Abstract
Telomerase-negative cancer cells maintain their telomeres via the Alternative Lengthening of Telomeres (ALT) pathway1–3. Although a growing body of evidence demonstrates that the ALT mechanism is a post-replicative telomere recombination process, molecular details of this pathway are largely unknown. Here we demonstrate that MUS81, a DNA structure–specific recombination endonuclease, plays a key role in the maintenance of telomeres in human ALT cells. We find that MUS81 specifically localizes to ALT-associated promyelocytic leukemia nuclear bodies (APBs) and associates with telomeric DNA in ALT cells, which is enriched during G2 phase of the cell cycle. Depletion of MUS81 results in reduction of ALT specific-telomere recombination and leads to proliferation arrest of ALT cells. In addition, the endonuclease activity of MUS81 is required for recombination-based ALT cell survival, and the interaction of MUS81 with TRF2 regulates this enzymatic activity to maintain telomere recombination. Thus, our results suggest that MUS81 is involved in the maintenance of ALT cell survival at least in part by telomere-HR process.
Keywords: ALT, APBs, MUS81, telomere recombination
Telomere homologous recombination (HR) is implicated in the ALT pathway1–4. The increased telomere recombination is ALT-specific and extends the proliferative life of ALT cells5. HR proteins and telomere associated proteins co-localize with telomeric DNA at ALT-associated promyelocytic leukemia (PML) nuclear bodies (APBs), which are present only in ALT cells6–9. Additionally, APBs are enriched during the G2 phase of the cell cycle when HR is most active and ALT activity is likely occurring10,11. MUS81 functions as an endonuclease by cleaving different DNA substrates12–17 and is required for the survival of cells undergoing aberrant replication and recombination12,14,17,18, suggesting that MUS81 might be a candidate “ALT protein” involved in abnormal telomere recombination of ALT cells. Here, we report that MUS81 specifically associates with telomeres in ALT cells and is essential for ALT cell viability through regulation of telomere recombination.
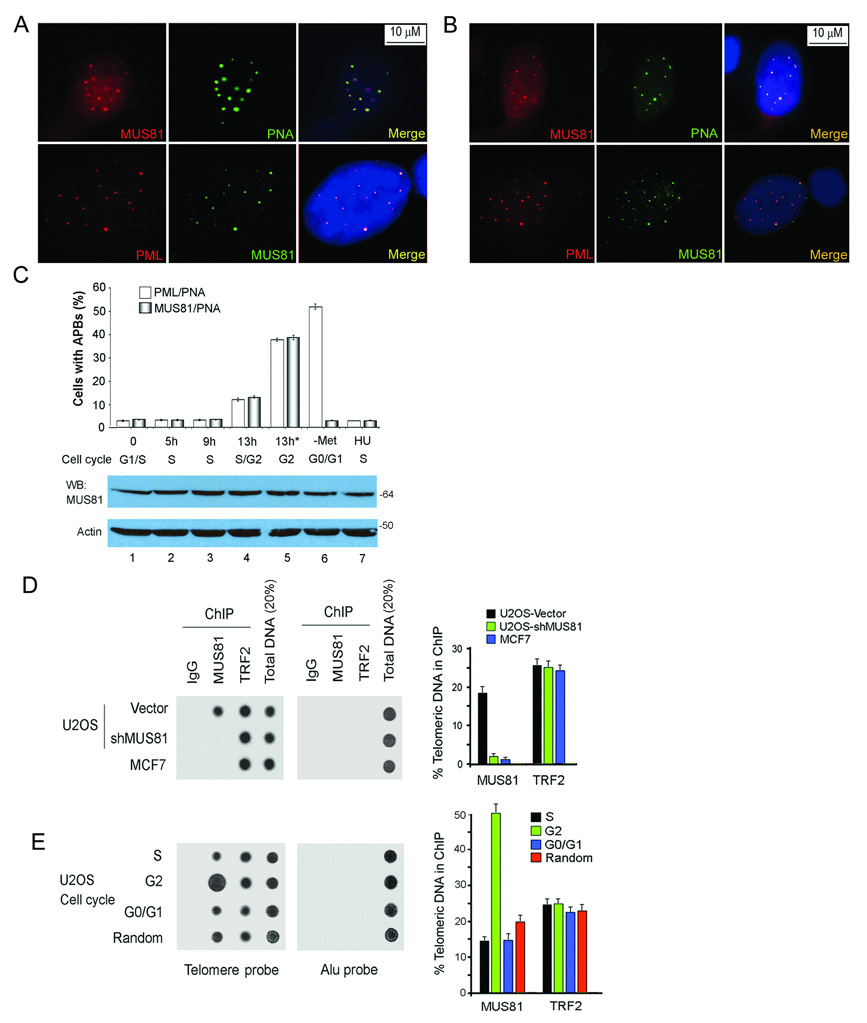
APBs are usually found in <5% of ALT cells growing asynchronously and are seen as large TRF1 or TRF2 foci that appear much brighter than signals from individual telomeres6–9. We confirmed the observation that TRF1, TRF2 and telomeric DNA co-localized with PML in APBs of GM847 cells (ALT positive)(Supplementary Information, Fig. S1A). We next examined the nuclear localization of MUS81 in an asynchronous culture of ALT cells and found that MUS81 specifically formed discrete nuclear foci in APB-positive cells and no MUS81 foci were seen in APB-negative cells. These MUS81 foci co-localized with telomeric DNA and PML in APBs in < 5% of GM847 cells (Fig. 1A). The presence of MUS81 in APBs was confirmed in another ALT cell line, U2OS cells (Fig. 1B). The foci were specific for MUS81 since depletion of MUS81 led to disappearance of the foci (Supplementary Information, Fig. S1B). These results indicate that MUS81 is an integral component of APBs in human ALT-positive cells.
Fig. 1.
MUS81 localizes to APBs in ALT cells. A. MUS81 co-localizes with telomeres at APBs in GM847 cells. Cells were processed for immunofluoresence and telomeric DNA-FISH or for double immunofluoresence with MUS81 (anti-mouse) and PML antibodies. B. MUS81 co-localizes with PML and telomeric DNA at APBs in U2OS cells (anti-rabbit MUS81 antibody). C. Cell cycle-dependent MUS81 foci formation in ALT cells. GM847 cells were fixed and processed for immunofluorescence at different time points following release from the double thymidine block (Lanes 1–5). The asterisk (*) shows an additional block by Hoechst 33342, in addition to the double thymidine treatment. Lanes 6 and 7: –Met: methionine restriction for 4 days; HU: 5 mM hydroxyurea treatment for 24 hours. The percentage of cells showing co-localization of PML or MUS81 foci with telomeric DNA (PNA probe) is shown in the upper panel. At least 200 cells were scored, and results represent three independent experiments (mean ± S.D.). The P value between S or G1 phase and G2 phase was <0.001, as determined by Student's t test. Western blot (WB) analysis was performed with indicated antibodies in the lower panel (Supplementary Information Fig. S9, Full scans). D. MUS81 binds to telomeric DNA in ALT cells, but not in non-ALT cells. ChIP of U2OS cells (with or without expression MUS81-shRNA-A) and MCF7 cells was conducted with indicated antibodies. Dot blots were probed for telomere or Alu repeats. The quantification of the data in the right panel represents the percentage of TTAGGG DNA recovered in each sample. Averaged signals obtained with total DNA samples were used as 100% value for the quantification and results were summarized from three independent experiments (mean ± S.D.). E. MUS81 associated with telomeres increases in G2 phase cells. ChIP assays were conducted from U2OS cells with double thymidine block (S and G2 phases) or methionine restriction for 4 days (G0/G1 phases). Random: asynchronous cells. The quantification of the data represents three independent experiments (mean ± S.D.). The P value between S or G0/G1 phase and G2 phase in the MUS81 group was <0.001, as determined by Student's t test.
To determine whether the association of MUS81 with APBs is cell-cycle dependent, we examined MUS81 foci formation in the different phases of cell cycle. GM847 cells synchronized at the G1/S boundary by a double thymidine block were released into the cell cycle and then fixed at specific time points post-release. FACS analysis confirmed cell cycle distributions (Supplementary Information, Fig. S1C). Consistent with previous reports10,11, <5% of GM847 cells displayed APBs during G1/S and S phases, and MUS81 foci were only observed in APBs (Fig. 1C). At G2 phase, ~40% of cells showed MUS81 foci that co-localized with APBs. We also observed less than 5% of cells with MUS81 foci when ALT cells were arrested at S phase with HU treatment. All together, our results demonstrate that the association of MUS81 with APBs is preferentially enriched at G2 phase.
GM847 cells arrested in G0/G1 phases by methionine restriction were accompanied by an induction of APBs in 50–60% of the population9 (Fig. 1C). However, MUS81 only co-localized to the less than 5% of APBs (original APBs), not to the large population of APBs (induced APBs). Thus, we conclude that MUS81 foci formation was only enriched in APBs at G2 phase. The abundance of MUS81 was not increased in G2 phase of the ALT cells (Fig. 1C, the lower panel), indicating that MUS81 may be recruited to APBs in ALT cells.
Gao and coworkers19 demonstrated that MUS81 localizes to nucleoli in human telomerase-positive (non-ALT) cells. We observed diffuse staining of MUS81 throughout the nucleoli in non-ALT HT1080 cells and also in ALT cells (Supplementary Information, Figs. S1D & E), suggesting that MUS81 localizes to both APBs and nucleoli in ALT cells. Interestingly, co-localization of MUS81 with telomeric DNA was not observed in non-ALT cells (HT1080), suggesting that MUS81 may only contribute to telomere maintenance in ALT cells.
Chromatin immunoprecipitation (ChIP) assays were performed to determine the association of MUS81 with telomeric DNA. We observed an enrichment of telomeric DNA coimmunoprecipitated with the MUS81 antibody in ALT cells (Fig. 1D), suggesting that MUS81 binds to telomeres. MUS81 depletion decreased the telomeric DNA signal, indicating the specificity of the ChIP assay for MUS81. We did not detect telomeric DNA signal with the MUS81 antibody in non-ALT MCF7 cells, consistent with the immunostaining results that MUS81 associates with telomeres specifically in ALT cells. Immunostaining results point to localization of MUS81 to APBs specifically enriched during G2 phase. Telomere ChIP assays with the antibody to MUS81 in U2OS cells led to recovery of TTAGGG repeats in extracts from G2 phase cells (Fig. 1E). We observed a significant enrichment of telomeric DNA co-immunoprecipitated with the MUS81 antibody from G2 phase cells. These results confirm that association of MUS81 with telomeres in APBs is enriched in G2 phase.
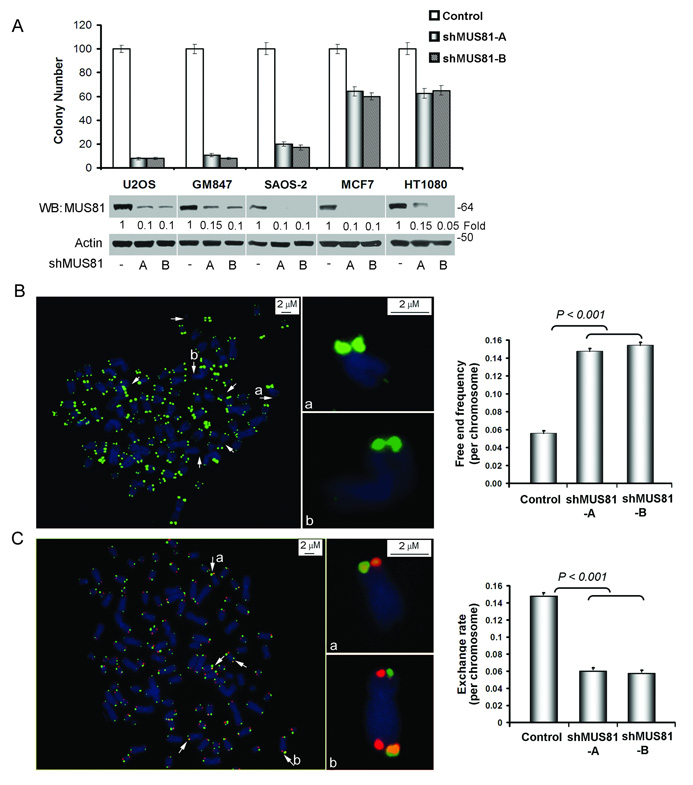
We next examined the role of MUS81 in ALT cell proliferation. Depletion of MUS81 caused growth arrest in the majority of ALT cells within 3–4 weeks. Colony formation assays showed that MUS81-shRNAs dramatically induced cell growth arrest in three ALT cell lines (GM847, U2OS and SAOS-2) (Fig. 2A and Supplementary Information, Fig. S2A). Inhibition of MUS81 expression in non-ALT cells (HT1080 and MCF7) only decreased the cell viability to ~60%, suggesting that the effect of MUS81 on cell viability is specific for ALT cells. Accordingly, a significant decrease in bromodeoxyuridine incorporation was associated with reduction of MUS81 levels in ALT cells, but not in non-ALT cells (Supplementary Information, Fig. S2B). Based on these results, we conclude that MUS81 plays a key role in the proliferation of ALT cells.
Fig. 2.
Depletion of MUS81 induces the cell growth arrest, telomere loss and decreases telomere recombination in ALT cells. A. Knockdown of MUS81 induces the cell growth arrest in ALT cells. Colony formation assays in three ALT cells (U2OS, GM847 and SAOS-2) and two non-ALT cells (MCF7, HT1080). Cells were equally seeded after infection with control lentiviral vector or MUS81-shRNAs in antibiotics selected medium for two weeks. Viable cell colonies were stained and counted. Depletion of MUS81 was determined by WB analysis (Supplementary Information Fig. S9, Full scans). Quantitative analysis of colony formation assay represents three independent experiments (mean ± S.D.). The P value between ALT+ and ALT− groups was <0.001. P values were determined by Student's t test. B. Knockdown of MUS81 induces telomere loss. Metaphases from GM847 cells with or without MUS81-shRNAs expression for one week were prepared and stained with Cy3-labled telomere PNA probe (red). DNA stained with DAPI (blue). The arrows pointed to the telomere loss. Quantitative data (the right panel) indicated that the free end telomere increased to ~15% in the shMUS81-A and shMUS81-B groups and 6% in the GFP-shRNA control group. The bar graph represents three independent experiments, showing the mean ± SD and Student t test were used for statistical analyses. C. Knockdown of MUS81 decreases the rate of T-SCE in GM847 cells. BrdU (30 µM) was incorporated into cells in one round of DNA replication (~26 hours), which were expressed MUS81-shRNAs or empty vector for one week. T-SCE was performed with telomeric G-strand PNA probe (red) and C-strand probe (green). Arrows showed telomere exchange signal (yellow). Quantitative analyses of telomere exchange signals were performed in GM847 cells expressing shMUS81-A, shMUS81-B or GFP-shRNA (the right panel). The bar graph represents three independent experiments, showing the mean ± SD and Student t test were used for statistical analyses.
The effect of MUS81 depletion on cell cycle progression was analyzed by FACS (Supplementary Information, Fig. S3A). Knockdown of MUS81 induced a diffused pattern of S and G2 phases, suggesting that depletion of MUS81 may induce a defective S phase. There was no significant change in HT1080 cells with expression of MUS81-shRNA. In addition, apoptotic cells did not increase in ALT and non-ALT cells upon depletion of MUS81 (Supplementary Information, Fig. S3B). These data suggest that MUS81 may mediate ALT cell proliferation by regulation of the cell cycle.
To further investigate the role of MUS81 on telomere maintenance, we conducted telomere FISH assay in ALT GM847 cells. An elevated frequency of telomere signal loss was observed in the MUS81-shRNA transduced cells (Fig. 2B). We detected 15% frequency of telomere signal loss per chromosome in the MUS81-shRNA transduced cells, which is approximately 3-fold higher than that in the control cells (5%, p < 0.001). Thus, depletion of MUS81 leads to increased loss of telomere signals.
Telomere sister chromatid exchange (T-SCE) is a hallmark of ALT cells associated with telomere recombination and ALT cell proliferation2,5,8. To test whether depletion of MUS81 hinders telomere recombination we monitored the frequency of T-SCEs in GM847 cells by performing CO-FISH. Consistent with previous reports2,5, we observed that a substantial proportion of metaphases had many chromosome extremities bearing “double signals (yellow)” in GM847 cells (Fig. 2C). Quantitative analysis showed that GM847 cells presented ~15% frequency of these exchange events. Importantly, depletion of MUS81 significantly decreased the frequency of T-SCE to ~5%. Depletion of MUS81 also decreased the same frequency of T-SCE in other ALT cells (U2OS and SAOS-2), demonstrating that MUS81 participates in T-SCE of ALT cells.
To investigate the effect of MUS81 on global telomere maintenance, we analyzed telomere loss and T-SCE in non-ALT cells. Knockdown of MUS81 in HT1080 and MCF7 cells did not induce telomere loss or affect T-SCE rate (Supplementary Information, Fig. S4). Moreover, Mus81-deficient MEFs did not show telomere loss and T-SCE phenotypes, confirming that MUS81 may not be involved in global telomere maintenance, but have a more restricted role in maintenance of telomeres by recombination in ALT cells.
To test whether depletion of MUS81 impacts on telomere length homeostasis, we performed Quantitative Fluorescence In Situ Hybridization (Q-FISH) on metaphase nuclei using a telomere specific PNA probe. We did not observe significant telomere length changes in the MUS81-shRNA transduced GM847 cells (Supplementary Information, Fig. S5A), including average telomere length, the frequency of shorter telomeres (<5 kb) or longer telomeres (>50 kb). Telomere restricted fragment (TRF) analysis verified these results as there was no detectable change in telomere length upon depletion of MUS81 in ALT or non-ALT cells (Supplementary Information, Fig. S5B). We did not observe obvious chromosome end-to-end fusion in ALT cells after depletion of MUS81 (data not shown). These results were consistent with the fact that knockdown of MUS81 did not induce the formation of telomere dysfunctional induced-foci (γH2AX foci formation on telomeres, Supplementary Information, Fig. S6), suggesting that MUS81 does not affect telomere end protection in ALT cells.
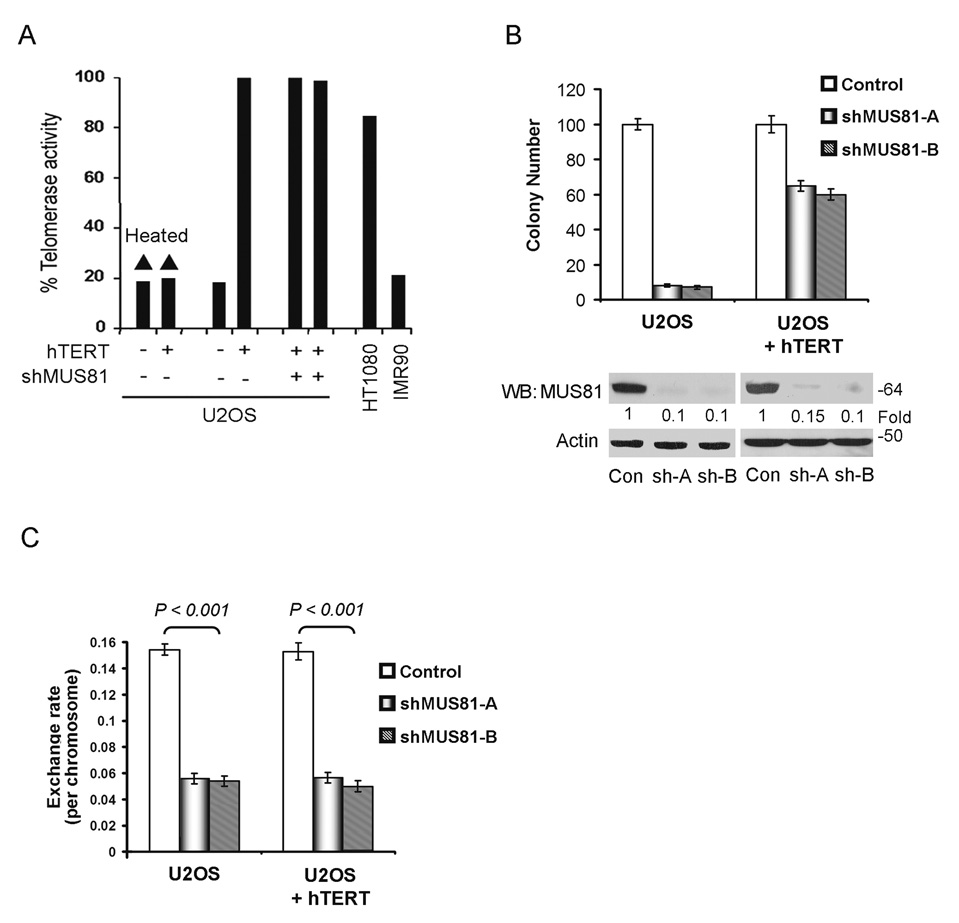
Expression of exogenous hTERT in ALT cells can induce telomerase activity, but does not abolish the ALT mechanism20,21, suggesting coexistence of ALT and telomerase in hTERT-transfected ALT cells. To investigate the relationship between MUS81 mediated-ALT cell proliferation and telomere maintenance, we stably expressed hTERT in U2OS cells. Telomerase was activated in these cells, as detected by the telomerase TRAP assay (Fig. 3A). The colony formation results showed that expression of hTERT partially rescued the arrested cell growth upon depletion of MUS81 (Fig. 3B). We also confirmed the results in another ALT cell line (SAOS-2). Expression of hTERT in SAOS-2 rescued the cell growth arrest and about 70% of the cells survived upon depletion of MUS81. U2OS cells expressing telomerase still showed a high rate of T-SCE and knockdown of MUS81 decreased the T-SCE rate in these cells (Fig. 3C), suggesting that rescue of MUS81-mediated ALT cell survival by telomerase is independent of the telomere recombination pathway. Thus, the results support that MUS81 regulates the growth of ALT cells mainly through maintenance of telomere by recombination.
Fig. 3.
Expression of hTERT rescues the MUS81 depletion-mediated cell growth arrest in ALT cells. A. hTERT was stably expressed in U2OS cells and the TRAP assay was performed to evaluate the telomerase activity. Heated U2OS cell lysates and lysates from primary fibroblasts IMR90 were used as negative controls. Lysates from HT1080 cells were used as a positive control. B. U2OS cells with or without stably expressing hTERT were transduced with shMUS81-A, shMUS81-B or empty vector for colony formation assays. After two weeks with drug-selection, cell colonies were counted and depletion of MUS81 was determined by WB analysis (Supplementary Information Fig. S9, Full scans). Quantitative analysis represents three independent experiments (mean ± S.D.). The P value between U2OS and U2OS+hTERT groups with expression of MUS81-shRNAs was <0.001. P values were determined by Student's t test. C. U2OS cells with or without stably expressing hTERT were transduced with shMUS81-A, shMUS81-B or empty vector for T-SCE assays. Quantitative analysis represents three independent experiments (mean ± S.D.).
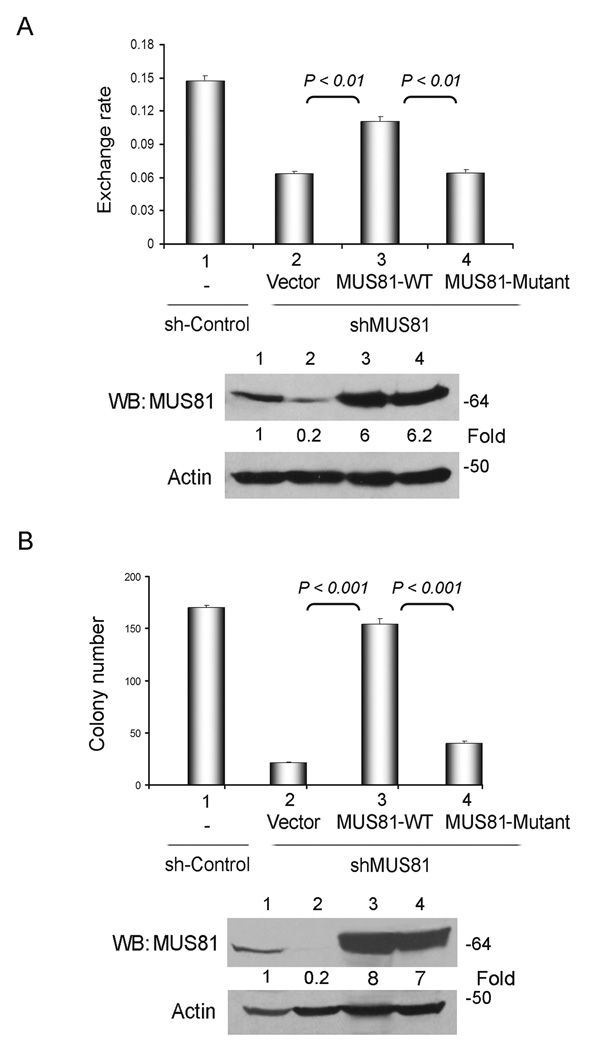
To investigate the role of endonuclease activity of MUS81 in the ALT pathway, we used a MUS81 construct in which the aspartic acid residues at 338 and 339 in VERK domain were substituted by alanine residues. Consistent with a previous report15, no endonuclease activity was detected in HA immunoprecipitates when this construct was tested (Supplementary Information, Fig. S7A). We observed that expression of wild-type MUS81 was sufficient to rescue the incidence of T-SCE in MUS81-depleted GM847 cells (Fig. 4A). Importantly, expression of the mutant MUS81 did not significantly affect T-SCE frequency, indicating that the endonuclease activity is key for the function of MUS81 on telomere recombination in ALT cells. Furthermore, we hypothesized that endonuclease activity of MUS81 is required for the viability of ALT cells. U2OS cells transduced with shMUS81-B (targeting in 3’UTR region) were infected with lentiviral particles containing MUS81 wild-type or mutant protein. Expression of wild-type MUS81 entirely restored viable cell colonies to the levels seen in transduced control vector cultures (Fig. 4B and Supplementary Information, Fig. S7C), demonstrating that the expression of wild-type MUS81 rescues the cell growth arrest induced by MUS81-shRNA. In contrast, the MUS81 endonuclease dead mutant did not significantly rescue the cell growth arrest upon MUS81 depletion. Thus, these results indicate that MUS81 enzymatic activity is required for the ALT cell survival and telomere recombination.
Fig. 4.
MUS81 endonuclease activity is required for the recombination-based ALT cell survival. A. MUS81 endonuclease activity is required for the telomere recombination. GM847 cells were expressed shMUS81-B (in the 3’UTR region) for three days and then were overexpressed wild-type or an endonuclease dead mutant MUS81. After three days, T-SCE was conducted. Quantitative analysis represents three independent experiments (mean ± S.D.). Equal amounts of wild-type or mutant MUS81 were determined by WB analysis (Supplementary Information Fig. S9, Full scans). B. MUS81 wild-type, but not the mutant, rescues the cell growth in ALT cells expressing shMUS81-B. shMUS81-B (in the 3’UTR region) were expressed in U2OS cells for three days and then wild-type or mutant MUS81 were over-expressed in these cells. After two weeks with drug-selection, cell colonies were counted. Depletion of MUS81 and equal amounts of wild-type or mutant MUS81 were determined by WB analysis (Supplementary Information Fig. S9, Full scans). The quantitative data represent three independent experiments (mean ± S.D.).
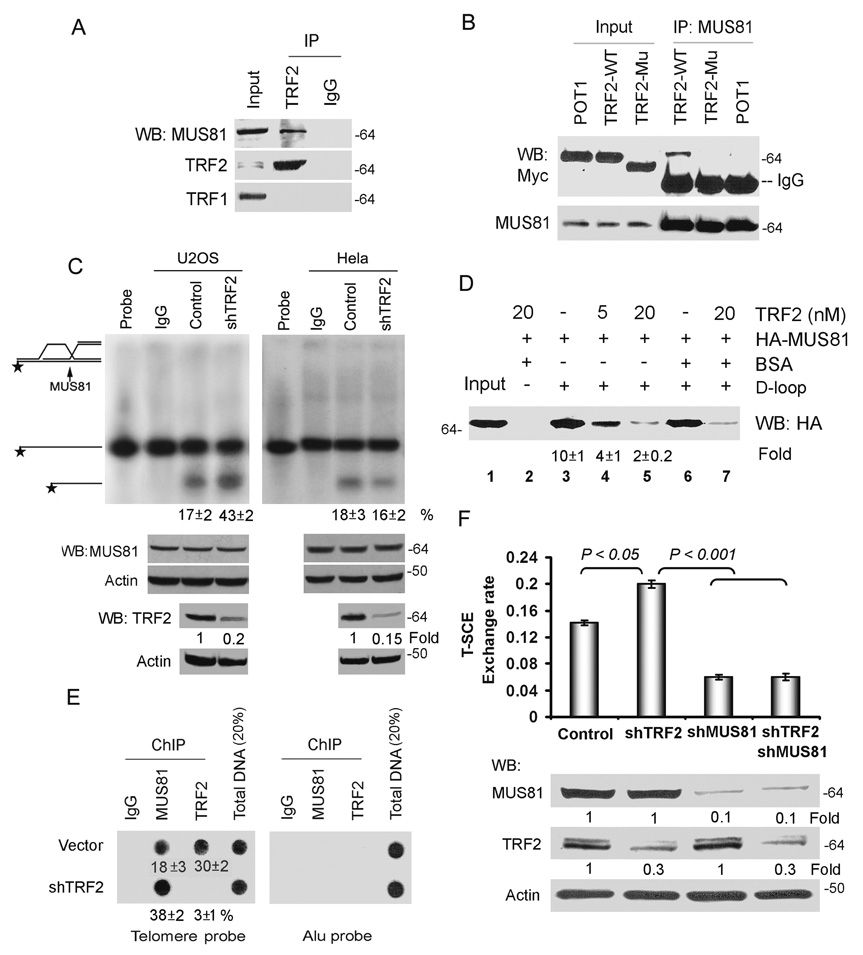
We used a proteomics approach to detect MUS81 interacting partners in the ALT pathway. A stable U2OS cell line expressing His- and Flag-tagged MUS81 was grown. After purification by using the His- and Flag-tag with chromatography columns, the MUS81 immunoprecipitates were analyzed by LC–MS/MS. MUS81 binding partner Eme1 and TRF2 were found in the MUS81 complex. Furthermore, we found that endogenous MUS81 immunoprecipitated with endogenous TRF2 (Fig. 5A). Treatment with DNase I did not affect interaction of TRF2-MUS81, excluding bridging effects of nucleic acids. Moreover, the MUS81 immunocomplex contained TRF2, but not TRF2ΔB, a dominant negative mutant of TRF2 lacking the N-terminal domain (Fig. 5B), confirming the specificity of the association of MUS81 with TRF2.
Fig. 5.
MUS81 physically and functionally interacts with TRF2. A. Endogenous MUS81 binds to TRF2 in U2OS cells by coimmunoprecipitation. Nuclear extracts (0.5 mg) were treated with DNase I and then subjected to immunoprecipitation with the anti-TRF2 antibody (Oncogene), followed by WB analysis with indicated antibodies (Supplementary Information Fig. S9, Full scans). Fifty micrograms of nuclear extract was loaded as input. B. Wild-type TRF2, but not POT1 or TRF2ΔB mutant, binds to MUS81 in U2OS cells. Myc-TRF2, myc-POT1 and myc-TRF2ΔB (TRF2-Mu) were transfected into U2OS cells. MUS81 was coimmunoprecipitated with an anti-MUS81 antibody. Immunoprecipitated proteins were visualized by WB analysis with anti-myc antibody. C. Depletion of TRF2 increases the endonuclease activity of MUS81 in ALT cells. The endonuclease activity assay was carried out by incubating the 32P D-loop substrate with immunoprecipitates of either anti-MUS81 or IgG from U2OS cell lysates (the left panel) or HeLa cell lysates (the right panel) in the presence or absence of TRF2-shRNA. The quantitative data represent three independent experiments (mean ± S.D.). TRF2-shRNA decreased expression level of TRF2 protein by 80%, determined by WB with the anti-TRF2 antibody (Upstate). D, Recombinant TRF2 inhibits MUS81 binding to the D-loop DNA substrate. The biotin-D-loop DNA was incubated with in vitro translated HA-MUS81 in the absence or presence of various amounts of recombinant TRF2 (5 and 20 nmol). The D-loop DNA and its bound proteins on the beads were analysed by SDS-PAGE gel. The quantitative data from three independent experiments (mean ± S.D.). HA-MUS81 was detected by WB with the anti-HA antibody. Input, 50% of the total HA-MUS81 used in each reaction. E. Depletion of TRF2 increases MUS81 binding to telomeres by ChIP assay. ChIP of U2OS cells with or without expression TRF2-shRNA was conducted with indicated antibodies. Dot blots were probed for telomere or Alu repeats. The quantification of the data represents three independent experiments (mean ± S.D.). F. Inhibition of T-SCE by TRF2 requires MUS81. T-SCE was performed with telomeric PNA probes in GM847 cells with expressing empty vector, shTRF2 and/or shMUS81-A. The bar graph represents three independent experiments, showing the mean ± SD and Student t test were used for statistical analyses.
We then determined whether TRF2 regulates MUS81 enzymatic activity. Immunoprecipitates of MUS81 from U2OS cells alone could cleave approximately 17% of the total D-loop substrates (Fig. 5C). However, depletion of TRF2 by shRNA significantly enhanced MUS81-mediated cleavage in ALT cells, but not in non-ALT cells. More than 43% of the total substrates were cleaved upon transduction of TRF2-shRNA in U2OS cells. To exclude that the putative endonuclease activity detected in the nuclease assays derives from another protein co-immunoprecipitating with endogenous MUS81, HA-tagged MUS81 wild-type or nuclease dead construct was expressed and immunoprecipitations and nuclease assays were performed by using a HA antibody. We observed that knockdown of TRF2 also increased the nuclease activity of HA-MUS81 (Supplementary Information, Fig. S7A). Thus, our results suggest that TRF2 may inhibit the MUS81 enzymatic activity. In addition, depletion of TRF2 increased MUS81 mediated-cleavage of 3’ flap substrate made by telomere sequences or non-telomere sequences (Supplementary Information, Fig. S7B), indicating that inhibition of the MUS81 enzymatic activity by TRF2 is not sequence specific. Quantitative Western analysis showed equivalent amounts of immunoprecipitated MUS81 from cells with or without TRF2-shRNA (Fig. 5C), suggesting that TRF2 does not affect the expression levels of MUS81.
Next, we tested whether recombinant TRF2 regulated the binding of HA-MUS81 to DNA structures. Immunoblot analysis with anti-HA antibody showed that in the absence of TRF2, HA-MUS81 was associated with biotinylated-DNA substrates, as retrieved by streptavidin beads (Fig. 5D; Supplementary Information, Fig. S7D). The addition of TRF2 inhibited the binding of HA-MUS81 to the DNA in a dose-dependent manner. These data indicate that TRF2 may suppress MUS81 endonuclease activity through inhibition of MUS81 binding to the DNA substrates. To confirm the results in cells, ChIP assays were performed to determine whether TRF2 regulates the ability of MUS81 to bind to telomeric DNA. Knockdown of TRF2 increased the telomere-bound levels of MUS81 (Fig. 5E). Thus, our results suggest that TRF2 inhibits the MUS81 enzymatic activity by regulation of MUS81 loading onto telomeres during telomere recombination process.
To investigate the functional relationship between TRF2 and MUS81, we examined their effects on telomere recombination. T-SCE rate increased with 20% frequency in GM847 cells upon depletion of TRF2 (Fig. 5F), suggesting that TRF2 suppresses telomere recombination in ALT cells, consistent with a previous model22,23. Knockdown of MUS81 decreased the frequency of T-SCE in cells expressing TRF2-shRNA (Fig. 5F), demonstrating that TRF2-mediated telomere recombination requires MUS81 in ALT cells.
We observed that there was no significant change of telomere circle (t-circle formation) after depletion of MUS81 (Supplementary Information, Fig. S8), suggesting that t-circles may be not involved in MUS81-mediated ALT cell survival. However, because a lower level of t-circles may be insufficient to detect, we can not rule out a role for MUS81 on t-circle formation. TRF2 mediates t-loop formation and disruption of t-loop by TRF2ΔB mutant generates t-circles3,24,25. MUS81 does not interact with TRF2ΔB mutant, implicating that the MUS81 endonuclease may be involved in the disruption of t-loop, which is mediated by TRF2.
In summary, our results demonstrate that MUS81 endonuclease is specifically involved in telomere recombination and in the maintenance of ALT cell survival. We propose a model where telomeres move into APBs for processing recombination based telomere maintenance during G2 phase. MUS81 is recruited into recombination sites to cleave the recombination structure. TRF2 negatively regulates MUS81 enzymatic activity to balance this process. In this manner, ALT cells maintain their telomeres and extend the proliferative lifespan.
METHODS
Cell culture, Western blot analyses and immunoprecipitation assays
ALT cell lines U2OS, GM847, SAOS-2, and non-ALT cell lines MCF7, HT1080, HeLa were cultured in respective mediums (Clotech, Mountain View, CA,). Mouse embryonic fibroblasts (MEFs) of Mus81+/+ and Mus81−/− with expression of human papillomavirus type 16 E6 were obtained from Clare H. McGowan26. Cells synchronized at the G1/S boundary by a double thymidine block were released into the cell cycle and then fixed at specific time points post-release. An additional block by Hoechst 33342, in addition to the double thymidine treatment, enriched cells at G2 phase10. Methionine restriction were performed as described9. To prepare arrested cells for FACS analysis, cells were made as single-cell suspension, fixed in ice-cold 70% ethanol and incubated in 1 ml PI buffer (Propidium iodide 25 µg/ml, 0.1% NP-40, 1 mg/ml RNAse) for FACS analysis. Western blotting analyses and immunoprecipitation were essentially as described previously27.
Lentiviral shRNA constructs and antibodies
MUS81 and TRF2 shRNAs were designed and cloned to the lentiviral construct (Sigma, Saint Louis, MO). shRNA MUS81-A at MUS81 cDNA (NM_025128), positions 973 to 993 (GAGTTGGTACTGGATCACATT); shMUS81-B at positions 1930 to 1950 (TGTCACCAGTTGGTCCTCATC); shRNA TRF2 at TRF2 cDNA (NM_005652), positions 615 to 635 (CCATCCTGTTATCCAGAACTT). The following antibodies were used: anti-mouse MUS81 (abCAM, Cambridge, MD), anti-rabbit MUS81 (gift from Clare H. McGowan), anti-PML, anti-nucleoli/C23 (Santa Cruz, Santa Cruz, CA), anti-TRF1 (Sigma), anti-BrdU (BD, San Jose, CA) and anti-TRF2 (Oncogene, Cambridge, MA; Upstate, Atlanta, GA).
Colony formation, BrdU incorporation and TUNEL assays
All the lentiviral infected cells for colony formation assay were equally seeded into 60 mm dishes (duplicate) and cell colonies were stained with 0.05% crystal violet after two weeks and counted. For BrdU incorporation analysis, cells were cultured in 4-well chamber slides, 100 µM BrdU was added to culture medium for 30 min before fixed with 4% paraformaldehyde. DNA was denatured by two consecutive treatments with 4 M HCl for 15 min. After washing, slides were incubated with anti-BrdU antibody at RT for 30 min, and with secondary antibody at RT for 30 min. Slides were covered with mounting medium and evaluated by fluorescence microscope. The terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay was performed with the TUNEL assay kit from Roche (Indianapolis, IN).
Immunofluresence and telomeric DNA-FISH assays
Cell were cultured on 4-well chamber slides and immunoflurescence staining was performed as describe9. We conducted pre-extract in 0.5% Triton/PBS for 1 minute on ice. For combined telomeric DNA-FISH, after finished immunostaining, slides were fixed, treated with 0.5 µg/ml RNase A in 2×SSC for 45 min at 37°C, and dehydrated in 70%, 80% and 100% ethanol. After denatured at 80°C for 3 min, slides were hybridized for 2 hours in hybridization solution contained 0.3 µg/ml Tel-C PNA telomere probe (Applied Biosystems, Forst City, CA) with 70% formamidine, 1% BSA, and 10 mM Tris, pH 7.2. Then, slides were washed, dehydrated and covered with anti-fading DAPI mounting media.
Telomere Q-FISH and T-SCE (CO-FISH) analyses
Cells were seeded to 40% confluency and cultured 26 hours with 30 εM BrdU (CO-FISH) or without BrdU (Q-FISH). For the metaphases preparation, cells were incubated with 0.1 µg/ml colcemid for 3 hours, followed by hypotonic shock in 0.065% KCl for 10 min at 37°C, and fixed in methanol/acetic acid (3:1) for 10 min twice at RT. Metaphases spread were prepared by dropping fixed cells onto cleared glass slides. Telomere Q-FISH procedure was performed as described28.
CO-FISH was performed as described29. Briefly, after stained with 0.5 µg/ml Hoechst 33258 (Sigma) and exposured UV light for 30 min, newly synthesized DNA strands were degraded by treating slides with 3 U/µl Exo III at RT for 30 min. Slides were washed and digested in 0.05% pepsin (dissolve in 0.005 N HCL) at 37°C for 10 min. FITC-OO-(CCCTAA)3 PNA probe was used as first staining in hybridization buffer (0.3 µg/ml probe, 1% blocking buffer, 10 mM Tris, pH7.2) at RT for 2 hours, then slides were washed and fixed again, followed by Cy3-OO-(TTAGGG)3 complement staining and covered with anti-fading DAPI mounting.
ChIP assays
Cells were cultured in 15 cm plates and ChIP assay was performed by using ChIP assay kit (Upstate) according to the manufacturer. ChIP samples passed through Bio-Dot apparatus by vacuum and loaded on NY+ membrane. The blots were fixed by UV crosslink and hybridized with 32P labeled telomere DNA probe27. Alu probe was used to re-hybridize with the blots after striped previous probe.
TRF and 2-D gel assays
Genomic DNA was isolated following standard procedure30 and digested with Hinf I. Samples were separated by electrophoresis on 0.5% agarose gel at 1v/cm for 18–20 hours. For TRF assay, alkaline transfer was performed. For 2-D gel assay, gels with DNA sample were cut and put into 1% agarose gel, and run for the secondary dimension at 5v/cm for 2.5 hours. Then the alkaline transfer was conducted. Membranes were hybridized with 32P labeled telomere probe.
MUS81 endonuclease assay
Oligonucleotides were labeled by using T4 PNK end labeling method with 32P-γ-ATP. DNA structures were generated by annealing labeled oligonucleotide to their complimentary sequence in annealing buffer (50 mM Tris-HCl, pH 7.5, 50 mM NaCl, 10 mM MgCl2) at 95°C for 4 min, 65°C for 10 min, 37°C for 10 min, 26°C for 10 min and 4°C for 10 min. The substrate products were purified by polyacrylamide gel. Non-telomere 3’-flap sequence was used as described31. Telomere 3’-flap sequences are available upon request. D-loop structure was used and endonuclease assay was performed as described15,32 with modifications. Briefly, 2 µg antibodies were used in each immunoprecipitation with 1 mg cell lysates. MUS81 immune precipitates were washed three times in the lysis buffer (50 mM Tris-HCl, pH 8.0; 120 mM NaCl, 0.05% NP-40) and two times in the reaction buffer (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 10 mM MgCl2, 0.2 mM DTT, 5% Glycerol). Reactions were initiated by adding 1 nM labeled DNA substrates in 25 µl reaction volume then incubate at 37°C for 30 min and stopped in 5x stop solution (10 mg/ml proteinse K, 2.5% SDS, 50 mM Tris-HCl, pH 7.5, 25 mM EDTA) at 37°C for 15 min. Products were separated by electrophoresis in 1×TBE 10% polyacrylamide gel (3’-flap substrate), or with 7M Urea (D-loop substrate).
MUS81-TRF2 in vitro DNA binding assay
The D-loop and 3' flap telomeric DNA substrates were biotinylated (BioServer Biotechnologies, Laurel, MD). In vitro translated HA-MUS81 (2 µL; Promega, Madison, WI) and biotinylated DNA substrates (0.1 pmol) were incubated in the absence or presence of various amounts of recombinant TRF2 for 30 minutes. The biotinylated DNA substrates were retrieved with streptavidin beads, and the unbound proteins were washed away with the binding buffer. Buffers and binding procedures were described previously33. HA-MUS81 bound to the biotinylated A was detected by WB.
Affinity purification and LC-MS/MS analysis of MUS81 associated proteins
A stable cell line expressing His- and flag-tagged MUS81 was grown. After purification by using the His-tag and flag-tag with chromatography columns, approximately 1 mg of protein was obtained from 125 mL of lysates. LC–MS/MS was carried out by nanoflow reverse-phase liquid chromatography (RPLC) coupled online with a quadrupole-orthogonal-time-of-flight tandem mass spectrometer (QSTAR XL, Applied Biosystems/MDS Sciex). The MS/MS spectra were subjected to an in-house MASCOT server for database searching, and the results were complied, validated, and summarized using in-house software.
Supplementary Material
ACKNOWLEDGMENTS
We thank Roger R. Reddel for providing the ALT cells, and Clare H. McGowan for providing the wild-type and mutant MUS81 constructs, MUS81 antibody and Mus81+/+ and Mus81−/− MEFs. We thank Ian Hickson and Joseph Roti Roti for proofreading this manuscript. This work is supported in part by grants from Concern Foundation (QY), NIH CA 10445 and CA 123232 (TKP). This research was also supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
References
- 1.Dunham MA, Neumann AA, Fasching CL, Reddel RR. Telomere maintenance by recombination in human cells. Nat. Genet. 2000;26:447–450. doi: 10.1038/82586. [DOI] [PubMed] [Google Scholar]
- 2.Londono-Vallejo JA, Der-Sarkissian H, Cazes L, Bacchetti S, Reddel RR. Alternative lengthening of telomeres is characterized by high rates of telomeric exchange. Cancer Res. 2004;64:2324–2327. doi: 10.1158/0008-5472.can-03-4035. [DOI] [PubMed] [Google Scholar]
- 3.Wang RC, Smogorzewska A, de Lange T. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell. 2004;119:355–368. doi: 10.1016/j.cell.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 4.Cesare AJ, Reddel RR. Telomere uncapping and alternative lengthening of telomeres. Mech. Ageing Dev. 2008;129:99–108. doi: 10.1016/j.mad.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 5.Bailey SM, Brenneman MA, Goodwin EH. Frequent recombination in telomeric DNA may extend the proliferative life of telomerase-negative cells. Nucleic Acids Res. 2004;32:3743–3751. doi: 10.1093/nar/gkh691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yeager TR, et al. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 1999;59:4175–4179. [PubMed] [Google Scholar]
- 7.Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddel RR. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 1995;14:4240–4248. doi: 10.1002/j.1460-2075.1995.tb00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muntoni A, Reddel RR. The first molecular details of ALT in human tumor cells. Hum. Mol. Genet. 2005;14(Spec No. 2):R191–R196. doi: 10.1093/hmg/ddi266. [DOI] [PubMed] [Google Scholar]
- 9.Jiang WQ, Zhong ZH, Henson JD, Reddel RR. Identification of candidate alternative lengthening of telomeres genes by methionine restriction and RNA interference. Oncogene. 2007;26:4635–4647. doi: 10.1038/sj.onc.1210260. [DOI] [PubMed] [Google Scholar]
- 10.Wu G, Lee WH, Chen PL. NBS1 and TRF1 colocalize at promyelocytic leukemia bodies during late S/G2 phases in immortalized telomerase-negative cells. Implication of NBS1 in alternative lengthening of telomeres. J Biol Chem. 2000;275:30618–30622. doi: 10.1074/jbc.C000390200. [DOI] [PubMed] [Google Scholar]
- 11.Grobelny JV, Godwin AK, Broccoli D. ALT-associated PML bodies are present in viable cells and are enriched in cells in the G(2)/M phase of the cell cycle. J. Cell Sci. 2000;113(Pt 24):4577–4585. doi: 10.1242/jcs.113.24.4577. [DOI] [PubMed] [Google Scholar]
- 12.Blais V, et al. RNA interference inhibition of Mus81 reduces mitotic recombination in human cells. Mol. Biol. Cell. 2004;15:552–562. doi: 10.1091/mbc.E03-08-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ehmsen KT, Heyer WD. Saccharomyces cerevisiae Mus81-Mms4 is a catalytic, DNA structure-selective endonuclease. Nucleic Acids Res. 2008;36:2182–2195. doi: 10.1093/nar/gkm1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Osman F, Whitby MC. Exploring the roles of Mus81-Eme1/Mms4 at perturbed replication forks. DNA Repair (Amst) 2007;6:1004–1017. doi: 10.1016/j.dnarep.2007.02.019. [DOI] [PubMed] [Google Scholar]
- 15.Chen XB, et al. Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol. Cell. 2001;8:1117–1127. doi: 10.1016/s1097-2765(01)00375-6. [DOI] [PubMed] [Google Scholar]
- 16.Boddy MN, et al. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107:537–548. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- 17.Hollingsworth NM, Brill SJ. The Mus81 solution to resolution: generating meiotic crossovers without Holliday junctions. Genes Dev. 2004;18:117–125. doi: 10.1101/gad.1165904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heyer WD. Recombination: Holliday junction resolution and crossover formation. Curr. Biol. 2004;14:R56–R58. doi: 10.1016/j.cub.2003.12.043. [DOI] [PubMed] [Google Scholar]
- 19.Gao H, Chen XB, McGowan CH. Mus81 endonuclease localizes to nucleoli and to regions of DNA damage in human S-phase cells. Mol. Biol. Cell. 2003;14:4826–4834. doi: 10.1091/mbc.E03-05-0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perrem K, Colgin LM, Neumann AA, Yeager TR, Reddel RR. Coexistence of alternative lengthening of telomeres and telomerase in hTERT-transfected GM847 cells. Mol. Cell Biol. 2001;21:3862–3875. doi: 10.1128/MCB.21.12.3862-3875.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cerone MA, Londono-Vallejo JA, Bacchetti S. Telomere maintenance by telomerase and by recombination can coexist in human cells. Hum. Mol. Genet. 2001;10:1945–1952. doi: 10.1093/hmg/10.18.1945. [DOI] [PubMed] [Google Scholar]
- 22.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 23.Palm W, de Lange T. How Shelterin Protects Mammalian Telomeres. Annu. Rev. Genet. 2008 doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 24.Griffith JD, et al. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 25.Stansel RM, de Lange T, Griffith JD. T-loop assembly in vitro involves binding of TRF2 near the 3' telomeric overhang. EMBO J. 2001;20:5532–5540. doi: 10.1093/emboj/20.19.5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dendouga N, et al. Disruption of murine Mus81 increases genomic instability and DNA damage sensitivity but does not promote tumorigenesis. Mol. Cell Biol. 2005;25:7569–7579. doi: 10.1128/MCB.25.17.7569-7579.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Q, Zheng YL, Harris CC. POT1 and TRF2 cooperate to maintain telomeric integrity. Mol. Cell Biol. 2005;25:1070–1080. doi: 10.1128/MCB.25.3.1070-1080.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia-Cao M, Gonzalo S, Dean D, Blasco MA. A role for the Rb family of proteins in controlling telomere length. Nat. Genet. 2002;32:415–419. doi: 10.1038/ng1011. [DOI] [PubMed] [Google Scholar]
- 29.Gonzalo S, et al. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat. Cell Biol. 2006;8:416–424. doi: 10.1038/ncb1386. [DOI] [PubMed] [Google Scholar]
- 30.Karlseder J, Smogorzewska A, de Lange T. Senescence induced by altered telomere state, not telomere loss. Science. 2002;295:2446–2449. doi: 10.1126/science.1069523. [DOI] [PubMed] [Google Scholar]
- 31.Kaliraman V, Mullen JR, Fricke WM, Bastin-Shanower SA, Brill SJ. Functional overlap between Sgs1-Top3 and the Mms4-Mus81 endonuclease. Genes Dev. 2001;15:2730–2740. doi: 10.1101/gad.932201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang R, et al. BLM helicase facilitates Mus81 endonuclease activity in human cells. Cancer Res. 2005;65:2526–2531. doi: 10.1158/0008-5472.CAN-04-2421. [DOI] [PubMed] [Google Scholar]
- 33.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.