treatment duration from two to four weeks and also permitting the foci to grow for two weeks longer than with F344 rats used in recent experiments (Roebuck et al., 2003; Yates et al., 2006). Throughout the experiments, both food and water were available ad libitum with only food being withdrawn for 12 h prior to autopsy to reduce the glycogen accumulation in the livers. At approximately 5 weeks of age and about 125 g body weight, AFB1 treatment began. AFB1 (25 μg/rat) was gavaged 5 days per week for 4 successive weeks and rats were autopsied 6 weeks later. The cumulative dose of AFB1 was 500 μg AFB1 per rat and the duration of the entire protocol was 10 weeks. From the left lateral hepatic lobe, multiple 2-mm-thick sections were cut by hand, fixed in acetone at 4°C, and embedded in paraffin. Hepatic sections (5 μm thick) were stained by standard immunohistochemical methods for expression of GST-placental isoform (GST-P) foci and the foci were identified and analyzed by light microscopy. As with previous analyses (Roebuck et al., 2003; Yates et al., 2006), the observed focal data of number of foci per unit tissue area examined and their focal transactional areas were first subjected to morphometric transformation resulting in the volume percent of liver occupied by GST-P positive foci, a parameter analogous to tumor burden. Details of this procedure have been published previously (Kensler et al., 1992; Maxuitenko et al., 1996; Yates et al., 2006).
Statistical analyses.
Statistical analyses of the in vivo metabolism data were performed with Sigma-Plot software (SYSTAT Software, Inc., San Jose, CA) using one-way analysis of variance (ANOVA). The Holm-Sidak post hoc method was used for multiple pairwise comparisons. Focal data (volume %) and bile duct cell labeling indices were statistically analyzed using a two-way ANOVA (Stata Corp., College Station, TX).
RESULTS
Characterization of Transgenic Rat Lines
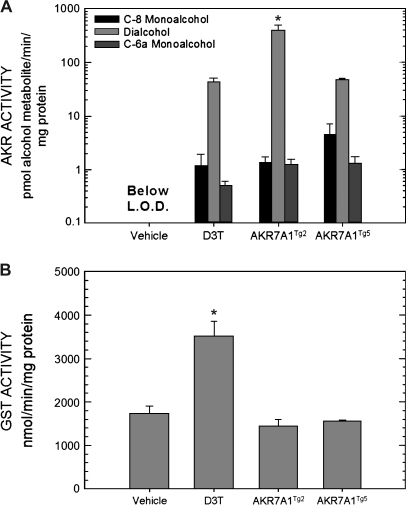
Four transgenic founder rats were produced using the APOE-AKR7A1 DNA construct to drive expression of AKR7A1 in the liver (Fig. 2A). Protein levels of AKR7A1 in hepatic cytosols prepared from the progeny of each founder are shown in Figure 2B. These immunoblots clearly show that lines AKR7A1Tg3 and AKR 7A1Tg4 have less immunoreactive protein than lines AKR7A1Tg2 and AKR7A1Tg5. The latter two transgenic lines exhibited similar or greater intensity for AKR protein than did the cytosols from the D3T-pretreated nontransgenic rats and were 18-fold and 8-fold greater than nontransgenic genetic control rats. Expression of AKR7A1 was similar between male and female rats for all transgenic lines. Measurement of 9,10-phenanthrene quinone reductase activity, a substrate for AKR7A1 (Knight et al., 1999), was used for initial functional characterization of the four transgenic lines. This analysis indicated that the AKR7A1Tg2 and AKR7A1Tg5 lines exhibited elevated activities compared to nontransgenic rats; whereas, the AKR7A1Tg3 and AKR 7A1Tg4 lines did not (data not shown). No discernable differences between male and female rats were seen in either the transgenic or nontransgenic lines with this assay. The copy number of AKR7A1 per genome was determined to be 25 for AKR7A1Tg2 and 8 for AKR7A1Tg5. More rigorous characterization of the catalytic capacity of the hepatic enzymes from the four rat lines was determined using AFB1 dialdehyde as substrate. Shown in Figure 3A, hepatic cytosols from lines AKR7A1Tg2 and AKR7A1Tg5, as well as from D3T-treated animals, produced C-6a monoalcohol, C-8 monoalcohol, and dialcohol. However, dialcohol represented between 90 and 95% of the product formed in all cases. None of the dialdehyde-derived alcohol metabolites were detected in hepatic cytosols from vehicle-treated, nontransgenic rats under the incubation conditions employed. Summing all alcohol metabolites, the specific activities (pmol alcohol metabolite/min/mg protein) were 418 ± 100, 49 ± 3, 42 ± 9 (mean ± SE, N = 3) for AKR7A1Tg2, AKR7A1Tg5 and D3T-treated nontransgenic animals, respectively. As a point of comparison, Figure 3B indicates that GST activity was not affected by transgene expression in either line, but was elevated twofold by D3T treatment, as has been observed previously (Roebuck et al., 2003). Collectively, the estimates of copy number, hepatic protein expression by Western blot, and assessment of specific activity indicate that AKR7A1 expression in line AKR7A1Tg5 is comparable to that achievable with a pharmacologic intervention (i.e., treatment with D3T), while that in line AKR7A1Tg2 is several fold higher.
FIG. 3.
(A) Formation of the C-6a monoalcohol, C-8 monoalcohol and dialcohol following incubation of hepatic cytosols with aflatoxin dialdehyde. Incubations were conducted for 3 min with AFB1 dialdehyde susbtrate (7μM). All three AFB1 alcohols could be detected in nontransgene hepatic cytosols under nonlinear incubation conditions (10μM substrate, 10 min). Metabolites were quantified by isotope dilution mass spectrometry. *p < 0.05, AKR7A1Tg2 compared to AKR7A1Tg5 by ANOVA. (B) GST activities in hepatic cytosols using chlorodinitrobenzene as substrate. *p < 0.05, D3T compared to vehicle treated by ANOVA. Values are mean ± SE (N = 3).
In Vivo Metabolism of AFB1 Dialdehyde
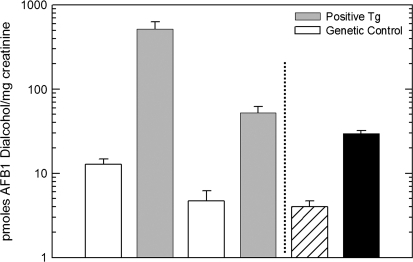
Of the three alcohols that could be formed from the AFB1 dialdehyde intermediate (Fig. 1), only C-6a monoalcohol and the dialcohol were detected in rat urine samples. No substantial differences (data not shown) were observed for the urinary excretion of C-6a monoalcohol as a function of either transgene or D3T treatment, suggesting the in vivo formation of this metabolite is principally affected by other enzymes and/or extrahepatic tissues. Shown in Figure 4 are the levels of AFB1-dialcohol, the major AKR7A1-derived metabolite from the C-6a monoalcohol, excreted during the 24 h following dosing with AFB1. Line AKR7A1Tg2 excreted 40-fold more dialcohol than their nontransgenic litter mates, while line AKR7A1Tg5 excreted 12-fold more dialcohol. D3T-treated nontransgenic animals excreted 7.5-fold more of the dialcohol metabolite. These results are consistent with the characterization of AKR7A1 protein expression (see Fig. 2B) and AFB1 alcohols produced in vitro by the hepatic cytosols preparations. Collectively, these results suggest the two transgenic lines provide a robust model to examine the impact of AKR7A1 on aflatoxin toxicity and carcinogenicity in vivo.
FIG. 4.
Urinary excretion of AFB1 dialcohol levels in AKR7A1 transgenic and D3T-treated nontransgenic rats. AFB1 dialcohol was quantified by isotope dilution mass spectrometry as described in “Materials and Methods” and in reference 24. Values are mean ± SE (N = 3).
AFB1 Toxicity
Without aflatoxin damage to the parenchymal cells, bile duct cells have a very low turnover and labeling index. The transgenic rats and their genetic controls had a labeling index of 0.3 ± 0.2 (mean ± SE, N = 7) which agrees well with historic controls (Maxuitenko et al., 1996). As with the in vivo metabolism of AFB1 described above, there were no statistically significant differences noted between male and female rats of either the transgene phenotype or their genetic controls; thus, these data were combined. For the AKR7A1Tg2 line, there was no difference between the bile duct labeling index of the positive (26.9 ± 1.7, N = 11) and genetic control litter mates (25.6 ± 1.2, N = 11). A similar observation was made with the AKR7A1Tg5 line, namely, the labeling indices of the transgenic positive rats (34.1 ± 2.2, N = 16) and genetic controls (32.4 ± 1.8, N = 14) did not differ statistically. The clear differences in extent of labeling between the two transgenic lines resulted because the experiments were undertaken at different times with different preparations of AFB1 and BrdU. In response to induced hepatotoxicity by AFB1, bile duct proliferation ensued and was not modulated by the presence of the additional AKR7A1 enzymatic activity in these rats. These results are in marked contrast to the protective effects from administration of dithiolethiones on this manifestation of AFB1 toxicity (Maxuitenko et al., 1996).
AFB1 Carcinogenicity
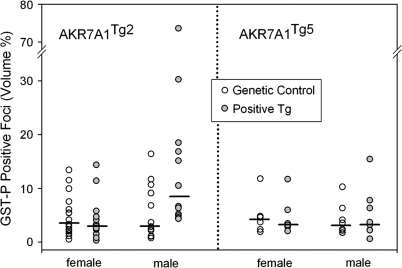
Using our well-characterized hepatic carcinogenesis model (Kensler et al., 1992; Roebuck et al., 2003; Yates et al., 2006), AKR7A1 transgene afforded no protection against AFB1-induced carcinogenesis (Fig. 5). With the exception of one rat (see below), there were no differences among any of the groups in the observed microscopic appearance of the GST-P positive foci. Irrespective of the sex of the rats or the transgenic line, AKR7A1Tg2 and AKR7A1Tg5, the focal burden (volume %) of GST-P foci did not differ statistically between groups carrying the transgene and groups of their genetic control, litter mates. The less robust, morphometrically derived data of foci number per unit volume of liver and mean focal diameter showed no statistically significant difference between groups (data not shown). At variance with all other rats in these groups, one of 12 male, AKR7A1Tg2 positive rats had an unusually high focal burden (74%) and mean focal diameter what was twice that of other rats, thus, largely accounting for the large variation in the male AKR7A1Tg2 group. In the AKR7A1Tg2 genetic controls, the focal volume was 5.50 ± 4.99 (mean ± SD); whereas, in the positive Tg2 group, the focal volume was 16.40 ± 19.64. There was not a statistically significant difference between these two groups (p = 0.065, t-test) and additionally, the higher volume percent was in the opposite direction for support of our initial hypothesis. An underlying explanation of the susceptibility of this one rat to AFB1 is not obvious.
FIG. 5.
Evaluation of the AKR7A1 transgene on AFB1-induced GST-P positive foci formation. At 5 weeks of age, litter mates expressing AKR7A1 and genetic controls were orally gavaged with 25 μg AFB1 5 days per week for 4 successive weeks. The livers were removed 6 weeks later, fixed in acetone, and immunohistochemically stained using an antibody recognizing GST-P. The number and size of the GST-P positive foci were measured by light microscopy and the volume % of GST-P positive foci (analogous to tumor burden) calculated. There were no statistically significant differences between AKR7A1 positive and genetic control. One of 12 rats with an unusually high focal burden (74%) accounted for the high group mean and large SE in the male AKR7A1Tg2 group. These scatter plots are accompanied with median bars (N ranged from 12 to 16 rats per group for the AKR7A1Tg2 and six to eight rats per group for the AKR7A1Tg5 rats).
AFB1 DNA and Protein Adducts
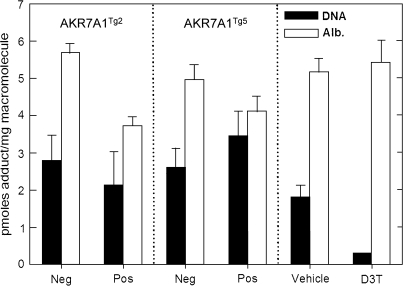
Further evaluation of the effects of transgene expression on aflatoxin toxicodynamics were assessed by quantification of biomarkers reflecting the internal dose and the biologically effective dose of aflatoxin, that is, aflatoxin-lysine adducts in serum and aflatoxin-N7-guanine adducts excreted in urine, respectively (Fig. 6). Neither of the AKR7A1 transgenic rat lines in vivo showed alterations in excretion of the DNA adduct biomarker; an outcome concordant with earlier observations in which the gene was overexpressed in cells in culture without impact on DNA adduct burden (Bodreddigari et al., 2008). By contrast, D3T, which is known to induce AKR7A1 in concert with many other Nrf2-regulated genes including GSTs, produced an 83% reduction in the urinary excretion of aflatoxin-N7-guanine adducts. Line AKR7A1Tg2 showed a significant 35% decrease in levels of aflatoxin-albumin adducts in serum following acute exposure to AFB1. Line AKR7A1Tg5, which has a several fold lower expression of the transgene, did not exhibit a statistically significant reduction in serum levels of the protein adduct. Additionally, no reductions in levels of this protein adduct biomarker were observed in the D3T-treated rats; however, D3T is known to reduce levels of aflatoxin-albumin adducts during chronic exposure to AFB1 (Groopman et al., 1992).
FIG. 6.
Influence of AKR transgene on AFB1 hepatic DNA and serum albumin protein adduct levels. AFARTg2 positive animals had statistically significant lower levels of AFB1-lysine adducts compared to their genetic control littermates (p < 0.05). D3T-treated animals had statistically significant lower levels of AFB1 N7-guanine adducts compared to vehicle treated (*p < 0.05). Quantification for data shown was by isotope dilution mass spectrometry. Statistical comparisons were achieved by ANOVA. Values are mean ± SE (N = 3).
DISCUSSION
Since their discovery in the early 1960s, the hepatotoxicity and carcinogenicity of the aflatoxins have been extensively explored (Busby and Wogan, 1984; Eaton et al., 1994; Kensler et al., 1999; Roebuck and Maxuitenko, 1994). Generally, experimental hepatic carcinogenesis of aflatoxins in rats has involved either chronic exposure over 50–80 weeks in the diet (Wogan, 1974; Wogan et al., 1974) or shorter exposures of usually 2–5 weeks by daily oral gavage (Kensler et al., 1997; Roebuck et al., 1991). With cumulative doses of approximately 100–170 μg AFB1 per rat, long-term feeding resulted in a 100% incidence of hepatocellular carcinomas (HCC); whereas, the shorter exposure protocols have yielded fewer HCC in spite of the cumulative doses being greater. Central to this process is the metabolic activation of AFB1 by CYP1A2 and CYP3A4 to yield two chemically reactive epoxides: AFB1-8,9-exo and -8,9-endo epoxides (Gallagher et al., 1996; Johnson and Guengerich, 1997). Only the AFB1-8,9-exo isomer reacts readily with DNA, forming the N7-guanine and its derivative AFB1 formamidopyrimidine adducts (Johnson and Guengerich, 1997). If not repaired, these DNA adducts or the abasic sites that remain from the spontaneous depurination of the N7-guanine adduct can rapidly lead to DNA mutations (Smela et al., 2001). Several studies have shown strong correlations between N7-guanine adducts and the putative preneoplastic GST-P positive foci (Kensler et al., 1986; Kensler et al., 1992; Roebuck et al., 1991; Yates et al., 2006). Kensler et al. (1986) observed that the first of a series of doses yielded more total DNA adducts than did subsequent doses. Possible explanations include that the ensuing toxicity from the initial doses of AFB1 may induce protective mechanisms such as GSTs that result in reduced genetic damage from subsequent exposures or the inherent toxicity of AFB1 may kill cells that have accumulated adducts thereby reducing the hepatic risk. There is an abundance of literature documenting AFB1 as a genetic toxin, a generalized growth inhibitor of the liver, and a hepatotoxin able to engender regenerative hyperplasia (Busby and Wogan, 1984; Newberne, 1973; Roebuck, 2004).
In studies of aflatoxin carcinogenesis, the incidence of HCC and timing of their occurrence are both modulated by the dithiolethione oltipraz (Kensler et al., 1997; Roebuck et al., 1991). In short-term studies, protection against the development of hepatic foci have been demonstrated with a variety of chemicals: antioxidants (Kensler et al., 1986), dithiolethiones (Kensler et al., 1992; Roebuck et al., 1991), and triterpenoids (Yates et al., 2006). These same agents protect against the acute toxic effects of AFB1 as adjudged by prevention of weight loss or maintenance of normal rates of growth, reduction in histological evidence of cellular damage, and mortality from liver failure. These chemoprotective agents all induce the enzymatic activities of both GST and AKR (Kensler et al., 1986, 1992; Roebuck et al., 2003; Yates et al., 2006). Interestingly, the mouse is highly resistance to AFB1 toxicity (Roebuck and Maxuitenko, 1994), an outcome that is considered to be accounted for predominantly by the high constitutive expression of GST in mouse liver (Eaton et al., 1994).
While the role of AFB1 N7-guanine adducts in the initiation of cancer is well documented, the contribution of hepatotoxicity to this process is less clear. Overall, the mechanisms of cytotoxicity by AFB1 are poorly understood. AFB1 can form protein adducts particularly with lysine through a Schiff base reaction of the AFB1 dialdehyde (Fig. 1), and several investigators have hypothesized that such protein damage may lead to hepatotoxicity (Ellis et al., 1993; Guengerich et al., 2002). However, several unknowns remain, including the critical protein targets that trigger cell death, the dose-response for these important reactions, and the mechanisms of cell protection against such attack. Several investigators have hypothesized that AKR7A1 activity is an important factor in this modulation and that elevated levels of AKR activity should prevent cytotoxicity and thereby attenuate carcinogenesis (Bodreddigari et al., 2008; Ellis et al., 1993; Guengerich et al., 2002).
In a recent study (Bodreddigari et al., 2008), we showed that recombinant in vitro expression of AKR7A1 afforded protection against AFB1 dialdehyde cytotoxicity, resulting in a sixfold increase in the LC50 of the dialdehyde accompanied by a marked decrease in cellular protein adducts. To address the role of the dialdehyde metabolite in the cytotoxicity of AFB1 and its modulation by AKR7A1 activity in vivo, a rat transgenic for AKR7A1 was constructed and characterized. Two transgenic lines, AKR7A1Tg2 and AKR7A1Tg5, with 25 and 8 inserted genomic copies, respectively, expressed enzymatically active AFR7A1. Furthermore, in the intact rats, the expected AFB1 alcohols were formed in relation to the extent of AKR7A1 expression in the liver. However, only the highest expression line, AKR7A1Tg2, exhibited reduced levels of serum albumin adducts. Importantly, the GST activity in the transgenic rat was at basal levels in these two transgenic lines, as were the level of N7-guanine adducts, unlike what occurs with the use of chemoprotective agents. Together, these data confirm appropriate expression of active liver AKR7A1 protein at levels comparable or exceeding those found in response to treatment with chemoprotective agents. Unexpectedly, this metabolic phenotype did not translate into either protection against AFB1-induced hepatic toxicity or diminution of foci. The high expression of AKR7A1 appears not to be relevant to levels achieved by treatment of rats with chemoprotective agents, indicating that in vivo neither the production of the AFB1 dialdehyde nor its metabolism by AKR7A1 are critical for cytotoxicity or carcinogenicity.
Of interest to these observations are the results of another in vitro experiment where we expressed either human CYP1A2 or human CYP1A2 and rat AKR7A1 in AHH-1 lymphoblastoid cells (Bodreddigari et al., 2008). In this experiment AFB1 was metabolized by CYP1A2 within a cell that has negligible levels of GST activity. Coexpression of AKR7A1 significantly increased cell survival, but did not alter the level of DNA adducts formed at a dose of 3 ng/ml of AFB1. Thus, at this dose, it appeared that cytotoxicity was being driven by the AFB1 dialdehyde and not by the CYP1A2-derived epoxide or other oxidative metabolites such as AFM1. At higher concentrations of AFB1, the AKR7A1-expressing cells exhibited a concentration-dependent decrease in the proportion of cells protected against cytotoxicity. This observation suggests that other factors are contributing to AFB1-mediated cytotoxicity. Of interest to this discussion, independent studies have demonstrated the apparent saturation of mutation induced by exposure to AFB1 in human lymphoblast cell lines (Crespi et al., 1990; Kaden et al., 1987). This plateau in mutant fraction is observed despite linear increases in the amount of AFB1 adducts formed (Crispi et al., 1990; Kaden et al., 1987) and in both studies, it occurred at concentrations of AFB1 exceeding 6 ng/ml. Having controlled for cell cycle and the potential for different subpopulations of cells, these studies support the concept of a disproportionate loss of cells exceeding some threshold of DNA damage. For example, apoptotic mechanisms activated at elevated levels of DNA damage could account for the concentration-dependent loss of protection afforded by AKR7A1. This idea is supported by an earlier in vitro study by Fields et al. (1999) where they expressed in hamster V79 cells either rat CYP2B1 or rat CYP2B1 and murine GST A3-3. In their study, coexpression of the GST resulted in a more than a 70% reduction in DNA and RNA adducts, and a 4.6-fold resistance to cytotoxicity (Fields et al., 1999). These data indicate a role of the epoxide in mediating cytotoxicity. However, this study and those described above cannot be directly compared, as the cells expressing rat CYP2B1 had to be treated with much higher concentrations of AFB1. Nonetheless, at doses of AFB1 required to produce foci in Sprague-Dawley rats, it seems likely that the role of the epoxide is much greater than the role of the dialdehyde in the observed hepatotoxicity. In the past, the available analytical techniques were not sufficiently sensitive to carefully document the mass balance of the dose of AFB1 to the liver and its metabolic fate. However, perhaps with the current techniques of HPLC with isotope dilution mass spectrometry (McCoy et al., 2005), this important question can be more fully elaborated.
The rationale for using AKR7A1 for the development of hepatocyte-specific transgenic rats was predicted on the basis of the extraordinary levels of induction of this enzyme in rodent liver by chemoprotective agents and the observed protection against AFB1 toxicity afforded by forced overexpression in cells in culture. Despite this strong evidence in support of the hypothesis that AKR7A1 could modulate susceptibility to aflatoxin carcinogenesis, especially when coupled with the pronounced phenotype of enhanced AFB1 dialdehyde metabolism in the transgenic rats, no protective effect was observed. Clearly, in vitro and in vivo observations are not always concordant. This study again sounds a cautionary note against total reliance on reductionist approaches to complex systems. Whether hepatic GST induction is solely responsible for protection by inducers from hepatic cytotoxicity, or more likely multiple aspects of the Keap1-Nrf2 transcriptional program work in concert to protect the liver from AFB1 is not known. However, with the hindsight afforded by this study, the focus of future studies on mechanisms of cancer chemoprevention using systems approaches might be more fruitful.
FUNDING
National Institutes of Health, National Cancer Institute (R01 CA 39416); and National Institute of Environmental Health Sciences (P30 ES 03819); and the W. Harry Feinstone Center for Genomic Research to T.R.S.
Acknowledgments
We thank Dr John Taylor, UCSF, for the liver-specific expression vector pLiv.7 and Mr Eric York, Department of Pathology, Dartmouth Medical School for superb histological services.
References
- Bodreddigari S, Jones LK, Egner PA, Groopman JD, Sutter CH, Roebuck BD, Guengerich FP, Kensler TW, Sutter TR. Protection against aflatoxin B1-induced cytotoxicity by expression of the cloned aflatoxin B1-aldehyde reductases rat AKR7A1 and human AKR7A3. Chem. Res. Toxicol. 2008;21:1134–1142. doi: 10.1021/tx7004458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby WF, Wogan GN. Aflatoxins. In: Searle CE, editor. Chemical Carcinogens. Washington, D.C.: American Chemical Society; 1984. pp. 945–1136. [Google Scholar]
- Crespi CL, Steimel DT, Aoyama T, Gelboin HV, Gonzalez FJ. Stable expression of human cytochrome P4501A2 cDNA in a human lymphoblastoid cell line: Role of the enzyme in the metabolic activation of aflatoxin B1. Mol. Carcinogenesis. 1990;3:5–8. doi: 10.1002/mc.2940030104. [DOI] [PubMed] [Google Scholar]
- Eaton DL, Bammler TK. Concise review of the glutathione S-transferases and their significance to toxicology. Toxicol. Sci. 1999;49:156–164. doi: 10.1093/toxsci/49.2.156. [DOI] [PubMed] [Google Scholar]
- Eaton DL, Ramsdell HS, Neal GE. Biotransformation of aflatoxins. In: Eaton DL, Groopman JD, editors. The Toxicology of Aflatoxins: Human Health, Veterinary, and Agricultural Significance. New York: Academic Press; 1994. pp. 45–72. [Google Scholar]
- Egner PA, Groopman JD, Wang J, Kensler TW, Friesen MD. Quantification of aflatoxin B1-N7-guaine in human urine by high-performance liquid chromatography and isotope dilution tandem mass spectrometry. Chem. Res. Toxicol. 2006;19:1191–1195. doi: 10.1021/tx060108d. [DOI] [PubMed] [Google Scholar]
- Ellis EM, Judah DJ, Neal GE, Hayes JD. An ethoxyquin-inducible aldehyde reductase from rat liver that metabolizes aflatoxin B1 defines a subfamily of aldo-keto reductases. Proc. Natl. Acad. Sci. U. S. A. 1993;90:10350–10354. doi: 10.1073/pnas.90.21.10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Wang J, Bensadoun A, Lauer SJ, Dang Q, Mahley RW, Taylor JM. Overexpression of hepatic lipase in transgenic rabbits leading to a marked reduction of plasma high density lipoproteins and intermediate density lipoproteins. Proc. Natl. Acad. Sci. U. S. A. 1994;91:8724–8728. doi: 10.1073/pnas.91.18.8724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields WR, Morrow CS, Doehmer J, Townsend AJ. Expression of stably transfected murine glutathione S-transferase A3-3 protects against nucleic acid alkylation and cytotoxicity by aflatoxin B1 in hamster V79 cells expressing rat cytochrome P450-2B1. Carcinogenesis. 1999;20:1121–1125. doi: 10.1093/carcin/20.6.1121. [DOI] [PubMed] [Google Scholar]
- Gallagher EP, Kunze KL, Stapleton PL, Eaton DL. The kinetics of aflatoxin B1 oxidation by human cDNA—Expressed and human liver microsomal cytochromes P450 1A2 and 3A4. Toxicol. Appl. Pharmacol. 1996;141:595–606. doi: 10.1006/taap.1996.0326. [DOI] [PubMed] [Google Scholar]
- Groopman JD, Busby WF, Jr, Wogan GN. Nuclear distribution of aflatoxin B1 and its interaction with histones in rat liver in vivo. Cancer Res. 1980;40:4343–4351. [PubMed] [Google Scholar]
- Groopman JD, DeMatos P, Egner PA, Love-hunt A, Kensler TW. Molecular dosimetry of urinary aflatoxin-N7-guanine and serum aflatoxin-albumin adducts predicts chemoprotection by 1,2-dithiole-3-thione in rats. Carcinogenesis. 1992;13:101–106. doi: 10.1093/carcin/13.1.101. [DOI] [PubMed] [Google Scholar]
- Guengerich FP, Arneson KO, Williams KM, Deng Z, Harris TM. Reaction of aflatoxin B1 oxidation products with lysine. Chem. Res. Toxicol. 2002;15:780–792. doi: 10.1021/tx010156s. [DOI] [PubMed] [Google Scholar]
- Guengerich FP, Cai H, McMahon M, Hayes JD, Sutter TR, Groopman JD, Deng Z, Harris TM. Reduction of aflatoxin B1 dialdehyde by rat and human aldo-keto reductases. Chem. Res. Toxicol. 2001;14:727–737. doi: 10.1021/tx010005p. [DOI] [PubMed] [Google Scholar]
- Habig WH, Pabst MJ, Jakoby WB. Glutathione S-transferase. The first enzymatic step in mercapturic acid formation. J. Biol. Chem. 1974;219:7130–7139. [PubMed] [Google Scholar]
- Hayes JD, Judah DJ, Neal GE. Resistance to aflatoxin B1 is associated with the expression of a novel aldo-keto reductase which has catalytic activity towards a cytotoxic aldehyde-containing metabolite of the toxin. Cancer Res. 1993;53:3887–3894. [PubMed] [Google Scholar]
- Hinshelwood A, McGarvie G, Ellis EM. Substrate specificity of mouse aldo-keto reductase AKR7A5. Chem. Biol. Interact. 2003;144:263–269. doi: 10.1016/s0009-2797(02)00173-4. [DOI] [PubMed] [Google Scholar]
- Ireland LS, Harrision DJ, Neal GE, Hayes JD. Molecular cloning, expression and catalytic activity of a hum n AKR7 member of the aldo-keto reductase superfamily: Evidence that the major 2-carboxybenzaldehyde reductase from human liver is a homologue of rat aflatoxin B1-aldehyde reductase. Biochem. J. 1998;332:21–34. doi: 10.1042/bj3320021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DN, Egner PA, OBrian G, Glassbrook N, Roebuck BD, Sutter TR, Payne GA, Kensler TW, Groopman JD. Quantification of urinary aflatoxin B1 dialdehyde metabolites formed by aflatoxin aldehyde reductase using isotopic dilution tandem mass spectrometry. Chem. Res. Toxicol. 2008;21:752–760. doi: 10.1021/tx700397n. [DOI] [PubMed] [Google Scholar]
- Johnson WW, Guengerich FP. Reaction of aflatoxin B1 exo-epoxide with DNA: Kinetic analysis of covalent binding and DNA-induced hydrolysis. Proc. Natl. Acad. Sci. U. S. A. 1997;94:6121–25. doi: 10.1073/pnas.94.12.6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WW, Harris TM, Guengerich FP. Kinetics and mechanism of hydrolysis of aflatoxin B1 exo-8,9-oxide and rearrangement of the dihyrodiol. J. Am. Chem. Soc. 1996;118:8213–8220. [Google Scholar]
- Kaden DA, Call KM, Leong P-M, Komives EA, Thilly WG. Killing and mutation of human lymphoblast cells by aflatoxin B1: Evidence for and inducible repair response. Cancer Res. 1987;47:1993–2001. [PubMed] [Google Scholar]
- Kensler TW, Egner PA, Davidson NE, Roebuck BD, Pikul A, Groopman JD. Modulation of aflatoxin metabolism, aflatoxin-N7-guanine formation, and hepatic tumorigenesis in rats fed ethoxyquin: Role of induction of glutathione S-transferases. Cancer Res. 1986;46:3924–3931. [PubMed] [Google Scholar]
- Kensler TW, Gange SJ, Egner PA, Dolan PM, Muñoz A, Groopman JD, Rogers AE, Roebuck BD. Predictive value of molecular dosimetry: Individual versus group effects of oltipraz on aflatoxin-albumin adducts and risk of liver cancer. Cancer Epidemiol. Biomarkers Prev. 1997;6:603–610. [PubMed] [Google Scholar]
- Kensler TW, Groopman JD, Eaton DL, Curphey TJ, Roebuck BD. Potent inhibition of aflatoxin-induced hepatic tumorigenesis by the monofunctional enzyme inducer 1,2-dithiole-3-thione. Carcinogenesis. 1992;13:95–100. doi: 10.1093/carcin/13.1.95. [DOI] [PubMed] [Google Scholar]
- Kensler TW, Groopman JD, Sutter TR, Curphey TJ, Roebuck BD. Development of cancer chemopreventive agents: Oltipraz as a paradigm. Chem. Res. Toxicol. 1999;12:113–126. doi: 10.1021/tx980185b. [DOI] [PubMed] [Google Scholar]
- Knight LP, Primiano T, Groopman JD, Kensler TW, Sutter TR. cDNA cloning, expression and activity of a second human aflatoxin B1-metabolizing member of the aldo-keto reductase superfamily, AKR7A3. Carcinogenesis. 1999;20:1215–1223. doi: 10.1093/carcin/20.7.1215. [DOI] [PubMed] [Google Scholar]
- Liu YL, Roebuck BD, Yager JD, Groopman JD, Kensler TM. Protection by 5-(2-pyrazinyl)-4-methyl-1,2-dithiol-3-thione (oltipraz) against the hepatotoxicity of aflatoxin B1 in the rat. Toxicol. Appl. Pharmacol. 1988;93:442–451. doi: 10.1016/0041-008x(88)90047-6. [DOI] [PubMed] [Google Scholar]
- Maxuitenko YY, Curphey TJ, Kensler TW, Roebuck BD. Protection against aflatoxin B1-induced hepatic toxicity as a short term screen of cancer chemopreventive dithiolethiones. Fundam. Appl. Toxicol. 1996;32:250–259. doi: 10.1006/faat.1996.0128. [DOI] [PubMed] [Google Scholar]
- McCoy LF, Scholl PF, Schleicher RL, Groopman JD, Powers CD, Pfieffer CC. Analysis of aflatoxin B1-lysine adduct in serum using isotope dilution liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2005;19:2203–2210. doi: 10.1002/rcm.2045. [DOI] [PubMed] [Google Scholar]
- Newberne PM. Chronic aflatoxicosis. J. Am. Vet. Med. Assoc. 1973;163:1262–1269. [PubMed] [Google Scholar]
- Roebuck BD. Hyperplasia, partial hepatectomy, and the carcinogenicity of aflatoxin B1. J. Cell. Biochem. 2004;91:243–249. doi: 10.1002/jcb.10758. [DOI] [PubMed] [Google Scholar]
- Roebuck BD, Curphey TJ, Li Y, Baumgartner KJ, Bodreddigari S, Yan J, Gange SJ, Kensler TW, Sutter TR. Evaluation of the cancer chemopreventive potency of dithiolethione analogs of oltipraz. Carcinogenesis. 2003;24:1919–28. doi: 10.1093/carcin/bgg173. [DOI] [PubMed] [Google Scholar]
- Roebuck BD, Liu Y-L, Rogers AE, Groopman JD, Kensler TW. Protection against aflatoxin B1-induced hepatocarcinogenesis in F344 rats by 5-(2-pyrazinyl)-4-methyl-1,2-dithiole-3-thione (Oltipraz): predictive role for short-term molecular dosimetry. Cancer Res. 1991;51:5501–5506. [PubMed] [Google Scholar]
- Roebuck BD, Maxuitenko Y. Biochemical mechanisms and biological implications of the toxicity of aflatoxins as related to aflatoxin carcinogenesis. In: Eaton DL, Groopman JD, editors. The Toxicology of Aflatoxins: Human Health, Veterinary, and Agricultural Significance. New York: Academic Press; 1994. pp. 27–43. [Google Scholar]
- Sabbioni G, Skipper PL, Buchi G, Tannenbaum SR. Isolation and characterization of the major serum albumin adduct formed by aflatoxin B1 in vivo in rats. Carcinogenesis. 1987;8:819–824. doi: 10.1093/carcin/8.6.819. [DOI] [PubMed] [Google Scholar]
- Smela ME, Currier SS, Bailey EA, Essigmann JM. The chemistry and biology of aflatoxin B1: from mutational spectrometry to carcinogenesis. Carcinogenesis. 2001;22:535–545. doi: 10.1093/carcin/22.4.535. [DOI] [PubMed] [Google Scholar]
- Wogan GN. Naturally occurring carcinogens. In: Shubik P, editor. The Physiopathology of Cancer. Biology and Biochemistry. Vol. 1. Basel: Karger; 1974. pp. 64–109. [Google Scholar]
- Wogan GN, Paglialunga S, Newberne PM. Carcinogenic effects of low dietary levels of aflatoxin B1 in rats. Fd Cosmet. Toxicol. 1974;121:681–685. doi: 10.1016/0015-6264(74)90239-9. [DOI] [PubMed] [Google Scholar]
- Yamanaka S, Balestra M, Ferrell LD, Fan J, Arnold KS, Taylor S, Taylor JM, Innerarity TL. Apolipoprotein B mRNA-editing protein induces hepatocellular carcinoma and dysplasia in transgenic animas. Proc. Natl. Acad. Sci. U. S. A. 1995;92:8483–8487. doi: 10.1073/pnas.92.18.8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates MS, Kwak M-K, Egner PA, Groopman JD, Bodreddigari S, Sutter TR, Baumgartner KJ, Roebuck BD, Liby KT, Yore MM, et al. Potent protection against aflatoxin-induced tumorigenesis through induction of Nrf2-regulated pathways by the synthetic triterpenoid, 1-[2-cyano-3-,12-dioxooleana-1,9(11)-dien-28-oly]imidazole. Cancer Res. 2006;66:2488–2494. doi: 10.1158/0008-5472.CAN-05-3823. [DOI] [PubMed] [Google Scholar]