Abstract
The virescent3 (v3) and stripe1 (st1) mutants in rice (Oryza sativa) produce chlorotic leaves in a growth stage-dependent manner under field conditions. They are temperature-conditional mutants that produce bleached leaves at a constant 20°C or 30°C but almost green leaves under diurnal 30°C/20°C conditions. Here, we show V3 and St1, which encode the large and small subunits of ribonucleotide reductase (RNR), RNRL1, and RNRS1, respectively. RNR regulates the rate of deoxyribonucleotide production for DNA synthesis and repair. RNRL1 and RNRS1 are highly expressed in the shoot base and in young leaves, and the expression of the genes that function in plastid transcription/translation and in photosynthesis is altered in v3 and st1 mutants, indicating that a threshold activity of RNR is required for chloroplast biogenesis in developing leaves. There are additional RNR homologs in rice, RNRL2 and RNRS2, and eukaryotic RNRs comprise α2β2 heterodimers. In yeast, RNRL1 interacts with RNRS1 (RNRL1:RNRS1) and RNRL2:RNRS2, but no interaction occurs between other combinations of the large and small subunits. The interacting activities are RNRL1:RNRS1 > RNRL1:rnrs1(st1) > rnrl1(v3):RNRS1 > rnrl1(v3):rnrs1(st1), which correlate with the degree of chlorosis for each genotype. This suggests that missense mutations in rnrl1(v3) and rnrs1(st1) attenuate the first αβ dimerization. Moreover, wild-type plants exposed to a low concentration of an RNR inhibitor, hydroxyurea, produce chlorotic leaves without growth retardation, reminiscent of v3 and st1 mutants. We thus propose that upon insufficient activity of RNR, plastid DNA synthesis is preferentially arrested to allow nuclear genome replication in developing leaves, leading to continuous plant growth.
Plastid development from proplastids to photosynthetically active chloroplasts is one of the most important metabolic processes during plant growth and is coordinately regulated by both plastid and nuclear genes. Chloroplast development is largely under nuclear control, because the coding capacity of plastids is very limited and nuclear genes encode more than 95% of the chloroplast proteins. Thus, the precise coordination of gene expression through two-way signaling between plastids and the nucleus is essential for chloroplast biogenesis in plant cells (Mandel et al., 1996; Koussevitzky et al., 2007).
A number of chlorophyll (Chl)- and chloroplast-associated mutations that affect leaf coloration and/or seedling viability have been identified and are referred to as virescent (v), stripe (st), albino, chlorina, zebra, and yellow variegated depending on their diverse phenotypes. Among these mutants, v plants suffer from Chl deficiency in the leaves that develop during the early growth stages and produce mostly green leaves during the late growth stages (Archer and Bonnett, 1987). This developmental phenotype suggests that some of the key factors required for Chl synthesis and/or chloroplast development are absent or insufficient at the earlier developmental stages but are present at adequate levels as the mutant plants mature. Three temperature-conditional v mutants (v1, v2, and v3) have been reported in rice (Oryza sativa; Iba et al., 1991). These mutants produce chlorotic leaves at a restrictive temperature (constant 20°C) but develop nearly green leaves at a permissive temperature (constant 30°C). It has been proposed that the V1 gene products should be involved in the timing of constructing the plastid transcription/translation apparatus, which is needed for chloroplast differentiation in developing leaves (Kusumi et al., 1997). Indeed, the V2 gene was recently identified to encode a new type of plastid/mitochondrial guanylate kinase (pt/mt GK) that regulates guanine nucleotide pools in developing leaves (Sugimoto et al., 2007).
In this study, we report our findings for two temperature-conditional Chl-deficient rice mutants, v3 and st1, which harbor mutations in the open reading frames (ORFs) of the V3 and St1 genes that encode the large and small subunits of ribonucleotide reductase (RNR), respectively. RNR is an essential enzyme for DNA replication and damage repair in all living organisms, because it provides the DNA precursors by catalyzing the de novo synthesis of deoxyribonucleotide diphosphates from their corresponding ribonucleotide diphosphates (Elledge et al., 1992; Eriksson et al., 1997). RNR also plays a key role in maintaining genomic stability in mammals, fungi, and plants, and its activity is precisely regulated at the transcriptional and posttranslational levels. These regulatory mechanisms include the control of RNR mRNA stability (Saitoh et al., 2002), the repression of RNR transcription (Huang et al., 1998), the inhibition of RNR activity (Chabes et al., 1999; Zhao et al., 2000), and the control of RNR degradation (Chabes et al., 2003). The existence of these multiple regulatory mechanisms for RNR activity underlines the importance of a proper dNTP pool for the fitness and survival of an organism.
In contrast to the many studies of RNR functions in yeast and mammals, there are few reports describing the roles of this enzyme in higher plants. R1 and R2 genes have been identified in tobacco (Nicotiana tabacum) and Arabidopsis (Arabidopsis thaliana; Philipps et al., 1995; Chabouté et al., 1998). In Arabidopsis, the large and small subunits of RNR were isolated to investigate their functions (Elleingand et al., 1998; Sauge-Merle et al., 1999). The large subunit (RNR1) in Arabidopsis was found to be encoded by a single-copy gene, and the small subunit (RNR2) was encoded by three genes (Philipps et al., 1995), whereas tobacco has two large RNR1 genes and one small RNR2 gene (Chabouté et al., 1998). Recently in Arabidopsis, mutations in the small subunit RNR2 genes such as TSO2, RNR2A, and RNR2B were characterized (Wang and Liu, 2006). These gene products display a degree of functional redundancy, although TSO2 is sufficient to provide enough RNR activity for maintaining normal organ development. The tso2 mutant exhibits several developmental defects, including callus-like floral organs and a fasciated shoot apical meristem. The Arabidopsis crinkled leaves8 (cls8) mutant is defective in the large subunit of RNR1 and exhibits a developmental phenotype of abnormal leaf and flower morphology, reduced root growth, and bleached leaf sectors, which is quite similar to the developmental phenotype of tso2 (Garton et al., 2007). These results indicate that RNR activity is critical for proper organ development in the Arabidopsis dicot model plant.
Here, we reveal a critical function for plant RNR activity in chloroplast biogenesis through the phenotypic and molecular characterization of two weak rnr mutants, v3 and st1, in rice as a monocot model plant. In contrast to the Arabidopsis rnr mutants (cls8 and tso2), the growth stage-specific and temperature-conditional development of chlorotic leaves in v3 and st1 mutants indicates that a threshold level of RNR activity plays an important role in balancing chloroplast biogenesis and cell division at a certain stage of leaf development. The interaction between wild-type and mutated RNR subunits in yeast two-hybrid analysis provided an understanding of how a possible dimerization among rice RNR homologs occurs in vivo. Furthermore, we show that under weak genotoxic stress conditions, including the introduction of missense mutations in the RNR subunits, chloroplast biogenesis is preferentially arrested without affecting nuclear genome replication during cell division in developing leaves. This leads to the production of chlorotic leaves without retardation of the whole plant growth. Thus, our analysis of v3 and st1 mutants provides new insights into the mechanisms underlying chloroplast biogenesis during cell division in plants.
RESULTS
Phenotypic Characterization of v3 and st1 Mutants
Under paddy field conditions, v3 plants largely exhibit a normal green phenotype up to the L3 stage, in which the third leaf blade is well expanded, and produce almost fully chlorotic leaves until the maximum tillering stage, but then they develop nearly normal green leaves after heading. Similarly, st1 plants produce green leaves until the L4 or L5 stage, in which the fourth or fifth leaf blade fully emerges from the sheath, then develop chlorotic leaves with a few longitudinal green stripes throughout the tillering stage and recover to generate mostly green leaves after heading. Thus, both v3 and st1 plants display a mixed phenotype in which there are more white leaves and fewer green leaves during the log phase (or tillering stage) of rice growth (Fig. 1A). This observation indicates that the mutant phenotype is expressed in a growth stage-dependent manner during the regular growing season. However, when mutant plants were transferred to growth chambers under 12-h-light (300 μmol m−2 s−1)/12-h-dark conditions at a constant 25°C in the middle of the tillering stage, they continued to produce entirely chlorotic leaves and were never restored (Fig. 1B).
Figure 1.
Phenotypic characteristics of v3 and st1 mutants. A, Phenotypes of wild-type and mutant rice plants grown in a paddy field. Wild-type and v3 plants were photographed at 50 DAG, and st1 plants were photographed at 60 DAG during the tillering stage. WT, Wild-type japonica Seolakbyeo rice. B, Leaf phenotypes of v3 and st1 mutants according to temperature conditions. After heading, mutant plants produced almost fully green leaves with some thin white stripes when grown only in a paddy field (F), whereas they continued to develop chlorotic leaves when transferred into a growth chamber under C25 conditions (12-h-light [300 μmol m−2 s−1]/12-h-dark conditions at a constant 25°C) at the maximum tillering stage. C, Phenotypic changes in wild-type and mutant plants under different temperature conditions were photographed at the L4 stage (left panels), and the Chl content for each leaf was also measured (right panels). The numbers indicate the order of leaf blades. Black and white boxes indicate Chl a and Chl b, respectively. Mean and sd values were obtained from three replicates. FW, Fresh weight.
In rice, v3 has been classified as a temperature-conditional mutant along with v1 and v2 (Iba et al., 1991). The leaves of these three mutants accumulate only a trace amount of Chls at a restrictive temperature (20°C [C20]). To characterize the developmental phenotypes of v3 and st1 mutants in more detail, we examined the expressivity of these phenotypes and the Chl contents according to temperature conditions (Fig. 1C). Under C20 conditions, both v3 and st1 mutants exhibited chlorosis from the L2 stage and contained only a trace amount of Chls in their leaves (Fig. 1C, top). These plants then withered to death at approximately 30 d after germination (DAG). Under 30°C (C30) conditions, these mutant plants produced chlorotic leaves starting from the L3 and L4 stages, respectively (Fig. 1C, middle) and showed a milder expression of the mutant phenotype and a greater accumulation of Chls than under C20. Under an alternative light/dark cycle (12 h light at 30°C/12 h of dark at 20°C [L30/D20]), however, these mutant plants developed almost normal green leaves with much higher Chl concentrations (Fig. 1C, bottom) and produced a few fine white-striped green leaves throughout all growth stages (data not shown). Although wild-type plants appeared to produce normal green leaves irrespective of the temperature conditions, the leaf Chl contents were significantly affected by temperature, indicating that the optimum growth conditions for rice plants in descending order are L30/D20 > C30 > C20. Based on the phenotypic development and Chl contents of these mutant plants, v3 mutants always exhibited more severe symptoms than st1 mutants, indicating that the v3 mutation is much more sensitive to all temperature conditions. Overall, our results demonstrate that the phenotypic development of v3 and st1 mutants is closely associated with the ambient temperature in a gradient of growth stages.
Next, we further examined whether the expressivity of the mutant phenotype is also affected by other external conditions. The observed mutant phenotypes became far more severe under low light (20 μmol m−2 s−1) compared with normal light (300 μmol m−2 s−1) under L30/D20 and more severe under 12 h of light at 20°C/12 h of dark at 30°C (L20/D30) than under L30/D20. Furthermore, phenotypic development was much delayed when these plants were grown in a Murashige and Skoog (MS) medium compared with MS-free agar medium under C20 (data not shown). These results suggest that the phenotypic expression of v3 and st1 mutants is also affected by several external factors, such as light intensity, day/night temperature, and nutrition. Thus, we conclude from our analyses that the defects associated with v3 and st1 mutants are more restored when these plants are grown under conditions that are more favorable to rice growth.
The Chlorotic Leaves of v3 and st1 Mutants Contain Fewer and Smaller Undifferentiated Chloroplasts
To investigate the effects of temperature on the v3 and st1 phenotypes, we examined the density of Chl-containing cells and the ultrastructure of the chloroplasts in these mutant plants. Confocal microscopic observations indicated that under L30/D20, the green leaf cells of mutant plants had an almost normal density and distribution of Chl-containing cells, except for a detectable sporadic deficiency in chloroplasts (Supplemental Fig. S1, B and D). Under C20, the mutant leaves had a few Chl-containing cells but only adjacent to the stomata (Supplemental Fig. S1, C and E). We next examined the ultrastructure of the chloroplasts in the mesophyll cells (Fig. 2). Under L30/D20, the chloroplasts in mutant plants displayed well-developed lamellar structures equipped with normally stacked grana and thylakoid membranes, which is comparable to those of wild-type plants (Fig. 2, A, B, and D). Under C20, however, these chlorotic leaf cells were found to contain undifferentiated chloroplasts that were fewer and smaller (Fig. 2, C and E). These results demonstrate that the Chl-free cells of v3 and st1 mutants are due to a complete arrest of chloroplast biogenesis under restrictive conditions.
Figure 2.
Ultrastructures of chloroplasts in the mesophyll cells of mutant plants. Shown are transmission electron microscopy images of chloroplasts of the wild type (WT) under L30/D20 (A), v3 mutant under L30/D20 (B) and C20 (C), and st1 mutant under L30/D20 (D) and C20 (E). All transmission electron microscopy samples were obtained from the third or fourth leaf blade of each plant. Through more than 50 observed cell images for each genotype, there were no differences found between wild-type and mutant plants in the sizes or numbers of nuclei and chloroplasts in the cells of green leaves under L30/D20. Under C20, mutant plants had almost equivalently sized nuclei to the wild type but much fewer and smaller undifferentiated chloroplasts compared with wild-type plants (data not shown). C, Chloroplast; G, grana; M, mitochondria; N, nucleus; OG, osmiophilic plastoglobuli; S, starch granule; TM, thylakoid membranes.
Map-Based Cloning of V3 and St1 Genes
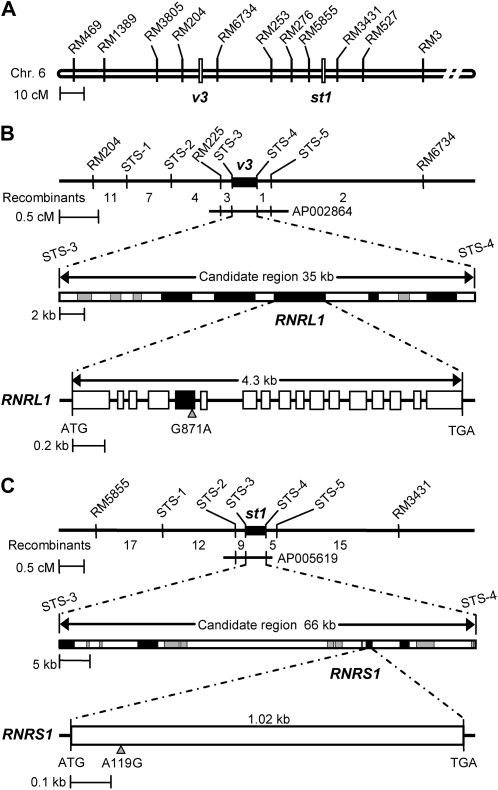
Genetic mapping of the V3 gene was performed using F2, F3, and F4 plants that were generated from a cross of v3 (japonica) mutant and ‘Milyang23’ (a tongil-type indica/japonica hybrid cultivar). Using PCR-based markers, the v3 locus was initially mapped to within 4.7 centimorgan (cM) between two simple sequence repeat (SSR) markers, RM204 and RM6734, on the short arm of chromosome 6 (Fig. 3A). To generate a fine physical map, sequence-tagged site (STS) markers were developed by comparing the genomic sequences of indica and japonica between the SSR markers, and the v3 locus was finally narrowed to a 35-kb interval between STS3 (AP002864-48 kb) and STS4 (AP002864-83 kb) in AP002864 (GenBank accession number; Fig. 3B, top). There were nine hypothetical and expressed genes in this region that were already identified by the Rice Genome Research Program (http://rgp.dna.affrc.go.jp/). Among these candidate genes, five expressed genes had full-length or partial mRNA sequences (Fig. 3B, middle, black boxes). We cloned five genes by reverse transcription (RT)-PCR with total RNA from the young leaves of wild-type and v3 plants, which revealed that in the RNR large subunit gene (designated RNRL1; LOC_Os06g07210), a single nucleotide change (G871A) in exon 5 of the v3 allele caused a missense mutation, Gly to Ser (G291S; Fig. 3B, bottom).
Figure 3.
Map-based cloning of the V3 and St1 genes. A, Genetic mapping of the V3 and St1 genes using SSR and STS markers. The loci of V3 and St1 were initially mapped on the short arm of chromosome 6 to within a 4.7-centimorgan (cM) region between the RM204 and RM6734 markers and a 9.8-cM region between the RM5855 and RM3431 markers, respectively. B, Physical mapping of the V3 gene narrowed to a 35-kb interval between the STS3 and STS4 markers using 28 recombinants of F2 individuals. The genomic region contains five expressed genes (black boxes) and four hypothetical genes (gray boxes). The genomic structure of RNRL1, comprising 17 exons and 16 introns, is indicated. A point mutation (G871A) was found in exon 5 of RNRL1 in the v3 mutant. C, Physical mapping of St1 to within a 66-kb interval between STS3 and STS4 using 58 recombinants of F2 individuals. The genomic region contains four expressed genes (black boxes). RNRS1 comprises one exon in which a point mutation (A119G) occurs in the st1 mutant.
To identify St1, a mapping population was generated from a cross of st1 mutant (japonica) and Milyang23. Genetic mapping with SSR markers located the st1 locus to within 7.5 cM between two SSR markers, RM5855 and RM3431, on the short arm of chromosome 6 (Fig. 3A). Utilizing 58 F2 recombinants from both directions, st1 was further delimited to a 66-kb interval between the STS markers STS3 (AP005619-85 kb) and STS4 (AP005619-151 kb) in AP005619 (Fig. 3C, top). Four of 13 candidate genes within this region had either full-length or partial mRNA sequences deposited in the Rice Genome Research Program (Fig. 3C, middle). RT-PCR and DNA sequencing of four expressed genes in the st1 mutant revealed that in the RNR small subunit gene (designated RNRS1; LOC_Os06g14620) comprising only one exon, a single-base change (A119G) occurred in the st1 allele, which caused a missense mutation, Lys to Arg (K40R; Fig. 3C, bottom). The point mutations in the v3 and st1 alleles were visually verified using the dCAPS method (Supplemental Fig. S2).
A National Center for Biotechnology Information BLAST search revealed the existence of two homologs of RNRL (designated RNRL1 and RNRL2) and RNRS (RNRS1 and RNRS2) in the rice genome. The full-length mRNAs of RNRL1 (GenBank accession no. EU602344) and RNRS1 (EU602345) contain 2,445- and 1,020-bp ORFs, respectively. RNRL1 comprises 17 exons, whereas RNRS1 has only one exon (Fig. 3, B and C). RNRL1 and RNRS1 encode 814 and 339 amino acids with predicted molecular masses of 92 and 38 kD, respectively. Similar to RNRL1 and RNRS1, RNRL2 (LOC_Os02g56100; EU602346) and RNRS2 (LOC_Os06g03720; EU602347) comprise 17 exons (2,433 bp) and one exon (1,038 bp), which encode 810- and 345-amino acid proteins with molecular masses of 91 and 39 kD, respectively. RNRL1 shares 89% homology with RNRL2, and RNRS1 shares 94% homology with RNRS2. A search for conserved domains in the National Center for Biotechnology Information database indicates that RNRL1 contains two conserved domains: the first domain (amino acids 1–90) is an ATP cone domain and the second (amino acids 167–760) is an RNR1 domain (class I RNR). RNRS1 contains only one conserved domain (amino acids 25–300), referred to as an RNR2 domain (Supplemental Fig. S2, C and D). Protein sequence alignments revealed that the substituted Ser-291 residue of rnrl1(v3) is largely conserved as Gly in plants or as Asn in animals (Supplemental Fig. S3A). The substituted Arg-40 residue of rnrs1(st1) is absolutely conserved as Lys in all living organisms (Supplemental Fig. S3B).
To perform the complementation tests for v3 and st1 mutations, 35S:RNRL1 and 35S:RNRS1 constructs (Supplemental Fig. S4A) were introduced into calli generated from the mature embryos of v3 and st1 mutant kernels, respectively, using Agrobacterium tumefaciens-mediated transformation (Jeon et al., 2000; Toki et al., 2006). We obtained 10 transgenic plants from the 35S:RNRL1 and nine from the 35S:RNRS1 transformations. All transgenic plants exhibited a normal green phenotype under C20 (Supplemental Fig. S4B). To confirm whether the regenerants were true transformants, PCR primers were designed between the cauliflower mosaic virus 35S promoter and the first exons of RNRL1 and RNRS1 (Supplemental Fig. S4A). All of the regenerated plants yielded one PCR product with the expected size (Supplemental Fig. S4C). Moreover, we confirmed by RT-PCR that the transgenes were highly expressed in all regenerants via the constitutive 35S promoter (Supplemental Fig. S4D).
Yeast Complementation Test for Rice RNR Activity
A yeast complementation test was performed to evaluate the activities of rice RNRL1, rnrl1(v3), and RNRL2 under the permissive (30°C) and restrictive (20°C) temperatures (Fig. 4). The yeast genome contains two homologs that encode the RNR large subunits, RNR1 (ScRNR1) and RNR3 (ScRNR3). The knockout of ScRNR1 is lethal in yeast, whereas the ScRNR3 knockout is viable (Elledge and Davis, 1990). Hence, we constructed the rnr1 rnr3 double mutant to examine cell survival in the presence of rice RNRL1, rnrl1(v3), or RNRL2. To avoid the lethality caused by the loss of ScRNR1 during genetic manipulation, we used the URA3 knockout system. The haploid Saccharomyces cerevisiae strain YPH499 (MATa, ura3-52, lys2-801amber, ade2-101ochre, trp1-Δ63, his3-Δ200, leu2-Δ1) was used for this analysis. To disrupt the ScRNR3 gene in yeast, the genomic ScRNR3 locus was replaced with the HIS3 gene. This haploid strain is referred to as rnr3∷HIS3. The rnr3∷HIS3 strain was transformed with a URA3 plasmid containing the ScRNR1 gene, and the genomic ScRNR1 locus was then replaced with the Leu-2 gene. This haploid strain is referred to as rnr1 rnr3/RNR1. Next, the rnr1 rnr3/RNR1 strain was transformed with constructs expressing the yeast ScRNR1 and ScRNR3 genes as the positive controls and the rice RNRL1, rnrl1(v3), and RNRL2 genes, which are under the control of the Gal-inducible GAL10 promoter.
Figure 4.
Yeast complementation test for the rnr1 rnr3 null mutation in rice RNRL1, rnrl1(v3), and RNRL2. Cells of the yeast rnr1 rnr3 null strain were transformed with plasmids expressing the yeast ScRNR1 and ScRNR3 genes and the rice RNRL1, rnrl1(v3), and RNRL2 genes under the control of a GAL-inducible promoter at a constant 30°C (top) or 20°C (bottom). Cultures were then grown to saturation, serially diluted, and spotted onto an SG plate lacking His (H), uracil (U), and Trp (W; SG-HUW; left) or a 5-FOA-containing SG plate without Trp [5-FOA (SG-W); right]. Growth on the control plate (SG-HUW) indicates that a similar number of cells were spotted for each sample. The rnr1 rnr3/RNR1 cells transformed with empty vector (construct 1) were used as a negative control, whereas the rnr1 rnr3/RNR1 cells transformed with ScRNR1 (construct 2) and ScRNR3 (construct 3) were used as positive controls. On 5-FOA (SG-W) plates, the rnr1 rnr3/RNR1 cells expressing the rice RNRL1 were able to complement the lethal phenotype at 30°C (construct 4). By contrast, the yeast cells expressing the rice rnrl1(v3) (construct 5) and rice RNRL2 (construct 6) were unable to grow. The complementation efficiencies of ScRNR1, ScRNR2, and rice RNRL1 in the rnr1 rnr3/RNR1 cells were much lower at 20°C than at 30°C.
5-Fluoro-orotic acid (5-FOA) is a pyrimidine analog that reacts with the orotidine-5′-phosphate decarboxylase (the URA3 gene product) to produce a toxic product, 5-fluorouracil, and provides a strong selection pressure in favor of cells that have lost the URA3 plasmid. As expected from a previous study (Elledge and Davis, 1990), the rnr1 rnr3/RNR1 yeast cells that were transformed with an empty expression vector failed to grow on the Gal medium containing 5-FOA (Fig. 4, construct 1), whereas cell viability was restored in the rnr1 rnr3/RNR1 cells with the ScRNR1 and ScRNR3 expression vectors (Fig. 4, constructs 2 and 3). We found that the expression of rice RNRL1 rescued the lethality of the yeast rnr1 rnr3/RNR1 cells at 30°C but not at 20°C (Fig. 4, construct 4). By contrast, the expression of neither rice rnrl1(v3) nor RNRL2 could rescue the lethal phenotype of the yeast rnr1 rnr3/RNR1 cells (Fig. 4, constructs 5 and 6). These results strongly suggest that rice RNRL1 encodes the functional large subunit of RNR and that neither rnrl1(v3) nor RNRL2 has sufficient RNR activity to rescue the lethal phenotype of yeast rnr1 rnr3/RNR1 cells under C30 conditions.
Relative dNTP Levels, Chloroplast-to-Nuclear DNA Copy Ratios, and RNR Expression in Mutant Plants
We speculated that reduced RNR activity might negatively affect plastid genome replication during cell division due to an insufficient and/or unbalanced supply of dNTPs, ultimately resulting in the arrest of chloroplast DNA synthesis in the mutant leaf cells during early development. To examine this possibility, we first measured the dNTP levels in the leaves of v3 and st1 mutants grown under C20 conditions using a polymerase-based assay as described previously (Roy et al., 1999; Wang and Liu, 2006). In general, the levels of the nucleotides were slightly reduced in mutant plants (Fig. 5A), although the levels of dATP and dCTP did not show statistically significant differences between wild-type and mutant plants. This strongly suggests that the v3 and st1 alleles are weak mutations in terms of RNR activity. This may explain why v3 and st1 mutants produce chlorotic leaves in either a temperature-conditional or a growth stage-dependent manner in contrast to the Arabidopsis cls8 and tso2 mutants, which exhibit constitutive developmental phenotypes (Wang and Liu, 2006; Garton et al., 2007).
Figure 5.
Relative dNTP levels, chloroplast-to-nuclear DNA ratios, and expression profiles of RNR genes. A, Relative dNTP levels in the leaves of v3 and st1 mutants. The fully expanded third leaf blades of 4-week-old plants grown under C20 were examined (A and B) and the values were normalized to the wild-type (WT) dTTP levels. Mean and sd values were obtained from three replicates (A and B). This experiment was performed four times independently with similar patterns. B, Relative ratios of chloroplast-to-nuclear DNA copy in v3 and st1 mutants. The relative ratios of chloroplast DNA (rps12) to nuclear DNA (SGR) copies were normalized to the wild-type values. Both plastid rps12 and nuclear SGR genes exist as single copies in their respective genomes. C, Expression levels of RNR genes according to the leaf stages in wild-type plants. Semiquantitative RT-PCR was performed over 27 cycles unless otherwise stated. The expression levels of Ubiquitin (Ub) were used as a loading control (C and D). Three-week-old plants grown under L30/D20 were used. Total RNA was treated with RNase-free DNase I to remove any possible contamination of genomic DNA for RT-PCR (C and D). This experiment was repeated at least three times with similar results. C, Coleoptile; L1 to L3, first to third leaves; L4, not yet emerged fourth leaf in the leaf sheath; L3L, the lower half of the third leaf blade; L3LS, the leaf sheath of the third leaf; L3U, the upper half of the third leaf blade; SB, shoot base (1.0 cm above the bottom). The diagram at right represents a 3-week-old rice plant with a fully emerged third leaf. D, Expression levels of RNR genes under C30 and C20 conditions evaluated using semiquantitative RT-PCR. The third leaf blades of 3-week-old plants were used. The expression levels of Ubiquitin were used as a loading control. This experiment was repeated three times with similar results.
To analyze for differences in the copy numbers of chloroplast DNA relative to nuclear DNA between wild-type and mutant plants, quantitative PCR was performed with Stay-green (SGR; Park et al., 2007) and rps12 (Sugimoto et al., 2007), which exist as single-copy genes in the rice nuclear and plastid genomes, respectively. The results revealed that the rps12-SGR ratios in the chlorotic leaves of mutant plants were much lower than in the green leaves of wild-type plants (Fig. 5B). This suggested that plastid DNA synthesis during chloroplast biogenesis is severely inhibited in the developing leaves of mutant plants. In particular, this ratio in the v3 mutant was lower than that of the st1 mutant, consistent with the fact that v3 always develops a more severe phenotype than st1 under the same restrictive conditions.
We next tested whether the expression levels of the four RNR genes are regulated during leaf development (Fig. 5C). Total RNA samples were prepared according to the leaf developmental stage as described previously (Kusumi et al., 1997). The tissue-specific expression results showed that RNRL1, RNRL2, and RNRS1 were highly expressed in the shoot base, whereas RNRS2 was slightly expressed in only mature leaf tissues, such as the fully elongated third leaf blade and sheath. The spatial and temporal expression profiles for RNRL1 and RNRS1 were quite similar, and the transcripts for these genes were observed throughout leaf development. In addition, RNRL2 mRNA was strongly detected only in the shoot base but was barely evident at other developmental stages in the rice leaves.
Finally, the expression patterns of four RNR genes were investigated in wild-type and mutant plants under the restrictive C20 and C30 conditions using a semiquantitative RT-RCR method (Fig. 5D). The expression of RNRL1 and RNRS1 in the leaves of v3 and st1 mutants exhibited the same pattern in response to restrictive temperatures. The expression of RNRL1 and RNRS1 was significantly up-regulated in mutant plants regardless of the temperature conditions, whereas the expression of RNRL2 was slightly increased in the st1 mutant only. By contrast, the expression of RNRS2 was either unaltered or somewhat decreased in mutant plants when compared with wild-type plants. The overall transcript levels of RNRL2 and RNRS2 were also relatively lower than those of RNRL1 and RNRS1. Furthermore, all four RNR transcripts were higher at C20 than at C30, suggesting that an increased activity of RNR is needed for developing leaves under low temperature. Taken together, the expression of RNRL1 and RNRS1 was found to be coordinately regulated by the developmental stage as well as the temperature conditions, and their expression was higher in mutant plants than in wild-type plants under the same restrictive conditions (Fig. 5D). These results indicate that insufficient RNR activity in the dividing leaf cells of mutant plants activates the transcription of RNRL1 and RNRS1 simultaneously, possibly through a feedback-like mechanism.
Expression Profiles of Genes Associated with Chloroplast Development in Mutant Plants
Chloroplast development is tightly regulated by the coordinated expression of plastid and nuclear genes during leaf development (Mandel et al., 1996). To investigate whether the regulation of nuclear and plastid genes was altered by the reduced activity of RNR in v3 and st1 mutants, semiquantitative RT-PCR was performed using total RNA extracts from the fully expanded third leaf blades of wild-type (green) and mutant (chlorotic) plants under C20 (Fig. 6). The transcripts of photosynthetic genes encoded in the plastid (psbA) and nucleus (LhcpII and RbcS) were found to be reduced in the mutant leaves. By contrast, the plastid genes encoding the transcription/translation apparatus (rpoA, rpoB, and rps7) were highly expressed in mutant plants. In wild-type plants, the plastid genes encoding the transcription/translation apparatus are transcribed by the nucleus-encoded plastid RNA polymerase during the early stages of chloroplast development, and the plastid genes encoding the photosynthetic apparatus are transcribed by the plastid-encoded plastid RNA polymerase at later stages of chloroplast development during early leaf development (Kusumi et al., 1997). In v1, the stage-specific expression of plastid genes involved in transcriptional regulation was revealed to be disrupted, because they were expressed in the later stages of leaf development. Our results indicate that the expression patterns of these genes in the v3 mutant are basically the same as in v1, as described previously (Kusumi et al., 1997). In addition, the chlorotic leaves of v3 and st1 mutants lacked plastid-encoded ribosomal RNA (16S rRNA). These results suggest that the reduced activity of RNR causes the delayed expression of plastid transcription/translation genes and almost completely blocks the expression of photosynthetic genes in the later stages of leaf development.
Figure 6.
Transcription analysis of chloroplast development-related genes in mutant plants. For semiquantitative RT-PCR, RNase-free DNase I-treated total RNA was used. The fully emerged third leaf blades of 3-week-old rice plants grown under C20 were used. The following numbers of PCR cycles were used for the specific genes under analysis: 20 (RbcS), 24 (LhcpII, psbA, and 16S rRNA), 25 (rpoA and Ub), 26 (rps7), and 29 (rpoB). Ubiquitin (Ub) was amplified as a loading control. This experiment was repeated more than three times with similar results. Primer information is listed in Supplemental Table S1. WT, Wild type.
RNRL1 Strongly Interacts with RNRS1
A yeast two-hybrid assay was performed to identify the two large and two small subunit pairs of RNRs as well as to examine the effects of the missense mutations in rnrl1(v3) and rnrs1(st1) on the interactions with their counterparts during the α2β2 heterodimerization of RNR. Prior to conducting the yeast two-hybrid assay, we examined the expression and stability of RNR subunits in yeast (Fig. 7A) and found that they are stably expressed at sufficient levels to perform this analysis. To investigate the interaction specificity between the subunits, α-galactosidase activity was measured (Fig. 7B) and revealed that binding occurs between the large and small subunits, RNRL1:RNRS1 and RNRL2:RNRS2, but not RNRL1:RNRS2 or RNRL2:RNRS1. Interestingly, no interaction was found between the large subunits (RNRL1:RNRL1, RNRL2:RNRL2, and RNRL1:RNRL2) or between the small subunits (RNRS1:RNRS1, RNRS2:RNRS2, and RNRS1:RNRS2). These findings strongly suggest that an RNR αβ dimer is first assembled and that two αβ dimers then interact with each other to establish a functional RNR heterodimer (α2β2). RNRL1 interacted most strongly with RNRS1 (RNRL1:RNRS1), and less interaction of RNRL1:rnrs1(st1) may be the situation in the st1 mutant. Next, rnrl1(v3) had much less interaction with RNRS1 [rnrl1(v3):RNRS1], which may underlie the defects in the v3 mutant. The rnrl1(v3):rnrs1(st1) interaction did not occur at all, possibly explaining why the v3 st1 double mutant produces albino leaves and exhibits seedling lethality even under the permissive L30/D20 conditions (data not shown). These results further suggest that missense mutations attenuate the first αβ dimerization. In addition, the interacting activity of rnrl1(v3):RNRS1was significantly lower than that of RNRL1:rnrs1(st1), possibly explaining why the v3 mutant exhibits a more severe chlorotic phenotype and a lower chloroplast-to-nuclear DNA copy ratio than st1 under the same growth conditions (Figs. 1 and 5B).
Figure 7.
Analysis of the interactions between RNR subunits in yeast two-hybrid assays. A, Verification of the protein expression and stability of wild-type and mutated RNR subunits in yeast. Each protein fraction was isolated from different yeast strains expressing a recombinant pGBKT7 plasmid containing RNRL1 (lane 1), rnrl1(v3) (lane 2), RNRL2 (lane 3), RNRS1 (lane 4), rnrs1(st1) (lane 5), or RNRS2 (lane 6). The large and small subunits of RNR are about 112 and 58 kD (asterisks) in size, respectively, due to the presence of the N-terminal fusion protein (about 20 kD) in the pGBKT7 plasmid. An anti-myc antibody (α-myc) was used to detect each subunit using an ECL system. After immunoblotting, the membrane was stained with Coomassie Brilliant Blue (CBB; Sigma) to check for equal loading. The additional 75-kD proteins reacting with α-myc in the left three lanes are possibly the superfluous RNRL subunits that are inactivated by the removal of the C-terminal region (about 37 kD) by an unknown mechanism(s). B, α-Galactosidase activities in yeast two-hybrid assays with all possible combinations of RNR subunits. Three colonies were selected from minimal medium lacking Trp and Leu and independently cultivated in liquid medium until the midexponential phase. The supernatants were then collected to determine the α-galactosidase activity. Mean and sd values were obtained from three replicates. BK, Empty pGBK7 plasmid; AD, empty pGADT7 plasmid; L1, RNRL1; v3, rnrl1(v3); L2, RNRL2; S1, RNRS1; st1, rnrs1(st1); S2, RNRS2.
Effects of an RNR Inhibitor on Chloroplast Biogenesis and Plant Growth
In Arabidopsis, strong mutations in RNR subunits generally cause severe abnormality and retardation, with partial deficiency of chloroplasts throughout development (Wang and Liu, 2006; Garton et al., 2007). However, unlike the constitutive defectiveness of the Arabidopsis rnr mutants, v3 and st1 mutants exhibited the chlorotic phenotype mainly during the tillering stage without significant organ or growth defects under field conditions, although lethality was observed for these mutations under restrictive conditions (C20 and C30). Hydroxyurea (HU) inhibits RNR activity by quenching the tyrosyl radical in the small subunit of RNR, which leads to a reduction in the dNTP pools and stalling of the DNA replication forks (Elleingand et al., 1998). To examine the effects of HU on the rate of seedling growth and Chl synthesis, both wild-type and mutant plants were grown in HU-treated MS under C30. The growth retardation became more severe as the concentration of HU increased. Wild-type plants displayed a rapid decrease of Chl concentration without growth retardation under a low concentration of HU (2 mm; Fig. 8), reminiscent of the developmental phenotypes of v3 and st1 mutants (Fig. 1). In the v3 mutant in particular, Chl synthesis was almost fully blocked by 2 mm HU, whereas the fresh weight was only slightly reduced. On the other hand, the Chl content of mutant plants was found to be similar to or slightly lower than that of the wild type under C30, but it was decreased more rapidly by exposure to 2 mm HU. These data suggest that mutant plants are much more sensitive to HU than wild-type plants, possibly because mutated RNRs are more easily deactivated than wild-type RNRs.
Figure 8.
Responses of wild-type and mutant plants to HU exposure. A, Phenotypic changes in wild-type (WT), v3, and st1 mutant plants upon exposure to increased concentrations of HU (0, 2, 4, 8, and 16 mm). Seedlings were grown for 2 weeks in MS agar medium under C30 conditions. B, Changes in the fresh weights (FW) and Chl a contents of wild-type and mutant plants in response to treatments with increasing concentrations of HU.
DISCUSSION
We have previously characterized three v (v1, v2, and v3) mutants in rice (Iba et al., 1991; Kusumi et al., 1997; Sugimoto et al., 2004) and recently identified the V2 gene that encodes pt/mt GK (Sugimoto et al., 2007). In this study, we have further identified the V3 and St1 genes that encode the large and small subunits of RNR, RNRL1 and RNRS1, respectively. Our finding that the chlorotic phenotypes of v3 and st1 mutants are conditionally developed by missense mutations in the RNR subunits (Fig. 3) demonstrates that chloroplast biogenesis is arrested (Fig. 2, C and E) when plants fail to satisfy an increased requirement of RNR activity, particularly under restrictive temperatures or during the most active growth period (tillering stage). This is supported by our evidence that the expression of RNRL1 and RNRS1 is more up-regulated at C20 than at C30 (Fig. 5D) and that the heterologous RNRL1 is able to rescue the yeast rnr1 rnr3 knockout line at 30°C but not at 20°C (Fig. 4, construct 4). Since the optimal temperature for yeast growth is 28°C to 30°C, 20°C can be considered as a restrictive low temperature for yeast growth and cell division. In addition, the single amino acid substitutions in rnrl1(v3) and rnrs1(st1) may attenuate the in vivo interactions with their wild-type counterparts (Fig. 7), which results in a failure to satisfy an increased requirement of RNR activity under the restrictive temperatures. Finally, the expression of RNRL1 and RNRS1 was significantly increased in actively growing tissues, such as the shoot base and young leaf tissues (Fig. 5C), and the mutant phenotype become more severe at the maximum tillering stage (Fig. 1A).
The Arabidopsis rnr mutants (tso2 and cls8) exhibit severe abnormality and retardation in growth and development, and they also display chloroplast deficiency in some parts of their organs (Wang and Liu, 2006; Garton et al., 2007). Garton et al. (2007) have suggested in their previous report that under a limited dNTP supply, the inhibition of chloroplast DNA replication may be a primary cause of the aberrant development of rnr mutants. In this study, the v3 and st1 alleles can be assumed to be weak mutations because their phenotypic development is conditional. We speculate also that under a limited dNTP supply during early leaf development, plastid DNA synthesis is preferentially blocked, rather than nuclear DNA replication, probably through a specific regulatory mechanism for proper plant growth. In support of this contention, we obtained two significant results from this study. First, mutant plants have lower copy numbers of chloroplast DNA compared with wild-type plants (Fig. 5B). Second, the chloroplast biogenesis of wild-type plants is arrested without growth retardation under a low concentration of an RNR inhibitor (2 mm HU; Fig. 8), reminiscent of the v3 and st1 phenotypes under restrictive temperature conditions (Fig. 1). This suggests that plants can arrest chloroplast biogenesis as a defense mechanism, since temporary deficiency of chloroplast activity at the tillering stage does not severely inhibit the growth of mutant plants under field conditions. Taken together, we propose from our current data that higher plants first establish the nuclear genome, which is critical for cell maintenance and division during growth and development. In other words, plastid DNA synthesis for chloroplast biogenesis is relatively less critical for plant survival, especially during the active growth of rice plants. Thus, under insufficient dNTP levels, the plants can make sacrifices and become heterotrophic in order to maintain their rates of growth and development and finally reach the reproductive stage in the correct season.
The interaction between the large and small subunits of RNR was previously studied in human and shown to establish a functional holoenzyme (Qiu et al., 2006). Although two small subunits, hRRM2 and p53R2, were shown to interact with the large subunit, hRRM1, to form a functional RNR holoenzyme, the hRRM1-hRRM2 pair had approximately twice as much kinetic activity as the hRRM1-p53R2 pair. RNR expression has also been shown to be closely linked to the rate of DNA synthesis during active mitosis by a feedback-like regulatory mechanism (Chabouté et al., 1998). Our results also indicate that for DNA synthesis, the expression of RNRL1 and RNRS1 is regulated in a spatial and temporal manner, most likely by forming a functional RNR heterodimer complex. We provide evidence for this from our yeast two-hybrid analyses, which show that in vivo, RNRL1 and RNRS1 form a functional RNR enzyme in rice (Fig. 7). Moreover, the severity of the mutant phenotype (v3 > st1) was found to be closely related to the activities of the first αβ dimerization between RNR subunits [rnrl1(v3):RNRS1 < RNRL1:rnrs1(st1); Figs. 1 and 7B]. In addition, RNRL1 and RNRS1 were highly expressed in the rice plants grown under a low temperature (C20) rather than a high temperature (C30), and this was more pronounced in mutant plants (Fig. 5D). This strongly suggests that under the restrictive C20 conditions, RNR activity may exhibit a lower level, as demonstrated by the yeast complementation test (Fig. 4), resulting in a lower dNTP pool in the dividing cells of actively growing leaves. Consequently, the cells try to increase the dNTP levels by up-regulating the transcription of RNRL1 and RNRS1 through a feedback regulation.
In rice plants, the shoot always harbors four immature leaves at different developmental stages, when a leaf begins to emerge from the leaf sheath (Yamazaki, 1963; Kusumi et al., 1997). These unemerged immature leaves show a series of successive stages of leaf development, termed P1 (leaf primordium), P2, P3, and P4, which are early leaf developmental stages. The emerging and well-expanded leaves were designated P5 and P6, respectively, which are late stages of leaf development. An as yet unidentified V1 gene is speculated to control the timing of expression of the key plastid genes of the transcription/translation apparatus that are essential for the subsequent activation of other plastid genes (Kusumi et al., 1997). Usually, the rpo and rps genes of the plastid transcription/translation apparatus are highly expressed at the P4 stage of leaf development in the wild type, but their expression was much more delayed in the v1 mutant. On the other hand, transcripts for the plastid transcription/translation apparatus were observed to accumulate at wild-type levels at the P4 stage of v2, but their translation is blocked at an early stage of chloroplast differentiation (Sugimoto et al., 2004). In v3 and st1 mutants, the expression of plastid genes is highly suppressed at the P4 stage (data not shown) while abnormally increased at a later stage (P6) of leaf development (Fig. 6), as observed previously for the v1 mutant (Kusumi et al., 1997). During leaf development in monocot plants, the events accompanying chloroplast differentiation can be divided into three steps (Chory, 1991; Mullet, 1993). The first and second of these occur at the leaf developmental stages from P1 to P4, and the third proceeds at stages P5 and P6. It has been speculated that the V1 gene product functions at the beginning of the second step of chloroplast differentiation (Kusumi et al., 1997), whereas V2 (pt/mt GK) plays a critical role in the second and third steps (Sugimoto et al., 2004, 2007). Plastids in the chlorotic leaves of v1 and v2 were shown to be normal in size and DNA copy number (Iba et al., 1991). However, v3 and st1 mutants exhibit a significant decrease in the size and number of their chloroplasts (Fig. 2, C and E) and a reduced chloroplast-nuclear genome ratio (Fig. 5A), indicating that RNR function is critical in the first step of chloroplast differentiation. Interestingly, all of these mutants are temperature conditional and have the same temperature-sensitive leaf stage (P4). The developing leaf at the P4 stage is still kept inside the leaf sheath. Just before the emergence of the leaf blade from the sheath, many genes are expressed that convert proplastids to chloroplasts, including those involved in chloroplast DNA synthesis, the plastid transcription/translation apparatus, and the photosynthetic system. At this critical stage, chloroplast development can be arrested if the activities of essential proteins or their products do not reach certain threshold levels. In v2, the insufficient activity of pt/mt GK probably inhibits the assembly and function of the plastid translation machinery, leading to a temperature-conditional arrest in chloroplast differentiation. In the case of v3 and st1 mutants, the reduced activity of RNR impairs chloroplast DNA replication in developing leaves, most markedly at restrictive temperatures or during the active growth phase.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
v3 and st1 mutants were spontaneously induced from a Japanese japonica rice variety (Oryza sativa ‘Kinmaze’; Omura et al., 1978) and the japonica rice strain ‘FL176’ (Nagamatsu and Omura, 1962), respectively. These plants were then crossed with a the Korean japonica rice ‘Seolakbyeo’ that was allowed to progress to the F7 generation. Thus, Seolakbyeo was used as the parental wild-type plant in this study. Plants were grown in a paddy field during the rice growing season or in a growth chamber. The chamber conditions were as follows: 12 h of light (300 μmol m−2 s−1), 30°C/12 h of dark, 20°C for diurnal conditions (L30/D20); 12 h of light, 20°C/12 h of dark, 30°C for temperature-reverse diurnal conditions (L20/D30); 12 h of light/12 h of dark at a constant 30°C (C30) or 20°C (C20); and 12 h of light (20 μmol m−2 s−1), 30°C/12 h of dark, 20°C for low-light diurnal conditions.
Measurement of Photosynthetic Pigments
For Chl extraction, equal fresh weights of rice leaf tissues at the fourth leaf stage were homogenized and resuspended in ice-cold 80% (v/v) acetone under dim green light. Residual plant debris was removed by centrifugation. The supernatants of these samples were then analyzed with a visible spectrophotometer, and the concentrations of Chls and carotenoids were measured at 663.2, 646.8, and 470 nm and determined using the method of Lichtenthaler (1987).
Confocal Microscopy and Transmission Electron Microscopy
To investigate the status of chloroplast development, leaf tissues of 3-week-old plants that were grown under C20 in growth chambers were harvested and observed with a confocal laser scanning microscope (Carl Zeiss). Transmission electron microscopy analysis was carried out as described previously (Inada et al., 1998) with minor modifications. Briefly, segments of leaf tissues were fixed with a modified Karnovsky's fixative (2% [v/v] paraformaldehyde, 2% [v/v] glutaraldehyde, and 50 mm sodium cacodylate buffer, pH 7.2) and washed three times with 50 mm sodium cacodylate buffer (pH 7.2) at 4°C for 10 min. The samples were then postfixed with 1% (w/v) osmium tetroxide in 50 mm sodium cacodylate buffer (pH 7.2) at 4°C for 2 h and briefly washed twice with distilled water at 25°C. Samples were en bloc stained in 0.5% (w/v) uranyl acetate at 4°C for 30 min, dehydrated in a gradient series of ethanol and propylene oxide, and finally embedded in Spurr's resin. After polymerization at 70°C for 24 h, ultrathin sections were prepared with a diamond knife on an ultramicrotome and then mounted onto Formvar-coated copper grids. The sections on the grids were next stained with 2% (w/v) uranyl acetate for 5 min and with Reynolds' lead citrate for 2 min at 25°C and then examined with a JEM-1010 EX electron microscope (JEOL).
Map-Based Cloning of V3 and St1
For map-based cloning of the V3 gene, a mapping population that consisted of 1,040 F2 individuals was generated by a cross between the v3 mutant (japonica) and the tongil-type ‘Milyang23’, which has a closer genetic makeup to that of indica. For the fine-mapping of the v3 locus, 1,810 F3 and 730 F4 progeny were further generated from F2 and F3 heterozygous lines, respectively. The st1 mapping population, which consisted of 595 F2 individuals, was generated by the cross of st1 and Milyang23. For the fine-mapping of the st1 locus, 2,809 F3 progeny were further generated from the F2 heterozygous lines. Based on the marker information in the GRAMENE (http://www.gramene.org/bd/markers/) and the Arizona Genomics Institute (http://www.genome.arizona.edu/fpc/rice/) Web sites, SSR markers and STS markers on chromosome 6 were used for the genetic and physical mapping. Five PCR-based STS markers were developed by nucleotide sequence alignments between japonica and indica in AB023482 and AP002864 and were used to map the v3 locus. For the physical mapping of the st1 locus, five STS markers in AP005107, AP005619, and AP005527 were used. The PCR primer information for these STS markers is listed in Supplemental Table S1.
dCAPS Analysis
For the dCAPS analysis of the v3 allele, primers (forward, 5′-ATGAAAGATGACAGCATTGAGGGAATC-3′; mismatched reverse, 5′-ATTGACCATAGAACCTACCCTTTCTCAAGC-3′) were designed using the Web server program dCAPS Finder 2.0 (http://helix.wustl.edu/dcaps/dcaps.html). These primers produce a 235-bp PCR product with a HindІII site specifically in the mutant allele of rnrl1(v3). For the st1 allele, a mismatched forward primer (5′-AGGTACCCGCAGATCTGGGAGTTCTCGA-3′) and a reverse primer (5′-GTTCTCGATGGCGATCTGGAATCCGTAG-3′) were designed to generate a 245-bp PCR product containing an XhoI site specific to the RNRS1 gene. PCR products of rnrl1(v3) and RNRS1 were digested with HindІII and XhoI, respectively, and separated on a 2.5% metaphore agarose gel that was then visualized and photographed under UV light.
Rice Transformation and Complementation Test
The recombinant pCAMLA plasmids that contain the 35S:RNRL1 and 35S:RNRS1 transgenes were used for the complementation testing of v3 and st1 mutations, respectively. These recombinant plasmids were introduced into an Agrobacterium tumefaciens strain, LBA4404, for transformation into the calli that were generated from the mature embryos of either v3 or st1 mutant seeds. The seed-based rice transformation (Toki et al., 2006) and Agrobacterium-mediated callus transformation (Jeon et al., 2000) methods were then used to obtain the transgenic plants. Transformants were confirmed by PCR amplification using primers designed between the CaMV35S promoter (5′-AGACCCTTCCTCTATATAAG-3′) and the first exon of four RNR genes, RNRL1 (5′-CTCTCGTTGAAATGTCCATAC-3′) and RNRS1 (5′-ATGTTCTCGATGGCGATCTG-3′) for the complementation test.
Yeast Strains, Growth Media, and Genetic Methods for the Yeast Complementation Test
The haploid Saccharomyces cerevisiae strain YPH499 (MATa, ura3-52, lys2-801anber, ade2-101ochre, trp1-Δ63, his3-Δ200, leu2-Δ1; Sikorski and Hieter, 1989) used in this study was obtained as described (Sugimoto et al., 2007). To generate the pSKRNR3HIS3 construct used for ScRNR3 disruption, the BamHI-digested HIS3 gene was inserted into the BamHI site of pBluescript II SK+ (Stratagene) to produce pSKHIS3. The 5′-end fragment of ScRNR3, which was obtained by PCR using the primers 5′-AGAGCTCGCCAAGTTATCTGCCTACGG-3′ and 5′-TACTAGTTTGTGTGGGAGTATTTGATTTAT-3′, was inserted into the pGEM-T Easy vector to produce pG-RNR3N. The 3′-end fragment of ScRNR3, which was obtained by the PCR primers 5′-AGAATTCAGAAGGATGCATCTCCAGTTCC-3′ and 5′-TGTCGACTTTGCGACGGTAAATCTGGA-3′, was inserted into the pGEM-T Easy vector to produce pG-RNR3C. The SacI-SpeI fragment of pG-RNR3N was then inserted into the SacI/SpeI sites of pSKHIS3 to produce pSKHIS3RNR3N. The EcoRI-SalI fragment of pG-RNR3C was inserted into the EcoRI/SalI sites of pSKHIS3RNR3N. To construct the plasmid vector pRS316RNR1 carrying ScRNR1 with a URA3 marker gene, a S. cerevisiae genomic fragment (4.34 kb) containing the ScRNR1 ORF was obtained using the PCR primers 5′-ACTCGAGGGGTTATGGAGAGTATGCTG-3′ and 5′-TGGATCCATAACTCTCCTCTGCCCGAA-3′. This fragment was then digested with BamHI and XhoI and inserted into the BamHI/XhoI sites of pRS316. For pGEMRNR1LEU2 used for ScRNR1 disruption, a 2.86-kb S. cerevisiae genomic fragment containing the ScRNR1 ORF was obtained using the PCR primers 5′-ACTGCAGAGATCTATTACATTATGGGTGGTATGTTG-3′ and 5′-TGCGGCCGCAGATCTGGTACTTTGGCCCC-3′ and inserted into the pGEM-T Easy vector to produce pG-LEU2. The 3′-end fragment of ScRNR1 that was obtained using the PCR primers 5′-ACTGCAGCGTTGATGATGAGGAAACCG-3′ and 5′-TGAGCTCGAACAATGTTGCCTAGACCC-3′ was inserted into the pGEM-T Easy vector to produce pG-RNR1C. The 5′-end fragment of ScRNR1 was obtained by PCR using the primers 5′-AGGGCCCTGTCGGGTAAGTTCATCCTC-3′ and 5′-TGCGGCCGCTTCGTTGGTGTCTCTCTTGC-3′ and inserted into the pGEM-T Easy vector to produce pG-RNR1N. The PstI-SacI fragment of pG-RNR1C was inserted into the PstI/SacI sites of pG-LEU2 to produce pG-LEU2RNR1C. The ApaI-NotI fragment of pG-RNR1N was then inserted into the ApaI/NotI sites of pG-LEU2RNR1C. To generate an rnr3 null allele in yeast, the SacI-SalI fragment of pSKRNR3HIS3 containing the disrupted ScRNR3 gene was introduced into YPH499, yielding the haploid rnr3∷HIS3 strain. To generate an rnr1 rnr3 null allele, the ApaI-SacI fragment of pGEMRNR1LEU2 containing the disrupted ScRNR1 gene was introduced into rnr3∷HIS3, which harbors pRS316RNR1, yielding the haploid rnr1 rnr3/RNR1 strain. For complementation assay using the rnr1 rnr3/RNR1 strain, we used YEpGAL112 as the expression vector (Saitoh et al., 2002). To produce YEpGAL112ScRNR1, a DNA fragment containing the ScRNR1 ORF obtained by PCR using the primers 5′-AAACATATGTACGTTTATAAAAGAGA-3′ and 5′-TTTGAGCTCTTAACCCGAACACATTTCAC-3′ was digested with NdeI and SacI and inserted into the corresponding sites of YEpGAL112. To produce YEpGAL112ScRNR3, a DNA fragment containing the ScRNR3 ORF was obtained using the PCR primers 5′-AAACATATGTACGTTATTAAAAGAGA-3′ and 5′-AAAGAGCTCTCAACCGGAACATGACTCAC-3′, digested with the NdeI and SacI enzymes, and then inserted into the NdeI/SacI sites of YEpGAL112. To construct YEpGAL112RNR1 and YEpGAL112rnrl1, a DNA fragment containing the rnrl1(v3) ORF was amplified using the primers 5′-AAACATATGTACGTGGTGAAGCGGGA-3′ and 5′-TTTGAATTCCTAACTTCCACACGCCAAGC-3′, digested with NdeI and SacI, and then inserted into the NdeI/SacI sites of YEpGAL112 to produce YEpGAL112RNRL1N and YEpGAL112rnrl1N. The DNA fragment containing the RNRL1 ORF, which was obtained by PCR as described above, was digested with EcoRI and SacI and then inserted into the EcoRI/SacI sites of YEpGAL112RNRL1N and YEpGAL112rnrl1N. To construct YEpGAL112RNRL2, a DNA fragment containing the RNRL2 ORF, which was obtained by PCR using the primers 5′-AAACATATGTACGTGGTGAAGCGCGA-3′ and 5′-TTTGAATTCCTAACTACCACATGCCAAGC-3′, was digested with NdeI and SacI and then inserted into the NdeI/SacI sites of YEpGAL112 to produce YEpGAL112RNRL2N. The DNA fragment containing the RNRL2 ORF that was also obtained using the PCR primers described above was digested with EcoRI and SacI and then inserted into the EcoRI/SacI sites of YEpGAL112RNRL2N. Yeast cells were grown and maintained in either a minimal synthetic medium containing either Glc (SD) or Gal (SG) or in complete medium containing either Glc or Gal. Strains carrying the URA3 plasmid were selected by culturing on solid synthetic medium containing 1 mg mL−1 5-FOA.
Semiquantitative RT-PCR and dNTP Measurements
Total RNA was extracted using the TRIzol Reagent (Invitrogen) from the leaf tissues of wild-type and mutant rice plants that were grown under three different growth conditions and then treated with the RNase-free DNase I (Ambion) in order to remove the DNA contamination for RT-PCR. Five micrograms of total RNA was used for each RT reaction. RT products that were equivalent to 50 ng of total RNA were used in the semiquantitative RT-PCR. The primer sets used for the four rice RNR genes, chloroplast development-related genes, and DNA sequences that exclusively existed in the nuclear or chloroplast genomes are listed in Supplemental Table S1. The dNTP levels were measured by a polymerase-based assay as described previously (Roy et al., 1999; Wang and Liu, 2006). Briefly, the leaf tissues of wild-type and mutant plants were harvested and ground to a fine powder in liquid nitrogen. Two hundred milligrams of ground tissues was then mixed with 60% ice-cold methanol by vigorous vortexing, incubated at 95°C for 5 min, and centrifuged at 17,000g for 20 min. The supernatants were dried in a SpeedVac (Hanil), resuspended in 0.1 mL of sterile distilled water, and stored at −20°C. Five microliters of each sample was then used in the assay. Commercial dNTPs (Promega) were used in parallel to establish a linear standard curve.
Yeast Protein Preparation
Total yeast protein extracts were prepared as described in the Clontech Yeast Protocol Handbook (PT3024-1; http://www.clontech.com/images/pt/PT3024-1.pdf). Briefly, yeast colonies were suspended in 5 mL of the appropriate SD liquid medium and cultured overnight at 30°C. A 2-mL aliquot of this culture was inoculated into 50 mL of appropriate SD liquid medium and grown until reaching an optical density at 600 nm of 0.7. The yeast cells were then treated with 25 μm MG-132 (Sigma) for 4 h. The media were then centrifuged, and the pellets were washed with ice-cold distilled water and resuspended in prewarmed complete cracking buffer (8 m urea, 5% SDS, 40 mm Tris-HCl [pH 6.8], 0.1 mm EDTA, 0.4 mg mL−1 bromphenol blue, 1% β-mercaptoethanol, 5 mm phenylmethylsulfonyl fluoride, and 1× Complete protease inhibitor cocktail [Roche]) containing 80 mg of acid-washed glass beads (G8772; Sigma-Aldrich). The solutions were next incubated at 70°C for 10 min and vortexed vigorously for 1 min. The samples were then centrifuged, and the supernatant was transferred to a new tube. The pellets were incubated at 100°C for 5 min, vortexed for 1 min, and centrifuged again. The first supernatant was then combined with the second supernatant. The samples were subsequently boiled briefly and fractionated by SDS-PAGE. For immunoblotting, N-terminal myc-tagged RNR subunits expressed from the recombinant pGBKT7 plasmid were detected by mouse monoclonal anti-myc antibody (Santa Cruz Biotechnology) using an enhanced chemiluminescence (ECL) detection system (WESTSAVE-Up; LabFrontier). After ECL detection, the membrane was stained with Coomassie Brilliant Blue (Sigma).
Yeast Two-Hybrid and α-Galactosidase Assays
Yeast two-hybrid assays were performed using the GAL4-based Matchmaker two-hybrid systems (Clontech) according to the manufacturer's protocols. Each of the bait and prey pairs was cotransformed into the yeast strain AH109. To eliminate false positives, the colonies obtained from the cotransformation were confirmed by separate streaking onto SD dropout medium. To monitor the interactions between the large and small subunits of RNR in the yeast two-hybrid samples, α-galactosidase activity was measured from liquid culture according to the manufacturer's instructions (Clontech).
Analysis of HU Sensitivity
To examine the effects of an RNR inhibitor, HU, on rice plants, a gradient of HU concentrations (0, 2, 4, 8, and 16 mm) was added to the MS phytoagar medium used to grow the seedlings. The kernels were sterilized as follows: seeds were washed with 70% (v/v) ethanol for 1 min, sterilized first with 2% (v/v) sodium hypochlorite containing Tween 20 (one drop in 50 mL) twice for 15 min and then with 2% (v/v) sodium hypochlorite for 10 min, followed by washing with sterile water five times. The seeds were then sown into the medium, which comprised 4.43 g L−1 MS basal medium (pH 5.6–5.8), 30 g L−1 Suc, and 3 g L−1 phytoagar. Plants were then grown in a growth chamber in C30 conditions.
The GenBank accession numbers of the rice RNR genes are as follows: RNRL1 (EU602344), RNRS1 (EU602345), RNRL2 (EU602346), and RNRS2 (EU602347).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Confocal microscopic analysis of the leaves of v3 and st1 mutants.
Supplemental Figure S2. Analysis of missense mutations in the v3 and st1 alleles via the dCAPS method.
Supplemental Figure S3. ClustalW alignments of the large and small subunits of RNR.
Supplemental Figure S4. Complementation tests of v3 and st1 mutants.
Supplemental Table S1. Primer information used in this study.
Supplementary Material
Acknowledgments
We thank Dr. Jong-Seong Jeon (Kyung Hee University) for technical assistance with rice transformation.
This work was supported by the Crop Functional Genomics Center of the 21C Frontier R&D Program (grant no. CG3131 from the Ministry of Science and Technology), by the Agricultural Plant Stress Research Center (grant no. R11–2001–092–05003–0 from the Korean Science and Engineering Foundation, Korea, to N.-C.P.), and by the Program for the Promotion of Basic and Applied Research for Innovations in Bio-oriented Industry, Japan (grant to K.I.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Nam-Chon Paek (ncpaek@snu.ac.kr).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Archer EK, Bonnett HT (1987) Characterization of a virescent chloroplast mutant of tobacco. Plant Physiol 83 920–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabes A, Domkin V, Thelander L (1999) Yeast Sml1, a protein inhibitor of ribonucleotide reductase. J Biol Chem 274 36679–36683 [DOI] [PubMed] [Google Scholar]
- Chabes AL, Pfleger CM, Kirschner MW, Thelander L (2003) Mouse ribonucleotide reductase R2 protein: a new target for anaphase-promoting complex-Cdh1-mediated proteolysis. Proc Natl Acad Sci USA 100 3925–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabouté ME, Combettes B, Clément B, Gigot C, Philipps G (1998) Molecular characterization of tobacco ribonucleotide reductase RNR1 and RNR2 cDNAs and cell cycle-regulated expression in synchronized plant cells. Plant Mol Biol 38 797–806 [DOI] [PubMed] [Google Scholar]
- Chory J (1991) Light signals in leaf and chloroplast development: photoreceptors and downstream responses in search of a transduction pathway. New Biol 3 538–548 [PubMed] [Google Scholar]
- Elledge SJ, Davis RW (1990) Two genes differentially regulated in the cell cycle and by DNA-damaging agents encode alternative regulatory subunits of ribonucleotide reductase. Genes Dev 4 740–751 [DOI] [PubMed] [Google Scholar]
- Elledge SJ, Zhou Z, Allen JB (1992) Ribonucleotide reductase: regulation, regulation, regulation. Trends Biochem Sci 17 119–123 [DOI] [PubMed] [Google Scholar]
- Elleingand E, Gerez C, Un S, Knüpling M, Lu G, Salem J, Rubin H, Sauge-Merle S, Laulhére JP, Fontecave M (1998) Reactivity studies of the tyrosyl radical in ribonucleotide reductase from Mycobacterium tuberculosis and Arabidopsis thaliana: comparison with Escherichia coli and mouse. Eur J Biochem 258 485–490 [DOI] [PubMed] [Google Scholar]
- Eriksson M, Uhlin U, Ramaswamy S, Ekberg M, Regnström K, Sjöberg BM, Eklund H (1997) Binding of allosteric effectors to ribonucleotide reductase protein R1: reduction of active-site cysteines promotes substrate binding. Structure 5 1077–1092 [DOI] [PubMed] [Google Scholar]
- Garton S, Knight H, Warren GJ, Knight MR, Thorlby GJ (2007) crinkled leaves 8—a mutation in the large subunit of ribonucleotide reductase—leads to defects in leaf development and chloroplast division in Arabidopsis thaliana. Plant J 50 118–127 [DOI] [PubMed] [Google Scholar]
- Huang M, Zhou Z, Elledge SJ (1998) The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 94 595–605 [DOI] [PubMed] [Google Scholar]
- Iba K, Takamiya KI, Toh Y, Satoh H, Nishimura M (1991) Formation of functionally active chloroplasts is determined at a limited stage of leaf development in virescent mutants of rice. Dev Genet 12 342–348 [Google Scholar]
- Inada N, Sakai A, Kuroiwa H, Kuroiwa T (1998) Three-dimensional analysis of the senescence program in rice (Oryza sativa L.) coleoptiles: investigations of tissues and cells by fluorescence microscopy. Planta 205 153–164 [DOI] [PubMed] [Google Scholar]
- Jeon JS, Lee S, Jung KH, Jun SH, Jeong DH, Lee J, Kim C, Jang S, Yang K, Nam J, et al (2000) T-DNA insertional mutagenesis for functional genomics in rice. Plant J 22 561–570 [DOI] [PubMed] [Google Scholar]
- Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J (2007) Signals from chloroplasts converge to regulate nuclear gene expression. Science 316 715–719 [PubMed] [Google Scholar]
- Kusumi K, Mizutani A, Nishimura M, Iba K (1997) A virescent gene V1 determines the expression timing of plastid genes for transcription/translation apparatus during early leaf development in rice. Plant J 12 1241–1250 [Google Scholar]
- Lichtenthaler H (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148 350–382 [Google Scholar]
- Mandel MA, Feldmann KA, Herrera-Estrella L, Rocha-Sosa M, León P (1996) CLA1, a novel gene required for chloroplast development, is highly conserved in evolution. Plant J 9 649–658 [DOI] [PubMed] [Google Scholar]
- Mullet JE (1993) Dynamic regulation of chloroplast transcription. Plant Physiol 103 309–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamatsu T, Omura T (1962) Linkage study of the genes belonging to the first chromosome in rice. Jpn J Breed 12 231–236 [Google Scholar]
- Omura T, Iwata N, Satoh H (1978) Linkage studies in rice (Oryza sativa L.): on some virescent and chlorina mutants. J Fac Agric Kyushu Univ 23 85–93 [Google Scholar]
- Park SY, Yu JW, Park JS, Li J, Yoo SC, Lee NY, Lee SK, Jeong SW, Seo HS, Koh HJ, et al (2007) The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell 19 1649–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipps G, Clément B, Gigot C (1995) Molecular characterization and cell cycle-regulated expression of a cDNA clone from Arabidopsis thaliana homologous to the small subunit of ribonucleotide reductase. FEBS Lett 358 67–70 [DOI] [PubMed] [Google Scholar]
- Qiu W, Zhou B, Darwish D, Shao J, Yen Y (2006) Characterization of enzymatic properties of human ribonucleotide reductase holoenzyme reconstituted in vitro from hRRM1, hRRM2, and p53R2 subunits. Biochem Biophys Res Commun 340 428–434 [DOI] [PubMed] [Google Scholar]
- Roy B, Beuneu C, Roux P, Buc H, Lemaire G, Lepoivre M (1999) Simultaneous determination of pyrimidine or purine deoxynucleoside triphosphates using a polymerase assay. Anal Biochem 269 403–409 [DOI] [PubMed] [Google Scholar]
- Saitoh S, Chabes A, McDonald WH, Thelander L, Yates JR, Russell P (2002) Cid13 is a cytoplasmic poly(A) polymerase that regulates ribonucleotide reductase mRNA. Cell 109 563–573 [DOI] [PubMed] [Google Scholar]
- Sauge-Merle S, Falconet D, Fontecave M (1999) An active ribonucleotide reductase from Arabidopsis thaliana cloning, expression and characterization of the large subunit. Eur J Biochem 266 62–69 [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto H, Kusumi K, Noguchi K, Yano M, Yoshimura A, Iba K (2007) The rice nuclear gene, VIRESCENT 2, is essential for chloroplast development and encodes a novel type of guanylate kinase targeted to plastids and mitochondria. Plant J 52 512–527 [DOI] [PubMed] [Google Scholar]
- Sugimoto H, Kusumi K, Tozawa Y, Yazaki J, Kishimoto N, Kikuchi S, Iba K (2004) The virescent-2 mutation inhibits translation of plastid transcripts for the plastid genetic system at an early stage of chloroplast differentiation. Plant Cell Physiol 45 985–996 [DOI] [PubMed] [Google Scholar]
- Toki S, Hara N, Ono K, Onodera H, Tagiri A, Oka S, Tanaka H (2006) Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J 47 969–976 [DOI] [PubMed] [Google Scholar]
- Wang C, Liu Z (2006) Arabidopsis ribonucleotide reductases are critical for cell cycle progression, DNA damage repair, and plant development. Plant Cell 18 350–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki K (1963) Studies on the leaf formation in rice plants. I. Observation on the successive development of the leaf. Jpn J Crop Sci 31 371–378 [Google Scholar]
- Zhao X, Georgieva B, Chabes A, Domkin V, Ippel JH, Schleucher J, Wijmenga S, Thelander L, Rothstein R (2000) Mutational and structural analyses of the ribonucleotide reductase inhibitor Sml1 define its RNR1 interaction domain whose inactivation allows suppression of mec1 and rad53 lethality. Mol Cell Biol 20 9076–9083 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.