Abstract
Cytosine methylation patterns are essential for the proper control of gene expression in higher vertebrates. Although alterations in methylation patterns are frequently observed in human tumors, neither the mechanisms for establishing methylation patterns during normal development nor the mechanisms leading to pathological alterations of methylation patterns are currently known. While epidemiological studies have implicated inflammation in cancer etiology, a mechanistic link has yet to be established. Investigations of inflammation-mediated DNA damage may have provided important new insights. Our in vitro studies revealed that the inflammation-mediated DNA damage product, 5-chlorocytosine, could direct fraudulent methylation of previously unmethylated CpG sites. The purpose of this study was to recapitulate our in vitro findings by introducing 5-chlorocytosine residues into the DNA of replicating mammalian cells and to examine its impact on gene expression and cytosine methylation patterns. CHO-K1 cells hemizygous for the hprt gene were electroporated with the triphosphates of cytosine [2′-deoxycytidine-5′-triphosphate (dCTP)], 5-methylcytosine [5-methyl-2′-deoxycytidine-5′-triphosphate (MedCTP)] and 5′-chloro-2′-deoxycytidine-5′-triphosphate (CldCTP), and then selected with 6-thioguanine for silencing the hprt gene. Both modified nucleotides, MedCTP and CldCTP, but not unmodified dCTP, silenced hprt gene expression. Subsequent bisulfite pyrosequencing of CpG sites within the hprt promoter region of the selected cells confirmed hypermethylation, although global methylation levels as measured by gas chromatography–mass spectrometry did not change. Modified nucleotide-induced gene silencing could be reversed with 5-aza-2′-deoxycytidine indicating an epigenetic rather than mutagenic alteration. These results provide further evidence that the inflammation damage product 5-chlorocytosine could be a link between inflammation and cancer development.
Introduction
Cytosine methylation patterns in DNA, in concert with covalent modifications to histone proteins, comprise an epigenetic code for controlling gene activity that is established during normal cellular development, but is frequently altered in human tumors (1–4). Methylation changes in human tumors are associated with inappropriate activation of tumor promoter genes or aberrant silencing of tumor suppressors (5–7). A renewed focus on the potential role of epigenetic alterations in the development of human disease has been highlighted recently in the Roadmap to Epigenomics Program from the National Institutes of Health (8).
DNA damage, inflammation and alterations in cytosine methylation patterns have all been associated with the development of cancer in humans (9,10); however, a mechanistic link has yet to be established among them. Recent studies on inflammation-mediated DNA damage may have provided important new clues. Although it has been long known that reactive species generated by activated neutrophils and eosinophils are potent antimicrobial agents, only recently the extent of collateral damage to host DNA and proteins has been revealed. The protein damage product, 3-chlorotyrosine, has been shown to be a specific marker of neutrophil-derived HOCl damage, and it is detected in diseased human tissues associated with atherosclerosis, sepsis and lung disease (11–13). Reactive inflammatory molecules can also cause an array of oxidized and halogenated damage products in the DNA of living cells and among these is a particularly sinister damage product, 5-chlorocytosine (14–16). In in vitro biochemical studies, 5-chlorocytosine has been shown to mimic 5-methylcytosine in directing the binding of methylation-sensitive DNA-binding proteins (17) and in misdirecting the human maintenance methylase to fraudulently methylate previously unmethylated CpG sites (18). While in vitro studies with 5-chlorocytosine provide a sound chemical rationale for linking inflammation, DNA damage and epigenetic changes observed in human tumors, studies with living cells are required to strengthen these links.
In order to investigate the behavior of 5-chlorocytosine in a cell culture system with respect to its ability to cause epigenetic alterations, 5-chlorocytosine was introduced into dividing mammalian cells as a 2′-deoxynucleoside triphosphate. Direct exposure of cells to HOCl would generate 5-chlorocytosine in DNA. However, it would also result in a complex array of other potentially cytotoxic and mutagenic damage products allowing for results that would be very difficult to interpret. Although mammalian cells will metabolize and incorporate 5-substituted 2′-deoxyuridine analogs into DNA, 5-substituted 2′-deoxycytidine analogs are deaminated much more efficiently than they are phosphorylated, rendering this method ineffective for introducing 5-chlorocytosine into the DNA of replicating cells (19,20). Triphosphate analogs in the growth medium, including 5-chloro-2′-deoxycytidine-5′-triphosphate (CldCTP), would not passively enter cells, requiring an alternative tactic. The approach utilized here was based upon previous studies that demonstrated that exogenous nucleotides can be electroporated into living cells, and specifically that the triphosphate analog of 5-methyl-2′-deoxycytidine (MedCTP) could be introduced into cells, incorporated into genomic DNA and heritably alter methylation patterns and gene expression (21–24).
In this study, we have synthesized CldCTP, examined its properties with DNA polymerase, and examined its capacity to alter methylation patterns and gene expression in replicating mammalian cells. We have also measured total 5-methylcytosine levels by gas chromatography–mass spectrometry (GC–MS).
Materials and methods
Synthesis, purification and confirmation of 5-chloro-2′-deoxycytidine-5′-triphosphate
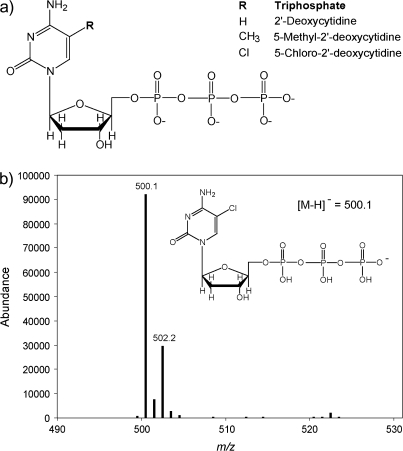
5-Chloro-2′-deoxycytidine (Figure 1a) was synthesized according to methods developed by this laboratory (25). The triphosphate was synthesized according to methods published by Ludwig (26). Briefly, 5-chloro-2′-deoxycytidine was reacted with phosphorus oxychloride under argon on ice. Tributylammonium pyrophosphate in anhydrous dimethylformamide was added, followed by tributylamine. The reaction was monitored using high-performance liquid chromatography on a MonoQ column with a gradient of triethylamonium acetate in water. The final product was purified on a MonoQ column with a gradient of triethylammonium bicarbonate in water. The identity of the desired product, CldCTP, was confirmed by electrospray ionization-mass spectrometry in the negative ion mode with a ThermoFinnigan LCQ Deca XP mass spectrometer (Figure 1b).
Fig. 1.
Triphosphates used for CHO-K1 electroporation. (a) R represents the 5-substituent of the pyrimidine ring. When R is H, then structure is 2′-deoxycytidine. When R is CH3, the structure is 5-methyl-2′-deoxycytidine. When R is Cl the structure is 5-chloro-2′-deoxycytidine. (b) Electrospray mass spectrometry of CldCTP. Predominant ion M-H seen at 500.1 m/z in the negative ion mode with an envelope consistent with a chlorine-containing compound.
Oligonucleotide synthesis and purification
Oligonucleotide 22 and 30mers containing standard, unmodified bases for polymerase studies were prepared by standard solid-phase synthesis (Figure 2). The duplex sequence used in this study was taken from a region of the hamster hypoxanthine-guanine phosphoribosyltransferase (hprt) promoter region. The 5-chlorocytosine phosphoramidite was prepared according to the methods developed by this laboratory (25). All other phosphoramidites used were obtained from Glen Research (Sterling, VA).
Fig. 2.
Klenow exo minus fragment is able to incorporate CldCTP into the newly synthesized strand as well as synthesize past ClC in the template strand. (a) Comparable polymerase incorporation of dCTP and CldCTP into the newly synthesized strand. Bottom band in each lane represents the 32P-labeled 22mer primer. Top band in represents the 30mer full-length extension product. (b) Comparable polymerase extension past C and ClC in template strand. Bottom most band in each lane represents the 32P-labeled 23mer primer. Top most band represents the 30mer full-length extension product.
Polymerase incorporation and extension assays
Exonuclease-deficient Klenow fragment and appropriate reaction buffers and cofactors were obtained from New England Biolabs. The deoxynucleoside triphosphates (dNTPs) other than CldCTP were obtained from Roche.
For the polymerase incorporation assays, the 22mer primer, derived from the hamster hprt promoter, was 5′-32P-end labeled by T4 polynucleotide kinase (New England Biolabs, Ipswich, MA) with [γ-32P]adenosine triphosphate (MP Biomedicals, Costa Mesa, CA) under conditions recommended by the enzyme supplier and subsequently purified using G50 Sephadex columns (Roche, Indianapolis, IN). The labeled 22mer was mixed with a 2-fold excess of the complementary 30mer strand in 100 mM Tris–HCl pH 7.0, incubated at 95°C for 5 min and allowed to cool to room temperature slowly to create a duplex with a 5′ overhang that provides a template for Klenow exo minus fragment (Figure 2a). A time course of dCTP and CldCTP incorporation was conducted. Briefly, the labeled substrate (2.5 pmol) was incubated with 0.025 U Klenow fragment in the presence of 100 μM dNTPs and 1 μM dCTP or CldCTP in 1× NEBuffer 2 (50 mM NaCl, 10 mM Tris–HCl, 10 mM MgCl2, 1 mM dithiothreitol, pH 7.9 at 25°C) in a total volume of 20 μl at 37°C. One unit of Klenow fragment is defined as the amount of enzyme required to convert 10 nM of dNTPs to an acid-insoluble material in 30 min at 37°C. The reactions were allowed to proceed for 0, 0.25, 0.5, 1, 2, 5, 10 and 20 min and stopped using equal volumes of Maxam–Gilbert loading buffer (98% formamide, 0.01 M ethylenediaminetetraacetic acid, 1 mg/ml xylene cyanol and 1 mg/ml bromophenol blue). The reaction products were electrophoresed on 20% (vol/vol) denaturing polyacrylamide gels and visualized and quantified using a phosphorimager (Molecular Dynamics, Sunnyvale, CA) and the ImageQuant 5.0 software (Molecular Dynamics). The same duplexes were used to determine the Km and Vmax of dCTP and CldCTP incorporation. Extension assays were conducted with 0.01, 0.02, 0.05, 0.1, 0.5 and 1 μM dCTP or CldCTP, 100 μM dNTPs, 0.025 U Klenow fragment and 2.5 pmol of labeled substrate duplexes. The reactions were allowed to proceed for 2 min. Reactions were then stopped, visualized and quantified as above. Non-linear regression to the fit the Michealis–Menten equation was done using SigmaPlot 10 to determine Km and Vmax.
For the polymerase extension assays, the 23mer primer, derived from the hamster hprt promoter, was 5′-32P-end labeled by T4 polynucleotide kinase (New England Biolabs) with [γ-32P]adenosine triphosphate (MP Biomedicals) under conditions recommended by the enzyme supplier and subsequently purified using G50 Sephadex columns (Roche). The labeled 23mer primer was annealed with a 2-fold excess of the complementary 30mer strand in 100 mM Tris–HCl pH 7.0 and incubated at 95°C for 5 min to create a duplex with a 5′ overhang, providing a template for Klenow fragment (Figure 2b). A time course of 2′-deoxyguanosine-5′-triphosphate (dGTP) incorporation opposite cytosine or 5-chlorocytosine was conducted. Briefly, the labeled substrate (2.5 pmol) was incubated with 0.025 U Klenow fragment in the presence of 100 μM dNTPs and 1 μM dGTP in 1× NEBuffer 2 (50 mM NaCl, 10 mM Tris–HCl, 10 mM MgCl2, 1 mM dithiothreitol, pH 7.9 at 25°C) in a total volume of 20 μl at 37°C. The reactions were also allowed to proceed for 0, 0.5, 1, 2, 5, 10, 20 and 60 min then stopped using an equal volume of Maxam–Gilbert loading buffer (98% formamide, 0.01 M ethylenediaminetetraacetic acid, 1 mg/ml xylene cyanol and 1 mg/ml bromophenol blue). The reaction products were electrophoresed on 20% (vol/vol) denaturing polyacrylamide gels and visualized and quantified as described above.
Electroporation of CHO-K1 cells with triphosphates and selection of hprt-silenced cells
Electroporation was done under conditions described previously by Nyce (22). CHO-K1 cells were obtained from American Type Culture Collection, Manassas, VA (product number CCL-61) and routinely grown at 37°C with 5% CO2 in Ham's F12 with 2 mM L-glutamine supplemented with 10% fetal bovine serum, 10 000 U/ml streptomycin and 10 000 μg/ml penicillin, which will from this point forward be referred to as Ham's F12 complete medium. Trypsinized cells were obtained from a log-phase culture and washed twice with phosphate-buffered saline pH 7.4, and 2.5 × 106 cells were resuspended in 0.5 ml buffer containing 1 mM of either MedCTP (Amersham Biosciences, Piscataway, NJ), CldCTP or dCTP (Sigma, St. Louis, MO). After the cells were incubated on ice for 10 min, electroporation was executed using 0.4 cm cuvettes with a BioRad Gene Pulser II with Capacitance Extender II Module set at 950 μF and 200 V. Post-treatment, cells were again incubated on ice for 10 min. Cells were recovered in Ham's F12 complete medium for ∼5–7 days before plating 1.0 × 10-5 cells in selective medium, Ham's F12 complete medium containing 15 μg/ml 6-thioguanine (6TG) (MB Biomedicals). Cells were grown in selective media for ∼15 days, when cells electroporated with MedCTP were confluent. The length of selection on average was 15 days. Viable colonies would generally appear at 7 days. These experiments were conducted in triplicate.
Reversion of hprt silenced CHO-K1 cells with 5-aza-2′-deoxycytidine
Cells that survived the selective medium with 6TG were subcultured before plating on Ham's F12 complete medium containing 5 μM 5-aza-2′-deoxycytidine (DAC) (Sigma) for 48 h, after that the medium was replaced with Ham's F12 complete medium and allowed to recover. Cells were trypsinized and then plated (3 × 105) in the following selective medium: hypoxantine aminopterin thymidine (HAT) (Ham's F12 complete medium with 100 μM hypoxanthine, 0.4 μM aminopterin and 16 μM thymidine), Ham's F 12 complete medium with 2 μg/ml 6TG and Ham's F12 complete medium. The selection process proceeded on average for 10 days; essentially, cells that survived were grown until confluence. Most cells grown in HAT media and in both DAC treated and untreated cells were plated in this manner in order to assay for reversion of hprt silencing. These experiments were repeated in triplicate.
Bisulfite pyrosequencing
Total DNA from the CHO-K1 cells after dCTP, MedCTP and CldCTP electroporation, recovery and 6TG selection were extracted using a Qiagen DNeasy Kit according to the manufacturer's protocol. Total DNA was also isolated from 6TG-selected cells after treatment with DAC and growth in selective HAT medium. These samples were then sent for bisulfite pyrosequencing (Biotage, Charlottesville, VA) to assay for methylation status within a 290 base region (−264 to +75, where +1 is A of ATG transcription start site, National Center for Biotechnology Information nucleotide X53073), containing 38 CpG sites that includes the transcription start site and portions of exon 1 and intron 1.
One microgram of total DNA was bisulfite converted using the EZ DNA Methylation kit (Zymo Research, Orange, CA) according to the manufacturer's protocol. A minimum of 100 ng of DNA before bisulfite treatment was used for the DNA control.
The following primers were used for polymerase chain reaction (PCR) amplification of the region of interest: Forward—5′-GTATTTGGTTTTAGGAGTTATTTAG-3′ and Reverse with 5′ biotin modification—5′-ACCYCCTAAACCTAACTATCC-3′. Briefly, in 50 μl reactions, 100 ng of DNA was used along with 10 pmol of forward primer as well as 10 pmol of the reverse 5′ biotin modified primer, 1× PCR buffer II (Qiagen, Valencia, CA), 1.5 mM MgCl2, 1.5 U Qiagen Hotstar Taq DNA polymerase (Qiagen) and 200 μM of each dNTP with the following PCR conditions: 94°C 15 min; 45 times (95°C 30 s; 51°C 30 s; 72°C 30 s); 72°C 5 min; 4°C infinity.
Solid-phase single-stranded DNA (ssDNA) templates were prepared according to the standard pyrosequencing sample preparation protocol at Biotage. Briefly, PCR products (10 μl of a 50 μl reaction) were captured on streptavidin sepharose high performance beads (GE Healthcare, Piscataway, NJ), washed with 70% EtOH then subsequently denatured with 0.2 N NaOH to produce ssDNA that was washed once more. The beads were then released with the ssDNA in to an annealing solution that contained 0.5 μM of the pyrosequencing primer. Samples were annealed at 80°C for 2 min. Samples were then analyzed via pyrosequencing preformed on PSQ96 HS system. Six separate pyrosequencing reactions were run in this assay to determine the methylation status for all 38 CpG sites. The following primers were used for pyrosequencing of the forward strand: CpG sites 1–7, 5′-TTGGTTTTAGGAGTTATTTA-3′; CpG sites 8–16, 5′-GTTTATTTTYGTYGTTAGGG-3′; CpG sites17–21, 5′-GGTAGYGGYGTTTGTGAT-3′; CpG sites 22–30, 5′-AGTTAGTTGGGTTTATTTTA-3′; CpG sites 31–34, 5′-GAGGGTTTTTTTTTTATA-3′ and CpG sites 35–38, 5′-GTTATGGAGATTAGTAGTTTTA-3′. Methylation at selected sites in the reverse strand was obtained using the previously mentioned PCR amplification primers and the following primers for pyrosequencing of the reverse strand: CpG sites 1–8, 5′-GAGGGTTTTTTTTTTATA-3′; CpG sites 9–15, 5′-AGTTAGTTGGGTTTATTTTA-3′; CpG sites13–20, 5′-GGTAGYGGYGTTTGTGAT-3′; CpG sites 21–25, 5′-GTTTATTTTYGTYGTTAGGG-3′ and CpG sites 26–36, 5′-TTGGTTTTAGGAGTTATTTA-3′.
Isotope dilution mass spectrometry
The global methylation status was measured with isotope dilution mass spectrometry using isotope-enriched cytosine and 5-methylcytosine standards (27). Total DNA was extracted from CHO-K1 parental cells as well as the MedCTP- and CldCTP-electroporated cells using the DNeasy Blood and Tissue Kit (Qiagen). Briefly, a total of 1 μg of DNA was used with a mixture of stable isotope-labeled internal standards consisting of 5-methylcytosine (15N2) and cytosine (15N2). The mixture was dried under reduced pressure, hydrolyzed with 88% formic acid at 140°C for 40 min and then derivatized in 20 μl anhydrous acetonitrile and 20 μl N-methyl-N(tert-butyldimethylsilyl)trifluoroacetamide with 1% tert-buthyldiamethylchlorosilane (Pierce, Rockford, IL) at 140°C for 40 min. The samples were then analyzed via GC–MS in the selected ion mode. The 5-methylcytosine to total cytosine ratios were measured in triplicate.
Results
The studies presented here were designed to test whether 5-chlorocytosine could mimic 5-methylcytosine in the DNA of replicating cells, as has been observed in in vitro systems (17,18). To accomplish these goals, the 5′-triphosphate analog of 5-chloro-2′-deoxycytidine (CldCTP) was chemically synthesized, and the product was verified by standard analytical methods including electrospray mass spectrometry (Figure 1). 5-Methyl-2′-deoxycytidine (MedCTP) and 2′-deoxycytidine (dCTP) were obtained from Amersham Biosciences and Sigma, respectively.
It was first established that CldCTP could serve as a substrate for DNA polymerase in an in vitro model system. We show that exonuclease-deficient Klenow polymerase incorporated dCTP and CldCTP opposite a template guanine residue with virtually indistinguishable kinetic parameters (Figure 2a). The apparent Km's for the substrates tested were 0.13 ± 0.01, 0.12 ± 0.02 (0.025 Units) with Vmax of 14.4 ± 0.03 and 13.2 ± 0.6.
In order to determine if 5-chlorocytosine could serve as a template for Klenow exo minus polymerase, an oligonucleotide was synthesized containing a 5-chlorocytosine residue and tested as a template. The data show that the polymerase inserts guanine opposite template 5-chlorocytosine residue and does not distinguish between 5-chlorocytosine (ClC) and cytosine in the template strand (Figure 2b). These in vitro results are not unexpected as it has long been established that the halogenated uracil analog, 5-bromouracil, could be substituted for thymine in DNA–protein interactions (28,29) and the replacement of cytosine by a 5-chlorocytosine residue has negligible impact on oligonucleotide melting temperatures (30).
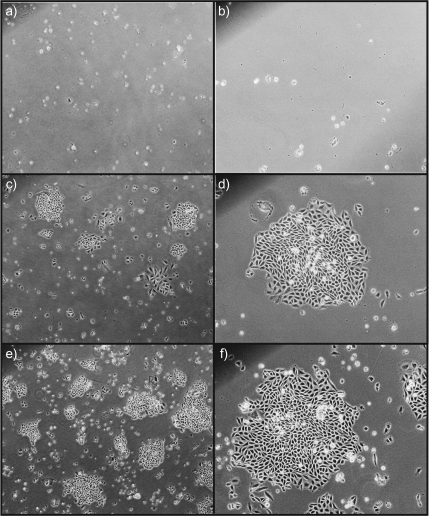
To test the capacity of 5-chlorocytosine to mimic 5-methylcytosine in the DNA of dividing vertebrate cells, CHO-K1 cells were electroporated with either dCTP, MedCTP or CldCTP. Following electroporation, cells were grown in medium containing the purine analog, 6TG, which is metabolized by HPRT to the nucleotide and subsequently incorporated into nucleic acids. The presence of 6TG interferes with the structure and function of nucleic acids, resulting in cell toxicity. If the hprt gene has been silenced, 6TG is not metabolized and cells survive. The results of this study reveal that only cells electroporated with either MedCTP or CldCTP survived 6TG selection, whereas all cells electroporated with dCTP did not (Figure 3). The results presented here are consistent with previous electroporation studies with MedCTP and dCTP (22–24). Attempts were made to quantify the amount of modified triphosphate incorporated into the cells after electroporation; however, as the electroporation process itself kills off the vast majority of cells, the amount of DNA available for analysis is below our limit of detection by GC–MS. Although the remaining viable cells after electroporation undergo cell division, the initial amount of 5-chlorocytosine that was incorporated will be continuously diluted by semi-conservative DNA replication and therefore cannot be measured by GC–MS methods.
Fig. 3.
HPRT silenced CHO-K1 cells after electroporation and selection with 6TG. dCTP-electroporated cells after 6TG selection, (a) ×4 magnification with no colonies seen and (b) ×10 magnification with no colonies seen. MedCTP-electroporated cells after 6TG selection, (c) ×4 magnification with several colonies seen and (d) ×10 magnification of one colony. CldCTP-electroporated cells after 6TG selection, (e) ×4 magnification with several colonies seen and (f) ×10 magnification of one colony.
Having established that the incorporation of 5-chlorocytosine could result in the silencing of the hprt gene, we sought to determine if expression could be reactivated by using the methyltransferase inhibitor DAC. Cells that survived growth in the 6TG-containing media were treated with the methyltransferase inhibitor DAC. DAC is a nucleoside analog that is metabolized to the nucleotide and incorporated into DNA where it forms a covalent-suicide complex with DNA methyltransferase (31,32). If the hprt gene had been silenced in the surviving cells by methylation, as opposed to mutation or gene disruption, then treatment with DAC would result in demethylation and gene reactivation. Subsequent selective HAT media allow only for survival of cells that have re-expressed HPRT. HAT media inhibits de novo nucleotide synthesis as it contains aminopterin, a dihydrofolate reductase inhibitor. For survival in HAT media, the hprt gene must be active, allowing for a functioning nucleotide salvage pathway to uptake the hypoxanthine present in the HAT media. If the hprt gene was silenced due to mutation or gene disruption, then the cells would not survive HAT selection. The results show that cells rendered resistant to 6TG by either MedCTP or CldCTP electroporation survived HAT selection following treatment with DAC, confirming that hypermethylation was the mechanism of hprt gene silencing in these cells.
If MedCTP and CldCTP indeed silenced the hprt gene by promoter methylation, methylation should be detectable in the promoter region by bisulfite sequencing. In order to confirm that changes in methylation in the hprt promoter occurred in parallel with the changes in HPRT expression, the methylation status of a 290 bp region within the promoter of the hprt gene was assessed for the CHO-K1 cells not subject to electroporation, as well as all cells surviving selection in the 6TG media and cells surviving HAT media after treatment with DAC. In the bisulfite sequencing method, cytosine residues in DNA are converted to uracil, whereas 5-methylcytosine residues are inert. Bisulfite pyrosequencing allows quantitative determination of the extent of methylation at a particular sequence position. The results reveal that prior to electroporation only four of the 38 methylation sites studied in the promoter region of the hprt gene were methylated, and on average at a low level of <1% (Figure 4). After electroporation with MedCTP and 6TG selection, the average methylation across the 38 CpG sites was 78% with a standard deviation between sites of 19%. The results obtained from the DNA of cells after electroporation with CldCTP and 6TG selection show an increase in methylation that was comparable with that found with the MedCTP-electroporated cells. Across the 38 CpG sites, the average methylation was 83% with a standard deviation between sites of 16%. These data show that, like 5-methylcytosine, 5-chlorocytosine in DNA can promote increased methylation in progeny cells.
Fig. 4.
Bisulfite pyrosequencing reveals increased promoter methylation after electroporation of CldCTP and MedCTP that is reversible with DAC treatment. The methylation status of CpG sites within the 290 bases within the promoter region of the hprt gene for CHO-K1 cells without treatment (white), CHO-K1 cells electroporated with MedCTP post-6TG selection (gray), CHO-K1 cells electroporated with CldCTP post-6TG selection (black), MedCTP-electroporated CHO-K1 cells after 6TG selection, DAC treatment and HAT selection (white with stripes) and CldCTP-electroporated CHO-K1 cells after 6TG selection, DAC treatment and HAT selection (gray with stripes) is shown. The percent methylation (y-axis) is shown for 38 CpG sites. The first CpG is located −236 bases upstream of the transcription start site. The last CpG assayed is 54 bases downstream of the transcription start site. All CpGs in between were assayed and are numbered consecutively from 1 to 38. Asterisk represents first CpG after transcription start site. Note that methylation levels for CHO-K1 cells without treatment are on average <1%, therefore may blend into the x-axis in the figure.
Bisulfite pyrosequencing data obtained from the promoter region of DAC-treated, HAT-selected cells show a drastic decrease in methylation of the hprt promoter region (Figure 4). The average percent methylation across the 38 CpG sites decreased from 78% to 42% in the MedCTP-electroporated cells and from 83% to 52% in the CldCTP-electroporated cells. These data are in accord with the phenotypic change in HPRT expression leading to survival in HAT media.
The results from our assay of the global methylation level (5-methylcytosine to total cytosine ratio) of CHO-K1 cells prior to electroporation with MedCTP and CldCTP show a baseline of 1.74 ± 0.03% within the genome of the parental cells. Global 5-methylcytosine levels were measured following electroporation with MedCTP or CldCTP and selection with 6TG were found to be 1.78 ± 0.15% and 1.59 ± 0.06%, respectively. Electroporation and selection therefore did not increase global 5-methylcytosine levels. However, after treatment with DAC, the global methylation is reduced to 0.61 ± 0.04% and 0.95 ± 0.01% for MedCTP- and CldCTP-electroporated cells, respectively.
Discussion
Methylation of cytosine residues in promoter regions has long been correlated with transcriptional silencing. A cause and effect relationship between methylation and gene silencing was substantially strengthened by studies demonstrating that MedCTP, introduced into cells, could result in gene silencing and that methyltransferase inhibitors could cause demethylation, resulting in gene reactivation (21–24). Emerging evidence now is providing a more complete, but more complicated picture of the role of methylation in gene silencing. Methylation of cytosine residues within CpG dinucleotides substantially increases the affinity of methylation-sensitive DNA-binding proteins (33–35). Methylation-sensitive DNA-binding proteins then recruit histone-modifying enzymes, which covalently modify histone proteins, generating compact chromatin, less accessible to transcription (3,4). As yet unanswered questions include how the initial methylation patterns are created in the process of cell differentiation, how these patterns are dynamically modified and how epigenetic patterns are perturbed in tumor cells. Of special interest are mechanisms that would result in inappropriate methylation of previously unmethylated sites, resulting in transcriptional silencing of tumor suppressor genes.
In parallel with these epigenetic studies, substantial efforts have been directed toward understanding pathways for DNA damage and repair. An important focus in cancer research has been on damage from endogenous reactive molecules including reactive oxygen species, reactive nitrogen species and most recently reactive halogen species such as hypochlorous acid that can cause a multitude of DNA damage products (36–43). Among the many alkylation, oxidation, hydrolysis and halogenation damage products examined to date, several have the potential to interfere with the binding of methylation-sensitive DNA-binding proteins or to interfere with DNA methyltransferases (17,18,33,44). The only endogenous damage products yet to emerge that might cause aberrant methylation are the 5-halocytosine analogs, 5-chlorocytosine and 5-bromocytosine generated by activated neutrophils and eosinophils, respectively.
In in vitro studies, it has been demonstrated that methylation-sensitive DNA-binding proteins cannot distinguish 5-halocytosines from 5-methylcytosine (17,18). In cells, this molecular mimicry could potentially result in the recruitment of histone-modifiying enzymes. In vitro studies have also demonstrated that the hemimethylated CpG dinucleotide, a substrate resembling a methylated region following DNA replication, is the preferred substrate for the mammalian maintenance methyltransferase (18,45,46). Like the methylation-sensitive DNA-binding proteins, the human maintenance methylase cannot distinguish halogenated from methylated cytosine residues (18).
In this study, the incorporation of 5-chlorocytosine in DNA is shown to cause changes in gene expression in replicating mammalian cells. The studies described here parallel the pioneering studies on the impact of electroporation of MedCTP on gene activity (22,23). In those studies, it was shown that MedCTP introduced into cells is incorporated into genomic DNA and that its incorporation alters methylation patterns and a population of cells could be selected with 6TG that had silenced the hprt gene secondary to methylation of the promoter. In the studies reported here, we observed that only cells electroporated with MedCTP or CldCTP, not dCTP, were able to survive 6TG selection. This result with replicating cells is consistent with the capacity of 5-chlorocytosine to mimic 5-methylcytosine as established in in vitro biochemical assays.
The incorporation of 5-chlorocytosine into DNA was shown to alter gene expression by epigenetic mechanisms. The capacity of nucleotide analogs to alter gene expression could result from either methylation changes or by mutagenic mechanisms. If the changes result from methylation-induced silencing, the silenced gene should be reactivated by exposure of cells to the demethylating agent DAC. In accord with previous studies, we confirmed here that MedCTP-induced silencing of the hprt gene could be reversed by treatment with DAC and selection in HAT media, indicating that the MedCTP-induced gene silencing resulted from increased methylation. Identical results were obtained with CldCTP, indicating that, like MedCTP, the incorporation of CldCTP results in increased local DNA methylation. If either of the analogs examined here had diminished expression of functional HPRT activity by mutagenic mechanisms, the silenced genes could not have been reactivated by treatment with DAC.
The incorporation of 5-chlorocytosine into DNA resulted in increased levels of methylation within the hprt gene promoter. In parallel with the functional assays described above, we sought to establish that incorporation of the nucleotide analogs increased methylation in the promoter of the hprt gene after only one electroporation event. Recently developed bisulfite pyrosequencing methods allow the extent of methylation at target cytosine residues to be measured (47). We observed that methylation of cytosine residues in the hprt gene indeed results from both MedCTP and CldCTP electroporation. In both cases, the increased extent and distribution of local methylation, followed by the decline in methylation following DAC treatment are indistinguishable. Upon the basis of the data reported here, however, we cannot determine which cytosine residues must be methylated in order to silence the hprt gene.
Although incorporation of both CldCTP and MedCTP resulted in silencing of the hprt gene and associated with increased methylation of the hprt promoter, we did not observe overall increases in total 5-methylcytosine content in the DNA. DAC treatment resulted in reactivation of the hprt gene associated with decreased methylation of the hprt promoter and significant decline of total 5-methylcytosine levels, in both cases.
In the experiments described here, electroporated nucleotide analogs would become part of the intracellular nucleotide pools and become available for incorporation into the replicating DNA. The cytosine analogs 5-chlorocytosine and 5-methylcytosine are incorporated into random positions along the DNA, potentially increasing DNA methylation by two mechanisms. In the first mechanism, DNA containing the analogs would bind with higher affinity methylation-sensitive DNA-binding proteins, and this complex could then recruit histone-modifying enzymes. In subsequent rounds of cell replication, the chromatin regions with the modified histones might provide preferred targets for DNA methyltransferases, thus propagating the initial modifications.
In a second potential mechanism, the 5-chloro or 5-methylcytosine residue incorporated into DNA during the initial cell replication would serve as the parental strand in a subsequent round of DNA and cell replication. The hemimethylated or hemi-chlorinated parental strand would then serve as a high-affinity target for the maintenance methyltransferase, generating a symmetrically modified CpG site. Having established aberrant methylation at this site, the pattern would be propagated at each cell replication. Both mechanisms are consistent with our previous in vitro studies (17,18). The studies described here cannot determine if one or both mechanisms account for the increased gene methylation observed in the progeny cells.
The formation of 5-chlorocytosine in DNA may provide a link between inflammation and cancer development. The development of cancer is a complex process involving multiple DNA changes in any given tumor. Agents that damage DNA tend to promote cancer development, and it is traditionally thought that damage-related changes result from damage-induced genetic mutations or chromosomal rearrangements. Growing evidence indicates that most human tumors also harbor epigenetic methylation changes that result in increased expression of tumor promoters or, more frequently, decreased expression of tumor suppressors (6,7).
The mechanisms by which pathologic epigenetic changes occur, as well as how DNA damage may be related to this process has yet to be fully elucidated. Among the various types of DNA damage, many would be predicted to interfere with DNA–protein interactions, potentially resulting in loss of methylation signals in progeny cells and inappropriate gene activation. In contrast, only one type of DNA damage identified to date, the 5-halocytosines, can mimic the endogenous methylation signal associated with gene silencing. Results of previous in vitro studies with 5-chlorocytosine have been recapitulated here in replicating mammalian cells, confirming the hypothesis that aberrant chlorination of cytosine residues in DNA could result in increased cytosine methylation and gene silencing that can be heritably transmitted to progeny cells.
Inflammation is one of the factors often associated with cancer development (9,10), although a mechanistic link has not been previously proposed. Reactive species from activated neutrophils could result in substantial DNA damage, depending upon local concentrations and the duration of exposure. Of the many damage products formed, most would be potentially mutagenic if not repaired. Although most of the promutagenic HOCl-mediated damage products would be efficiently repaired in normal cells, no repair activity has been identified for 5-chlorocytosine (25,30), perhaps because it is such good mimic of 5-methylcytosine, as shown by this series of experiments. The 5-chlorocytosine formed in DNA, either from direct modification of cytosine or incorporation of a modified nucleotide, could thus accumulate in a given cell with repeated exposure to inflammation. The accumulated halogenation product would then be predicted to cause increased methylation and ultimately gene silencing. Cell replication in tissues exposed to inflammation could create selective pressure for cells with altered patterns of gene expression.
We show here that the inflammation-mediated DNA damage product 5-chlorocytosine could have harmful and heritable biological consequences. 5-Chlorocytosine in DNA is recognized by cellular machinery as 5-methylcytosine, allowing this damage product to alter methylation patterns. Furthermore, because it cannot be distinguished from 5-methylcytosine, it is probably to accumulate in cellular DNA over time.
The studies here demonstrate that the incorporation of 5-chlorocytosine into DNA can increase methylation and associated transcriptional silencing of a target gene. We did not observe significant increases in global 5-methylcytosine levels, suggesting that selective pressure could influence the genomic location of methylation changes in progeny cells. In the studies reported here, 5-chlorocytosine-induced methylation changes occur acutely, whereas the role of ClC in the development of human cancer is probably to be the result of its accumulation overtime in areas exposed to increased inflammatory changes. It has recently been determined that ClC is the most likely product to form in double-stranded DNA as a result of HOCl attack (14). Furthermore, there is evidence that ClC occurs more frequently at CpG sites as compared with non-CpG sites (14). The incorporation of CldCTP from a damaged triphosphate pool may also contribute to the formation of ClC in DNA; however, this pathway has yet to be fully investigated. The results of the studies described here further establish a role for inflammation in epigenetic gene silencing and will hopefully lead to future studies that may help further understand the relationship between inflammation and cancer development.
Funding
Grant support: National Institutes of Health and National Cancer Institute. V.V.Lao is supported in part by Loma Linda University School of Medicine Medical Scientist Training Program.
Acknowledgments
The authors thank Ana Dumitrescu for her technical assistance. Funding to pay the Open Access publication charges for this article was provided by Loma Linda University School of Medicine.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- CldCTP
5-chloro-2′-deoxycytidine-5′-triphosphate
- DAC
5-aza-2′-deoxycytidine
- dCTP
2′-deoxycytidine-5′-triphosphate
- dNTP
deoxynucleoside triphosphate
- GC–MS
gas chromatography–mass spectrometry
- HAT
hypoxantine aminopterin thymidine
- hprt
hypoxanthine-guanine phosphoribosyltransferase
- MedCTP
5-methyl-2′-deoxycytidine-5′-triphosphate
- PCR
polymerase chain reaction
- 6TG
6-thioguanine
References
- 1.Feinberg AP, et al. The history of cancer epigenetics. Nat. Rev. Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 2.Esteller M. Aberrant DNA methylation as a cancer-inducing mechanism. Annu. Rev. Pharmacol. Toxicol. 2005;45:629–656. doi: 10.1146/annurev.pharmtox.45.120403.095832. [DOI] [PubMed] [Google Scholar]
- 3.Klose RJ, et al. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 4.Grønbaek K, et al. Epigenetic changes in cancer. APMIS. 2007;115:1039–1059. doi: 10.1111/j.1600-0463.2007.apm_636.xml.x. [DOI] [PubMed] [Google Scholar]
- 5.Herman JG, et al. Gene silencing in cancer in association with promoter hypermethylation. N. Engl. J. Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 6.Feinberg AP, et al. Hypomethylation of ras oncogenes in primary human cancers. Biochem. Biophys. Res. Commun. 1983;111:47–54. doi: 10.1016/s0006-291x(83)80115-6. [DOI] [PubMed] [Google Scholar]
- 7.Ehrlich M. DNA hypomethylation, cancer, the immunodeficiency, centromeric region instability, facial anomalies syndrome and chromosomal rearrangements. J. Nutr. 2002;132:2424S–2429S. doi: 10.1093/jn/132.8.2424S. [DOI] [PubMed] [Google Scholar]
- 8.Office of Portfolio Analysis and Strategic Initiatives National Institutes of Health. [homepage on the internet]. NIH Roadmap for Medical Research. http://nihroadmap.nih.gov/epigenomics/ (18 April 2008, date last accessed) [Google Scholar]
- 9.Ames BN, et al. The causes and prevention of cancer. Proc. Natl Acad. Sci. USA. 1995;92:5258–5265. doi: 10.1073/pnas.92.12.5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marnett LJ. Oxyradicals and DNA damage. Carcinogenesis. 2000;21:361–370. doi: 10.1093/carcin/21.3.361. [DOI] [PubMed] [Google Scholar]
- 11.Buss IH, et al. 3-Chlorotyrosine as a marker of protein damage by myeloperoxidase in tracheal aspirates from preterm infants: association with adverse respiratory outcome. Pediatr. Res. 2003;53:455–462. doi: 10.1203/01.PDR.0000050655.25689.CE. [DOI] [PubMed] [Google Scholar]
- 12.Gaut JP, et al. Neutrophils employ the myeloperoxidase system to generate antimicrobial brominating and chlorinating oxidants during sepsis. Proc. Natl Acad. Sci. USA. 2001;98:11961–11966. doi: 10.1073/pnas.211190298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamb NJ, et al. Oxidative damage to proteins of bronchoalveolar lavage fluid in patients with acute respiratory distress syndrome: evidence for neutrophil-mediated hydroxylation, nitration, and chlorination. Crit. Care Med. 1999;27:1738–1744. doi: 10.1097/00003246-199909000-00007. [DOI] [PubMed] [Google Scholar]
- 14.Kang JI, Jr, et al. Examination of hypochlorous acid-induced damage to cytosine residues in a CpG dinucleotide in DNA. Chem. Res. Toxicol. 2008;6:1211–1218. doi: 10.1021/tx800037h. [DOI] [PubMed] [Google Scholar]
- 15.Badouard C, et al. Detection of chlorinated DNA and RNA nucleosides by HPLC coupled to tandem mass spectrometry as potential biomarkers of inflammation. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2005;827:26–31. doi: 10.1016/j.jchromb.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 16.Whiteman M, et al. Hypochlorous acid-induced base modifications in isolated calf thymus DNA. Chem. Res. Toxicol. 1997;10:1240–1246. doi: 10.1021/tx970086i. [DOI] [PubMed] [Google Scholar]
- 17.Valinluck V, et al. 5-Halogenated pyrimidine lesions within a CpG sequence context mimic 5-methylcytosine by enhancing the binding of the methyl-CpG-binding domain of methyl-CpG-binding protein 2 (MeCP2) Nucleic Acids Res. 2005;33:3057–3064. doi: 10.1093/nar/gki612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valinluck V, et al. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Res. 2007;67:946–950. doi: 10.1158/0008-5472.CAN-06-3123. [DOI] [PubMed] [Google Scholar]
- 19.Jekunen A, et al. 5-Methyl-2′-deoxycytidine. Metabolism and effects on cell lethality studied with human leukemic cells in vitro. Mol. Pharmacol. 1983;25:431–435. [PubMed] [Google Scholar]
- 20.Jekunen A, et al. Exclusion of exogenous 5-methyl-2′-deoxycytidine from DNA in human leukemic cells. A study with [2(-14)C]- and [methyl-14C]5-methyl-2′-deoxycytidine. Biochem. Pharmacol. 1983;32:1165–1168. doi: 10.1016/0006-2952(83)90265-4. [DOI] [PubMed] [Google Scholar]
- 21.Chen J, et al. Direct induction of DNA hypermethylation in sea urchin embryos by microinjection of 5-methyl dCTP stimulates early histone gene expression and leads to developmental arrest. Dev. Biol. 1993;155:75–86. doi: 10.1006/dbio.1993.1008. [DOI] [PubMed] [Google Scholar]
- 22.Nyce J. Gene silencing in mammalian cells by direct incorporation of electroporated 5-methyl-2′-deoxycytidine 5′-triphosphate. Somat. Cell Mol. Genet. 1991;17:543–550. doi: 10.1007/BF01233619. [DOI] [PubMed] [Google Scholar]
- 23.Holliday R, et al. Gene silencing in mammalian cells by uptake of 5-methyl deoxycytidine-5′-triphosphate. Somat. Cell Mol. Genet. 1991;17:537–542. doi: 10.1007/BF01233618. [DOI] [PubMed] [Google Scholar]
- 24.Holliday R, et al. DNA methylation and epigenetic inheritance. Methods. 2002;27:179–183. doi: 10.1016/s1046-2023(02)00072-5. [DOI] [PubMed] [Google Scholar]
- 25.Kang JI, Jr, et al. Synthesis and characterization of oligonucleotides containing 5-chlorocytosine. Chem. Res. Toxicol. 2004;17:1236–1244. doi: 10.1021/tx0498962. [DOI] [PubMed] [Google Scholar]
- 26.Ludwig J. A new route to nucleoside 5′-triphosphates. Acta Biochim. Biophys. Acad. Sci. Hung. 1981;16:131–133. [PubMed] [Google Scholar]
- 27.Burdzy A, et al. Synthesis of stable-isotope enriched 5-methylpyrimidines and their use as probes of base reactivity in DNA. Nucleic Acids Res. 2002;18:4068–4074. doi: 10.1093/nar/gkf520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zamenhof S, et al. Studies on thymine-5-bromouracil “exchange” in deoxyribonucleic acids of Escherichia coli. J. Biol. Chem. 1959;234:2960–2964. [PubMed] [Google Scholar]
- 29.Kanner L, et al. Efficiency of utilization of thymine and 5-bromouracil for normal and repair DNA synthesis in bacteria. Biochim. Biophys. Acta. 1968;157:532–545. doi: 10.1016/0005-2787(68)90151-2. [DOI] [PubMed] [Google Scholar]
- 30.Valinluck V, et al. Impact of cytosine 5-halogens on the interaction of DNA with restriction endonucleases and methyltransferase. Chem. Res. Toxicol. 2006;19:556–562. doi: 10.1021/tx050341w. [DOI] [PubMed] [Google Scholar]
- 31.Creusot F, et al. Inhibition of DNA methyltransferase and induction of Friend erythroleukemia cell differentiation by 5-azacytidine and 5-aza-2′-deoxycytidine. J. Biol. Chem. 1982;257:2041–2048. [PubMed] [Google Scholar]
- 32.Taylor SM, et al. Mechanism of action of eukaryotic DNA methyltransferase. Use of 5-azacytosine-containing DNA. J. Mol. Biol. 1982;162:679–692. doi: 10.1016/0022-2836(82)90395-3. [DOI] [PubMed] [Google Scholar]
- 33.Valinluck V, et al. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of MeCP2. Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nan X, et al. Dissection of the methyl-CpG binding domain from the chromosomal protein MeCP2. Nucleic Acids Res. 1993;21:4886–4892. doi: 10.1093/nar/21.21.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Free A, et al. DNA recognition by the methyl-CpG binding domain of MeCP2. J. Biol. Chem. 2001;276:3353–3360. doi: 10.1074/jbc.M007224200. [DOI] [PubMed] [Google Scholar]
- 36.Henderson JP, et al. Molecular chlorine generated by the myeloperoxidase-hydrogen peroxide-chloride system of phagocytes produces 5-chlorocytosine in bacterial RNA. J. Biol. Chem. 1999;274:33440–33448. doi: 10.1074/jbc.274.47.33440. [DOI] [PubMed] [Google Scholar]
- 37.Henderson JP, et al. Production of brominating intermediates by myeloperoxidase. A transhalogenation pathway for generating mutagenic nucleobases during inflammation. J. Biol. Chem. 2001;276:7867–7875. doi: 10.1074/jbc.M005379200. [DOI] [PubMed] [Google Scholar]
- 38.Whiteman M, et al. Hypochlorous acid-induced base modification: potentiation by nitrite: biomarkers of DNA damage by reactive oxygen species. Biochem. Biophys. Res. Commun. 1999;257:572–576. doi: 10.1006/bbrc.1999.0448. [DOI] [PubMed] [Google Scholar]
- 39.Harrison JE, et al. Studies on the chlorinating activity of myeloperoxidase. J. Biol. Chem. 1976;251:1371–1374. [PubMed] [Google Scholar]
- 40.Foote CS, et al. Assessment of chlorination by human neutrophils. Nature. 1983;301:715–716. doi: 10.1038/301715a0. [DOI] [PubMed] [Google Scholar]
- 41.Kettle AJ. Neutrophils convert tyrosyl residues in albumin to chlorotyrosine. FEBS Lett. 1996;379:103–106. doi: 10.1016/0014-5793(95)01494-2. [DOI] [PubMed] [Google Scholar]
- 42.Klebanoff SJ. Myeloperoxidase: friend and foe. J. Leukoc. Biol. 2005;77:598–625. doi: 10.1189/jlb.1204697. [DOI] [PubMed] [Google Scholar]
- 43.King CC, et al. Secretion and inactivation of myeloperoxidase by isolated neutrophils. J. Leukoc. Biol. 2002;61:239–302. doi: 10.1002/jlb.61.3.293. [DOI] [PubMed] [Google Scholar]
- 44.Turk PW, et al. DNA adduct 8-hydroxyl-2′-deoxyguanosine (8-hydroxyguanine) affects function of human DNA methyltransferase. Carcinogenesis. 1995;16:1253–1255. doi: 10.1093/carcin/16.5.1253. [DOI] [PubMed] [Google Scholar]
- 45.Pradhan S, et al. Recombinant human DNA (cytosine-5) methyltransferase. I. Expression, purification, and comparison of de novo and maintenance methylation. J. Biol. Chem. 1999;274:33002–33010. doi: 10.1074/jbc.274.46.33002. [DOI] [PubMed] [Google Scholar]
- 46.Bacolla A, et al. Recombinant human DNA (cytosine-5) methyltransferase.II. Steady-state kinetics reveal allosteric activation by methylated dna. J. Biol. Chem. 1999;274:33011–33019. doi: 10.1074/jbc.274.46.33011. [DOI] [PubMed] [Google Scholar]
- 47.Tost J, et al. DNA methylation analysis by pyrosequencing. Nat. Protoc. 2007;2:2265–2275. doi: 10.1038/nprot.2007.314. [DOI] [PubMed] [Google Scholar]