Summary
Objective
An apparent database error in the sequence underlying the Helix-II cartilage biomarker immunoassay was investigated at the protein level.
Methods and Results
Tandem mass spectrometry established the peptide sequence ERGETGPP*GPA in human type II collagen, not ERGETGPP*GTS used to generate the antibody for the Helix-II assay.
Conclusions
Recent reports in which the Helix-II assay was applied to urine or serum as a marker of cartilage collagen degradation need to be re-evaluated since the epitope does not occur in cartilage type II collagen. Based on collagen sequences and Helix-II epitope properties, type III collagen is one of several candidate sources of the cross-reacting signal in body fluids, but not type II collagen. The findings highlight the need for more stringent scrutiny of the origins and validation of molecular markers in body fluid assays in general.
Keywords: cartilage, collagen, biomarker
Introduction
There is a pressing need to develop reliable biochemical markers that can inform on the process of joint destruction in osteoarthritis (OA), a disorder of essentially silent onset1. Such markers could aid in drug development by identifying fast progressors and detecting early response to therapy and so reducing patient numbers and time required for clinical trials. The NIH-sponsored Osteoarthritis Initiative (web link: http://www.oai.ucsf.edu/) is a nationwide research study and clinical sample resource that recognizes the need to promote biomarker development. Body fluid markers of cartilage metabolism have received much research attention, in particular type II collagen breakdown products2. Since type II collagen is largely restricted to hyaline cartilages it presents an attractive target as a potential monitor of joint erosion processes. Besides face validity, however, it would seem wise to establish any candidate marker by unequivocal molecular identification in biological samples as a prelude to wide-ranging, potentially misleading clinical applications. It is notable that of the several collagen type II peptide-based immunoassays in clinical research, none have been validated by definitive direct methods as the primary signal generator in adult serum or urine. These include CTX-II, TIINE and Helix-II as intended collagen degradation markers3.
With this in mind in reviewing the status of collagen II biomarkers actively promoted through publications in the clinical research literature, we noticed a curious anomaly in the origin of the marker Helix-II originally described by Charni et al.4. The authors reported selecting this target peptide immunogen by database searching for a unique occurrence in the α1(II) chain among the different collagen gene products. On inspection of DNA and Protein databases maintained by NCBI (http://www.ncbi.nlm.nihgov/) however, it is apparent that the target peptide sequence may owe its uniqueness to a sequencing error in the original 1989 submission to the Swiss Protein database (P02458)5. The genomic entry for the Helix-II epitope sequence in COL2A1 encodes ERGETGPPGPA not ERGETGPPGTS, the latter being the basis of the immunogen for the Helix-II assay. To confirm beyond any doubt the nature of this collagen domain at the protein level, we isolated the appropriate fragment from human cartilage type II collagen and established its structure directly by mass spectrometry.
A growing number of reports based on the Helix-II assay have appeared in the clinical research literature with results interpreted in terms of its usefulness as a monitor of joint cartilage degradation4, 6-8. Here we confirm at the protein level that the Helix-II epitope is based on a non-existent sequence and predict that a competing epitope, possibly from type III collagen, is responsible for the clinical findings.
Methods
Consensus protein sequences were taken from the NCBI database using human genomic sequences for COL1A1, COL1A2, COL2A1 and COL3A1, and compared with the source sequence (Swiss Prot. P02458) for the Helix-II immunogen4. Collagen type II CNBr-derived fragments were prepared from multiple samples of fetal human cartilage and adult human articular cartilage. The fragment α1(II)CB10 containing the putative Helix-II sequence was resolved from individual samples by SDS-PAGE, located by Coomassie Blue staining, excised and digested in-gel with trypsin9 prior to protein mass spectrometry. Resulting peptides were subjected to microbore column liquid chromatography (Vydac C8 mass spectrometry 0.3mm × 15cm) interfaced directly to an ion trap tandem mass spectrometer (Thermo Finnigan LCQ Deca XP) equipped with a micro-electrospray ionization source. Peptide sequences were identified by comparison with the NCBI non-redundant protein database using Sequest, an automated database search algorithm designed for use with tandem mass spectrometry data, and by manual interpretation of the MS/MS fragmentation results.
Results and Discussion
Fig. 1 compares homologous protein sequences from the Human Genome database at NCBI for α1(II), α1(I) and α1(III) collagen chains with that of the Helix-II sequence as submitted in the original SwissProt Accession No. P02458. As shown, a cDNA sequencing error in the original submission and report5 misidentified TSGI in place of the sequence PAGF, as subsequently established by the human genomic sequence. This error was corrected in later P02458 database entries and in other COL2A1 cDNA submissions but apparently overlooked in Helix-II assay design4.
Fig. 1.
Comparison of the Helix-II target epitope sequence (based on original Swiss Protein uncorrected entry Accession No. P02458) to homologous protein sequences at position 623-633 of the α1(II) helix from the human genome database for collagen chains α1(II), α1(I) and α1(III).
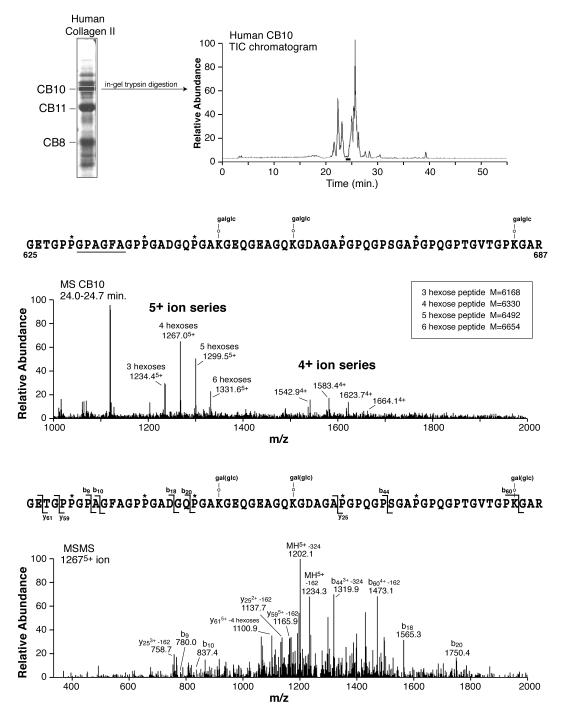
To be completely certain that this was wholly an artifactual error of cDNA sequencing, and not for example the result of a natural polymorphism, we screened more than 10 individual collagen type II protein samples from our bank of control fetal and adult human tissues. The same tryptic peptide was recovered and identified by mass spectrometry from them all. Its sequence is shown in Fig. 2. The peptide mass and MS/MS fragmentation data showed that PAGF not TSGI is the correct protein sequence at the Helix-II locus. We could not find a TSGI version in any sample examined.
Fig. 2.
Direct confirmation by tandem mass spectrometry that the α1(II) collagen sequence from human cartilage at helix residues 631-636 is PAGF not TSGI. The upper panel shows the tryptic peptide profile by total ion current (TIC) on HPLC from CNBr-fragment α1(II)CB10. The middle panel shows the mass spectrum from the peptide encompassing the Helix-II epitope of mass that fits the internal sequence PAGF (not TSGI) and the lower panel the MS/MS fragmentation spectrum of the 12675+ ion confirming the structure shown.
What then is the source of the signal given by urine and serum in the Helix-II immunoassay? Clearly human urine and serum give an inhibitory signal and statistically significant differences are evident between patients and control subjects in several clinical studies4,6-8. The Helix-II epitope can also be generated from human cartilage in vitro by cathepsin digestion10, shows diurnal variability in urinary excretion11 and no response to estrogen in post-menopausal women or experimental rats12.
The simplest explanation would be cross-reaction of the antiserum raised against ERGETGPP*GTS, with peptides of the correct collagen type II peptide sequence, ERGETGPP*GPA, from this locus in the triple-helix. However, the control experiments of Charni et al. ruled this out4. They synthesized the appropriate homologous peptides for α1(I) and α1(III) chains, which showed no cross-reactivity in the Helix-II inhibition assay. The α1(III) sequence, ERGETGPP*GPA, is identical to that of α1(II), so neither α1(II), α1(I) nor α1(III) can be the source of the inhibition at least from peptides originating at this helical locus. Rat type II collagen has the same sequence, ERGETGPPGPA, as human, so the Helix-II assay results reported for experimental rats12 have a similar problem.
In their original assay validation, Charni et al.4 used various synthetic peptides to establish the requirement of a free C-terminal GTS sequence and hence proteolytic neoepitope recognition. In addition, hydroxyproline at the P* site of GPP*GTS gave a >10-fold higher affinity than proline. The helical domains of human α1(I), α2(I) and α1(II) lack a GTS triplet, but the human α1(III) chain contains two, one of which is in the context GPP*GTS (at res 16-21 of the αI(III) triple-helical domain) as in the α1(II) artifactual sequence. One possibility, therefore, is that the Helix-II immunoassay is measuring collagen type III breakdown products in body fluids. If so, it is not unreasonable that in cross-sectional clinical studies comparing patients and control subjects, significantly elevated levels of Helix-II signal will be found in keeping with observations using other generalized inflammatory markers (e.g., serum C-reactive protein and hyaluronic acid) in OA and RA. Collagen type III turnover in the body is high and indeed as assessed by the serum type III N-propeptide assay, PIIINP, significantly higher levels have been reported for knee OA patients13. Etanercept, an anti-inflammatory, is also likely to suppress type III collagen turnover14, which would explain Helix-II findings with spondylarthropathy patients8. Adult human articular cartilage contains significant amounts of type III collagen15 which could explain the in vitro findings10. The GPPGTS motif is also present in human α2(XI) and α5(IV) collagen (http://www.ncbi.nlm.nih.gov/) so various alternative origins are possible provided that proteolysis occurs after GTS. Whatever the origin of the competing antigen(s), however, it seems unlikely that the differences observed in clinical studies are due primarily to metabolic processes in cartilage or even joint tissues.
There are important lessons from this cautionary tale. The first is the obvious need for care and oversight when translating basic science into clinical application. The second is the valuable insight from the clinical studies on the dangers of cross-reactivity when antibodies raised against short collagen sequences are applied directly to biological fluids. Significant clinical differences can be misleading. In short, the pitfalls are clear for all biomarker validations that rely on findings from peptide-based immunoassays in the absence of rigorous scientific evidence that the intended peptide targets indeed are responsible for the assay signal. Even when the authentic peptide sequence is used for antibody production, the only convincing and reliable proof would be to immunopurify the binding partner(s) from the body fluid using the assay antibody and identify the peptide(s) and hence their molecular origin by definitive methods, ideally mass spectrometry. Finally, the important message is that the scientific bar needs to be raised in the development and validation of molecular markers in general for osteoarthritis research.
Acknowledgements
This study was supported by grants AR37318 and AR36794 from the National Institute of Arthritis, Musculoskeletal and Skin Diseases (NIAMS) of the National Institutes of Health (USA), and the Ernest M. Burgess endowed chair of the University of Washington.
Footnotes
Conflict of interest statement: The authors report no conflicts of interest with this study.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lohmander LS, Eyre DR. Biochemical Markers as Surrogate End Points of Joint Disease. In: Reid DM, Miller CG, editors. Clinical Trials in Rheumatoid Arthritis and Osteoarthritis. Springer; 2008. pp. 249–274. [Google Scholar]
- 2.Birmingham JD, Vilim V, Kraus VB. Collagen biomarkers for arthritis applications. Biomarker Insights. 2006;2:61–76. [PMC free article] [PubMed] [Google Scholar]
- 3.Garnero P. New biochemical markers of cartilage turnover in osteoarthritis: Recent developments and remaining challenges. http://www.bonekey-ibms.org/cgi/content/full/ibmske;4/1/7.
- 4.Charni N, Juillet F, Garnero P. Urinary type II collagen helical peptide (HELIX-II) as a new biochemical marker of cartilage degradation in patients with osteoarthritis and rheumatoid arthritis. Arth. Rheum. 2005;52:1081–90. doi: 10.1002/art.20930. [DOI] [PubMed] [Google Scholar]
- 5.Su MW, Lee B, Ramirez F, Machado MA, Horton WA. Nucleotide sequence of the full-length cDNA encoding for human type II procollagen. Nucleic Acids Res. 1989;17:9473. doi: 10.1093/nar/17.22.9473. (abstract) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garnero P, Charni N, Juillet F, Conrozier T, Vignon E. Increased urinary type II collagen helical and C-telopeptide levels are independently associated with a rapidly destructive hip osteoarthritis. Ann. Rheum. Dis. 2006;65:1639–44. doi: 10.1136/ard.2006.052621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charni N, Juillet F, Garnero P. A novel serum ELISA for II collagen helical domain degradation (S-helix-II) to assess cartilage damage in patients with rheumatoid arthritis. Ann. Rheum. Dis. 2006;65(Suppl 2):145. [Google Scholar]
- 8.Briot K, Roux C, Gossec L, Charni N, Kolta S, Dougados M, et al. Effects of etanercept on serum biochemical markers of cartilage metabolism in patients with spondyloarthropathy. J. Rheumatol. 2008;35(2):310–4. [PubMed] [Google Scholar]
- 9.Kinter M, Sherman NE. In: Protein Sequencing and Identification Using Tandem Mass Spectrometry. Desiderio DM, Nibbering NMM, editors. John Wiley & Sons; New York, NY: pp. 152–7. [Google Scholar]
- 10.Tabassi N Charni-Ben, Desmarais S, Bay-Jensen A-C, Delaissé JM, Percival MD, Garnero P. The type II collagen fragments Helix-II and CTX-II reveal different enzymatic pathways of human cartilage collagen degradation. Osteoarth. Cart. 2008;16:1183–91. doi: 10.1016/j.joca.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 11.Quintana DJ, Garnero P, Huebner MS, Tabassi N Charni-Ben, Kraus VB. Brief Report: PIIANP and HELIXII diurnal variation. Osteoarth. Cart. 2008;16:1192–5. doi: 10.1016/j.joca.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bay-Jensen AC, Tabassi NC, Sondergaard LV, Andersen TL, Dagnaes-Hansen F, Garnero P, et al. The response to estrogen deprivation of the cartilage collagen degradation marker, CTX-II, is unique compared to other markers of collagen turnover. Arth. Res. Ther. 2009 doi: 10.1186/ar2596. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garnero P, Piperno M, Gineyts E, Christgau S, Delmas PD, Vignon E. Cross sectional evaluation of biochemical markers of bone, cartilage, and synovial tissue metabolism in patients with knee osteoarthritis: relations with disease activity and joint damage. Ann. Rheum. Dis. 2001;60:619–26. doi: 10.1136/ard.60.6.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Radauceanu A, Ducki C, Virion J-M, Rossignol P, Mallat Z, McMurray J, et al. Extracellular matrix turnover and inflammatory markers independently predict functional status and outcome in chronic heart failure. J. Cardiac Failure. 2008;14:467–74. doi: 10.1016/j.cardfail.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 15.Eyre DR, Weis MA, Wu JJ. Articular cartilage collagen: An irreplaceable framework? Eur. Cell Mater. 2006;12:57–63. doi: 10.22203/ecm.v012a07. [DOI] [PubMed] [Google Scholar]