Abstract
The bacterial Nramp family protein MntH is a divalent metal transporter, but mntH mutants have little or no phenotype in organisms where it has been studied. Here, we identify the mntH homolog of Bradyrhizobium japonicum, and demonstrate that it is essential for Mn2+ transport and for maintenance of cellular manganese homeostasis. Transport activity was induced under manganese deficiency, and Fe2+ did not compete with 54Mn2+ for uptake by cells. The steady state level of mntH mRNA was negatively regulated by manganese, but was unaffected by iron. Control of mntH expression and Mn2+ transport by manganese was lost in a fur strain, resulting in constitutively high activity. Fur protected a 35 bp region of the mntH promoter in DNase I footprinting analysis that includes three imperfect direct repeat hexamers that are needed for full occupancy. Mn2+ increased the affinity of Fur for the mntH promoter by over 50-fold, with a Kd value of 2.2 nM in the presence of metal. The findings identify MntH as the major Mn2+ transporter in B. japonicum, and show that Fur is a manganese-responsive regulator in that organism. Furthermore, Fe2+ is neither a substrate for MntH nor a regulator of mntH expression in vivo.
Keywords: bradyrhizobium, manganese, metal homeostasis
INTRODUCTION
Manganese is required for many biological processes as an enzyme cofactor, and serves as a protectant from oxidative stress. However, manganese can be toxic at high concentrations (Kehres and Maguire, 2003; Moore and Helmann, 2005; Papp-Wallace and Maguire, 2006; Que and Helmann, 2000), and thus manganese homeostasis is maintained in part by regulating acquisition of the metal from the environment. Manganese uptake by bacterial cells proceeds by the divalent metal transporters SitABCD (MntABCD) and MntH. SitABCD is an ABC-type transport complex that was initially described as an Fe2+ transporter (Bearden et al., 1998; Zhou et al., 1999), but is now known to transport Mn2+ as well. The high affinity of Salmonella enterica serovar Typhimurum SitABCD for Mn2+ compared to Fe2+ supports the argument that it functions physiologically as only a Mn2+ transporter (Kehres et al., 2002a). However, abrogation of Fe2+ transport in that organism requires mutation of sitABCD as well as the ferrous iron transporter gene feoB (Boyer et al., 2002; Perry et al., 2007; Sabri et al., 2008). In addition, iron-related phenotypes of sitABCD mutants in other bacteria have been described (Bearden et al., 1998; Runyen-Janecky et al., 2003; Sabri et al., 2008; Zhou et al., 1999).
MntH is a bacterial member of the Nramp family of divalent cation transporters originally described in eukaryotes (Horsburgh et al., 2002; Kehres et al., 2000; Makui et al., 2000; Que and Helmann, 2000). Like SitABCD, MntH has preferential affinity for Mn2+, but can also take up Fe2+ (Kehres et al., 2000; Makui et al., 2000). Bacterial mntH mutants generally do not have a growth defect or other phenotypes associated with manganese deficiency (Kehres et al., 2000; Makui et al., 2000), and characterization is carried out in strains where mntH is overexpressed from a plasmid. The lack of strong phenotypes is due in some cases to the presence of sitABCD, which carries out a similar function (Boyer et al., 2002; Horsburgh et al., 2002).
In numerous bacterial species, the sitABCD and mntH genes are regulated by Mn2+ via MntR, a transcriptional repressor that binds to target promoters when complexed with the metal (Horsburgh et al., 2002; Kehres et al., 2002b; Patzer and Hantke, 2001; Que and Helmann, 2000). These metal transporter genes are also repressed in the presence of Fe2+ due to the activity of the Fur repressor (Guedon et al., 2003; Kehres et al., 2002b; Patzer and Hantke, 2001). Whereas the physiological significance of Fe2+ transport by MntH or SitABCD has been questioned (Kehres et al., 2000; Kehres et al., 2002a), the regulation of mntH or sitABCD by Fe2+ is specific in the model systems E. coli, S. enterica and B. subtilis, and underscores the integration of iron and manganese metabolism in those bacteria.
We are interested in understanding manganese homeostasis in Bradyrhizobium japonicum, a bacterium that lives as a free-living soil organism, or as the endosymbiont of soybean. B. japonicum belongs to the α-Proteobacteria, a diverse taxonomic group of gram negative bacteria containing numerous members that form close or intracellular associations with eukaryotic hosts in a symbiotic or pathogenic context. The sitABCD operon has been described in Sinorhizobium meliloti (Chao et al., 2004; Davies and Walker, 2007; Platero et al., 2004; Platero et al., 2003). A sitA mutant has a growth deficiency in metal-deplete media and other phenotypes that are rescued by manganese supplementation. Thus, SitABCD is likely to be the major Mn2+ transporter in those species. Interestingly, Fur mediates Mn2+ control of the sitABCD operon in S. meliloti (Chao et al., 2004; Platero et al., 2004; Platero et al., 2007), Rhizobium leguminosarum (Diaz-Mireles et al., 2004) and Agrobacterium tumefaciens (Kitphati et al., 2007). The Fur homolog has been renamed Mur in those rhizobial species, but it shows iron responsiveness in A. tumefaciens (Kitphati et al., 2007). The Fur homolog in B. japonicum has thus far been characterized only as an iron responsive regulator (Friedman and O’Brian, 2004; Yang et al., 2006b), and was originally identified by complementation of an E. coli Fur mutant (Hamza et al., 1999).
In the present study, we show the mntH gene is essential for Mn2+ transport, growth in manganese-deficient media and for maintenance of manganese homeostasis. Furthermore, Fur mediates Mn2+-dependent control of mntH gene expression, thus a new role for B. japonicum Fur as a Mn2+-responsive regulator is described.
RESULTS
B. japonicum has a manganese-regulated Mn2+ transport activity
Most studies that characterize bacterial Mn2+ transport directly in detail are carried out in cells in which a transport gene is expressed from on a high copy plasmid (Kehres et al., 2000; Makui et al., 2000). Although informative, that work cannot assess the physiological Mn2+ transport activity, its regulation, or the contribution of a particular transporter to that activity. Therefore, we wanted to ask the simple question of whether B. japonicum has a high affinity Mn2+ transport activity, and whether it is affected by cellular exposure to manganese. Mn2+ uptake was measured in cells grown in high manganese (50 μM MnCl2) or low manganese (1 μM MnCl2) media using 5 nM 54Mn2+ as the initial substrate concentration (Fig. 1A). Cells grown in high manganese medium had very low Mn2+ uptake activity. However, uptake was observed in cells grown in low manganese medium, showing that B. japonicum has a manganese-dependent Mn2+ uptake activity.
Fig. 1. Mn2+ uptake by B. japonicum.
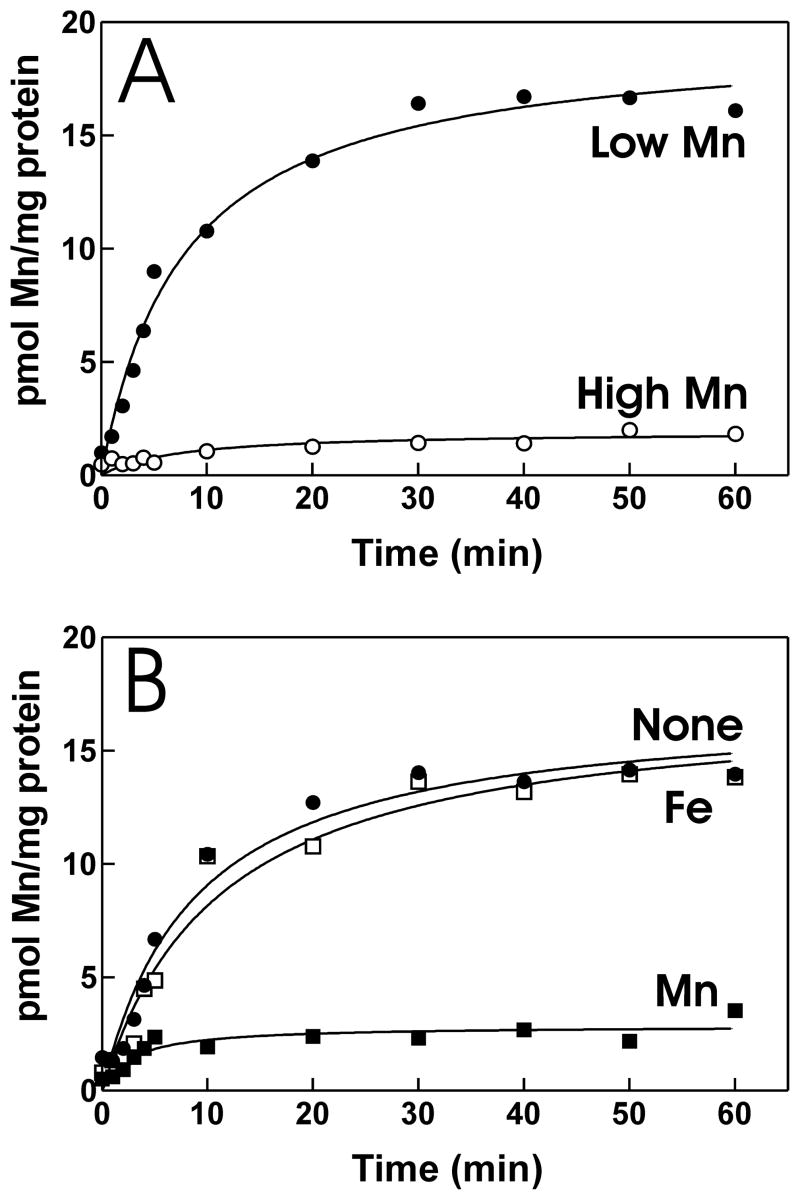
A. 54Mn2+ uptake was measured in parent strain USDA110 in cells grown in media containing 1 μM (closed circles) or 50 μM MnCl2 (open circles). At time zero, 5 nM 54Mn2+ was added to the cells in assay medium, and aliquots were subsequently taken at various time points and counted. B. Competition of 54Mn2+ uptake with unlabeled Mn2+ or Fe2+. Cells of the parent strain were grown in media containing no added manganese to induce Mn2+ transport activity. The assay medium containing cells was supplemented with either no metal (closed circles) or with 1 μM MnCl2 (closed squares) or 1 μM FeSO4 (open squares) immediately prior to addition of 5 nM 54Mn.
The Mn2+ uptake system does not transport Fe2+
Transport systems that take up Mn2+ also take up Fe2+, and can be regulated by both metals. We examined whether 54Mn2+ was taken up by as system that also transports Fe2+ by competition experiments with unlabeled metals (Fig. 1B). 1 μM unlabeled Mn2+ inhibited uptake of 5 nM 54Mn2+, but 1 μM Fe2+ did not. This suggests that Mn2+ and Fe2+ are not taken up by a common transport system.
The mntH homolog bll5044 is essential for high affinity Mn2+ transport activity
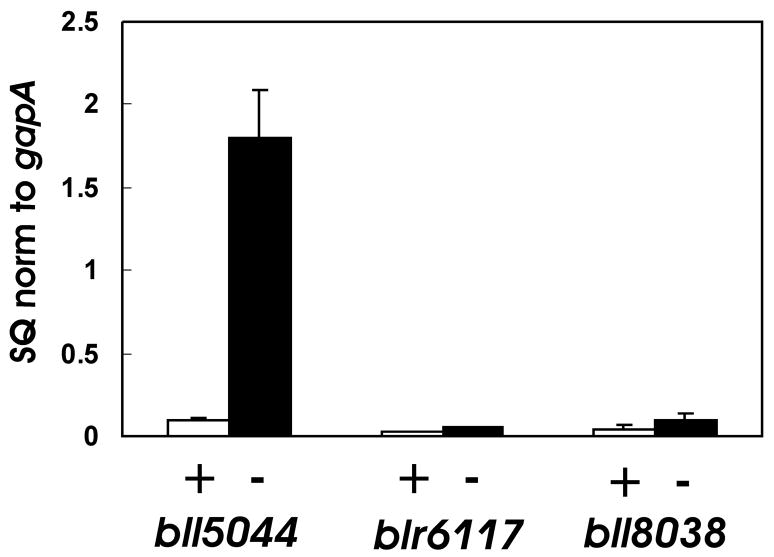
Mn2+ is likely to be transported by SitABCD in Sinorhizobium meliloti as judged by the rescue of growth and other phenotypes of sitA mutants with Mn2+ (Chao et al., 2004; Davies and Walker, 2007; Platero et al., 2003). However, B. japonicum genome does not contain an obvious sitABCD operon homolog (but see Discussion). A BLAST search using E. coli MntH identified Bll5044 as a putative protein with 40% identity to it, and Blr6117 and Bll8038 with 25% identity. To further narrow down a candidate Mn2+ transporter gene, we examined mRNA levels of bll5044, blr6117 and bll8038 in cells grown in high or low Mn2+ media by quantitative real time PCR (qPCR) (Fig. 2). Whereas blr6117 and bll8038 mRNA levels were low independent of the Mn2+ status, bll5044 message was high in cells grown in low manganese media, and very low in the presence of the metal. Thus, bll5044 is a manganese-regulated gene, and its expression correlated with Mn2+ transport activity (Fig. 1A).
Fig. 2. Manganese-dependent expression of putative mntH homologs.
mRNAs were analyzed by quantitative real-time PCR from cells grown in media supplemented with no manganese (solid bars) or with 50 μM MnCl2 (open bars). The data are expressed as the relative starting quantity (SQ) of the respective mRNAs normalized to the housekeeping gene gapA. The data are expressed as the average of three replicates with error bars representing the standard deviation.
We constructed a mutant strain defective in the bll5044 gene such that the open reading frame was deleted and replaced with an Ω cassette. We measured high affinity Mn2+ transport activity in the mutant and the parent strain grown in low manganese media (1 μM MnCl2) (Fig. 3A), since this was the lowest manganese concentration in which the mutant grew (see below). The transport activity observed in the parent strain was almost completely abolished in the mutant. Thus, bll5044 is required for Mn2+ uptake, and is a structural and functional homolog of mntH. Accordingly, bll5044 was designated mntH.
Fig. 3. Mn2+ uptake by B. japonicum mutants.
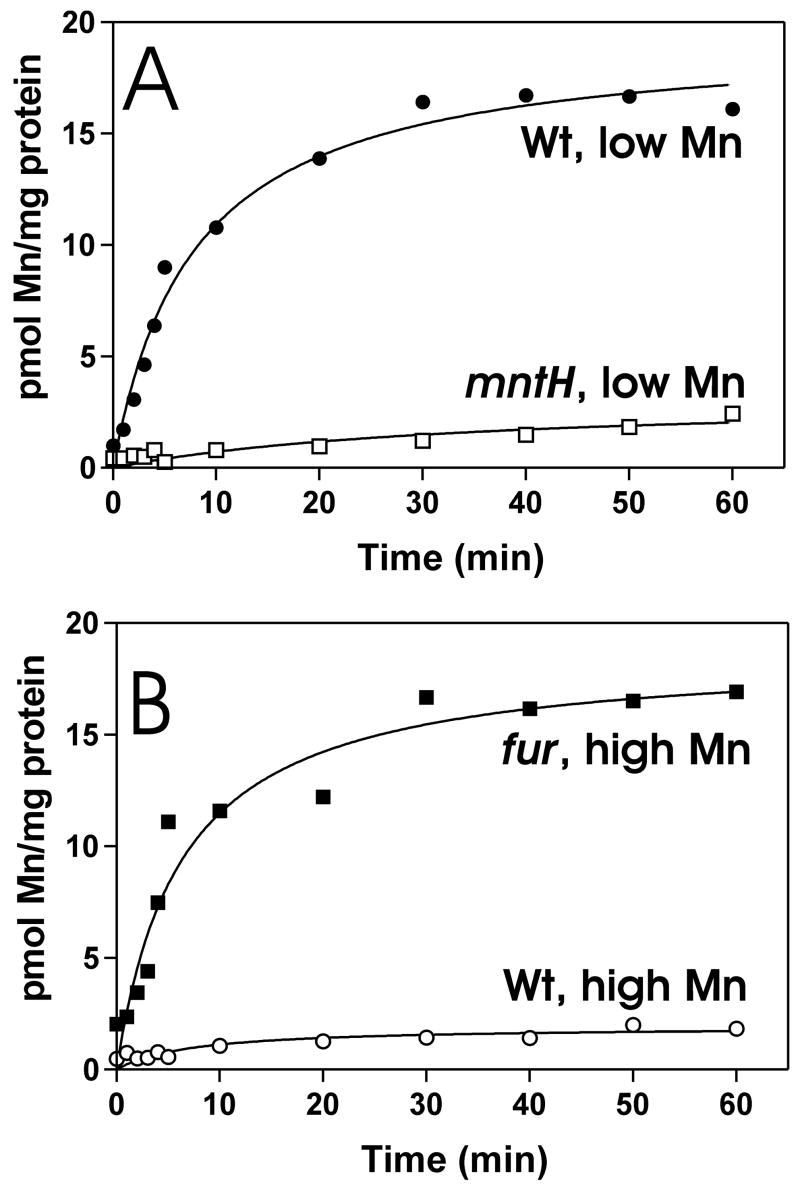
A. Cells of the parent strain (closed circles) or the mntH mutant strain (open squares) were grown in media containing 1 μM MnCl2. At time zero, 5 nM 54Mn was added to the assay medium and aliquots were subsequently taken at various time points and counted. The uptake data of the parent strain cells grown in low manganese are the same as in Fig. 1A, and are shown in two different panels for clarity of presentation.
B. Cells of the parent strain (open circles) or the fur mutant (closed squares) were grown in media containing 50 μM MnCl2. At time zero, 5 nM 54Mn was added to the assay medium and aliquots were subsequently taken at various time points and counted. The uptake data of the parent strain cells grown in high manganese media are the same as in Fig. 1A, and are shown in two different panels for clarity of presentation.
The mntH gene is required for growth and maintenance of manganese homeostasis under manganese deficiency
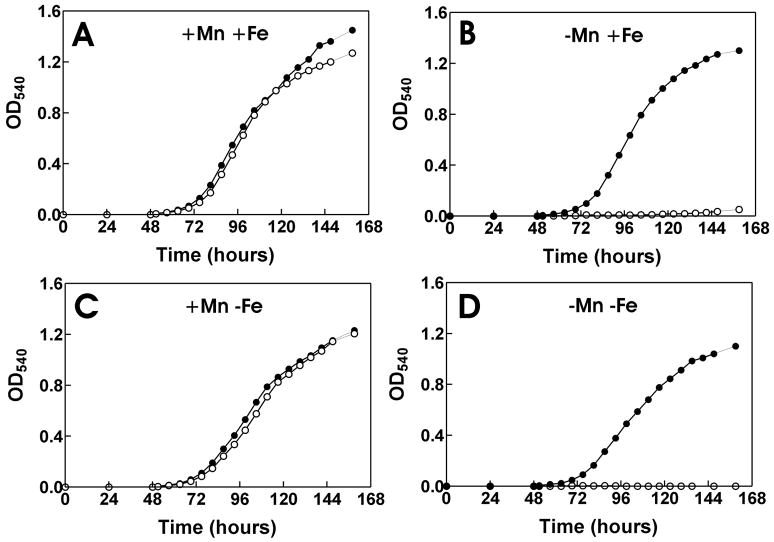
The data indicate that MntH is responsible for most of the observed Mn2+ transport activity in cells under the growth conditions tested (Fig. 3A), and therefore we predict that the mntH strain would be manganese-deficient. To test this, we examined the growth of the parent and mutant strains in manganese-deficient and –replete media (Fig. 4). The mntH mutant grew well in media supplemented with 50 μM MnCl2, but did not grow in media with no added manganese (0.2 μM final concentration). The parent strain grew well under either media condition, and thus mntH is necessary for growth in manganese-deficient medium. In other bacteria, MntH is reported to transport iron as well as manganese. To verify that that rescue of the mntH strain by 50 μM MnCl2 was not due to iron contamination, we carried out the growth studies in low and high iron media (Fig. 4). Iron did not rescue the growth phenotype of the mntH strain in low manganese medium, and the mutant grew in high or low iron conditions as long as manganese was present. FeCl3 was used in Fig. 4, and FeSO4 yielded the same results. We conclude that the growth phenotype of the mntH strain is due to a manganese deficiency.
Fig. 4. Dependence on manganese or iron supplementation for growth of B. japonicum parent or mntH strains.
Growth media were inoculated with 5 × 105 cells/ml of parent strain (closed circles) or the mntH mutant (open circles) and grown in media containing (A) 50 μM MnCl2 and 20 μM FeCl3, (B) no exogenous manganese and 20 μM FeCl3, (C) 50 μM MnCl2 and no exogenous iron, or (D) no exogenous manganese and no exogenous iron. Unsupplemented media contains 0.2 μM and 0.3 μM manganese and iron, respectively. Aliquots were taken at the indicated time points and the optical density was measured at 540 nm (OD540).
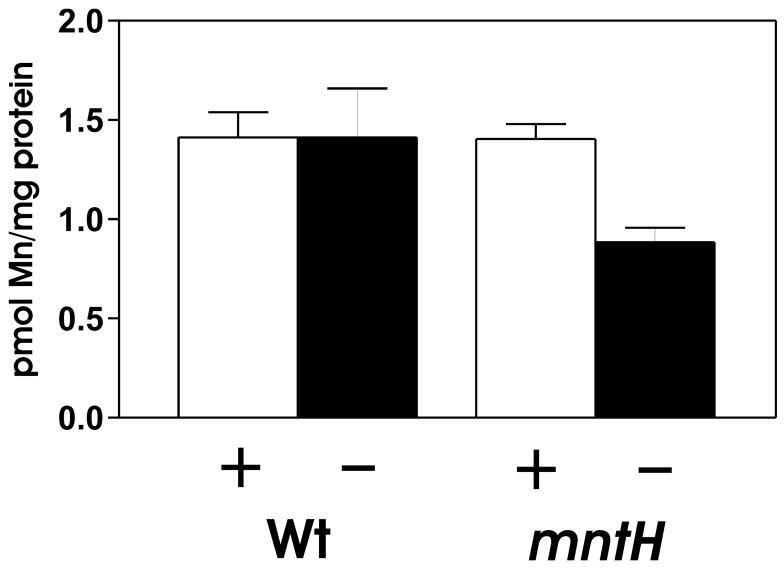
The intracellular manganese content was determined for the parent and mntH strains grown in high (50 μM) or low (1 μM) manganese media. The manganese content in the parent strain was approximately the same in high or low manganese media (Fig. 5), indicating that B. japonicum is able to regulate its cellular manganese content over at least a 50-fold range of extracellular manganese. However, the cellular manganese level of the mntH mutant was only about 60% of the wild type when grown in low manganese media. The experimental design, by necessity, underestimates the importance of mntH on manganese homeostasis because we must grow the cells at a manganese concentration (1 μM) that allows some MntH-independent uptake of Mn2+. This lower affinity uptake activity is not observed in assays (Fig. 3) because 5 nM of 54Mn2+ is used as the initial substrate concentration. Nevertheless, these observations, along with the growth phenotype of the mutant, shows that mntH is required to maintain manganese homeostasis under low manganese conditions.
Fig. 5. Cellular manganese content of the parent strain and mntH mutant grown in low or high manganese media.
Cells were grown in media supplemented with 1 μM MnCl2 (−, solid bars) or 50 μM MnCl2 (+, open bars). Manganese content of whole cells was determined by absorption spectroscopy. The data are based on triplicate samples with error bars representing the standard deviation.
Mn2+ transport and mntH expression are deregulated in a fur mutant
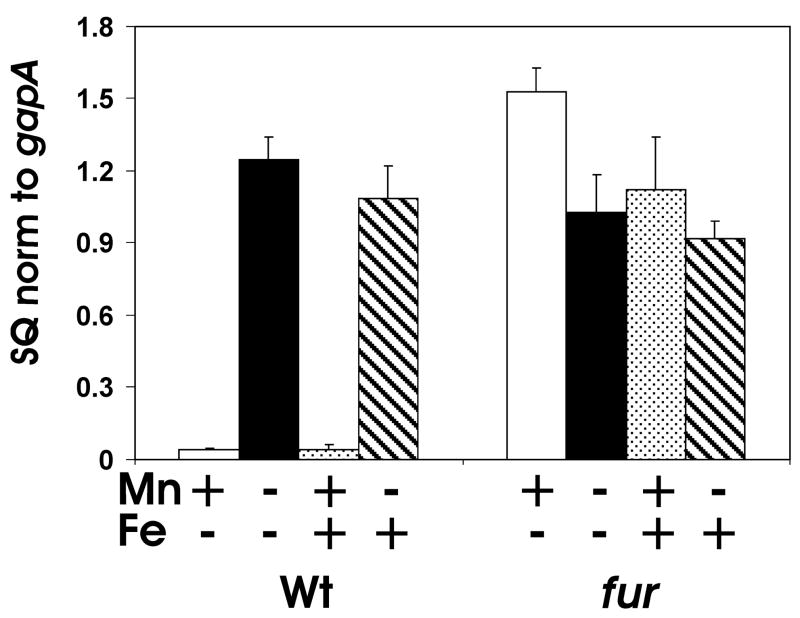
Manganese transport genes are regulated by Mn2+ via MntR in numerous bacterial species (Horsburgh et al., 2002; Kehres et al., 2002b; Patzer and Hantke, 2001; Que and Helmann, 2000), but B. japonicum and most rhizobial species do not have an mntR gene homolog in their genomes. However, microarray analysis revealed that bll5044 (mntH) in B. japonicum is strongly regulated by Fur (Yang et al., 2006b). Also, the Fur-like protein Mur mediates control of sitABCD in S. meliloti and R. leguminosarum (Chao et al., 2004; Diaz-Mireles et al., 2004; Platero et al., 2004). Thus, we examined control of mntH in a B. japonicum fur strain by qPCR (Fig. 6). The strong Mn2+-dependent expression of mntH transcript observed in the parent strain was lost in the fur mutant, showing high levels of transcript in the presence of Mn2+. Thus, Fur normally negatively affects mntH expression in the presence of Mn2+.
Fig. 6. Regulation of mntH gene mRNA by manganese and iron in the parent strain and a fur mutant.
mRNAs were analyzed by quantitative real-time PCR from cells grown in media supplemented with no metal (black bars), 50 μM MnCl2 (white bars), 20 μM FeCl3 (striped bars) or both MnCl2 and FeCl3 (stippled bars). The data are expressed as the relative starting quantity (SQ) of the respective mRNAs normalized to the housekeeping gene gapA, and presented as the average of triplicate samples ± the standard deviation.
To further address the role of Fur on manganese metabolism, we examined high affinity Mn2+ transport in a fur strain (Fig. 3B). Transport of 54Mn was very low in wild type cells grown in high manganese media, but was high in the fur strain, comparable to the activity of the parent strain grown in manganese-deficient media. Collectively, the observations show that Mn2+ transport is under Fur control via manganese-dependent regulation of the mntH gene.
The mntH gene is not regulated by iron
Although Fur mediates iron control of numerous B. japonicum genes (Friedman and O’Brian, 2004; Yang et al., 2006b), and mntH is iron regulated in numerous bacteria (Guedon et al., 2003; Kehres et al., 2002b; Patzer and Hantke, 2001), it was not controlled by iron in the parent strain (Fig. 6). Furthermore, its aberrant expression in the fur mutant is not substantially affected by the iron status (Fig. 6). We conclude that mntH is not an iron-responsive gene, and that Fur controls this gene only in response to manganese.
Fur binds directly to the mntH promoter in vitro with high affinity in a metal-dependent manner
To determine whether Fur is a direct regulator of mntH, we determined the transcription start site using 5′ RACE and then analyzed the region upstream of it. DNaseI footprinting analysis showed that recombinant Fur bound the mntH promoter in the presence of Mn2+, with maximum occupancy in the −56 to −22 region relative to the transcription start site (+1) (Fig. 7). In addition, a series of double-stranded oligonucleotides that collectively cover 171 bp upstream of the mntH transcription start site were analyzed by electrophoretic gel mobility shift assays (EMSA). The smallest DNA fragment tested that bound Fur was a 39 bp fragment that included −56 to −22 region, and no fragments lacking this region bound to Fur (data not shown).
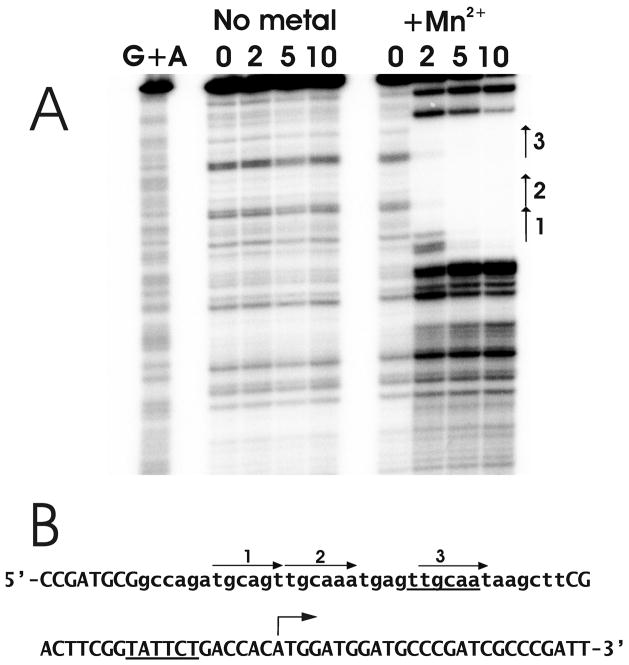
Fig. 7. DNase I Footprinting of the mntH promoter with B. japonicum Fur in the presence and absence of Mn2+.
A. Protection of DNA from DNase I digestion by Fur was carried out in the presence or absence of MnCl2 using 0, 2, 5 or 10 nM Fur. The DNA was radiolabeled at the 5′ end of the non-template strand with respect to the mntH gene, and thus the 3′ end is at the top of the gel. The arrows on the right denote the sites of the three hexamer repeats.
B. The sequence of the protected region of mntH and the direct repeats are shown. The lower case letters show the protected region. The bent arrow represents the transcription start site. The direct repeats are shown by the straight arrows. The underlined sequences show the putative −10 and −35 regions of the promoter.
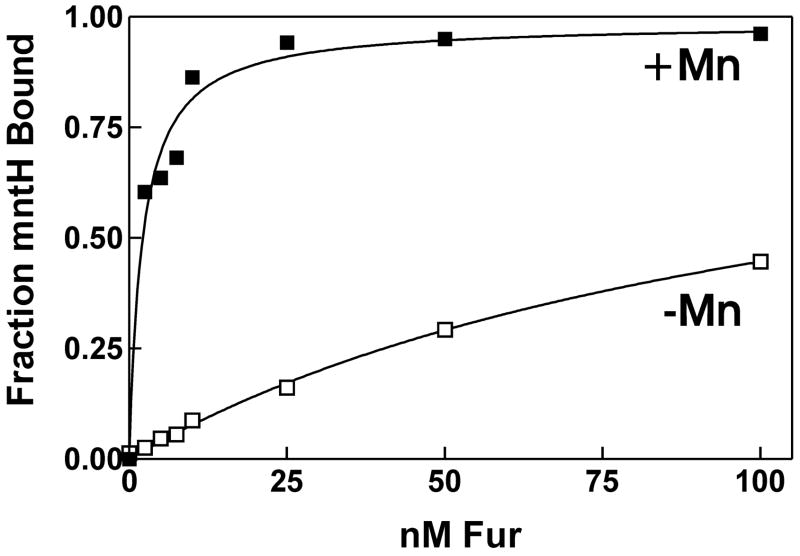
To determine the affinity of Fur for its binding site on the mntH promoter, binding reactions containing 0.1 nM radiolabeled DNA were titrated with Fur, and bound DNA was analyzed by EMSA (Fig. 8) The dissociation binding constant (Kd) was 2.2 nM in the presence of Mn2+, which is consistent with a role for Fur as a regulator of mntH.
Fig. 8. Effect of manganese on B. japonicum Fur binding to the mntH promoter.
EMSA was carried out using 100 pM 32P-labelled mntH promoter DNA titrated with various concentrations of Fur in the presence (closed squares) or absence (open squares) of 100 μM MnCl2. Bound and unbound DNA was resolved on a 5% non-denaturing polyacrylamide gel and visualized by autoradiography. Autoradiograms were scanned, and bands were quantified to determine bound and unbound DNA.
Mn2+ transport activity and mntH expressions are regulated by manganese. Based on the current findings and the present understanding of Fur family proteins, the observed control is likely due to sensing Mn2+ by Fur through direct interactions of the protein with the metal, which affects DNA binding activity. This was addressed in several ways. Using atomic absorption spectroscopy, we established that manganese binds Fur directly, with a stoichiometry of 0.96 ± 0.01 manganese atoms per Fur monomer. The effect of metal on mntH promoter occupancy by Fur was assessed by DNase footprinting (Fig. 7). No protection was observed in the absence of metal in the binding reaction, but Fur bound the promoter in the presence of Mn2+, showing that binding is metal-dependent. Furthermore, the affinity of Fur for mntH promoter DNA in the absence of metal was approximately 110 nM, over 50-fold weaker than in the presence of Mn2+ (Fig. 8). We conclude that Fur mediates Mn2+-dependent control of the mntH gene.
The Fur binding site on the mntH promoter contains three imperfect direct repeat hexamers
EMSA analysis using 50 nM Fur shows a high mobility complex (HMC) and a low mobility complex (LMC), but an HMC is predominant with 5 nM Fur (Fig. 9). This is similar to what was observed for Fur binding to the irr promoter, which corresponded to a dimer and a tetramer or two dimers (Friedman and O’Brian, 2003). Comparison of the mntH and irr promoters by EMSA using the DNA probes of equal size (39 bp) revealed that the HMC and LMC ran with the same mobilities, indicating that the mntH promoter is also occupied as a dimer at low Fur concentrations and two dimers or tetramer at high Fur concentrations (data not shown).
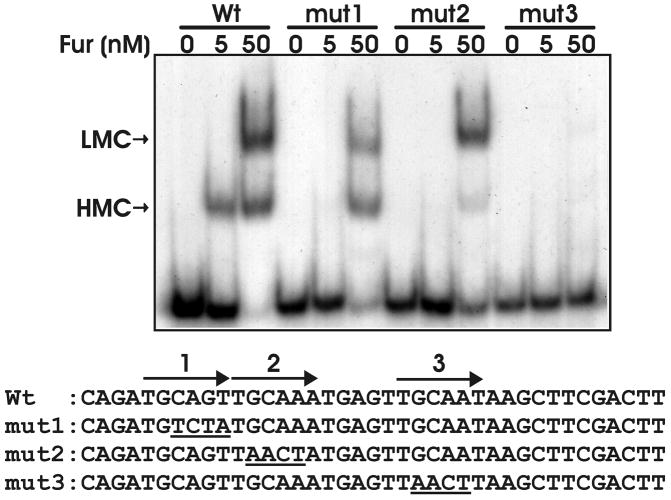
Fig 9. Effect of mutation of direct repeat sequences of the mntH promoter on formation of a high mobility complex (HMC) and low mobility complex (LMC) with Fur.
EMSAs were carried out using purified recombinant Fur (0, 5, or 50 nM) and either the wild type (Wt) 39-bp mntH promoter region DNA or DNA containing substitution mutations in one of the direct repeat sequence DNA. The complexes were resolved on a 5% non-denaturing polyacrylamide gel and visualized by autoradiography.
Examination of the protected region of DNA on the mntH promoter or the minimal binding DNA in EMSA shows that the sequence contains three imperfect direct repeat hexamers, similar to that observed within the irr promoter (Friedman and O’Brian, 2003). However, the Fur/Mur binding site of the S. meliloti or R. leguminosarum sitA promoter best fits a palindromic inverted repeat sequence (Diaz-Mireles et al., 2004; Platero et al., 2007) (Fig. 9), and a bioinformatic search for Fur/Mur binding sites in the rhizobia also defines the consensus as palindrome as well (Rodionov et al., 2006). Thus, we determined the effects of mutation of each repeat within the mntH promoter on Fur binding using EMSA (Fig. 9). Substitution mutations within direct repeat 1 or 2 (mut1 or mut2) abrogated binding with 5 nM Fur as observed by the loss of the HMC, but DNA was bound with 50 nM Fur. Mutation of the direct repeat 3 (mut3) abrogated Fur binding completely with 5 or 50 nM Fur in the binding reaction. These observations and previous work (Friedman and O’Brian, 2003) support the conclusion that the Fur binding site is best described as a three direct repeat sequence of hexamers in B. japonicum.
The mntH gene is not required for development of symbiosis with soybean
The parent strain and mntH mutant were used to inoculate soybean seedlings, and symbiotic properties of the resultant nodules from plants 26 days post-inoculation. The parent strain elicited an average of 49.3±6.4 nodules per plant, and the mutant 52.2±3.6 nodules per plant (n=5). Acetylene reduction activity, which is a measurement of nitrogen fixation activity, was 10.6±1.2 and 12.8±2.0 μmol ethylene produced/h/g nodule fresh weight for nodules produced from the parent strain and mntH mutant, respectively. Thus, mntH is not essential for the development of nodules or for symbiotic nitrogen fixation activity.
DISCUSSION
In the present study, we established that B. japonicum can maintain manganese homeostasis, as observed by an almost constant intracellular manganese level when grown in 1 μM or 50 μM manganese (Fig. 5). We identified bll5044 as the mntH homolog in B. japonicum, and demonstrate that it encodes the primary Mn2+ transporter in that organism. MntH contributes to homeostasis as observed by the lower manganese level in the mntH strain compared to the wild type (Fig. 5). Consistent with this, the mutant was unviable in media containing 0.2 μM manganese, a concentration sufficient for growth of the parent strain (Fig. 4). Mn2+ transport activity was induced under manganese limitation in the parent strain, and this activity was abrogated in the mntH strain (Figs. 1A, 3A). Regulation of this activity was found to be due to control by the Fur protein, which binds directly to the mntH promoter in the presence of Mn2+ to repress gene expression (Figs. 3B, 6, 7). Thus, Fur is a Mn2+-responsive regulator in B. japonicum.
Two additional weak mntH homologs, bll6117 and bll8038, were also identified based on sequence similarity. We are not aware of any bacterium that has multiple mntH genes. It is possible that they transport a different cation, but the genes were expressed at very low levels (Fig. 2), and therefore they were not further characterized.
The strong phenotypes observed for the B. japonicum mntH strain are in stark contrast to those described in other bacterial species. In those cases, mntH mutants show approximately wild type levels of growth in metal-limited media, cellular manganese content, Mn2+ transport activity, manganese toxicity and pathogenicity (Boyer et al., 2002; Domenech et al., 2002; Horsburgh et al., 2002; Kehres et al., 2000; Makui et al., 2000; Que and Helmann, 2000). The lack of phenotypes has made it necessary to study the properties of MntH by overexpression from high copy plasmids (Kehres et al., 2000; Makui et al., 2000). S. enterica, B. subtilis. and Staphylococcus aureus have sitABCD as well as mntH, which likely accounts for the lack of phenotypes for the mntH strain. However, sitABCD does not appear to be present in the E. coli K12 genome, and presumably Mn2+ can be taken up by a heretofore unidentified transporter.
Among the rhizobia, sitA mutants of S. meliloti have numerous phenotypes that can be rescued by addition of exogenous manganese (Chao et al., 2004; Davies and Walker, 2007; Platero et al., 2003), strongly supporting a role for SitABCD in manganese acquisition in that organism, and raises the possibility that it is the only Mn2+ transporter expressed. S. meliloti has a weak mntH gene homolog (sma1115) based on similarity to B. japonicum mntH, but it is not homologous over the entire length, and has not been studied. A modest sitA homolog is found in the B. japonicum genome (bll7769, 32% identity to S. meliloti sitA) adjacent to two additional putative ABC-type transporter genes. However ABC-type transporter proteins are similar to each other at the amino acid sequence level, and thus homology is usually not sufficient to determine the solute substrate. The Mn2+ transport defect of the mntH strain indicates that bll7769 is not required for uptake, and it may transport a different solute.
We found that B. japonicum mntH is not essential for establishing a symbiosis with soybean. This suggests either that the host provides a sufficiently rich manganese milieu that renders high affinity transport unnecessary, or else the bacterium has another mechanism for manganese acquisition that is not expressed in free living cells. It is intriguing that B. japonicum and S. meliloti use different systems to transport manganese into cells. In Salmonella enterica serovar Typhimurium, MntH activity has a pH optimum of 5.5–6.0, whereas the optimum for SitABCD is greater than 8.0 (Kehres et al., 2002a). This correlates with the optimal growth for soybean, the B. japonicum plant host, at moderately acidic soils (www.nsrl.uiuc.edu/aboutsoy/production02.html), whereas Medicago hosts of S. meliloti prefer more alkaline soils (Garau et al., 2005). Although B. japonicum mntH is not essential for symbiosis with soybean, it is nevertheless plausible that it has adapted to the same niche as its host. Further experiments are needed to test this idea.
Both SitABCD and MntH can utilize Fe2+ as well as Mn2+ in numerous bacterial species, and the genes that encode them are also regulated by iron at the mRNA level (Bearden and Perry, 1999; Boyer et al., 2002; Ikeda et al., 2005; Kehres et al., 2000; Kehres et al., 2002a, b; Makui et al., 2000; Patzer and Hantke, 2001; Runyen-Janecky et al., 2003). However, B. japonicum mntH gene was not regulated by iron (Fig. 6), nor was the growth phenotype of a mntH mutant rescued by iron (Fig. 4). In addition, Fe2+ did not compete with 54Mn2+ for transport of wild type cells even at 500-fold excess of the radiolabeled metal (Fig. 1B). These observations indicate that MntH does not have broad substrate specificity, but is specific for Mn2+.
We demonstrate that Fur mediates manganese-dependent expression of B. japonicum mntH. Fur also controls mntH expression in E. coli (Patzer and Hantke, 2001), S. enterica (Kehres et al., 2002b) and B. subtilis (Guedon et al., 2003), but in those systems Fur mediates regulation by iron, and control by manganese requires MntR. MntR appears to be absent in rhizobia based on homology, with the possible exception of Mesorhizobium loti. B. japonicum Fur has been described previously as an iron-responsive regulator (Friedman and O’Brian, 2004; Yang et al., 2006b), and therefore its role in manganese-dependent gene expression is a novel function. However, the Fur protein in S. meliloti and R. leguminosarum (also called Mur) mediates manganese control of the sitABCD operon (Chao et al., 2004; Diaz-Mireles et al., 2004; Platero et al., 2004; Platero et al., 2007), and is both manganese and iron responsive in Agrobacterium tumefaciens (Kitphati et al., 2007). In vitro characterization of the R. leguminosarum Fur/Mur protein shows that it binds Fe2+, Mn2+ and other divalent metals with similar affinities, and that binding of the holo-protein to DNA requires metal, but is not strongly dependent on which divalent metal is bound (Bellini and Hemmings, 2006). Thus, the basis of metal discrimination appears not to be at the level of binding affinity. It is plausible that metal chaperones play a role in metal discrimination, but there is no direct evidence for it. Although this could be unique to the rhizobia, a role for Fur in both Fe2+ and Mn2+ control of gene expression has been described in Yersinia pestis (Bearden et al., 1998). Guedon et al (Guedon et al., 2003) propose that the effects of Mn2+ on the Fur regulon in B. subtilis is an indirect consequence of disruption of the cellular iron pool. This is clearly not the case in B. japonicum with respect to mntH since the gene is not regulated by iron (Fig. 6).
B. japonicum Fur protected a three imperfect hexameric repeat sequence in footprinting assays (Fig. 7), and mutation of any of the repeats affected Fur binding (Fig. 9), similar to what was previously observed for the irr promoter (Friedman and O’Brian, 2003). Thus, this motif best represents the Fur-binding site in B. japonicum, which differs from the inverted repeat described for the sitA promoter of R. leguminosarum and S. meliloti (Diaz-Mireles et al., 2004; Platero et al., 2007), or predicted based on bioinformatic analysis (Rodionov et al., 2006).
EXPERIMENTAL PROCEDURES
Strains and media
B. japonicum USDA110 was the parent strain used in this study. Strains GEM4 (Hamza et al., 1999) and mntHΩΔ are mutant derivatives of the parent strain containing a DNA cassette encoding spectinomycin and streptomycin replacing the fur and mntH genes, respectively. B. japonicum strains were routinely grown at 29°C in glycerol-salts-yeast extract (GSY) medium as described previously (Frustaci et al., 1991). Strains GEM4 and mntHΩΔ were grown in the presence of 25 μg/ml streptomycin and 100 μg/ml spectinomycin. For low manganese conditions, modified GSY medium was used, containing 0.5 g/L yeast extract instead of 1 g/l, with either no exogenous manganese or 1 μM MnCl2 added prior to growth. For low iron conditions, the same modified GSY medium was used, with no exogenous iron. The actual concentrations of manganese and iron in the unsupplemented media are 0.2 μM and 0.3 μM, respectively, as determined by atomic absorption using a Perkin-Elmer model 1100B atomic absorption spectrometer. High metal medium was supplemented with either 50 μM MnCl2, 20 μM FeCl3, or both.
Manganese uptake assay
Cells were grown to mid-log phase (OD540 0.4–0.6) in low- or high- manganese media, harvested by centrifugation, washed twice and resuspended in uptake buffer (0.2 M MOPS and 2% (w/v) glycerol, pH 6.8) to a OD540 of 0.4. 30 mL of cell solution were placed into a 125-ml Erlenmeyer flask and preincubated 15 minutes at room temperature with shaking. At time zero, 5 nM 54Mn was added to the cell solution. 1-ml aliquots were removed at various time points and added to ice cold quench buffer (0.1 M Tris and 100 μM MnCl2, pH 6.0). The cells were collected immediately after quenching on 0.45μm filters presoaked in quench buffer. Cells were washed with 3 mL ice cold quench buffer and counted using a Wallac 1480 Wizard™ 3″ automatic gamma counter. Internalized 54Mn levels were normalized to protein levels in the cell. 54Mn Uptake Competition Assays were done as described above in the presence or absence of 1 μM MnCl2, or 1 μM FeSO4. The competing metal was added to the cell solution immediately prior to the start of the assay.
Construction of a B. japonicum mntH mutant
An mntH-deletion strain was constructed for this study. The open reading frame of mntH (bll5044) and 500 bp flanking DNA on each side of it, was amplified by PCR using USDA110 genomic DNA as the template, and ligated into pBluescriptSK+. The open reading frame was deleted using inverse PCR as described previously (Panek and O’Brian, 2004) and replaced with an Ω-cassette encoding for streptomycin and spectinomycin resistance (Prentki and Krisch, 1984). The construct was introduced into pLO1 (Lenz et al., 1994), mobilized into USDA110, and selected for double recombinant mutants as previously described (Panek and O’Brian, 2004). Mutants were confirmed using PCR and antibiotic resistance.
Bacterial growth studies
The parent strain and mntHΩΔ were grown in modified GSY medium under high (50 μM MnCl2) or low (0.2 μM) manganese conditions supplemented with or without 20 μM FeCl3 as described above. Growth rates were analyzed by measuring the optical density of cells at 540 nm every six hours until reaching stationary phase.
Determination of intracellular manganese content
Steady state levels of internalized manganese was determined using Atomic absorption spectroscopy. 40 ml of cultures grown to mid-log phase (OD540 0.4–0.6) were harvested by centrifugation. The pellet was washed twice with 40 ml ice cold 0.1 M Tris and once with double distilled metal free water to remove salts. Cells were lysed by resuspending the pellet in 100 μl of 70% HNO3 (J.T. Baker, AAS grade), lightly vortexed and incubated at 75°C for 5 minutes. 1 ml metal-free double distilled water was added to the lysed cells and mixed by vortex. Samples were centrifuged at 13 000 r.p.m. for 5 minutes and the supernatant was analyzed for Mn content. Atomic absorption was performed in the furnace mode on a Perkin Elmer Atomic Absorption Spectrometer model 1100B equipped with a model HGA 700 graphite furnace. All samples were diluted with metal free double distilled water to contain 1% HNO3. A Mn standard curve was set-up by diluting a stock solution of Mn (1mg/ml in 2% HNO3, Perkin Elmer) ranging from 0.1 ng/10 μl to 1.0 ng/10 μl in 1% HNO3. 10 μL of sample was used for each measurement, with each sample being performed in triplicate. Samples were run at 120°C for 40 seconds, 5 second ramp to 1000°C and held for 20 seconds, 2200°C for 5 seconds, and 2300°C for 5 seconds. Mn content was normalized to protein level of the cells. The manganese content of modified GSY medium, containing no exogenous manganese, in 1% HNO3 was measured on a Perkin Elmer Atomic Absorption Spectrometer model 1100B using the protocol above.
Analysis of RNA
Expression levels of selected genes were determined by quantitative real real time PCR (qPCR) with iQ™ SYBR green supermix (Bio-Rad) using iCycler thermal cycler (Bio-Rad). RNA was isolated from B. japonicum cells using a hot phenol method as described previously (Yang et al., 2006a). cDNA was synthesized from 2 μg total RNA using iScript™ cDNA Synthesis Kit (Bio-Rad). Each PCR reaction contained 10 μl 2 × SYBR green supermix, 0.2 μM primers (IDT DNA Technology) and 25ng cDNA in a 20 μl volume. PCR reactions were heated to 95°C for 3 minutes, followed by 40 cycles with steps of 95°C, 56°C, and 72°C for 30 seconds each. The generation of specific PCR products was confirmed using melting curve analysis. RNA samples in which the iScript reverse transcriptase was omitted were used as negative controls. The standard curve method was used for relative quantification of the template, with gapA being employed as a housekeeping gene control. Genomic DNA from B. japonicum USDA110 was used as PCR templates to generate a standard curve for each gene. Relative starting quantities (SQ) of the mRNAs for the genes of interest and gapA were calculated from the corresponding standard curves. Quantity of the interested genes was normalized to the quantity of gapA for each respective condition. The results were based on average of triplicates and the standard deviation is shown as the error bar in the results.
The transcriptional start site of mntH was determined utilizing 5′ RACE System kit (Invitrogen) as per manufacturer’s protocol. RNA was isolated from B. japonicum USDA110 as described above. 5 μg total RNA was used for 5′ RACE.
DNase I Footprinting Analysis
DNase I Footprinting analyses examined the DNA region protected by Fur. Fur was incubated for 30 minutes at room temperature in a 50 μl volume of EMSA binding buffer containing 125 ng dI.dC, 5 μg of bovine serum albumin, and 1nM radiolabeled mntH probe in the presence or absence of 100 μM MnCl2. The labeled DNA template was synthesized by PCR. The 5′ end of the forward primer was radiolabeled using [γ-32P]dATP (6000Ci/mmol) (Perkin Elmer) and Polynucleotide Kinase (Promega). 50 μl of room temperature solution of 5mM CaCl2 and 10mM MgCl2 was added to the binding reaction and allowed to incubate for 1 min before the addition of 0.45 units of RQ1 RNase-free DNase (Promega) in 5 μl of 40mM Tris-HCl, pH 7.0. Reactions were incubated for 1 minute at room temperature before being stopped with the addition of 90 μl stop solution (200mM NaCl, 30mM EDTA, 1% SDS, 100 μg/ml yeast RNA). DNA was extracted from the reaction using phenol:chloroform (1:1) followed by ethanol precipitation. G + A ladders were produced as described (Maxam and Gilbert, 1980). DNase I digested products were separated on an 8% denaturing polyacrylamide gel containing 7 M urea in Tris borate EDTA electrophoresis buffer. After electrophoresis, gel was exposed on Imaging Screen K, 35 × 45cm (Bio-Rad), and scanned using a Personal Molecular Imager FX (Bio-Rad).
Electrophorectic mobility shift assay
EMSA analysis was employed to study Fur binding to DNA using a protocol modified from de Lorenzo et al (de Lorenzo et al., 1988). Fur was incubated 30 minutes at room temperature in a 20-μl volume of EMSA buffer, consisting of 50 ng dI.dC, 2 μg bovine serum albumin, 100 μM MnCl2, and 1 nM radiolabeled DNA probe. Double stranded DNA probes were produced by boiling and slowly cooling synthetic DNA oligonucleotides (Integrated DNA technologies) in annealing buffer (150 mM NaCl2, 10 mM Tris-HCl, 1 mM EDTA, pH 8.0) and filled in with [α-32P]dCTP (3000 Ci/mmol) (Perkin Elmer) with the klenow fragment of DNA polymerase (Promega). Following the incubation, EMSA reactions were analyzed on a 5% non-denaturing polyacrylamide gel in electrophoresis buffer (20 mM bis-Tris borate, 100μM MnCl2, pH 7.5) that were prerun for 30 minutes at a constant 200 V at 4°C. After electrophoresis at 4°C for 60 minutes at 200 V, gels were dried and autoradiographed. Autoradiograms were developed on BioMax film (Eastman Kodak Co.) and scanned using a GS-700 densitometer (Bio-Rad). Signal intensities were detected and quantified using Quantity One (Bio-Rad). To determine the dissociation constant (Kd), binding reactions were titrated with a varying amount of Fur. Bound and unbound DNA was quantified by comparing relative signal intensities and analyzed using Graphpad Prism (Graphpad Software Inc.). DNA probes used to represent the Fur binding site in the mntH promoter were synthesized as overlapping fragments and compared by EMSA.
Plant growth, infection and assays
Soybeans (Glycine max cv. Essex) were inoculated with the parent strain or mntHΔΩ and grown in an environmental growth chamber under a 16 h day/8 h dark regime as described previously (Frustaci and O’Brian, 1992). Nitrogen fixation in root nodules from plants 26 days post-inoculation was assessed as the reduction of acetylene to ethylene as described previously (Frustaci and O’Brian, 1992).
Acknowledgments
This work was supported by NIH grant R01GM067966 to M.R.O’B. T.H.H. was supported by NIH training grant T32 AI707614.
References
- Bearden SW, Staggs TM, Perry RD. An ABC transporter system of Yersinia pestis allows utilization of chelated iron by Escherichia coli SAB11. J Bacteriol. 1998;180:1135–1147. doi: 10.1128/jb.180.5.1135-1147.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearden SW, Perry RD. The Yfe system of Yersinia pestis transports iron and manganese and is required for full virulence of plague. Mol Microbiol. 1999;32:403–414. doi: 10.1046/j.1365-2958.1999.01360.x. [DOI] [PubMed] [Google Scholar]
- Bellini P, Hemmings AM. In vitro characterization of a bacterial manganese uptake regulator of the fur superfamily. Biochemistry. 2006;45:2686–2698. doi: 10.1021/bi052081n. [DOI] [PubMed] [Google Scholar]
- Boyer E, Bergevin I, Malo D, Gros P, Cellier MF. Acquisition of Mn(II) in addition to Fe(II) is required for full virulence of Salmonella enterica serovar Typhimurium. Infect Immun. 2002;70:6032–6042. doi: 10.1128/IAI.70.11.6032-6042.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao TC, Becker A, Buhrmester J, Pühler A, Weidner S. The Sinorhizobium meliloti fur gene regulates, with dependence on Mn(II), transcription of the sitABCD operon, encoding a metal-type transporter. J Bacteriol. 2004;186:3609–3620. doi: 10.1128/JB.186.11.3609-3620.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies BW, Walker GC. Disruption of sitA compromises Sinorhizobium meliloti for manganese uptake required for protection against oxidative stress. J Bacteriol. 2007;189:2101–2109. doi: 10.1128/JB.01377-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorenzo V, Giovannini F, Herrero M, Neilands JB. Metal ion regulation of gene expression. Fur repressor-operator interaction at the promoter region of the aerobactin system of pColV-K30. J Mol Biol. 1988;203:875–884. doi: 10.1016/0022-2836(88)90113-1. [DOI] [PubMed] [Google Scholar]
- Diaz-Mireles E, Wexler M, Sawers G, Bellini D, Todd JD, Johnston AW. The Fur-like protein Mur of Rhizobium leguminosarum is a Mn2+-responsive transcriptional regulator. Microbiology. 2004;150:1447–1456. doi: 10.1099/mic.0.26961-0. [DOI] [PubMed] [Google Scholar]
- Domenech P, Pym AS, Cellier M, Barry CE, 3rd, Cole ST. Inactivation of the Mycobacterium tuberculosis Nramp orthologue (mntH) does not affect virulence in a mouse model of tuberculosis. FEMS Microbiol Lett. 2002;207:81–86. doi: 10.1111/j.1574-6968.2002.tb11032.x. [DOI] [PubMed] [Google Scholar]
- Friedman YE, O’Brian MR. A novel DNA-binding site for the ferric uptake regulator (Fur) protein from Bradyrhizobium japonicum. J Biol Chem. 2003;278:38395–38401. doi: 10.1074/jbc.M306710200. [DOI] [PubMed] [Google Scholar]
- Friedman YE, O’Brian MR. The ferric uptake regulator (Fur) protein from Bradyrhizobium japonicum is an iron-responsive transcriptional repressor in vitro. J Biol Chem. 2004;279:32100–32105. doi: 10.1074/jbc.M404924200. [DOI] [PubMed] [Google Scholar]
- Frustaci JM, Sangwan I, O’Brian MR. Aerobic growth and respiration of a δ-aminolevulinic acid synthase (hemA) mutant of Bradyrhizobium japonicum. J Bacteriol. 1991;173:1145–1150. doi: 10.1128/jb.173.3.1145-1150.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frustaci JM, O’Brian MR. Characterization of a Bradyrhizobium japonicum ferrochelatase mutant and isolation of the hemH gene. J Bacteriol. 1992;174:4223–4229. doi: 10.1128/jb.174.13.4223-4229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garau G, Reeve WG, Brau L, Deiana P, Yates RJ, James D, Tiwari R, O’Hara GW, Howieson JG. The symbiotic requirements of different Medicago spp. suggest the evolution of Sinorhizobium meliloti and S. medicae with hosts differentially adapted to soil pH. Plant and Soil. 2005;276:263–277. [Google Scholar]
- Guedon E, Moore CM, Que Q, Wang T, Ye RW, Helmann JD. The global transcriptional response of Bacillus subtilis to manganese involves the MntR, Fur, TnrA and sigmaB regulons. Mol Microbiol. 2003;49:1477–1491. doi: 10.1046/j.1365-2958.2003.03648.x. [DOI] [PubMed] [Google Scholar]
- Hamza I, Hassett R, O’Brian MR. Identification of a functional fur gene in Bradyrhizobium japonicum. J Bacteriol. 1999;181:5843–5846. doi: 10.1128/jb.181.18.5843-5846.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsburgh MJ, Wharton SJ, Cox AG, Ingham E, Peacock S, Foster SJ. MntR modulates expression of the PerR regulon and superoxide resistance in Staphylococcus aureus through control of manganese uptake. Mol Microbiol. 2002;44:1269–1286. doi: 10.1046/j.1365-2958.2002.02944.x. [DOI] [PubMed] [Google Scholar]
- Ikeda JS, Janakiraman A, Kehres DG, Maguire ME, Slauch JM. Transcriptional regulation of sitABCD of Salmonella enterica serovar Typhimurium by MntR and Fur. J Bacteriol. 2005;187:912–922. doi: 10.1128/JB.187.3.912-922.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehres DG, Zaharik ML, Finlay BB, Maguire ME. The NRAMP proteins of Salmonella typhimurium and Escherichia coli are selective manganese transporters involved in the response to reactive oxygen. Mol Microbiol. 2000;36:1085–1100. doi: 10.1046/j.1365-2958.2000.01922.x. [DOI] [PubMed] [Google Scholar]
- Kehres DG, Janakiraman A, Slauch JM, Maguire ME. SitABCD is the alkaline Mn(2+) transporter of Salmonella enterica serovar Typhimurium. J Bacteriol. 2002a;184:3159–3166. doi: 10.1128/JB.184.12.3159-3166.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehres DG, Janakiraman A, Slauch JM, Maguire ME. Regulation of Salmonella enterica serovar Typhimurium mntH transcription by H(2)O(2), Fe(2+), and Mn(2+) J Bacteriol. 2002b;184:3151–3158. doi: 10.1128/JB.184.12.3151-3158.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehres DG, Maguire ME. Emerging themes in manganese transport, biochemistry and pathogenesis in bacteria. FEMS Microbiol Rev. 2003;27:263–290. doi: 10.1016/S0168-6445(03)00052-4. [DOI] [PubMed] [Google Scholar]
- Kitphati W, Ngok-Ngam P, Suwanmaneerat S, Sukchawalit R, Mongkolsuk S. Agrobacterium tumefaciens fur has important physiological roles in iron and manganese homeostasis, the oxidative stress response, and full virulence. Appl Environ Microbiol. 2007;73:4760–4768. doi: 10.1128/AEM.00531-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz O, Schwartz E, Dernedde J, Eitinger M, Friedrich B. The Alcaligenes eutrophus H16 hoxX gene participates in hydrogenase regulation. J Bacteriol. 1994;176:4385–4393. doi: 10.1128/jb.176.14.4385-4393.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makui H, Roig E, Cole ST, Helmann JD, Gros P, Cellier MF. Identification of the Escherichia coli K-12 Nramp orthologue (MntH) as a selective divalent metal ion transporter. Mol Microbiol. 2000;35:1065–1078. doi: 10.1046/j.1365-2958.2000.01774.x. [DOI] [PubMed] [Google Scholar]
- Maxam AM, Gilbert W. Sequencing end-labeled DNA with base specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Moore CM, Helmann JD. Metal ion homeostasis in Bacillus subtilis. Curr Opin Microbiol. 2005;8:188–195. doi: 10.1016/j.mib.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Panek HR, O’Brian MR. KatG is the primary detoxifier of hydrogen peroxide produced by aerobic metabolism in Bradyrhizobium japonicum. J Bacteriol. 2004;186:7874–7880. doi: 10.1128/JB.186.23.7874-7880.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp-Wallace KM, Maguire ME. Manganese transport and the role of manganese in virulence. Annu Rev Microbiol. 2006;60:187–209. doi: 10.1146/annurev.micro.60.080805.142149. [DOI] [PubMed] [Google Scholar]
- Patzer SI, Hantke K. Dual repression by Fe2+-Fur and Mn2+-MntR of the mntH gene, encoding an NRAMP-like Mn2+ transporter in Escherichia coli. J Bacteriol. 2001;183:4806–4813. doi: 10.1128/JB.183.16.4806-4813.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry RD, Mier I, Jr, Fetherston JD. Roles of the Yfe and Feo transporters of Yersinia pestis in iron uptake and intracellular growth. Biometals. 2007;20:699–703. doi: 10.1007/s10534-006-9051-x. [DOI] [PubMed] [Google Scholar]
- Platero R, Peixoto L, O’Brian MR, Fabiano E. Fur is involved in manganese-dependent regulation of mntA (sitA) expression in Sinorhizobium meliloti. Appl Environ Microbiol. 2004;70:4349–4355. doi: 10.1128/AEM.70.7.4349-4355.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platero R, de Lorenzo V, Garat B, Fabiano E. Sinorhizobium meliloti fur-like (Mur) protein binds a fur box-like sequence present in the mntA promoter in a manganese-responsive manner. Appl Environ Microbiol. 2007;73:4832–4838. doi: 10.1128/AEM.00686-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platero RA, Jaureguy M, Battistoni FJ, Fabiano ER. Mutations in sitB and sitD genes affect manganese-growth requirements in Sinorhizobium meliloti. FEMS Microbiol Lett. 2003;218:65–70. doi: 10.1111/j.1574-6968.2003.tb11499.x. [DOI] [PubMed] [Google Scholar]
- Prentki P, Krisch HM. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;28:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- Que Q, Helmann JD. Manganese homeostasis in Bacillus subtilis is regulated by MntR, a bifunctional regulator related to the diphtheria toxin repressor family of proteins. Mol Microbiol. 2000;35:1454–1468. doi: 10.1046/j.1365-2958.2000.01811.x. [DOI] [PubMed] [Google Scholar]
- Rodionov DA, Gelfand MS, Todd JD, Curson AR, Johnston AW. Computational reconstruction of iron- and manganese-responsive transcriptional networks in alpha-Proteobacteria. PLoS Comput Biol. 2006;2:e163. doi: 10.1371/journal.pcbi.0020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runyen-Janecky LJ, Reeves SA, Gonzales EG, Payne SM. Contribution of the Shigella flexneri Sit, Iuc, and Feo iron acquisition systems to iron acquisition in vitro and in cultured cells. Infect Immun. 2003;71:1919–1928. doi: 10.1128/IAI.71.4.1919-1928.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabri M, Caza M, Proulx J, Lymberopoulos MH, Bree A, Moulin-Schouleur M, Curtiss R, 3rd, Dozois CM. Contribution of the SitABCD, MntH, and FeoB metal transporters to the virulence of avian pathogenic Escherichia coli O78 strain chi7122. Infect Immun. 2008;76:601–611. doi: 10.1128/IAI.00789-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Sangwan I, Lindemann A, Hauser F, Hennecke H, Fischer HM, O’Brian MR. Bradyrhizobium japonicum senses iron through the status of haem to regulate iron homeostasis and metabolism. Mol Microbiol. 2006a;60:427–437. doi: 10.1111/j.1365-2958.2006.05101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Sangwan I, O’Brian MR. The Bradyrhizobium japonicum Fur protein is an iron-responsive regulator in vivo. Mol Genet Genomics. 2006b;276:555–564. doi: 10.1007/s00438-006-0162-4. [DOI] [PubMed] [Google Scholar]
- Zhou D, Hardt WD, Galan JE. Salmonella typhimurium encodes a putative iron transport system within the centisome 63 pathogenicity island. Infect Immun. 1999;67:1974–1981. doi: 10.1128/iai.67.4.1974-1981.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]