Abstract
One of the most fascinating aspects of plant morphology is the regular geometric arrangement of leaves and flowers, called phyllotaxy. The shoot apical meristem (SAM) determines these patterns, which vary depending on species and developmental stage. Auxin acts as an instructive signal in leaf initiation, and its transport has been implicated in phyllotaxy regulation in Arabidopsis (Arabidopsis thaliana). Altered phyllotactic patterns are observed in a maize (Zea mays) mutant, aberrant phyllotaxy1 (abph1, also known as abphyl1), and ABPH1 encodes a cytokinin-inducible type A response regulator, suggesting that cytokinin signals are also involved in the mechanism by which phyllotactic patterns are established. Therefore, we investigated the interaction between auxin and cytokinin signaling in phyllotaxy. Treatment of maize shoots with a polar auxin transport inhibitor, 1-naphthylphthalamic acid, strongly reduced ABPH1 expression, suggesting that auxin or its polar transport is required for ABPH1 expression. Immunolocalization of the PINFORMED1 (PIN1) polar auxin transporter revealed that PIN1 expression marks leaf primordia in maize, similarly to Arabidopsis. Interestingly, maize PIN1 expression at the incipient leaf primordium was greatly reduced in abph1 mutants. Consistently, auxin levels were reduced in abph1, and the maize PIN1 homolog was induced not only by auxin but also by cytokinin treatments. Our results indicate distinct roles for ABPH1 as a negative regulator of SAM size and a positive regulator of PIN1 expression. These studies highlight a complex interaction between auxin and cytokinin signaling in the specification of phyllotactic patterns and suggest an alternative model for the generation of altered phyllotactic patterns in abph1 mutants. We propose that reduced auxin levels and PIN1 expression in abph1 mutant SAMs delay leaf initiation, contributing to the enlarged SAM and altered phyllotaxy of these mutants.
The shoot apical meristem (SAM) initiates lateral organs and determines their regular geometric spacing, or phyllotaxy. Different phyllotactic patterns are characteristic of specific species or developmental stages and include alternate, opposite and decussate, whorled, and spiral patterns. Although it has long drawn attention from biologists and mathematicians, until recently the molecular mechanism behind the generation of these fascinating patterns was not well understood. Molecular developmental studies suggest a phyllotaxy regulation model based on polar auxin transport by the PINFORMED1 (PIN1) auxin efflux carrier (Reinhardt et al., 2003; Petrasek et al., 2006; Wisniewska et al., 2006). Inhibition of polar auxin transport or mutations in the PIN1 auxin efflux carrier disrupt leaf or flower initiation in tomato (Solanum lycopersicum), Arabidopsis (Arabidopsis thaliana), and maize (Zea mays; Okada et al., 1991; Reinhardt et al., 2000; Scanlon, 2003). When auxin was applied locally to these pin-like SAMs, lateral organ outgrowth was restored at the application site. In Arabidopsis, PIN1-regulated polar auxin transport plays a crucial role in lateral organ positioning and initiation (Benkova et al., 2003; Reinhardt et al., 2003). PIN1 protein is expressed throughout the L1 layer of the SAM, and near the leaf initiation sites, its polar localization is directed toward the initiation sites (Reinhardt et al., 2003). As the subcellular localization of PIN directs the flow of auxin (Wisniewska et al., 2006), the hormone is transported in the L1 layer to leaf initiation sites, where auxin maxima are formed. Subsequently, the PIN1 expression domain extends inward to form a provascular stream toward the vascular tissues in the stem. These cells drain auxin away from the leaf primordium, causing depletion of auxin in the surrounding area. As a result of this sink, only regions distant from existing primordia are able to accumulate auxin and initiate new primordia. This model, therefore, explains the spacing component of phyllotaxy, but not how the patterns arise de novo. From these basic principles, several computational models of phyllotaxy have been developed (de Reuille et al., 2006; Jonsson et al., 2006; Smith et al., 2006). Despite the strong implication of auxin in phyllotaxy regulation, no auxin mutants in Arabidopsis display obvious changes in their phyllotactic pattern.
One of the very few mutants known to specifically affect phyllotaxy maps to the ABERRANT PHYLLOTAXY1 (ABPH1) locus in maize (Giulini et al., 2004). abph1 mutants develop decussate phyllotaxy, in which leaves are paired at 180°, whereas normal maize plants have a distichous phyllotaxy, with leaves developing in an alternating pattern (Jackson and Hake, 1999). ABPH1 encodes a cytokinin-inducible type A response regulator, suggesting that cytokinin signals are important for phyllotaxy regulation in maize (Giulini et al., 2004). Cytokinin signaling occurs through a two-component signaling pathway, involving a phosphorylation cascade from His kinase receptors at the cell surface to nuclear response regulators (Hwang and Sheen, 2001). The general importance of cytokinin signaling in the SAM is well established. Cytokinin overproduction results in increased transcript levels of KNOTTED-LIKE1 (KNAT1) and SHOOTMERISTEMLESS (STM), genes that are involved in the regulation of SAM function, while cytokinin-deficient plants or triple cytokinin receptor knockout mutants show reduced SAM development (Rupp et al., 1999; Werner et al., 2001, 2003; Higuchi et al., 2004; Nishimura et al., 2004). Cytokinins also appear to regulate the maintenance of a stem cell population in the SAM via a feedback loop between CLAVATA3 and WUSCHEL (WUS) genes (Brand et al., 2000; Schoof et al., 2000; Spradling et al., 2001; Weigel and Jurgens, 2002; Laux, 2003). Recently, WUS was shown to directly repress several cytokinin-inducible type A response regulators in the SAM (Leibfried et al., 2005). This result directly connects stem cell maintenance in the SAM to cytokinin signaling.
Auxins and cytokinins function together in many plant developmental processes. One classical example is in vitro plant culture, where commitment to shoot or root development is determined by the ratio of these hormones (Skoog and Miller, 1957). We hypothesized previously that the change in phyllotaxy in abph1 mutants results from a cytokinin-dependent increase in SAM size, which creates more available space for leaf initiation. However the observation that ABPH1 expression in the seedling SAM overlaps with the “plastochron zero” (P0) domain of leaf initiation suggests an alternative hypothesis that invokes a role for ABPH1 in the regulation of leaf initiation. This hypothesis is attractive in light of the fact that auxin transport by PIN1 functions in Arabidopsis phyllotaxy, and cytokinin regulation by ABPH1 in maize phyllotaxy, and suggests that these differences are not species specific but that cross talk between auxin and cytokinin signaling may be important in phyllotaxy regulation in diverse plant species. This hypothesis is supported by the observation that in maize both PIN1 and ABPH1 are expressed in what appear to be overlapping domains at the leaf initiation site or P0 (Giulini et al., 2004; Carraro et al., 2006). Cytokinins and auxins also appear to coordinately regulate root development. Cytokinins disrupt the formation of auxin maxima during Arabidopsis lateral root development by reducing the expression levels of PIN genes (Laplaze et al., 2007). Auxin-cytokinin cross talk also regulates development of the Arabidopsis root apical meristem (Dello Ioio et al., 2007) and the establishment of the embryonic root apical meristem (Müller and Sheen, 2008). Therefore, we investigated interactions between cytokinin and auxin signaling in the control of phyllotaxy in maize.
Here, we show that treatment with the polar auxin transport inhibitor 1-naphthylphthalamic acid (NPA) greatly reduced the expression of ABPH1, suggesting that ABPH1 expression is dependent on the accumulation of auxin at incipient leaf primordia. Surprisingly, we also found that maize PIN1 expression was strongly reduced in abph1 mutant meristems, indicating that ABPH1 is required for normal expression of PIN1. In addition, abph1 mutant embryos had lower concentrations of auxin compared with normal embryos, supporting the idea that ABPH1 is important for auxin accumulation. Taken together, these results imply a positive feedback loop involving polar auxin transport and maize PIN1, and ABPH1 expression at incipient leaf primordia. Therefore, ABPH1 acts both as a positive regulator of auxin levels and PIN1 signaling (this study) and as a negative regulator in cytokinin signaling in SAM size determination (Giulini et al., 2004). We propose that the reduced levels of PIN1 expression in abph1 shoot meristems may explain the observed delay in leaf initiation in abph1 mutants, which suggests an alternative model to explain the altered phyllotaxy in abph1 mutants.
RESULTS
ABPH1 Expression Is Reduced following NPA Treatment
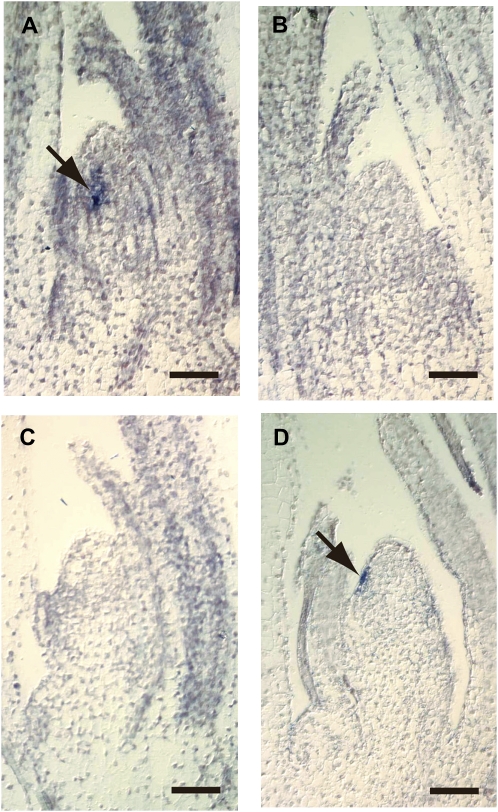
abph1 mutants initiate leaves in an altered phyllotaxy, in opposite pairs, in contrast to the normal alternate pattern in maize. During seedling development, ABPH1 is expressed in the leaf initiation site (P0) of the maize SAM, suggesting that it plays a role in leaf positioning (Fig. 1A; Giulini et al., 2004). As polar auxin transport and local auxin accumulation at the leaf initiation site are required for leaf initiation in Arabidopsis and maize (Reinhardt et al., 2003; Scanlon, 2003; Carraro et al., 2006), we examined ABPH1 expression after treatment with the polar auxin transport inhibitor NPA. Two-week-old shoot apices were cultured on control Murashige and Skoog (MS) medium or on medium with 30 μm NPA, as described (Scanlon, 2003). Apices were then fixed and subjected to in situ hybridization. While the expected ABPH1 expression domain (Giulini et al., 2004) was detected in the P0 or incipient leaf primordium in the control apices without NPA treatment (Fig. 1A), ABPH1 expression was absent (Fig. 1, B and C) or reduced and confined to the L1 layer of incipient leaf primordia (Fig. 1D) in apices treated with NPA for 5 to 14 d. Similar results were found in more than 12 treated apices.
Figure 1.
NPA treatment inhibits ABPH1 expression in the SAM. Longitudinal sections of maize shoot apices processed for ABPH1 in situ hybridization. Two-week-old maize seedlings were grown on MS medium without NPA for 10 d (A) or with 30 μm NPA for 5 d (B), 10 d (C), or 14 d (D). Arrows indicate ABPH1 expression (A) and residual weak expression (D). Bars = 100 μm.
To ask if this effect was likely to be due to a direct effect of auxin signaling or a downstream consequence of blocking leaf initiation by NPA treatment, we measured ABPH1 expression following shorter NPA treatments. After a 24-h treatment with NPA, we found no significant difference in ABPH1 expression in the SAMs of treated and control plants (ABPH1 levels were normalized relative to ubiquitin expression, and signal intensity in control treatments was arbitrarily set to 1; treated plants had a signal intensity of 0.96 ± 0.20 for the treated samples). As a control to confirm that a 24-h treatment with NPA is sufficient to affect auxin transport in the SAM, we measured ZmPIN1a expression in the SAMs of NPA-treated and control plants. We found that ZmPIN1a expression was increased in the SAMs of NPA-treated seedlings compared with control seedlings (ZmPIN1a levels were normalized relative to ubiquitin expression, and signal intensity in control treatments was arbitrarily set to 1; treated plants had a signal intensity of 2.90 ± 0.19). This observation is consistent with a previous report that enhanced expression of the auxin-responsive DR5 promoter-GFP reporter was observed in the Arabidopsis SAM after short-term NPA treatment (Benkova et al., 2003). Our result serves as a control to show that NPA influences auxin dynamics in the maize SAM within 24 h. The finding that 24 h of treatment with NPA does not alter ABPH1 expression suggests that the reduction in ABPH1 expression seen in SAMs treated for several days with NPA is unlikely to be a direct effect of reduced auxin transport and may be downstream of the block in leaf initiation. Therefore, the expression of ABPH1 is dependent on polar auxin transport, but this response is likely to be indirect.
An Arabidopsis PIN1 Antiserum Detects Maize PIN1 Expression
Here, we used an Arabidopsis PIN1 antiserum raised against a peptide (GPTPRPSNYEEDGGPA) located in the Gly-rich domain of the large intracellular loop of PIN1, which has previously been used to detect PIN1 proteins in Arabidopsis, maize, and Medicago (Boutte et al., 2006; Carraro et al., 2006; de Reuille et al., 2006). This antiserum detects PIN1 localization in the Arabidopsis SAM in the same pattern as in other studies using different PIN1 antisera or GFP fusions (Benkova et al., 2003; Reinhardt et al., 2003; Heisler et al., 2005; de Reuille et al., 2006). We first tested the specificity of the antiserum using maize protein extracts from shoot apices. A western blot detected a band of similar size (approximately 65 kD) to the predicted maize PIN1 proteins (see below; Supplemental Fig. S1). The band was weak in the soluble fraction and strong in the membrane fraction, as predicted for a membrane-localized protein.
An earlier report (Carraro et al., 2006) suggested that the pattern of PIN1 expression in maize was different, as they did not detect prominent PIN1 expression in the L1 layer like the reported expression in Arabidopsis. Here, we detected significant L1 expression of maize PIN1, and our results are described in detail below.
The Maize PIN1 Expression Pattern in the SAM Is Similar to That in Arabidopsis
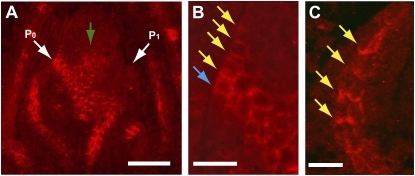
In Arabidopsis, PIN1 is preferentially expressed throughout the epidermis (L1 layer) of the SAM and, in most cells, is localized to the side of the cell that is nearest the tip of the SAM (Reinhardt et al., 2003). However, in cells near the incipient leaf primordium (P0), PIN1 is localized toward the P0, suggesting that auxin is transported in the L1 toward the P0 (Reinhardt et al., 2003). Immunolocalization with maize shoot apices also revealed polar localization patterns in the SAM (Fig. 2). Therefore, the Arabidopsis PIN1 antiserum appears able to detect maize PIN1 proteins in situ. In the maize SAM, PIN1 was not expressed strongly throughout the L1 layer but accumulated preferentially in epidermal cells near the P0 (Fig. 2, A and B). In epidermal cells that expressed maize PIN1, the protein was preferentially localized to the side of the cell nearest the P0, similar to the pattern described in Arabidopsis (Fig. 2B). Therefore, it appears likely that auxin is transported in the L1 toward the P0 in maize, as in Arabidopsis.
Figure 2.
Maize PIN1 is detected by an Arabidopsis PIN1 antiserum. A, Maize PIN1 immunolocalization in normal maize SAM. P0 and P1 leaf primordia are indicated with white arrows. The green arrow shows the maize PIN1 expression domain in the center of the SAM. Bar = 100 μm. B, Magnified incipient leaf initiation site (P0). Yellow arrows indicate basal localization of maize PIN1, and the blue arrow indicates apical localization of maize PIN1 in the L1 epidermal cells. Bar = 30 μm. C, Basal localization of maize PIN1 in provascular cells of a P1 leaf primordium. Yellow arrows show the basal localization of maize PIN1 protein. Bar = 25 μm.
We also observed that maize PIN1 expression extended inward from the P0 and young leaf primordia, presumably marking the provascular strands (Fig. 2, A and B). In these cells, the protein was polarly localized downward and toward the center of the shoot apex in two to three cell files. These cell files merged in the center of the stem (Fig. 2A). These patterns are also similar to those seen in Arabidopsis (Reinhardt et al., 2003). Thus, as proposed in Arabidopsis, the inward PIN1 localization from the P0 and young leaf primordia likely serves to form auxin sinks to deplete auxin close to existing primordia. Interestingly, we also observed an additional domain of maize PIN1 expression in the center of the SAM (Fig. 2A), as reported previously (Carraro et al., 2006). The importance of this domain is unknown, but it may be significant because PIN1 was also polarly localized within these cells.
Immunolocalization Shows That Maize PIN1 Expression Is Strongly Reduced in abph1 Plants
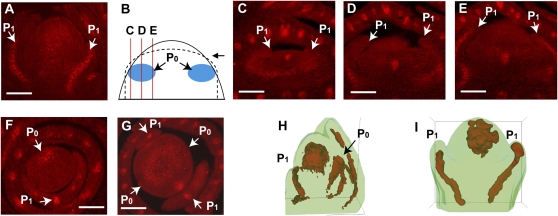
To investigate how PIN1 signaling might be affected in the abph1 phyllotaxy mutant, we also performed immunolocalization in the mutant background. As in normal maize shoot apices, PIN1 expression marked young leaf primordia in abph1, suggesting that the auxin sink function in these young primordia is not affected in abph1 mutants (Fig. 3A). However, the PIN1 expression that normally marks the P0 site was absent or very weak in abph1 (Fig. 3, B–E). As the P0 should be located at 90° to the P1 in abph1, rather than opposite as in the wild type, longitudinal median sections might miss the P0. Thus, we carried out immunolocalizations using transverse sections of abph1 apices and found the same results (Fig. 3, F and G). In abph1 mutants, leaf initiation itself is not defective, and older abph1 leaf primordia have normal PIN1 expression (Fig. 3, A and G); therefore, we interpret the weak expression in the P0 as a delay in the onset of PIN1 expression in abph1 mutants. We also observed maize PIN1 expression in a small domain in the center of the abph1 SAM, as seen in the wild type (Fig. 3A).
Figure 3.
Immunolocalization reveals PIN1 expression differences in abph1 mutants. A, Immunolocalization of PIN1 in abph1 SAM showing normal expression at P1 (arrows). B, Diagram to show the positions of longitudinal sections in C to E. The solid line indicates the SAM. The dotted line indicates the newly initiated (P1) leaf primordia. Red lines indicate sectioning planes. Letters correspond to each labeled panel. The arrow indicates the view point. P0 is labeled in blue. C to E, PIN1 immunolocalization in serial sections of the abph1 shoot apex. Bright signal in the center indicates provascular development. Arrows indicate P1. Note the lack of PIN1 expression in the presumed P0 position. F and G, PIN1 immunolocalization in transverse sections of normal (F) and abph1 (G) shoot apices. Arrows indicate each leaf position, as labeled. Note that the plane of the section in G is not precisely horizontal, hence the two P1 primordia appear slightly different sizes. Note the lack of PIN1 expression in the presumed P0 position. H and I, 3D reconstructions of PIN1 immunolocalization in normal maize (H) and abph1 (I) shoot apices. Arrows indicate each leaf primordium, as labeled. Bars = 100 μm.
To confirm the changes in maize PIN1 expression, we performed three-dimensional (3D) image reconstruction of a series of longitudinal immunolocalization sections. These reconstructions confirmed the maize PIN1 expression domains associated with leaf primordia (Fig. 3, H and I; Supplemental Videos S1 and S2) and again indicated clearly that PIN1 expression was absent from the P0 positions in abph1.
A Maize PIN1 Homolog Is Auxin Inducible in the SAM and Is Down-Regulated in abph1 Mutants
To determine whether our findings with the cross-reacting Arabidopsis PIN1 antiserum were also true for the endogenous maize PIN1 gene, and if regulation by ABPH1 might be at the transcriptional level, we identified maize PIN1 homologs. We screened a maize bacterial artificial chromosome (BAC) library (NSF B73; Clemson University Genomics Institute) using the first predicted exon sequence of the maize gene most similar to PIN1 (AZM5_9949; http://maize.tigr.org/). We identified two overlapping BAC clones and sequenced approximately 10 kb of genomic sequence of the locus, which we named ZmPIN1. This sequence corresponds to ZmPIN1a, as also described by Carraro et al. (2006). Shoot meristem-enriched cDNA libraries from immature ears and tassels were screened for maize PIN1 cDNAs, using the same probe used for BAC library screening. Seven clones were identified, and all of them corresponded exactly to the ZmPIN1a genomic clones. Comparison between ZmPIN1a genomic DNA and cDNA sequences revealed five introns, as in Arabidopsis PIN1.
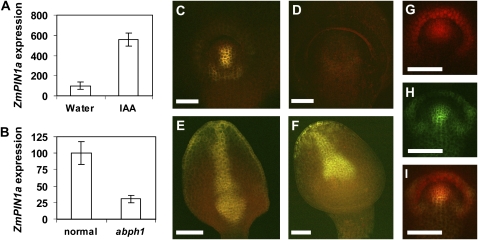
To determine if ZmPIN1a is functionally related to Arabidopsis PIN1, we first examined if it was expressed in the maize SAM. Using reverse transcription (RT)-PCR, we could indeed detect ZmPIN1a transcripts in RNA from dissected maize SAMs (data not shown). Another property of Arabidopsis PIN1 is its rapid induction by auxin (Heisler et al., 2005). To establish if ZmPIN1a is similarly induced, we performed semiquantitative RT-PCR using total RNA from 2-week-old maize apices treated with the auxin indole-3-acetic acid (IAA) for 1 h. This short treatment increased ZmPIN1a transcript levels significantly (Fig. 4A). In summary, ZmPIN1a is a maize homolog of Arabidopsis PIN1, and it is functionally related by its localization and auxin inducibility.
Figure 4.
ZmPIN1a is auxin induced, and its expression is reduced in abph1 seedlings and embryos. A, Quantification of ZmPIN1a expression by semiquantitative RT-PCR in shoot apices of seedlings treated with 100 μm IAA for 1 h. Expression levels (arbitrary units) were normalized based on ubiquitin expression levels, and control was adjusted to 100. Error bars indicate se (n = 3). B, Quantification by semiquantitative RT-PCR of ZmPIN1a expression levels in normal and abph1 SAMs. Expression levels (arbitrary units) were normalized based on ubiquitin expression levels. Levels of normal sample were arbitrarily adjusted to 100. Error bars indicate se (n = 3). C, ZmPIN1a-YFP expression in a normal embryo 11 d after pollination. The strongest ZmPIN1a-YFP expression is in the central and lower parts of the SAM. PIN1 is also expressed in the emerging coleoptile. D, An abph1 embryo 12 d after pollination. The embryo has an enlarged SAM, but no ZmPIN1a-YFP expression is visible. E, Deeper optical section through the scutellum of the normal embryo shown in C. F, Deeper optical section through the scutellum of the abph1 embryo. G, ABPH1-RFP expression in a normal embryo 11 d after pollination. Expression is seen in the upper part of the SAM and in the coleoptile. H, ZmPIN1a-YFP expression in the normal embryo shown in G. I, Colocalization of ABPH1-RFP and ZmPIN1a-YFP in the normal embryo shown in G and H. Bars = 100 μm.
To investigate if ZmPIN1a is regulated by ABPH1, as predicted by our immunolocalization results, we performed semiquantitative RT-PCR. Meristems from normal and abph1 seedlings were exposed by removing leaf primordia and then excised above the point of insertion of the P1 primordia, and total RNA was extracted. Semiquantitative RT-PCR results confirmed that ZmPIN1a transcript levels were significantly reduced in abph1 mutant meristems (Fig. 4B). These results are consistent with the reduced PIN1 expression detected by immunolocalization in abph1 mutants and suggest that this regulation of ZmPIN1a occurs at least in part at the transcriptional level.
Spatial Expression of ZmPIN1a-YFP Is Altered in abph1 Mutant Embryos
Since we observed reduced PIN1 expression in seedlings, we also investigated whether PIN1 expression was altered in abph1 embryos prior to leaf initiation. We used confocal microscopy to examine the expression of ZmPIN1a-yellow fluorescent protein (ZmPIN1a-YFP; Gallavotti et al., 2008) in embryos 11 to 14 d after pollination (Fig. 4, C–I). We found that ZmPIN1a-YFP expression was significantly weaker in the SAMs of abph1 embryos compared with the wild type. In wild-type embryos, ZmPIN1a-YFP was observed in the center and lower regions of the initiating SAM prior to initiation of the first leaf (Fig. 4C). A corresponding expression domain was never observed in the SAM of abph1 embryos (Fig. 4D). During the course of these observations, we also noticed that leaf initiation was delayed in abph1 embryos. Embryos were staged by observation of morphological landmarks, such as the appearance of the SAM, the coleoptilar ridge, or the first leaf primordium. We estimated that this delay was approximately 1 d. We also checked older abph1 embryos for ZmPIN1a-YFP expression. However, ZmPIN1a-YFP expression was never observed in the abph1 embryo SAM prior to leaf emergence. As in seedlings, ZmPIN1a-YFP was expressed in abph1 mutants in the provasculature of the first pair of leaf primordia after their initiation.
We also observed ZmPIN1a-YFP expression in deeper optical sections through the scutellum of both wild-type and abph1 embryos (Fig. 4, E and F). The expression patterns and levels of expression in the scutellum were similar in both backgrounds, indicating that the alterations in ZmPIN1a-YFP expression were specific to the SAM and that the abph1 embryos we observed were expressing the ZmPIN1a-YFP construct. Strong ZmPIN1a-YFP expression was observed in a patch beneath the SAM and in a central strand, presumably vasculature, that extended to the tip of the scutellum. ZmPIN1a-YFP expression was also seen at the upper edge of the scutellum.
ZmPIN1a-YFP and ABPH1-RFP Are Expressed in Partially Overlapping Domains in the Embryo SAM
Given the apparent effect of ABPH1 on PIN1 expression, we looked more closely to determine if their expression colocalized in the embryo during SAM initiation. (Fig. 4, G–I). We imaged embryos that were expressing ZmPIN1a-YFP and an ABPH1-red fluorescent protein (ABPH1-RFP) transgene that was also driven by its native regulatory elements. We found that these proteins were indeed expressed in partially overlapping domains in the embryo SAM. ABPH1-RFP was expressed in the upper part of the SAM and in the emerging coleoptile (Fig. 4G), in a pattern similar to that seen by in situ hybridization (Giulini et al., 2004). ZmPIN1a-YFP was expressed most strongly in the central and lower parts of the SAM (Fig. 4H). The domains of ZmPIN1a-YFP and ABPH1-RFP expression overlapped in the center of the SAM (Fig. 4I).
abph1 Mutant Embryos Have a Reduced Auxin Content
The decreased levels of ZmPIN1a expression in abph1 SAMs prompted us to ask whether auxin levels were altered in the mutant. In order to compare hormone levels, we used embryos similar to those shown in Figure 4 that had just started to initiate their first leaf primordia. This stage is enriched for SAM tissue (Jackson and Hake, 1999) and is preferred over isolated maize SAMs because they are too small for auxin quantification (H. Sakakibara, personal communication). ABPH1 is expressed specifically in the SAM region in the embryo (Giulini et al., 2004); therefore, the contributions of the abph1 mutation to the auxin differences should be mainly from the SAM, rather than from other regions of the embryo.
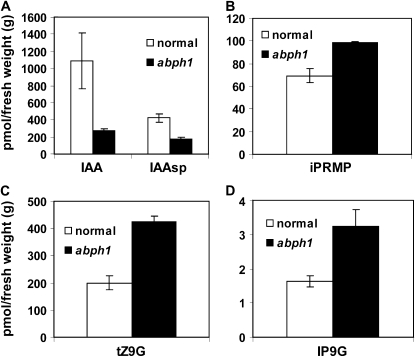
We found that the level of IAA was significantly lower in abph1 mutant embryos compared with normal siblings (Fig. 5A; Supplemental Table S1). The IAA conjugate, indole-3-acetyl aspartic acid (IAAsp), was also reduced in abph1 (Fig. 5A). The level of IAAsp is usually correlated with the levels of IAA (Ostin et al., 1998; Barratt et al., 1999; Barlier et al., 2000), consistent with the reduced IAA levels in abph1. Thus, the levels of IAA and an IAA conjugate were lower in abph1 embryos, a finding that is consistent with the reduced ZmPIN1a expression in the abph1 SAM.
Figure 5.
Hormone and hormone conjugate levels are altered in abph1 mutants. Hormone and hormone conjugates that showed significant differences between normal maize and abph1 are plotted. A, IAA and IAAsp. B, N6-(Δ2-Isopentenyl)adenine riboside 5′-monophosphate (iPRMP). C, Trans-zeatin-N9-glucoside (tZ9G). D, Isopentenyladenine-N9-glucoside (IP9G). Averages were calculated from three independent biological pools of approximately 10 embryos. Error bars indicate se.
We also analyzed cytokinin levels in abph1. Among the active cytokinins analyzed, cis-zeatin, dihydrozeatin, and N6-(Δ2-isopentenyl)adenine were below the detection limit and trans-zeatin did not display any significant difference. However, some cytokinin intermediates, such as N6-(Δ2-isopentenyl)adenine riboside 5′-monophosphate, and the inactivation reaction products isopentenyladenine-N9-glucoside and trans-zeatin-N9-glucoside levels were substantially increased in abph1 mutants (Fig. 5, B–D; Supplemental Table S1). These changes could reflect local alterations in cytokinin levels in abph1 mutants; however, this speculation is limited by our inability to measure cytokinins from very small tissue sources.
ZmPIN1a Is Rapidly Induced by Cytokinin
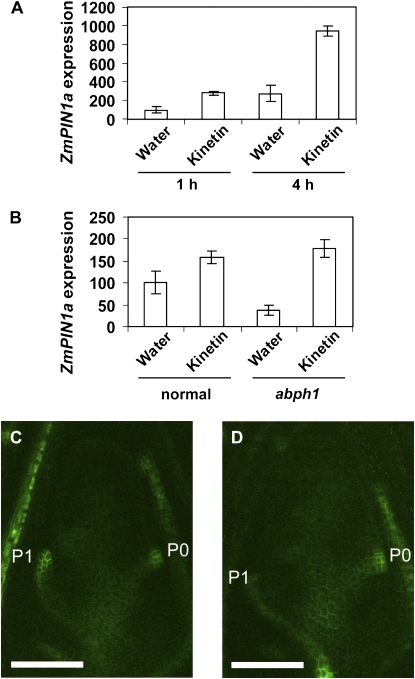
Since ABPH1 was required for normal PIN1 expression in the P0 of the SAM and ABPH1 is rapidly induced by cytokinin (Giulini et al., 2004), we asked if PIN1 expression was also regulated by cytokinins. We measured ZmPIN1a expression using semiquantitative RT-PCR of total RNA from dissected meristems. Indeed, following 1 h of treatment, ZmPIN1a expression was higher in the treated sample than in the water control, and a prolonged cytokinin treatment (4 h) showed further induction of ZmPIN1a in the cytokinin-treated SAM compared with the control (Fig. 6A). We also asked if the cytokinin induction of ZmPIN1a expression was altered in the abph1 mutant. As already shown, ZmPIN1a expression in untreated meristems was significantly lower in abph1 than in the wild type (Fig. 6B). However, cytokinin treatment induced ZmPIN1a expression to similar levels in abph1 and normal seedlings, suggesting that ZmPIN1a sensitivity to cytokinin is independent of abph1 (Fig. 6B).
Figure 6.
ZmPIN1a expression is rapidly induced by cytokinin in the SAM. A, Quantification of ZmPIN1a expression by semiquantitative RT-PCR using total RNA from single normal meristems treated with water or 100 μm kinetin for the indicated times. Expression levels (arbitrary units) were normalized based on ubiquitin expression levels. The 1-h water control levels were adjusted to 100 (n = 3; error bars indicate se). B, Quantification of ZmPIN1a by semiquantitative RT-PCR using total RNA from single normal or abph1 mutant meristems treated with 10 μm kinetin for 4 h. Expression levels (arbitrary units) were normalized based on ubiquitin expression levels. Levels of water control of normal samples were adjusted to 100 (n = 4; error bars indicate se). C, Confocal image of ZmPIN1a-YFP expression in a median longitudinal section through SAM of a seedling treated with control solution for 4 h. D, Confocal image of ZmPIN1a-YFP expression in a median longitudinal section through SAM of a seedling treated with 100 μm kinetin for 4 h. Bars = 100 μm. Additional images are shown in Supplemental Figure S3.
To determine if cytokinin treatment alters the level of ZmPIN1a expression throughout the SAM, or if the increased expression is limited to the P0, we imaged ZmPIN1a-YFP expression in the SAMs of cytokinin-treated and control plants (Fig. 6, C and D; Supplemental Fig. S3). We found that the fluorescence intensity in the P0 was greater in cytokinin-treated than in untreated plants (Fig. 6, C and D; Supplemental Table S2; Supplemental Fig. S3). When fluorescence intensity in the entire P0 was compared, control plants had a mean fluorescence intensity of 48 (se = 3), whereas treated plants had a mean value of 57 (se = 5) on a scale of 0 to 255. Measurements of fluorescence intensity in a set area of the P0 gave similar results. The mean area of ZmPIN1a-YFP-expressing cells also increased in cytokinin-treated plants from 579 μm2 (se = 31) to 703 μm2 (se = 33). We also calculated integrated density values (defined as the product of mean fluorescence intensity and area) in the P0 of control and treated plants as 27,143 (se = 2,009) and 40,344 (se = 4,156), respectively. ZmPIN1a-YFP expression in other regions of the SAM did not show a significant difference. These results indicate that cytokinin treatment specifically promotes expression of ZmPIN1a in the incipient leaf primordium.
In summary, we found a mutual requirement of auxin transport for ABPH1 expression and ABPH1 for ZmPIN1a expression, a finding that is consistent with reduced auxin levels in abph1 mutants. ZmPIN1a expression was also rapidly up-regulated by cytokinin. Together, these findings suggest that complex cross talk between auxin and cytokinin signaling is involved in the mechanism of phyllotaxy regulation. The reduced PIN expression in abph1 mutants also suggests an alternative model for the change in phyllotaxy that is observed in these mutants.
DISCUSSION
In this study, we investigated interactions between ABPH1-mediated cytokinin signaling and PIN1-dependent polar auxin transport in relation to phyllotaxy. The role of auxin transport by PIN1 and local auxin accumulation in determination of the leaf initiation site and leaf spacing are well established in Arabidopsis and other dicots (Reinhardt et al., 2000, 2003; Heisler et al., 2005; de Reuille et al., 2006; Jonsson et al., 2006; Smith et al., 2006). Polar auxin transport inhibitor studies (Scanlon, 2003) and PIN1 localization (Carraro et al., 2006; this study) strongly suggest that PIN1 also regulates leaf initiation and positioning in monocots such as maize. However, Arabidopsis auxin mutants do not show discrete changes of phyllotactic patterns, and one of the only mutants known to specifically change these patterns is the abph1 mutant of maize, which is mutated in a cytokinin-inducible response regulator. ABPH1 is expressed in the P0, in a region overlapping the domain of PIN1 accumulation. These observations prompted us to investigate interactions between auxin and cytokinin signaling in phyllotaxy.
The interaction between ABPH1 and auxin signaling was first examined by inhibition of polar auxin transport by NPA. This treatment leads to a loss of leaf initiation, presumably because auxin no longer accumulates at the P0 (Reinhardt et al., 2000; Scanlon, 2003). We found that NPA treatment of maize shoots for 5 to 10 d resulted in the loss of ABPH1 expression at the P0. However, shorter (24 h) treatments did not alter ABPH1 expression. Thus, the loss of ABPH1 expression may be a downstream consequence of the loss of leaf initiation in NPA-treated shoot apices, rather than a direct effect. We also tested whether ABPH1 was auxin inducible and found that it was not (Supplemental Fig. S2). This result is consistent with the hypothesis that ABPH1 is not a direct target of auxin signaling. Alternatively, ABPH1 expression might be saturated in normal physiological conditions, where auxin levels are maximal at the P0 (Reinhardt et al., 2003). Taken together, these results suggest that ABPH1 expression is dependent on a signal derived from the P0 but may not be a direct target of polar auxin transport or auxin accumulation. We also consider the possibility that other signal(s), in addition to auxin, may be involved, since the mechanism of NPA action is not specific to auxin (Geldner et al., 2001).
To elucidate interactions between ABPH1 and the PIN1 polar auxin transporter in maize, we investigated PIN1 expression in wild-type and abph1 seedling SAMs and embryos. Using a cross-reacting Arabidopsis PIN1 antiserum, we observed a similar PIN1 expression pattern in the maize SAM as found in Arabidopsis. In contrast to a previous report (Carraro et al., 2006), where it was suggested that PIN1 expression in the L1 layer may be less important in maize than in Arabidopsis, we found very clear PIN1 expression in the L1 cells, especially around the P0. The polar subcellular localization in these L1 cells and vascular localization patterns of maize PIN1 were similar to those seen in Arabidopsis. PIN1 protein in the L1 was most clearly detected close to the P0, and not throughout the epidermal layer of the maize SAM, unlike in Arabidopsis, where PIN1 accumulates throughout the L1. However, we suspect that this difference may be due to differences in expression levels between vegetative and inflorescence apices, rather than a fundamental species-specific difference. The published immunolocalization studies of Arabidopsis PIN1 are mostly in inflorescence meristems, and we also observed PIN1 localization throughout the entire L1 layer of inflorescence and floral meristems in maize (Gallavotti et al., 2008). Our observations provide evidence for polar auxin transport in the L1 layer toward the P0 and its removal through the developing provascular tissue. Hence, it appears likely that polar auxin transport in the L1 and auxin sinks contribute to leaf initiation and spacing in the maize shoot apex, as in Arabidopsis (Reinhardt et al., 2003). Interestingly, we also observed PIN1 expression in the center of the maize SAM by immunolocalization and by expression of the ZmPIN1a-YFP construct. Computer simulations based on PIN1 immunolabeling in Arabidopsis predicted an accumulation of auxin in the central zone, and higher IAA content in this zone has been experimentally confirmed and is proposed to play an important role in creating auxin maxima at incipient leaf initiation sites (de Reuille et al., 2006). Thus, our observations in the maize SAM are consistent with the computational model, even though PIN1 was not detected in the central zone of the Arabidopsis SAM (Reinhardt et al., 2003).
Surprisingly, we found significantly lower PIN1 expression in the P0 domain of abph1 seedlings and in the SAM of abph1 embryos. This difference was not an artifact of genetic background or experimental protocol, because normal PIN1 levels in abph1 mutants were found later in development, for example in P1 or older leaf primordia, and the PIN1 expression patterns in the scutellums of wild-type and abph1 embryos were similar. The reduced PIN1 expression in the SAM was confirmed by semiquantitative RT-PCR of a maize PIN1 homolog, suggesting that the regulation of PIN1 at the P0 by ABPH1 was at least in part at the transcriptional level.
The positive regulation of PIN1 by ABPH1 suggests a new model to explain the larger SAM and altered phyllotaxy phenotypes of abph1 mutants (Jackson and Hake, 1999; Fig. 7). According to this model, the lower level of PIN1 expression in the mutants could cause a delay in leaf initiation, allowing for extra growth of the meristem before leaves are initiated. Consistent with this model, we observed that initiation of the first leaf primordia in abph1 embryos occurs approximately 1 d later than in wild-type siblings (B.-h. Lee, R. Johnston, and D. Jackson, unpublished data). We propose that both the delayed leaf initiation and the enlarged SAM phenotypes contribute to the altered phyllotaxy observed in these mutants (Fig. 7). Given that the formation of auxin maxima is required for leaf initiation, our findings that auxin levels are lower in abph1 embryos, but that two leaves are initiated simultaneously in these mutants, seem contradictory. However, this may be explained if relative, rather than absolute, auxin levels are required to induce leaf initiation and if leaf initiation is triggered in cells with higher auxin levels relative to their neighbors. This model does not preclude a requirement for a basal auxin threshold for leaf initiation, and in this scenario, leaf initiation would be delayed until the threshold level is reached. There is already evidence for auxin thresholds, for example in vein formation in Arabidopsis flowers, where there is a correlation between YUCCA gene dosage and the number of veins formed (Cheng et al., 2006).
Figure 7.
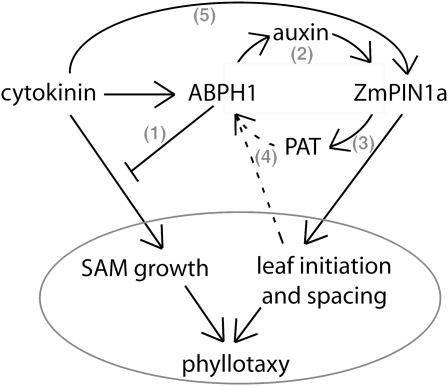
ABPH1 regulation of both cytokinin and auxin signaling appears to control phyllotaxy in maize. In step 1, ABPH1 negatively regulates cytokinin induction of SAM growth and therefore functions to maintain normal meristem size. ABPH1 also induces ZmPIN1a expression at the P0, possibly through facilitating auxin accumulation (step 2). ZmPIN1a likely functions in polar auxin transport (PAT) in maize to control leaf initiation and spacing (step 3), and PAT, or a downstream event in leaf initiation, is required for ABPH1 expression, although this effect is probably indirect (dotted lines; step 4). ZmPIN1a is also rapidly induced by cytokinin, although this induction appears to be ABPH1 independent (step 5). We propose that these complex interactions between auxin-mediated leaf initiation/spacing and cytokinin-mediated SAM growth contribute to the regulation of phyllotactic patterns, and our data suggest that ABPH1 performs a central function within this regulatory network.
Although it is generally thought that type A cytokinin response regulators act as negative regulators or repressors (Hwang and Sheen, 2001), we found that ABPH1 acts as a positive regulator of PIN1 expression. Other examples in which type A cytokinin response regulators act as positive regulators have been described. For example, overexpression of Arabidopsis ARR4 resulted in increased shoot regeneration in tissue culture (Osakabe et al., 2002) and ARR knockouts have reduced WUS expression (Leibfried et al., 2005), indicating a positive cytokinin-dependent role in shoot regeneration and WUS induction. ABPH1 is thought to act as a repressor of cytokinin signaling and a negative regulator of SAM size (Giulini et al., 2004). The data presented here indicate that ABPH1 also acts as a positive regulator of ZmPIN1a in the SAM of embryos and seedlings. Consistent with lower PIN1 expression levels in abph1, the mutant embryos also had lower auxin levels. ABPH1 is specifically expressed in the embryonic SAM (Giulini et al., 2004), so this reduction likely reflects a lower level of auxin in the embryonic SAM. This is supported by our observations of lower ZmPIN1a-YFP expression in the SAM, but not the scutellum, of abph1 embryos. In Arabidopsis, auxin increases PIN1 levels and facilitates its own efflux, leading to a presumed feed-forward positive reinforcement (Paciorek et al., 2005; Jonsson et al., 2006; Smith et al., 2006). Therefore, ABPH1 may regulate auxin levels directly, or the effect could be mediated through its positive regulation of PIN1.
Recently, the effects of cytokinins on PIN gene expression in Arabidopsis lateral root primordia were also investigated (Laplaze et al., 2007). In contrast to our findings in the shoot meristem, cytokinin treatment blocked the formation of auxin maxima at lateral root initiation sites by down-regulating PIN expression. Although we found the opposite effect, that cytokinin induced PIN1 expression in the shoot meristem, we are confident that our data are consistent. For example, cytokinins induce ABPH1 expression (Giulini et al., 2004), and ABPH1 is required for PIN1 expression in the P0. The different effects of cytokinins on PIN1 expression may reflect species-specific differences or, more likely, developmentally specific differences between root (lateral meristem initiation) and shoot (leaf initiation).
In summary, our results provide evidence for both negative and positive regulatory roles of ABPH1 (Fig. 7). While we cannot be certain that the interactions we found are direct, where we could measure kinetics (e.g. cytokinin induction of ABPH1 [Giulini et al., 2004] or of PIN1 [reported here]), these effects were rapid, occurring within 1 h, and certainly much faster than the length of a plastochron in maize, which is in the order of 24 h (Smith and Hake, 1992), suggesting that they are likely to be developmentally relevant. Together with the findings of cross talk between cytokinin and auxin signaling in initiation and regulation of the Arabidopsis root apical meristem or of lateral root primordia (Dello Ioio et al., 2007; Laplaze et al., 2007; Müller and Sheen, 2008), our results suggest a common theme of auxin-cytokinin cross talk in the regulation of plant apical meristems and organ initiation.
MATERIALS AND METHODS
Plant Treatments
For NPA treatment of cultured apices, we followed published methods (Scanlon, 2003). Briefly, shoot apices from 2-week-old maize (Zea mays) seedlings were dissected down to the SAM with four to five leaves attached and placed on MS/phytagel (Sigma) medium with or without 30 μm NPA (Chem Service).
For hormone and NPA treatments, 2-week-old maize seedlings were cut at the root-shoot junction and the shoot portion was placed in solution as described by Giulini et al. (2004). Kinetin or IAA was dissolved in 1 m KOH to make 10 mm stock solutions. The working solutions were diluted from the stock, and the pH was adjusted to 5.8 with HCl. The control solution was prepared from the same volume of 1 m KOH diluted with water, and pH for the control was also adjusted. NPA was dissolved in dimethyl sulfoxide and then diluted with water to make a 30 μm solution. A control solution was made by adding an equivalent quantity of dimethyl sulfoxide to water. After appropriate treatments, meristems were dissected and collected for further analyses.
In Situ Hybridization and Immunolocalization
Using the full-length cDNA of ABPH1, in situ hybridization was carried out on shoot apices following the method described by Jackson et al. (1994).
For immunolocalization, 2-week-old maize shoot apices were fixed in 5% acetic acid, 45% ethanol, and 10% formalin overnight. After dehydration through an ethanol series, the apices were infiltrated with Steedman's wax (polyethylene glycol 400 distearate:hexadecanol, 9:1) at 37°C (Steedman, 1957). Following embedding and sectioning, 10-μm sections were placed onto Chrom-Alum-coated slides (0.5% fish gelatin [Sigma] and 0.05% chromium potassium sulfate). For antibody reactions, Steedman's wax was removed in 100% ethanol. The sections were then rehydrated in an ethanol series and blocked by incubating in phosphate-buffered saline (PBS) buffer (130 mm NaCl and 10 mm PO4) with 1% bovine serum albumin. The slides were incubated with an Arabidopsis (Arabidopsis thaliana) PIN1 antiserum (Boutte et al., 2006; 1:300 dilution) in PBS with 1% bovine serum albumin for 2 h at room temperature. After two to three washes in PBS with 1% fish gelatin, the slides were incubated with a Cy3-conjugated anti-rabbit secondary antibody (1:800 dilution; Jackson ImmunoResearch Laboratory). Slides were washed five times in PBS with 1% fish gelatin, mounted in Citifluor AF1 (Electron Microscopy Sciences), and observed by fluorescence microscopy.
Molecular Biology
The ZmPIN1a-YFP construct has been described (Gallavotti et al., 2008). The ABPH1-RFP construct was generated using the fluorescent tagging of full-length proteins method (Tian et al., 2004). The construct includes 5′ upstream DNA sequence (3,558 bp) and 3′ downstream sequence (662 bp) of the ABPH1 open reading frame (842 bp) with a monomeric RFP (Campbell et al., 2002) internally inserted between nucleotides 830 and 831 of the ABPH1 open reading frame. This causes internal insertion of RFP between amino acids 132 and 133 of the ABPH1 protein. The primers used were AB1-P3.5F-P1 (5′-gctcgatccacctaggctCAGGGTGCCAAGATCTCTCC-3′), AB1-RFP-P2 (5′-tccacctccacctccaggccggccGCGGCTGCACAGGCGCGA-3′), AB1-RFP-P3 (5′-tggtgctgctgcggccgctggggccGTCCTGCGGTGAGCACGG-3′), and AB1-P4 (5′-cgtagcgagaccacaggaGCAACCGTGACCAAGATGAG-3′). Gene-specific sequences are shown in uppercase letters. Following the fluorescent tagging of full-length protein method, the fusion product was subcloned into the pTF101.1-derived pAM1006 vector containing a Gateway cassette.
For analysis of ZmPIN1a by semiquantitative RT-PCR, maize shoot meristems were isolated from 2-week-old normal and abph1 maize plants. Total RNA was extracted with Trizol (Invitrogen). Semiquantitative RT-PCR was carried out for ZmPIN1a using the One-Step RT-PCR kit (Qiagen) with primers UCSD-ZmPIN1-F1 (5′-ATAATCGCGTGCGGGAACAA-3′) and UCSD-ZmPIN1-R1 (5′-TCCTGCTCCACATCCCCATC-3′) and for ubiquitin with primers Ubi 5′ (5′-TAAGCTGCCGATGTGCCTGCGTCG-3′) and Ubi 3′ (5′-CTGAAAGACAGAACATAATGAGCACAG-3′). The ubiquitin gene was chosen as a loading control. The one-step RT-PCR was done as follows: 50°C for 30 min, 94°C for 15 min, seven cycles of 94°C for 15 s, 65°C (−1°C per cycle) for 15 s, and 72°C 20 s, and 18 cycles of 94°C for 15 s, 59°C for 15 s, and 72°C for 20 s. For analysis of ABPH1 expression by semiquantitative RT-PCR following NPA treatment, the SAM plus two leaf primordia were dissected from seedlings treated for 24 h with NPA or control solution. Two biological replicates were used for each treatment, and 10 SAMs were pooled for each replicate. Total RNA was extracted using Trizol, followed by RNAeasy columns (Qiagen). Semiquantitative RT-PCR was carried out using the SuperScript III One-Step RT-PCR kit (Invitrogen). PCR cycles were as follows: 94°C for 4 min, 10 cycles of 94°C for 30 s, 65°C (−1°C per cycle) for 20 s, and 75°C for 20 s, and 26 cycles of 94°C for 30 s, 55°C for 20 s, and 72°C for 30 s. The ABPH1 primer sequences were Fwd (5′-GATGGCGAGCCGCAAGTGT-3′) and Rev (5′-AATGCCGCTGCTACAGCTACCA-3′; Giulini et al., 2004). Ubiquitin was used as a loading control. After fractionation on a 1.2% agarose gel, the band intensity was quantified using a GelDoc system (Bio-Rad) by ethidium bromide fluorescence or a phosphoimager (Fuji) after DNA blot hybridization. Semiquantitative RT-PCR was performed using a cycle number low enough to ensure that amplification was in the linear phase. ZmPIN1a and ABPH1 signal levels were normalized by the ubiquitin signal intensity.
Protein Blot Analysis
Proteins were extracted from maize shoot apices by grinding in the protein extraction buffer (150 mm NaCl, 50 mm Tris-Cl [pH 7.5], 1 mm phenylmethylsulfonyl fluoride, 0.5 mm dithiothreitol, and Complete EDTA-free Protease Inhibitor Cocktail; Roche Applied Science).
The total fraction was obtained after centrifugation of the sample at 8,000g at 4°C for 10 min. This total fraction was centrifuged at 100,000g at 4°C for 1 h to separate soluble and membrane fractions; the supernatant fluid was taken as the soluble fraction, and the pellet was resuspended in extraction buffer containing 1% Triton X-100, incubated on ice for 30 min, and centrifuged at 100,000g at 4°C for 1 h. The resulting supernatant fluid was the membrane fraction. SDS-polyacrylamide gels (8%) were loaded with 16 μg of protein per lane. After running at 200 V for 45 min, they were blotted onto a polyvinylidene fluoride membrane (Millipore) using a Trans-Blot SD semidry transfer cell (Bio-Rad). Blocking was carried out by immersing the membrane in 5% nonfat milk in TBS-T buffer (140 mm NaCl, 20 mm Tris, pH 7.6, and 0.1% [v/v] Tween 20) for 1 h. The Arabidopsis PIN1 antiserum was diluted in the blocking solution to 1:2,000 and incubated with the blocked membrane for 1 h. After washing three times with TBS-T for 10 min, a horseradish peroxidase-labeled anti-mouse antibody (Amersham Bioscience) was diluted in the blocking solution to 1:5,000 and used as a secondary antibody. After 1 h of incubation, the membrane was washed four times for 10 min with TBS-T, visualized by the ECL Plus Western Blotting Detection System (Amersham Bioscience), and exposed to x-ray films for 30 s.
3D Image Reconstructions
A series of PIN1 immunolocalization images was collected using fluorescence microscopy from sequential sections of normal and abph1 SAMs (11 10-μm sections for normal SAM; 18 10-μm sections for abph1 SAM). Each series was used to construct 3D images following three steps: (1) preprocessing of the images in order to identify the areas of interest (Costa and Cesar, 2000) and to align each subsequent pair of slices; (2) morphing between subsequent pairs of images in order to obtain smoother transitions between slices, using Squrlz Morph application (http://www.xiberpix.com/SqirlzMorph.html); and (3) 3D reconstruction, using the whole set of images, of the structures of interest (Schroeder et al., 2003). Finally, the data were exported into Virtual Reality Modeling Language format.
Hormone Content Analysis
To achieve similar developmental stages with isogenic controls, pollen from normal maize or abph1 mutants was used to pollinate ears of abph1 mutants, generating abph1 heterozygous (“wild type”) and homozygous embryos, respectively. Embryos were harvested at about 10 d after pollination. For each extraction, 10 embryos (7–10 mg fresh weight) were harvested and soaked in 1 mL of extraction solvent (methanol:chloroform:water, 3:1:1). Hormones and related compounds were extracted using a TissueLyser (Qiagen) with zirconia beads, an HLB column (Waters), and an MCX column (Waters). In the solid-phase extraction, the cytokinin nucleotides were eluted with 0.35 m NH4OH, other cytokinin species were eluted with 0.35 m NH4OH in 60% methanol, and the auxins were eluted with methanol (Dobrev and Kaminek, 2002). The cytokinins were then measured with a liquid chromatography-mass chromatography system (UPLC/Quattro Ultima Pt; Waters) as described previously (Dobrev and Kaminek, 2002; Nakagawa et al., 2005). For measurement of IAA and IAAsp, the fraction eluted from the MCX column (Dobrev and Kaminek, 2002) was further purified using DEAE-cellulose. IAA and the amino acid conjugates were eluted with 0.5% and 3% formic acid, respectively. As the internal standards, stable isotope IAA (deuterium-labeled [2H5]IAA; OlchemIm) was added together with the internal standard of cytokinins. The recovery yield of IAAsp was calculated using that of IAA. The IAA and IAAsp were determined with a liquid chromatography-mass chromatography system. Cone voltage for detection of IAA and IAAsp was 53 V, collision energy was 13 to 19 eV, and capillary voltage was 3.12 kV.
Confocal Imaging of ZmPIN1a-YFP and ABPH1-RFP Expression in Embryos
Confocal images were taken with a Zeiss LSM510 confocal microscope, and images were acquired using the LSM software. For analysis of ZmPIN1a-YFP expression, wild-type (B73) and abph1 ears were pollinated with pollen from abph1 plants expressing the ZmPIN1a-YFP construct to generate normal and abph1 embryos, respectively. For colocalization of ZmPIN1a-YFP and ABPH1-RFP, pollen from plants expressing both constructs was used to pollinate wild-type ears. Embryos were dissected from kernels at 11 to 14 d after pollination, fixed for 10 min in 2.5% paraformaldehyde, mounted on glass microscope slides in FocusClear (CelExplorer Labs), and left to clear for 30 min.
Quantification of ZmPIN1a-YFP after Cytokinin Treatment
Seedlings expressing the ZmPIN1a-YFP construct were treated for 4 h with 100 μm kinetin or control solution. Median longitudinal hand sections were made of seedling apices and then mounted on glass microscope slides in 1× PBS buffer. Confocal settings were optimized so that fluorescence levels were below saturation and no fluorescence was seen in control (nonexpressing) SAMs. Quantification of fluorescence was done using ImageJ software (http://rsb.info.nih.gov/ij). For each section, the P0 site of leaf initiation, marked by high PIN1-YFP expression, was outlined using the polygon tool. Mean fluorescence intensity (on a scale of 0–255) and the area of the P0 were calculated for the selected region. Integrated density was calculated by multiplying the mean fluorescence intensity by the area. Mean fluorescence intensity was also quantified for a square with a set area of 225 μm2 (15 μm × 15 μm) that was centered on the P0 leaf primordium of each sample. The values obtained were compared using Student's t test.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Maize PIN1 is detected by an Arabidopsis PIN1 antiserum.
Supplemental Figure S2. ABPH1 is not induced by auxin.
Supplemental Figure S3. Cytokinin treatment induces ZmPIN1a-YFP in the P0 leaf primordium.
Supplemental Table S1. Contents of cytokinins, auxin, and related compounds in normal and abph1 embryos.
Supplemental Table S2. Quantification of ZmPIN1a-YFP fluorescence in the SAM of kinetin-treated and control seedlings.
Supplemental Video S1. 3D reconstruction of maize PIN1 immunolocalization in the shoot apex of normal maize.
Supplemental Video S2. 3D reconstruction of maize PIN1 immunolocalization in the shoot apex of abph1.
Supplementary Material
Acknowledgments
We thank Amitabh Mohanty for technical advice in ZmPIN1a-YFP and ABPH1-RFP construction, Tim Mulligan for plant care, Erin Jimenez for assistance with dissections and RT-PCR, and members of the Jackson laboratory for valuable advice.
This work was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (grant no. 308231/03–1 to L.d.F.C.), by the Fundação de Amparo à Pesquisa do Estado de São Paulo (grant no. 03/13072–8 to B.A.N.T.), by a Cold Spring Harbor Laboratory association fellowship to A.G., by a Special Research Grant of Sogang University to B.-h.L., and by the National Science Foundation (grant no. IOB 0642707 to B.-h.L., R.J., and D.J. and grant no. DBI–0501862 to Y.Y. and D.J.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: David Jackson (jacksond@cshl.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Barlier I, Kowalczyk M, Marchant A, Ljung K, Bhalerao R, Bennett M, Sandberg G, Bellini C (2000) The SUR2 gene of Arabidopsis thaliana encodes the cytochrome P450CYP83B1, a modulator of auxin homeostasis. Proc Natl Acad Sci USA 97 14819–14824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barratt NM, Dong WQ, Gage DA, Magnus V, Town CD (1999) Metabolism of exogenous auxin by Arabidopsis thaliana: identification of the conjugate N-alpha-(indol-3-ylacetyl)-glutamine and initiation of a mutant screen. Physiol Plant 105 207–217 [Google Scholar]
- Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, Jurgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115 591–602 [DOI] [PubMed] [Google Scholar]
- Boutte Y, Crosnier MT, Carraro N, Traas J, Satiat-Jeunemaitre B (2006) The plasma membrane recycling pathway and cell polarity in plants: studies on PIN proteins. J Cell Sci 119 1255–1265 [DOI] [PubMed] [Google Scholar]
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R (2000) Dependence of stem cell fate in Arabidopsis and a feedback loop regulated by CLV3 activity. Science 289 617–619 [DOI] [PubMed] [Google Scholar]
- Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY (2002) A monomeric red fluorescent protein. Proc Natl Acad Sci USA 99 7877–7882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraro N, Forestan C, Canova S, Traas J, Varotto S (2006) ZmPIN1a and ZmPIN1b encode two novel putative candidates for polar auxin transport and plant architecture determination of maize. Plant Physiol 142 254–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YF, Dai XH, Zhao YD (2006) Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 20 1790–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa LDF, Cesar RM Jr (2000) Shape Analysis and Classification: Theory and Practice. CRC Press, Boca Raton, FL
- Dello Ioio R, Linhares FS, Scacchi E, Casamitjana-Martinez E, Heidstra R, Costantino P, Sabatini S (2007) Cytokinins determine Arabidopsis root meristem size by controlling cell differentiation. Curr Biol 17 678–682 [DOI] [PubMed] [Google Scholar]
- de Reuille PB, Bohn-Courseau I, Ljung K, Morin H, Carraro N, Godin C, Traas J (2006) Computer simulations reveal properties of the cell-cell signaling network at the shoot apex in Arabidopsis. Proc Natl Acad Sci USA 103 1627–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrev PI, Kaminek M (2002) Fast and efficient separation of cytokinins from auxin and abscisic acid and their purification using mixed-mode solid-phase extraction. J Chromatogr A 950 21–29 [DOI] [PubMed] [Google Scholar]
- Gallavotti A, Yang Y, Schmidt RJ, Jackson D (2008) The relationship between auxin transport and maize branching. Plant Physiol 147 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N, Friml J, Stierhof YD, Jurgens G, Palme K (2001) Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413 425–428 [DOI] [PubMed] [Google Scholar]
- Giulini A, Wang J, Jackson D (2004) Control of phyllotaxy by the cytokinin-inducible response regulator homologue ABPHYL1. Nature 430 1031–1034 [DOI] [PubMed] [Google Scholar]
- Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, Long JA, Meyerowitz EM (2005) Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr Biol 15 1899–1911 [DOI] [PubMed] [Google Scholar]
- Higuchi M, Pischke MS, Mahonen AP, Miyawaki K, Hashimoto Y, Seki M, Kobayashi M, Shinozaki K, Kato T, Tabata S, et al (2004) In planta functions of the Arabidopsis cytokinin receptor family. Proc Natl Acad Sci USA 101 8821–8826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sheen J (2001) Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413 383–389 [DOI] [PubMed] [Google Scholar]
- Jackson D, Hake S (1999) Control of phyllotaxy in maize by the abphyl1 gene. Development 126 315–323 [DOI] [PubMed] [Google Scholar]
- Jackson D, Veit B, Hake S (1994) Expression of maize Knotted1-related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120 405–413 [Google Scholar]
- Jonsson H, Heisler MG, Shapiro BE, Meyerowitz EM, Mjolsness E (2006) An auxin-driven polarized transport model for phyllotaxis. Proc Natl Acad Sci USA 103 1633–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplaze L, Benkova E, Casimiro I, Maes L, Vanneste S, Swarup R, Weijers D, Calvo V, Parizot B, Herrera-Rodriguez MB, et al (2007) Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell 19 3889–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laux T (2003) The stem cell concept in plants: a matter of debate. Cell 113 281–283 [DOI] [PubMed] [Google Scholar]
- Leibfried A, To JPC, Busch W, Stehling S, Kehle A, Demar M, Kieber JJ, Lohmann JU (2005) WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438 1172–1175 [DOI] [PubMed] [Google Scholar]
- Müller B, Sheen J (2008) Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453 1094–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa H, Jiang CJ, Sakakibara H, Kojima M, Honda I, Ajisaka H, Nishijima T, Koshioka M, Homma T, Mander LN, et al (2005) Overexpression of a petunia zinc-finger gene alters cytokinin metabolism and plant forms. Plant J 41 512–523 [DOI] [PubMed] [Google Scholar]
- Nishimura C, Ohashi Y, Sato S, Kato T, Tabata S, Ueguchi C (2004) Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 16 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Ueda J, Komaki MK, Bell CJ, Shimura Y (1991) Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 3 677–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe Y, Miyata S, Urao T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2002) Overexpression of Arabidopsis response regulators, ARR4/ATRR1/IBC7 and ARR8/ATRR3, alters cytokinin responses differentially in the shoot and in callus formation. Biochem Biophys Res Commun 293 806–815 [DOI] [PubMed] [Google Scholar]
- Ostin A, Kowalyczk M, Bhalerao RP, Sandberg G (1998) Metabolism of indole-3-acetic acid in Arabidopsis. Plant Physiol 118 285–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciorek T, Zazimalova E, Ruthardt N, Petrasek J, Stierhof YD, Kleine-Vehn J, Morris DA, Emans N, Jurgens G, Geldner N, et al (2005) Auxin inhibits endocytosis and promotes its own efflux from cells. Nature 435 1251–1256 [DOI] [PubMed] [Google Scholar]
- Petrasek J, Mravec J, Bouchard R, Blakeslee JJ, Abas M, Seifertova D, Wisniewska J, Tadele Z, Kubes M, Covanova M, et al (2006) PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312 914–918 [DOI] [PubMed] [Google Scholar]
- Reinhardt D, Mandel T, Kuhlemeier C (2000) Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell 12 507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D, Pesce ER, Stieger P, Mandel T, Baltensperger K, Bennett M, Traas J, Friml J, Kuhlemeier C (2003) Regulation of phyllotaxis by polar auxin transport. Nature 426 255–260 [DOI] [PubMed] [Google Scholar]
- Rupp HM, Frank M, Werner T, Strnad M, Schmulling T (1999) Increased steady state mRNA levels of the STM and KNAT1 homeobox genes in cytokinin overproducing Arabidopsis thaliana indicate a role for cytokinins in the shoot apical meristem. Plant J 18 557–563 [DOI] [PubMed] [Google Scholar]
- Scanlon MJ (2003) The polar auxin transport inhibitor N-1-naphthylphthalamic acid disrupts leaf initiation, KNOX protein regulation, and formation of leaf margins in maize. Plant Physiol 133 597–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KFX, Jurgens G, Laux T (2000) The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100 635–644 [DOI] [PubMed] [Google Scholar]
- Schroeder WJ, Martin KM, Lorensen WE (2003) The Visualization Toolkit: An Object-Oriented Approach to 3D Graphics, Ed 3. Kitware, New York
- Skoog F, Miller CO (1957) Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp Soc Exp Biol 11 118–130 [PubMed] [Google Scholar]
- Smith LG, Hake S (1992) The initiation and determination of leaves. Plant Cell 4 1017–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RS, Guyomarc'h S, Mandel T, Reinhardt D, Kuhlemeier C, Prusinkiewicz P (2006) A plausible model of phyllotaxis. Proc Natl Acad Sci USA 103 1301–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A, Drummond-Barbosa D, Kai T (2001) Stem cells find their niche. Nature 414 98–104 [DOI] [PubMed] [Google Scholar]
- Steedman HF (1957) Polyester wax: a new ribboning embedding medium for histology. Nature 179 1345. [DOI] [PubMed] [Google Scholar]
- Tian GW, Mohanty A, Chary SN, Li SJ, Paap B, Drakakaki G, Kopec CD, Li JX, Ehrhardt D, Jackson D, et al (2004) High-throughput fluorescent tagging of full-length Arabidopsis gene products in planta. Plant Physiol 135 25–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Jurgens G (2002) Stem cells that make stems. Nature 415 751–754 [DOI] [PubMed] [Google Scholar]
- Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmulling T (2003) Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15 2532–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Motyka V, Strnad M, Schmulling T (2001) Regulation of plant growth by cytokinin. Proc Natl Acad Sci USA 98 10487–10492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewska J, Xu J, Seifertova D, Brewer PB, Ruzicka K, Blilou I, Rouquie D, Benkova E, Scheres B, Friml J (2006) Polar PIN localization directs auxin flow in plants. Science 312 883. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.