Abstract
We report that photoconversion is fairly common among orange and red fluorescent proteins, as a screen of 12 variants yielded 8 that exhibit photoconversion. Specifically, three red fluorescent proteins can be switched into a green state, and two orange variants can be photoconverted to the far red. The orange highlighters are ideal for dual-probe highlighter applications, and they exhibit the most red-shifted excitation of all fluorescent protein described to date.
Photoconvertable fluorescent proteins (pc-FPs) exhibit a change in fluorescence excitation and emission spectra after excitation at a specific wavelength, and are thus useful as optical highlighters. Since the first description of a photoactivatable probe, PA-GFP1, which switches from a dark to a fluorescent state, many fluorescent proteins have been developed that permit photoconversion rather than just photoactivation. These fluorescent proteins switch between two colors (typically from green to red), both of which can be visualized. The palette of currently useful pc-FPs includes PS-CFP2, Dendra3, 4, EosFP5, Kaede6, and KikGR7, all of which exhibit red-shifted fluorescence emission accompanied by a concomitant shift in the excitation maxima after irradiation with near-UV or deep blue light.
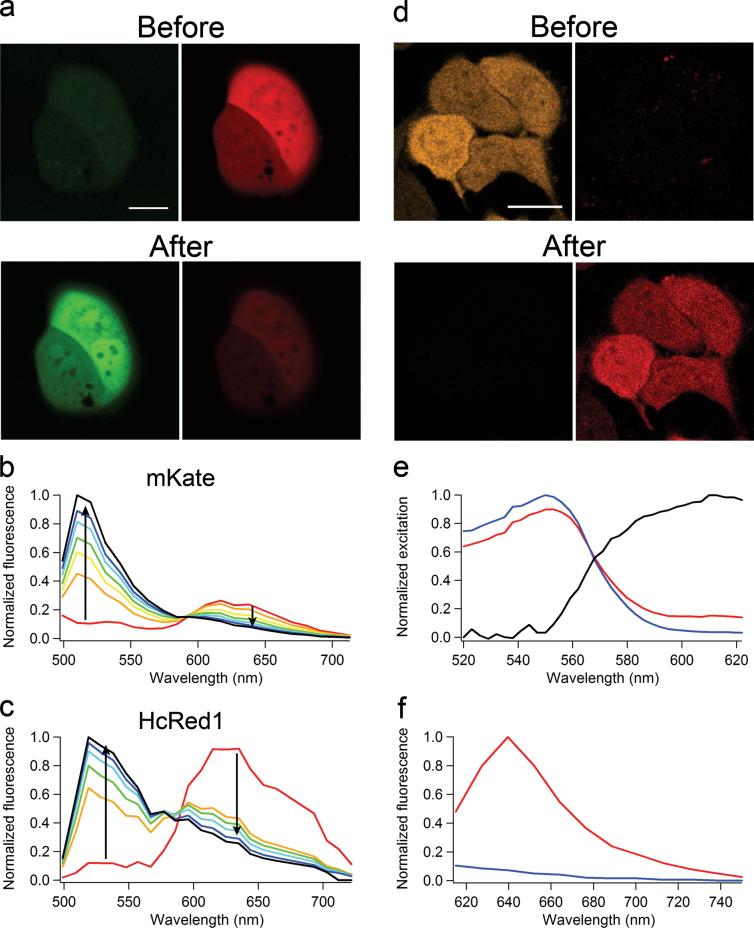
The dimeric red fluorescent protein (RFP) Katushka and its monomeric variant mKate yield the greatest levels of fluorescence emission above 650 nm of all presently characterized RFPs8. Cells expressing Katushka or mKate were fluorescent upon two-photon excitation at 750 nm, although the fluorescence decreased rapidly, apparently due to photobleaching. However, subsequent imaging of the cells through a GFP emission filter showed a novel green fluorescence (Fig. 1a,b), indicating a photoconversion of the red fluorescent proteins to a green state. Further investigation revealed that red-to-green photoconversion also occured upon single-photon laser excitation at 405 and 561 nm (Table 1), and that the green state had excitation and emission maxima of 495 and 518 nm, respectively. Katushka, mKate, mTagRFP9, and mTagRFP-T (a variant with increased photostability)10 are all derived from the same sea anemone protein, EqFP578. All four proteins share the same chromophore (Met-Tyr-Gly), but we observed no photoconversion of mTagRFP or mTagRFP-T at any excitation wavelength.
Figure 1.
Photoconversion of mKate, HcRed1, and mOrange1 in HeLa cells. a) Cells expressing mKate were imaged before and after photoconversion induced with 750 nm two-photon excitation. Green and red fluorescence was collected simultaneously using 488 nm excitation. b) mKate emission spectra (ex 488 nm) showing gradual red to green photoconversion. c) HcRed1 emission spectra (ex 488 nm) showing gradual red to green photoconversion using 561 nm excitation. d) Cells expressing mOrange1 before and after photoconversion induced at 488 nm. e) Excitation spectrum of the mOrange1 far-red species (black line) obtained by linear unmixing of the excitation spectra of a partially photoconverted cell (red line) and a non-converted cell (blue line). f) Emission spectra (ex 600 nm) of mOrange1 before and after photoconversion. Scale bars are 20 μm.
Table 1.
Photoconversion properties
| Color change | Photoactivation wavelength and power | Fold activationa | Residual fluorescenceb | Change in contrastc | |
|---|---|---|---|---|---|
| Katushka | Red to green | 750 nm 17.9 mW | 6 ± 2d | 0.22 ± 0.07 (0.30 ± 0.02)e | 27 (13) |
| 405 nm 1.4 mW | 7 ± 1 | 0.66 ± 0.03 (0.68 ± 0.03) | 10 (10) | ||
| 561 nm 0.32 mW | 5 ± 4 | 0.06 ± 0.03 | 82 | ||
| mKate | Red to green | 750 nm 17.9 mW | 11 ± 2 | 0.59 ± 0.08 (0.94 ± 0.09) | 19 (12) |
| 405 nm 1.4 mW | 10 ± 3 | 0.41 ± 0.03 (0.8 ± 0.6) | 26 (13) | ||
| 561 nm 0.32 mW | 2.0 ± 0.3 | 0.07 ± 0.03 | 29 | ||
| HcRed1 | Red to green | 561 nm 0.32 mW | 26 ± 18 | 0.22 ± 0.09 | 119 |
| mOrange1 | Orange to far-red | 488 nm 1.2 mW | 16 ± 4 | 0.10 ± 0.03 | 160 |
| mOrange2 | Orange to far-red | 488 nm 1.2 mW | 16 ± 5 | 0.10 ± 0.02 | 161 |
| Kaede | Green to red | 405 nm 0.38 mW | 28 ± 10 (1.8 ± 0.1) | 0.19 ± 0.02 | 148 (10) |
| Dendra2 | Green to red | 405 nm 0.25 mW | 47 ± 26 (1.2 ± 0.4) | 0.48 ± 0.16 | 97 (2) |
Fold increase in photoconverted fluorescence.
Fraction of original fluorescence remaining after photoconversion.
The change in contrast is determined by the increase in photoconverted fluorescence divided by the residual original fluorescence.
Values are mean ± s.d (n = 7).
Values between brackets are for single wavelength excitation (ex 488 nm).
HcRed1 is a far-red fluorescent protein and originally developed from a sea anemone chromoprotein11. We observed red-to-green photoconversion after illumination at 405, 543, or 750 nm two-photon excitation, but excitation at 561 nm produced the green fluorescent species with much better efficiency (Fig. 1c, Table 1 and Supplementary Fig. 1 online). This green species had an emission peak at ∼510 nm and was excited at 458 or 488 nm.
We also tested several red “mFruit” fluorescent proteins derived from DsRed 10, 12. Neither the widely used RFP, mCherry, nor its brighter variant, mApple110, exhibited any photoconversion upon one-photon excitation ranging from 405 nm to 633 nm or two-photon excitation at 750 nm. Both mPlum and mRaspberry showed a 10 nm blue-shift in the emission maximum after prolonged illumination with 405 nm or 543 nm light (Supplementary Fig. 2 online). These spectral changes were too small for use in optical highlighter applications and therefore we did not further characterize photoconversion of these proteins. In contrast, we observed considerable photoconversion for the orange fluorescent proteins, mOrange1 and mOrange2. We performed a complete characterization for both variants, and the photoconversion characteristics were practically identical, so we report the details only for mOrange1. Photoconversion occured at 458 nm or 488 nm laser excitation but is far less efficient at 405 nm excitation. Since most pc-FPs photoconvert upon near-UV excitation, which can be harmful to cells, this visible photoconversion may be advantageous for live cell applications. mOrange1 is not normally excited at 633 nm, but after photoconversion, we observed bright red fluorescence (Fig. 1d). Using a white-light continuum laser, we measured photoconverted excitation and emission spectra in live cells (Fig. 1e, f). The excitation and emission maxima of the far-red species were approximately 610 nm and 640 nm, respectively. To our knowledge, this species exhibits the most red-shifted absorption peak (610 nm) of any fluorescent protein thus far characterized.
Kusabira-Orange (mKO)13 has a similar chromophore and spectral properties to mOrange, although the fluorescent proteins are unrelated. Previously, mKO has been shown to photoconvert to a green fluorescent species under widefield illumination at 436 nm14, but we did not observe this blue-shift in fluorescence using laser excitation. However, excitation of mKO using the 514 nm or 543 nm laser resulted in a minor emission red-shift (Supplementary Fig. 3 online). This fluorescence was dim, and it remains unclear whether this is photoconversion or the result of selective photobleaching of an mKO subpopulation.
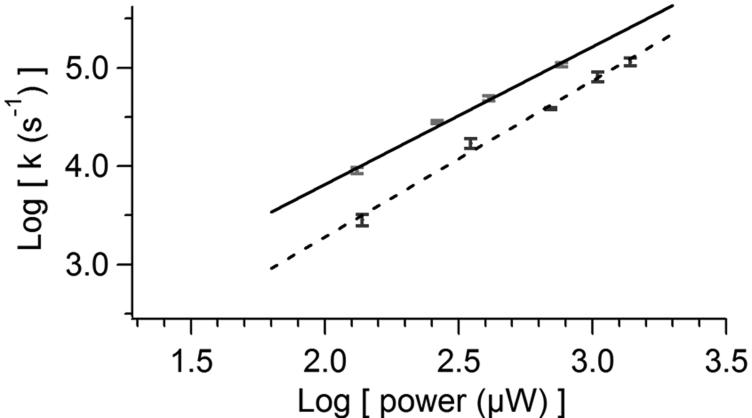
The dynamics of each of these photoconversion reactions were studied using micro-droplets of purified protein. Photoconversion of all pc-FPs, except Katushka and mKate, resulted in a mono-exponential increase in the photoconverted species. The photoconversion rate increased supralinearly with laser power (Fig. 2a), which indicated that these photoconversions were multi-photon phenomena. Photoconversion of the pc-FPs described here required more light than photoconversion of Kaede and Dendra2. Photoconversion was generally maximal at lower laser powers because of increased photobleaching at higher power (Supplementary Fig. 4 online). An exception was mOrange for which we observed the greatest photoconversion at the highest laser power. mOrange1 photoconversion was also not significantly affected by pH (Fig. 2b), which permits optical highlighting in a wide pH-range of cellular compartments. In comparison, Kaede photoconversion, like all other reported pc-FPs, was highly pH dependent, and Kaede was nonfluorescent at pH 5.45. Many of the photoconversion dynamics were complex (Supplementary Fig. 4 online). For example, Dendra2 showed an initial increase in green fluorescence upon photoconversion, and similar behavior has been observed for Kaede15. We also observed a novel blue fluorescent Kaede species during its photoconversion at high laser power. Finally, photoconversion of Katushka and mKate showed an initial lag in both the decrease of red fluorescence and increase of green fluorescence.
Figure 2.
mOrange1 and Kaede photoconversion rates. a) Log-log plot of laser power dependence of photoconversion for Kaede (solid) and mOrange1 (dashed). Linear fits yield a slope for Kaede of 1.40 ± 0.07 (r = 0.997) and for mOrange1 of 1.59 ± 0.08 (r = 0.996). b) pH-dependence of Kaede (solid) and mOrange1 (dashed) photoconversion rates. Data points are averages of at least 5 measurements. Error bars indicate standard deviations.
Most pc-FPs perform better under prolonged illumination at low intensities, but many applications require fast photoconversion (i.e. within seconds). For this purpose, we measured photoconversion efficiency of pc-FPs in HeLa cells with the excitation set to reach a maximum increase within ∼14 seconds. The new pc-FPs displayed a smaller increase in photoconverted fluorescence than Kaede and Dendra2 (Table 1). However, the contrast change for mOrange was comparable to Kaede. For applications using only 488 nm excitation, mKate outperformed Kaede in photoconverted fluorescence and contrast ratio.
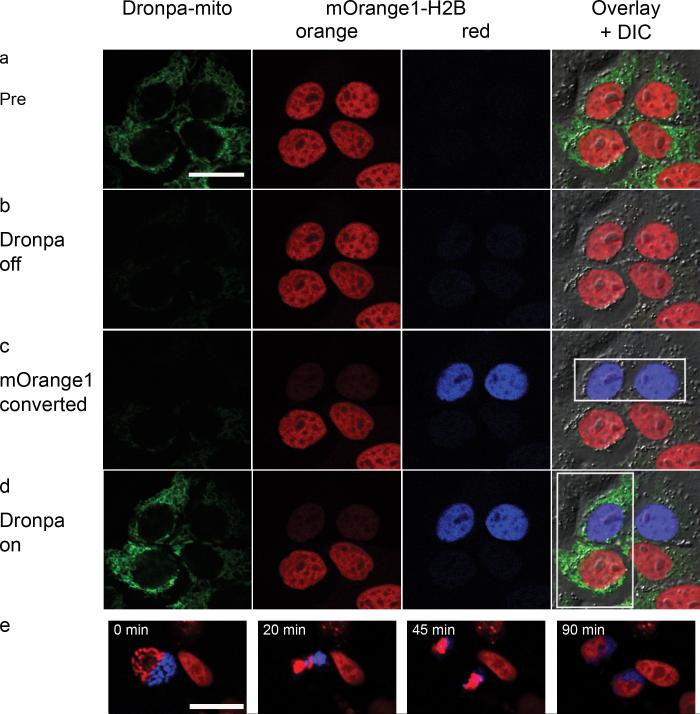
The mOrange fluorescent proteins have great potential as optical highlighters because of their monomeric nature, excellent photoconversion contrast, and relative pH-insensitivity of photoconversion. The red-shifted spectral properties also enabled their use in dual-probe optical highlighting together with the green photoswitchable fluorescent protein Dronpa16 to allow selective highlighting of 4 individual cell populations (Fig. 3a-d). We achieved this, by first switching off all Dronpa fluorescence, followed subsequently by photoconversion of mOrange and switching Dronpa on again in selected cells. To show cell viability, we followed photoconverted H2B-mOrange2 in cells for 140 minutes through mitosis (Fig 3e and Supplementary Movie online). While photoconverted mOrange was less photostable than Kaede and Dendra2 (Supplementary Note 1 online), the photostability was sufficient for time-lapse imaging. mOrange variants were also well-suited to laser scanning microscopy applications, because they are photoconverted at 488 nm and their contrast ratio was maximal at the highest excitation powers. This was in contrast to Kaede and Dendra2, for which the contrast ratio was maximal under low light conditions (Supplementary Fig. 4 online). In fact, we observed a smaller contrast ratio for Kaede after laser-based photoconversion (148-fold change; Table 1) compared to widefield photoconversion (∼2000-fold change)6.
Figure 3.
Dual-probe optical highlighting with H2B-mOrange1 and Dronpa-mito. a) Before photoconversion cells have green labeled mitochondria and orange labeled nuclei (pseudo-colored in red). b) Dronpa fluorescence is switched off with 74 μW 488 nm excitation; causing minimal photoconversion of mOrange1. c) mOrange1 is specifically photoconverted in the two upper nuclei with 742 μW 488 nm excitation (pseudo-colored blue). d) Finally, Dronpa fluorescence is switched on again in the two left-hand cells with 248 μW 405 nm illumination. Boxes indicate active regions of photoconversion. e) Time-lapse images from Supplementary Movie showing mitosis of a HeLa cell expressing H2B-mOrange2. Part of the nucleus was photoconverted during prophase (pseudo-colored in blue). The images in panel e are contrast-enhanced to help visualization. Scale bars are 20 μm.
Existing green-to-red optical highlighters are prone to spectral bleedthrough of emission from the original species into the photoconverted species detector. Thus, sequential two-channel imaging with multiple excitation wavelengths is required to quantify the photoconverted signal. In contrast, red-to-green optical highlighters do not suffer from such spectral overlap. Although the contrast ratio was similar for mKate and Kaede when using single wavelength excitation (488 nm), the increase in photoconverted fluorescence was 5 times higher for mKate (Table 1).
For the fluorescent proteins that exhibited red-to-green photoconversion, the spectral properties of the photoconverted green species resembled those of maturation intermediates. In fact, “greening” of DsRed fluorescence has been previously described17, but this effect was attributed to the presence of immature green DsRed chromophores within the DsRed tetramer. Our data suggest that this “greening” might be caused in part by photoconversion. If photoconversion yields an immature intermediate, then the photoconversion would be reversible as the immature intermediate matures again. Improving the maturation rate of these newly discovered pc-FPs could thus yield novel reversible highlighters. It has also been shown that DsRed can photoconvert to a red-shifted fluorescent species upon high intensity irradiation at 532nm18. That photoconversion occurs through a sequential two-photon absorption process that involves decarboxylation of a glutamic acid residue adjacent to the chromophore. Since mOrange is derived from DsRed, it is tempting to speculate that a similar conversion mechanism is involved in mOrange photoconversion as well. If this speculation proves correct, structural determination of photoconverted mOrange might reveal new design principles for far red fluorescent proteins.
Combined with previous reports, our data show that fluorescent protein photoconversion is fairly common, as eight out of twelve proteins investigated display this phenomenon. Our results demonstrate once more the complex photophysical characteristics of fluorescent proteins, and point toward the need for wariness in double labeling and fluorescence resonance energy transfer (FRET) experiments, where an unnoticed color-shift could lead to serious misinterpretations. Such a problem has already been reported for FRET experiments that use cyan and yellow fluorescent proteins and rely on acceptor photobleaching19. We also observed this blue-shifted photoconverted species for mVenus after laser-based bleaching (Supplementary Note 2 online). Further, the small spectral shifts observed for mPlum and mRaspberry would affect spectral unmixing experiments. Of course, detailed knowledge of the photophysics permits further uses for these fluorescent proteins. For instance, mOrange could also benefit superresolution microscopy approaches20 by leveraging the reduced autofluorescence in the red spectral region.
Supplementary Material
Acknowledgements
We thank D.M. Chudakov (Shemiakin-Ovchinnikov Institute of Bioorganic Chemistry, Moscow, Russia), A. Miyawaki (RIKEN, Wako, Saitama, Japan) and R.Y. Tsien (University of California at San Diego ) for providing plasmids encoding fluorescent protein variants. Measurements were performed in part at the Vanderbilt University Medical Center Cell Imaging Shared Resource (NIH grants CA68485, DK20593, DK58404) and at the QFM course at Mount Desert Island Biological Lab. Thanks to G. Daniels (Leica), J. Wailes (Zeiss), B. Burklow (Olympus), and S. Schwartz (Nikon) for assistance with their microscopes. Thanks to B. Livesay for help with the mVenus experiments. This work was supported by NIH Grant GM72048 (DWP).
Appendix
Methods
Cell culture
HeLa and Cos7 cells were cultured in 35 mm glass bottom dishes (MatTek Corp.) or Delta T imaging chambers (Bioptechs) using Dulbecco’s modified Eagle’s medium (Invitrogen) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin and 2 mM L-glutamine. Cell were transfected using a transfection mix consisting either of 100 ng DNA and 1.5 μl Lipofectamine2000 (Invitrogen) in 100 μl Optimem (Invitrogen) media or 1 μg DNA with Effectene (Qiagen) in growth medium. Cells were imaged after 24 hours. Fluorescent proteins were expressed without a fusion partner or as histone H2B fusion proteins. The vector for Dronpa-mito was generated by substituting Dronpa for EGFP in pEGFP-mito-7 (Clontech).
Microscopy
Several laser scanning confocal microscopy platforms were used for data collection, including Zeiss LSM510 Meta and LSM710 (Zeiss), Olympus FV1000 (Olympus), Leica TCS SP5 X (Leica), and Nikon C1Si (Nikon). All microscopes were equipped with a Fluor design 40× 1.3 numerical aperture (NA) oil immersion objective. Imaging and photoconversion were done at 4× optical zoom using a 8 μs pixel dwell time and a 155 nm pixel size (512 × 512 pixels). Red-to-green and green-to-red photoconversion was detected using the same filter settings. Green fluorescence was excited at 488 nm and detected through a 530-550 nm bandpass filter. Red fluorescence was excited at 543 nm and detected through a 560 nm longpass emission filter. For single wavelength excitation applications green and red fluorescence were excited at 488 nm and detected simultaneous. For orange-to-red photoswitching, orange fluorescence was excited at 543 nm and detected through a 555-625 nm bandpass emission filter. Far-red fluorescence was excited at 633 nm and detected through a 650 nm longpass emission filter. Dronpa fluorescence was detected using 488 nm excitation and a 500-530 nm bandpass emission filter.
Power measurements
Accurate power measurements of high NA oil immersion objectives are difficult to do directly. Therefore power measurements at the back aperture of the objective lens were done through a 10× 0.3NA air objective using a Coherent Fieldmaster light meter (Coherent). During the measurement, the microscope was set to scan at its highest speed and maximum zoom. The measured average power was corrected for the off-time of the laser during scanning as well as the previously measured transmission efficiency of the objective lenses (90% for a 10× 0.3NA air objective and 70% for a 40× 1.4NA oil immersion objective).
Photoconversion of protein purification
Fluorescent proteins were expressed as his-tagged proteins in E. coli XL1Blue. Protein was purified using Talon metal affinity resin (Clontech) and buffer exchanged into phosphate buffered saline pH 7.4 using PD10 columns (GE Healthcare). Micro-droplets containing fluorescent protein were generated by mixing aqueous buffered fluorescent protein solutions and octanol in a 1:10 volume ratio followed by sonication. Microscopy samples consisted of 4 μl emulsion between an object slide and a 22 mm square cover glass. Glassware was treated with methyltrimethoxysilane (Acros Organics) to prevent protein absorbance to the glass surface. The droplet size (∼10-50 μm in diameter) was such that 3 to 10 droplets could be photoconverted simultaneously. Photoconversion was followed over time by sequential time-lapse imaging. Separate tracks were used to scan the original and photoconverted fluorescence, followed by a single scan with the photoconversion laser. Photoconversion rates were determined by fitting the increase in photoconverted fluorescence to a single exponential function. The rate constants were divided by the pixel dwell time (in seconds) to convert the values from scan-1 to s-1.
pH dependence of photoconversion
A series of titration buffers was prepared using 100 mM citric acid / Na citrate (pH 5.45), 100 mM KH2PO4 / Na2HPO4 (pH 6.72, 7.46, and 8.20), and 100 mM glycin / NaOH (pH 9.00). A 5-fold dilution of purified protein in each pH buffer was used to prepare microdroplet samples. Photoconversion was done as described for purified protein. The wavelength and laser power used for photoconversion were: mOrange1, 488 nm 632 μW; Kaede 405 nm 251 μW.
Photoconversion in Mammalian cells
Cells with medium fluorescence after transfection with equal amounts of plasmid DNA were selected for photoconversion. Initial and photoconverted fluorescence was imaged with the gain for each detector set at 850 volts. Laser power and duration of photoconversion were adjusted to reach a maximum increase in photoconverted fluorescence within 5 scan iterations. Changes in fluorescence were calculated after background subtraction, by dividing the fluorescence immediately after photoconversion by the fluorescence before photoactivation. In the mitosis experiments, cells expressing a fusion of mOrange2 to human histone H2B (mOrange2-H2B) were scanned to find cells entering prophase. One-half of the prophase nucleus was photoconverted and subsequently observed at lower laser power for a period ranging from 2 to 6 h.
Photostability of photoconverted species in Mammalian cells
Cells were transfected with equal amounts of plasmid DNA and cells with medium fluorescence were selected for photoconversion. Laser power and duration of photoconversion were selected to reach the maximum increase in photoconverted fluorescence within 5 scan iterations. The initial and photoconverted fluorescence was detected with detector gains set at 750 volts and 850 volts, respectively. The laser power for imaging the initial fluorescence as well as the photoconverted fluorescence immediately after photoconversion was adjusted to yield ∼2000 counts. The wavelength and power used for imaging the photoconverted species were: Dendra2, 543 nm 5.57 μW; Kaede, 543 nm 14 μW; Katushka and mKate, 488 nm 133 μW; mOrange 633 nm 312 μW.
Photoconversion of mVenus
mVenus protein purification and sample preparation were performed as described. Experiments were performed on a Zeiss LSM510 laser scanning confocal microscope equipped with a Chameleon mode-locked titanium sapphire laser oscillator (Chameleon, Coherent). Imaging was done using a Fluar 40x 1.3NA oil immersion objective at 4x optical zoom using a 6.4 μs pixel dwell time and a 110 nm pixel size (512 × 512 pixels). Photoconversion in microdroplets was measured by time-lapse imaging. A single track was used to image the original and photoconverted fluorescence simultaneously upon two-photon excitation at 850 nm, followed by a single scan with the photoconversion laser at 514 nm. Yellow and photoconverted cyan fluorescence was detected through 530-550 nm and 470-510 nm bandpass emission filters, respectively. Photoconversion rates were determined by fitting the increase in photoconverted cyan fluorescence to a single exponential function. The rate constants were divided by the pixel dwell time (in s) to convert the values from scan-1 to s-1.
References
- 1.Patterson GH, Lippincott-Schwartz J. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- 2.Chudakov DM, et al. Nat Biotechnol. 2004;22:1435–1439. doi: 10.1038/nbt1025. [DOI] [PubMed] [Google Scholar]
- 3.Gurskaya NG, et al. Nat Biotechnol. 2006;24:461–465. doi: 10.1038/nbt1191. [DOI] [PubMed] [Google Scholar]
- 4.Chudakov DM, Lukyanov S, Lukyanov KA. Nat Protoc. 2007;2:2024–2032. doi: 10.1038/nprot.2007.291. [DOI] [PubMed] [Google Scholar]
- 5.Wiedenmann J, et al. Proc Natl Acad Sci U S A. 2004;101:15905–15910. doi: 10.1073/pnas.0403668101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ando R, Hama H, Yamamoto-Hino M, Mizuno H, Miyawaki A. Proc Natl Acad Sci U S A. 2002;99:12651–12656. doi: 10.1073/pnas.202320599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsutsui H, Karasawa S, Shimizu H, Nukina N, Miyawaki A. EMBO Rep. 2005;6:233–238. doi: 10.1038/sj.embor.7400361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shcherbo D, et al. Nat Methods. 2007;4:741–746. doi: 10.1038/nmeth1083. [DOI] [PubMed] [Google Scholar]
- 9.Merzlyak EM, et al. Nat Methods. 2007;4:555–557. doi: 10.1038/nmeth1062. [DOI] [PubMed] [Google Scholar]
- 10.Shaner NC, et al. Nat Methods. 2008;5:545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gurskaya NG, et al. FEBS Lett. 2001;507:16–20. doi: 10.1016/s0014-5793(01)02930-1. [DOI] [PubMed] [Google Scholar]
- 12.Shaner NC, et al. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 13.Karasawa S, Araki T, Nagai T, Mizuno H, Miyawaki A. Biochem J. 2004;381:307–312. doi: 10.1042/BJ20040321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goedhart J, Vermeer JE, Adjobo-Hermans MJ, van Weeren L, Gadella TW., Jr. PLoS ONE. 2007;2:e1011. doi: 10.1371/journal.pone.0001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dittrich PS, Schafer SP, Schwille P. Biophys J. 2005;89:3446–3455. doi: 10.1529/biophysj.105.061713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Habuchi S, et al. Proc Natl Acad Sci U S A. 2005;102:9511–9516. doi: 10.1073/pnas.0500489102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marchant JS, Stutzmann GE, Leissring MA, LaFerla FM, Parker I. Nat Biotechnol. 2001;19:645–649. doi: 10.1038/90249. [DOI] [PubMed] [Google Scholar]
- 18.Habuchi S, et al. J Am Chem Soc. 2005;127:8977–8984. doi: 10.1021/ja047023o. [DOI] [PubMed] [Google Scholar]
- 19.Valentin G, et al. Nat Methods. 2005;2:801. doi: 10.1038/nmeth1105-801. [DOI] [PubMed] [Google Scholar]
- 20.Betzig E, et al. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.