Bone morphogenetic protein (BMP) signaling activates transcription of the master iron regulator hepcidin in the liver. This study shows that a heterozygous mutation in the BMP-responsive element of the hepcidin gene promoter is associated with massive iron overload in a patient homozygous for the common HFE mutation, suggesting a new molecular mechanism of iron overload.
Keywords: hepcidin, BMP, hemochromatosis, iron, gene expression
Abstract
Low levels of hepcidin are responsible for the development of iron overload in p.Cys282Tyr HFE related hemochromatosis. Every genetic factor lowering the hepcidin gene expression could contribute to a more severe phenotype in HFE hemochromatosis. Based on this hypothesis, we identified a heterozygous nc.-153 C>T mutation in the hepcidin gene promoter sequence in a patient homozygous for the p.Cys282Tyr HFE mutation who presented massive iron overload, resisting to well conducted iron depletive treatment. Our results demonstrate that the nc.-153 C>T mutation, located within a BMP-RE (Bone Morphogenetic Protein-Responsive Element): i) decreases the transcriptional activity of the hepcidin promoter, ii) alters its IL-6 (Interleukin-6) total responsiveness, and iii) prevents the binding of the SMAD protein complex (1/5/8 and 4) to the BPM-RE. In conclusion, our results suggest that a mutation in the BMP-RE of hepcidin promoter may impact on human iron metabolism.
Introduction
Hepcidin, synthesized mainly by hepatocytes and secreted in plasma,1,2 plays a role in the control of iron metabolism3 by interacting with ferroportin,4 the major iron exporter expressed especially on macrophages and on basolateral membranes of enterocytes. Hepcidin induces degradation of ferroportin and thus iron sequestration within these cells.4,5 Therefore, hepcidin may modulate plasma iron concentration and transferrin saturation.
The homozygous p.Cys282Tyr mutation of HFE gene is the most frequent etiology of genetic hemochromatosis (GH)6 linked to an inaccurate low level of hepcidin regarding iron status.7,8 The molecular mechanisms linking the p.Cys282Tyr mutation and low hepcidin levels are not fully characterized. Mutations in other genes, including transferrin receptor 2 (TFR2), hemojuvelin (HJV) and ferroportin (FPN),9,10 as well as in coding and non-coding regions of hepcidin (HAMP),11–13 are known to induce phenotypic presentations similar to HFE related GH. HFE related GH is a disease with incomplete penetrance.6 This suggests that modifier genes modulate the expressivity of HFE hemochromatosis. Mutations in hepcidin (HAMP), TFR2 or HJV genes have been demonstrated to favor the development of iron overload in p.Cys282Tyr heterozygote patients14 and to increase iron burden in p.Cys282Tyr HFE homozygotes.15,16
In HFE-GH it can be hypothesized that every genetic factor lowering hepcidin gene expression could lead to a more severe phenotype. Therefore, we searched for mutations in iron metabolism regulatory genes in one patient with homozygous p.Cys282Tyr mutation presenting severe iron overload. We identified a heterozygous mutation located within the recently reported BMP-RE17 which could influence hepcidin gene expression. In this report, we demonstrate that the nc.-153 C>T hepcidin promoter mutation decreases its transcriptional activity thus underlining the potential impact of mutation in BMP-RE of hepcidin promoter on iron metabolism in humans.
Design and Methods
Patient and genetic studies
In 1994, the diagnosis of genetic hemochromatosis was established in a 37-year old man with massive hepatic iron overload (450 μmoL of iron/g dry liver weight, n<36) involving mainly hepatocytes and associated to liver fibrosis stage 2–3 in Metavir classification.18 Iron depletive treatment based on weekly 400 mL phlebotomies was initiated. In 1997, the patient was demonstrated to be homozygous for the p.Cys282Tyr mutation. In 2004 (47 years), 468 venesections had been performed, corresponding to the removal of 85g of iron. However, the patient was always exhibiting abnormal iron parameters with 40 μmol/L for serum iron, 100% for transferrin saturation, and 2,066 μg/L for serum ferritin. In addition, hepatic iron concentration remained very high (close to 300 μmol/g dry liver weight) at MRI. Serum hepcidin level quantified by Elisa19 was low (24 ng/L; 29<N< 254 for men) despite major iron overload with a decoupling between serum hepcidin and ferritin values compared to the correlation between both parameters reported by Ganz et al.19 Clinical iron burden consisted of asthenia, icthyosis, melanodermia, gonalgia, osteoporosis, and hypogonadism. There were no heart symptoms and serum glucose was normal. Prothrombin time was normal (90%). Serum transaminases and GGT activities were slightly increased (1.5-2N). Complete sequencing of the coding sequence of the HFE, HJV, TFR2, FPN and HAMP (including its promoter) genes was performed, as part of the diagnostic procedure, after having obtained a written informed consent from the patient. Unfortunately, the realization of a family study did not prove possible.
Sequencing of these genes was performed in one hundred subjects recruited from a health appraisal center with normal iron metabolism as judged on serum iron parameters and hemoglobin levels. These subjects had given their informed written consent for the study of iron metabolism genes after approval of the protocol by the local ethical committee (98/35-197).
Cell culture
Human hepatoma HepG2 cells were grown in Minimum Essential Medium α (invitrogen) supplemented with 10% fetal bovine serum (invitrogen), 100 UI/mL penicillin, 100 mg/mL streptomycin and 2 mM L-glutamine.
Plasmid constructions
The -1024 bp from the translation start site of the human hepcidin promoter were amplified from HuH7 genomic DNA and subcloned into pGL4.17 (Promega) to generate -1024/BMP-RE wt Hep/Luc plasmid construct. The internal mutations in the BMP responsive element or in the STAT3 DNA binding site were made using -1024 Hep/Luc as template and the internal following primers: GCCTTTTCGGTGCCACCACC; antisense GGTGGTGGCACCGAAAAGGC for BMP-RE or CACCTTCTTGGCCGTGAGAC, antisense GTCTCACGGCCAAGAAGGTG for STAT3 binding site. The nucleotide mutations are underlined. The identity of the constructs was confirmed by DNA sequencing.
Transfection, luciferase assay
Cells were transfected, using transfectine (Bio-Rad) with 100 ng of pGL4-hepcidin promoter vector with a normalization plasmid (pRL-TK, Promega). After 48 h Luciferase activities were measured in cell lysates using the Dual-Luciferase-Reporter assay system (Promega) and a Centro LB 960 luminometer (Berthold Technologies). For treatments, either 10 ng/mL of human BMP9 or 20 ng/mL of human BMP4 or 20 ng/mL IL-6 (R&D systems) were added to the cells for 48 h.
Electrophoretic mobility shift assay
HepG2 cells were incubated with either 10 ng/mL of human BMP9 or 20 ng/mL of human BMP4 for one hour. Then crude nuclear extracts were prepared as described.20 For gel retardation assays, nuclear extracts (10 μg) were preincubated with 250 ng of poly(dI-dC).poly(dI-dC), used as non-specific competitor DNA, in a binding buffer (10 mM Tris-HCl pH 7.5, 50mM NaCl, 1mM DTT, 1 mM EDTA, 5% glycerol) for 10 min on ice. This mixture was then added to the 32P-end-labeled probe either with or without specific competitor oligonucleotides, and the incubation was carried out for a further 30 min at room temperature. Gel shift assay with purified polyclonal rabbit IgG anti-SMAD4 or anti-SMAD1/5/8 (Santa Cruz) or non-relevant antibody were performed under the same conditions except that the antibodies were preincubated with nuclear extracts for one hour on ice. Complexes were resolved by electrophoresis on prerun Tris-Glycine 5% native polyacrylamide gels. The following chemically synthesized double-stranded oligonucleotides were used as probes and as specific competitors (nucleotide mutation is underlined): BMPRE-wt GCCTTTTCGGCGCCACCACC, BMPRE-mut GCCTTTTCGGTGCCACCACC.
Results and Discussion
The heterozygous nc -153 C>T was found in hepcidin promoter
In genetic hemochromatosis, iron loading results from abnormally low levels of hepcidin linked to mutations in the hepcidin gene itself or in genes involved in hepcidin expression control, such as the HFE, HJV and TfR2 genes.9,10 This prompted us to search for additional mutations in iron regulatory genes in a p.Cys282YTyr homozygous patient presenting a massive iron overload. The sequencing of the HFE gene confirmed the homozygous p.CysC282Tyr mutation and excluded another mutation. In addition, sequencing of HJV, TFR2 and FPN genes were normal. Finally, sequencing of the hepcidin gene led to the identification of a punctual mutation in position -153 from the translational start site. This mutation was localized within the BMP-RE as recently proposed,17 and was found neither in the web databases nor in 200 chromosomes issued from 100 healthy volunteers.
The heterozygous nc.-153 C>T mutation decreases hepcidin promoter transcriptional activity
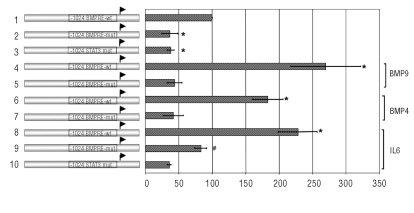
As described previously,17 the BMP-RE controls human hepcidin promoter transcriptional activity under steady state-condition. In order to assess the effect of the heterozygous nc.-153 C>T mutation on basal hepcidin gene expression, plasmid constructs containing the -1024 bp of the human hepcidin promoter with the wild type or the mutated BMP-RE inserted among the luciferase gene (-1024/BMP-RE wt Hep/Luc and -1024/BMP-RE mut Hep/Luc) were transfected in HepG2 cells. Luciferase activity from the -1024/BMP-RE mut Hep/Luc construct was about 2.4 fold reduced compared to the luciferase activity from the plasmid construct containing the wild type BMP-RE (Figure 1, lanes 1–2), demonstrating that this mutation decreased hepcidin transcriptional activity in basal condition. As this BMP-RE mediates the induction of hepcidin gene expression in response to BMP,17 HepG2 cells were transfected with the -1024/BMP-RE wt or mutated Hep/Luc plasmid constructs and were treated with either BMP9 or BMP4 in order to determine if the mutation impaired or not the response to these BMPs.21 As expected, for -1024/BMP-RE wt Hep/Luc construct, the BMP9 treatment increased luciferase activity by about 2.7 fold and BMP4 treatment by 1.8 fold (Figure 1, lanes 1,4,6). By contrast, with the -1024/BMP-RE mut Hep/Luc after BMP9 or BMP4 treatment, there was no change observed for luciferase activities compared to the basal condition (Figure 1, lanes 2,5,7). These results demonstrate that the heterozygous nc.-153 C>T mutation affects the basal hepcidin gene expression and impairs its BMP4/9 response.
Figure 1.
The heterozygous -153C/T mutation decreases hepcidin promoter transcription activity. HepG2 cells were transfected with the -1024/BMP-RE wt or -1024/BMP-RE mut Hep/Luc constructs and were treated with BMP9 (lanes 4,5), BMP4 (lanes 6,7), IL-6 (lanes 8,9,10) or not (lanes 1–2) for 48 h. Luciferase activity values represent Firefly/Renilla Luciferase activity ratios relative to that obtained with the -1024/BMP-RE wt Hep/Luc plasmid construct, which was arbitrarily set at 100%. Data represent means of at least three independent experiments (± SEM). *p<0.001 compared to the -1024/BMP-RE wt Hep/Luc construct and #p<0.001 compared to the -1024/BMP-RE mut determined by 1-way ANOVA and the Student-Newman-Keuls comparisons test.
Hepcidin gene responsiveness to IL-6 has been recorded decreasing when the BMP-RE or STAT3 binding sites are mutated17 or in SMAD4−/− mice.22 Accordingly, we found (Figure 1, lanes 8–9), by comparing the impact of IL-6 on wild type and mutated promoter, that the nc.153C>T was critical for IL-6 total responsiveness. However, the mutation did not affect the ability to respond in terms of fold induction, by comparing the ratio between basal and IL-6 stimulated mutated construct (Figure 1 lanes 2,9) to the ratio between basal and IL-6 stimulated wild type construct. (Figure 1, lanes 1,8).
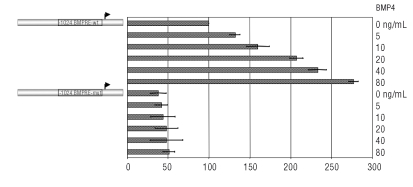
To test the severity of the mutation we performed a dose dependent assessment of the BMP4 effect (Figure 2). Whereas we found a dose dependent induction of Luciferase activity from the -1024/BMP-RE wt, no response was observed for the -1024/BMP-RE mut construct, thus demonstrating the critical impact of the mutation on the BMP related signaling. Taken together, these results demonstrate that the nc.-153 C>T mutation affects the basal hepcidin gene expression and impairs its response to BMP4/9 and the IL-6 total responsiveness.
Figure 2.
The -1024 BMPRE-wt Hep/Luc but not the mutated constructs responds in a dose-dependent manner to BMP4. HepG2 cells were transfected with the -1024 BMPRE wt or mutated and were treated with increasing amount of BMP4.
The heterozygous nc.-153 C>T mutation impairs the binding of SMAD1/5/8/4 to the BMP-RE
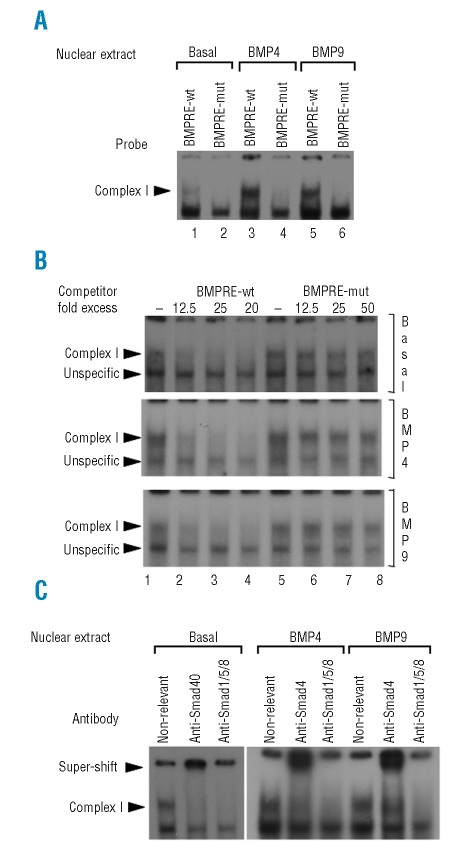
The BMP-RE DNA region is a potential binding site for SMAD1/5/8 transcription factors, which are phosphorylated upon BMP binding to their receptors and migrate to the nucleus associated to SMAD4, to activate transcription of target genes.23 In order to determine if the heterozygous nc.-153 C>T mutation impaired the formation of a nucleoprotein complex of SMAD1/5/8 with the BMP-RE, an EMSA was performed using nuclear extracts from HepG2 cells treated or not with BMP4 or BMP9 and probes containing the wild type or mutated BMP-RE. A complex was observed using the probe containing the wild type BMP-RE (Figure 3A, lanes 1,3,5; complex I). This complex I is present to a lower extent in nuclear extracts obtained from basal condition compared to those from BMP stimulated cells. The complex I formation was inhibited by using a probe which contained the mutated BMP-RE (Figure 3A, lanes 2,4,6). The specificity of formation of this complex with the probe containing the wild type BMP-RE was demonstrated using unlabeled oligonucleotides containing wild type or mutated BMP-RE in excess. Complex I binding was competed with the wild type BMP-RE used as cold competitor (Figure 3B, lanes 1–4) whereas competitor with the mutated BMP-RE had no effect (Figure 3B, lanes 5–8), thus demonstrating its binding specificity. The lower band was not affected by the wt competitor, demonstrating its unspecificity.
Figure 3.
The -153C/T mutation impairs the binding of SMAD1/5/8/4 to the BMP responsive element. (A) EMSA was performed using HepG2 cell nuclear extracts treated or not with either BMP4 or BMP9, which were incubated with radiolabeled probes containing the BMP-RE wt or mutated. The presence of a nucleoprotein complex (complex I) was found with the BMP-RE wt probe and not with the mutated. (B) Binding specificity of complex I was ascertained by EMSA using HepG2 cell nuclear extracts treated or not with either BMP4 or BMP9, which were incubated with the radiolabeled probe containing the BMP-RE wt and 12.5, 25 or 50 fold excess of cold competitor oligonucleotides containing the BMP-RE wt or mutated. The second arrowhead corresponds to an unspecific complex. (C) Protein content of complex I was analyzed by EMSA using HepG2 cell nuclear extracts treated or not with either BMP4 or BMP9, which were incubated with a non-relevant, SMAD4 or SMAD1/5/8 antibody.
To identify the protein content of complex I, HepG2 nuclear extracts were incubated with antibodies against a non-relevant protein as control, SMAD4 or SMAD1/5/8. Supershifts were observed with anti-SMAD4 antibody and anti-SMAD1/5/8 antibody specifically blocked the nucleoprotein complex I formation, thus inducing its disappearance. These results suggest that in HepG2 cells, a nucleoprotein complex containing the SMAD1/5/8/4 proteins is observed with the wild type BMP-RE DNA binding site and that this nucleoprotein complex formation is abrogated by the nc.-153 C>T mutation. Thus, the impairment of SMAD1/5/8/4 binding to the nc.-153 C>T mutated BMP-RE could reduce hepcidin gene expression.
Taken together, our data supports the deleterious impact of the nc.-153 C>T hepcidin promoter mutation. The iron depletive treatment (85g of iron subtracted, including 18g potentially absorbed iron during the whole venesection period, according to Olsson et al.24 was only partially successful as compared with what can be expected in classical HFE hemochromatosis as well as in juvelin hemochromatosis related to either hepcidin or hemojuvelin gene mutations. These data suggest that: i) the nc.-153 C>T mutation may have an additive effect to the homozygous p.Cys282Tyr mutation, which remains to be characterized and, ii) that the addition of an iron chelator treatment could be relevant both to potentiate iron elimination and to limit venesection-related iron absorption.
Our overall results suggest that mutations in BMP-RE of hepcidin promoter may alter iron homeostasis, contributing to the increase in the iron burden in HFE related hemochromatosis.
Acknowledgments:
Acknowledgments: we also wish to thank Dr. Jean-Michel Rotty (Ploermel hospital, France) for having referred the patient to us.
Footnotes
Authorship and Disclosures
MLI designed and performed experiments, analyzed data and wrote the manuscript. PB diagnosed the patient and participated in writing the manuscript. AMJ and AM performed sequencing and analyzed data. VD and YD coordinated the study in healthy volunteers and participated in writing the manuscript. OL initiated the study, analyzed data and wrote the paper.
The authors reported no potential conflicts of interest.
Funding: This work was supported by the LSHM-CT-2006-037296 European Community Grant (Euroiron1), the Association Fer et Foie, the French National Centre for Rare Genetic Iron Overload Disorders, and the Biological Ressources Centre of Rennes.
References
- 1.Pigeon C, Ilyin G, Courselaud B, Leroyer P, Turlin B, Brissot P, et al. A new mouse liver-specific gene, encoding a protein homologous to human antimicrobial peptide hepcidin, is overexpressed during iron overload. J Biol Chem. 2001;276:7811–9. doi: 10.1074/jbc.M008923200. [DOI] [PubMed] [Google Scholar]
- 2.Ganz T. Hepcidin, a urinary antimicrobial peptide synthesized in the liver. J Biol Chem. 2001;276:7806–10. doi: 10.1074/jbc.M008922200. [DOI] [PubMed] [Google Scholar]
- 3.Nicolas G, Bennoun M, Devaux I, Beaumont C, Grandchamp B, Kahn A, et al. Lack of hepcidin gene expression and severe tissue iron overload in upstream stimulatory factor 2 (USF2) knockout mice. Proc Natl Acad Sci USA. 2001;98:8780–5. doi: 10.1073/pnas.151179498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, Ward DM, et al. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306:2090–3. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- 5.De Domenico I, Ward DM, Langelier C, Vaughn MB, Nemeth E, Sundquist WI, et al. The molecular mechanism of hepcidin-mediated ferroportin down-regulation. Mol Biol Cell. 2007;18:2569–78. doi: 10.1091/mbc.E07-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brissot P, Troadec MB, Bardou-Jacquet E, Lan CL, Jouanolle AM, Deugnier Y, et al. Current approach to hemochromatosis. Blood Rev. 2008;22:195–210. doi: 10.1016/j.blre.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 7.Gehrke SG, Kulaksiz H, Herrmann T, Riedel HD, Bents K, Veltkamp C, et al. Expression of hepcidin in hereditary hemochromatosis: evidence for a regulation in response to serum transferrin saturation and non-transferrin-bound iron. Blood. 2003;102:371–6. doi: 10.1182/blood-2002-11-3610. [DOI] [PubMed] [Google Scholar]
- 8.Bridle KR, Frazer DM, Wilkins SJ, Dixon JL, Purdie DM, Crawford DH, et al. Disrupted hepcidin regulation in HFE-associated haemochromatosis and the liver as a regulator of body iron homoeostasis. Lancet. 2003;361:669–73. doi: 10.1016/S0140-6736(03)12602-5. [DOI] [PubMed] [Google Scholar]
- 9.Roetto A, Camaschella C. New insights into iron homeostasis through the study of non-HFE hereditary haemochromatosis. Best Pract Res Clin Haematol. 2005;18:235–50. doi: 10.1016/j.beha.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 10.Pietrangelo A. Non-HFE hemochromatosis. Hepatology. 2004;39:21–9. doi: 10.1002/hep.20007. [DOI] [PubMed] [Google Scholar]
- 11.Roetto A, Papanikolaou G, Politou M, Alberti F, Girelli D, Christakis J, et al. Mutant antimicrobial peptide hepcidin is associated with severe juvenile hemochromatosis. Nat Genet. 2003;33:21–2. doi: 10.1038/ng1053. [DOI] [PubMed] [Google Scholar]
- 12.Matthes T, Aguilar-Martinez P, Pizzi-Bosman L, Darbellay R, Rubbia-Brandt L, Giostra E, et al. Severe hemochromatosis in a Portuguese family associated with a new mutation in the 5'-UTR of the HAMP gene. Blood. 2004;104:2181–3. doi: 10.1182/blood-2004-01-0332. [DOI] [PubMed] [Google Scholar]
- 13.Porto G, Roetto A, Daraio F, Pinto JP, Almeida S, Bacelar C, et al. A Portuguese patient homozygous for the -25G>A mutation of the HAMP promoter shows evidence of steady-state transcription but fails to up-regulate hepcidin levels by iron. Blood. 2005;106:2922–3. doi: 10.1182/blood-2005-04-1630. [DOI] [PubMed] [Google Scholar]
- 14.Merryweather-Clarke AT, Cadet E, Bomford A, Capron D, Viprakasit V, Miller A, et al. Digenic inheritance of mutations in HAMP and HFE results in different types of haemochromatosis. Hum Mol Genet. 2003;12:2241–7. doi: 10.1093/hmg/ddg225. [DOI] [PubMed] [Google Scholar]
- 15.Le Gac G, Scotet V, Ka C, Gour-laouen I, Bryckaert L, Jacolot S, et al. The recently identified type 2A juvenile haemochromatosis gene (HJV), a second candidate modifier of the C282Y homozygous phenotype. Hum Mol Genet. 2004;13:1913–8. doi: 10.1093/hmg/ddh206. [DOI] [PubMed] [Google Scholar]
- 16.Jacolot S, Le Gac G, Scotet V, Quere I, Mura C, Ferec C. HAMP as a modifier gene that increases the phenotypic expression of the HFE pC282Y homozygous genotype. Blood. 2004;103:2835–40. doi: 10.1182/blood-2003-10-3366. [DOI] [PubMed] [Google Scholar]
- 17.Verga Falzacappa MV, Casanovas G, Hentze MW, Muckenthaler MU. A bone morphogenetic protein (BMP)-responsive element in the hepcidin promoter controls HFE2-mediated hepatic hepcidin expression and its response to IL-6 in cultured cells. J Mol Med. 2008;86:531–40. doi: 10.1007/s00109-008-0313-7. [DOI] [PubMed] [Google Scholar]
- 18.The French METAVIR Cooperative Study Group. TFMCS. Intraobserver and interobserver variations in liver biopsy interpretation in patients with chronic hepatitis C. Hepatology. 1994;20:15–20. [PubMed] [Google Scholar]
- 19.Ganz T, Olbina G, Girelli D, Nemeth E, Westerman M. Immunoassay for human serum hepcidin. Blood. 2008;112:4292–7. doi: 10.1182/blood-2008-02-139915. [DOI] [PubMed] [Google Scholar]
- 20.Therrien M, Drouin J. Cell-specific helix-loop-helix factor required for pituitary expression of the pro-opiomelanocortin gene. Mol Cell Biol. 1993;13:2342–53. doi: 10.1128/mcb.13.4.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Babitt JL, Huang FW, Wrighting DM, Xia Y, Sidis Y, Samad TA, et al. Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nat Genet. 2006;38:531–9. doi: 10.1038/ng1777. [DOI] [PubMed] [Google Scholar]
- 22.Wang RH, Li C, Xu X, Zheng Y, Xiao C, Zerfas P, et al. A role of SMAD4 in iron metabolism through the positive regulation of hepcidin expression. Cell Metab. 2005;2:399–409. doi: 10.1016/j.cmet.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 23.Kusanagi K, Inoue H, Ishidou Y, Mishima HK, Kawabata M, Miyazono K. Characterization of a bone morphogenetic protein-responsive Smad-binding element. Mol Biol Cell. 2000;11:555–65. doi: 10.1091/mbc.11.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olsson KS, Vaisanen M, Konar J, Bruce A. The effect of withdrawal of food iron fortification in Sweden as studied with phlebotomy in subjects with genetic hemochromatosis. Eur J Clin Nutr. 1997;51:782–6. doi: 10.1038/sj.ejcn.1600488. [DOI] [PubMed] [Google Scholar]