Abstract
Barley (Hordeum vulgare) grains synthesize starch as the main storage compound. However, some starch is degraded already during caryopsis development. We studied temporal and spatial expression patterns of genes coding for enzymes of starch synthesis and degradation. These profiles coupled with measurements of selected enzyme activities and metabolites have allowed us to propose a role for starch degradation in maternal and filial tissues of developing grains. Early maternal pericarp functions as a major short-term starch storage tissue, possibly ensuring sink strength of the young caryopsis. Gene expression patterns and enzyme activities suggest two different pathways for starch degradation in maternal tissues. One pathway possibly occurs via α-amylases 1 and 4 and β-amylase 1 in pericarp, nucellus, and nucellar projection, tissues that undergo programmed cell death. Another pathway is deducted for living pericarp and chlorenchyma cells, where transient starch breakdown correlates with expression of chloroplast-localized β-amylases 5, 6, and 7, glucan, water dikinase 1, phosphoglucan, water dikinase, isoamylase 3, and disproportionating enzyme. The suite of genes involved in starch synthesis in filial starchy endosperm is much more complex than in pericarp and involves several endosperm-specific genes. Transient starch turnover occurs in transfer cells, ensuring the maintenance of sink strength in filial tissues and the reallocation of sugars into more proximal regions of the starchy endosperm. Starch is temporally accumulated also in aleurone cells, where it is degraded during the seed filling period, to be replaced by storage proteins and lipids.
Starch accumulated in the cereal endosperm serves as a primary source of carbohydrate for the human and animal diet and also has numerous industrial applications. Much attention has been paid over the last decades to better understanding the process of starch synthesis in cereal grains (for review, see Smith, 1999; Emes et al., 2003; James et al., 2003; Tomlinson and Denyer, 2003; Morell and Myers, 2005). At least four classes of enzyme activities have been identified as being necessary for starch granule synthesis. Multiple isoforms of each enzyme are found in graminaceous plants, and specific roles have been proposed for each isoform on the basis of either mutant or transgenic phenotypes (Morell and Myers, 2005). The first key enzyme is ADP-glucose pyrophosphorylase (AGPase), as shown in the barley (Hordeum vulgare) Risø16 mutant, in which its activity is much compromised (Johnson et al., 2003; Rösti et al., 2006). AGPase is a heterotetramer consisting of two large (AGP-L) and two small (AGP-S) subunits, each being encoded by distinct genes (Emes et al., 2003). Two distinct AGP-S subunits in both barley and wheat (Triticum aestivum) arise through alternative splicing of the same gene (Thorbjørnsen et al., 1996; Burton et al., 2002b). One subunit is localized in the plastids and is the major subunit in leaves (Rösti et al., 2006), while the other is cytosolic and contributes to starch synthesis exclusively in the endosperm (Johnson et al., 2003). A third AGP-S isoform has been described as plastidial and is localized in endosperm (Johnson et al., 2003). ADP-Glc, produced by AGPase, is utilized in plastids as a Glc donor for starch synthesis by starch synthases. Granule-bound starch synthase (GBSS) is mainly involved in amylose synthesis (Patron et al., 2002). The synthesis pathway of the rather complex amylopectin molecule includes soluble starch synthase (SS), starch-branching enzyme (SBE), and starch-debranching enzyme (DBE). Each individual SS isoform is believed to have a unique function in amylopectin synthesis (James et al., 2003; Li et al., 2003; Morell et al., 2003; Morell and Myers, 2005). Distinct SBE isoforms are recruited for starch synthesis during particular stages of seed filling in barley (Mutisya et al., 2003). Plant DBEs are categorized into the isoamylase (ISA) and the pullanase (or β-limit dextrinase; PUL) types. The two enzymes differ with respect to their substrate specificity (James et al., 2003). Starch granule initiation and growth are altered in a barley mutant lacking ISA activity (Burton et al., 2002a). The PUL gene is expressed in both the developing barley endosperm and the germinating caryopsis, indicating its potential function for starch modification during synthesis and for starch hydrolysis during germination (Burton et al., 1999; Dinges et al., 2003).
Different pathways exist for starch breakdown in distinct plant tissues (Zeeman et al., 2007). In nonliving tissues of germinating cereal seeds, degradation of storage starch proceeds by the combined actions of α- and β-amylases (AMY and BAM), limit dextrinase (PUL), β-glucosidase, and possibly α-glucan phosphorylase (PHO), although direct evidence for the importance of some of these enzymes is still absent (Beck and Ziegler, 1989; Smith et al., 2005). AMY initiates the breakdown of starch in the germinating grain, and repression of its expression in rice (Oryza sativa) has been shown to delay caryopsis germination (Asatsuma et al., 2005). Breakdown of transient starch in living tissues (e.g. leaves of Arabidopsis [Arabidopsis thaliana]) was studied in more detail (for review, see Lloyd et al., 2005; Smith et al., 2005; Zeeman et al., 2007) and involves another set of enzymes. A prerequisite for transient starch mobilization is the phosphorylation of a portion of the glucosyl residues by glucan, water dikinase (GWD; Ritte et al., 2002) and phosphoglucan, water dikinase (PWD; Kötting et al., 2005). AMY, PUL, PHO, and α-glucosidase are established to play minor roles (if any) in transient starch breakdown (Lloyd et al., 2005; Yu et al., 2005), while BAM is a major enzyme (Fulton et al., 2008). Four BAM isoforms of Arabidopsis are plastid localized, and three of them are involved in degradation of transient starch to maltose (Fulton et al., 2008). ISA3 (Wattebled et al., 2005; Delatte et al., 2006) is also capable of degrading transient starch to maltooligosaccharides, which are further converted either to Glc by plastidial disproportionating enzyme 1 (DPE1; Critchley et al., 2001) or by BAM to maltose. Produced maltose is metabolized to Glc in cytosol by DPE2 (Lu and Sharkey, 2004).
The major starch storage organ of the cereal seed is the filial starchy endosperm, while the aleurone accumulates only transitory starch during grain development. In mature aleurone cells, only lipid and protein bodies but no starch granules are present (Duffus and Cochrane, 1993). Some starch is also synthesized in both embryo and pericarp. The maternal pericarp functions as a transitory storage organ during early grain development (Weschke et al., 2000; Sreenivasulu et al., 2004). By the time the caryopsis has reached its maximum length and the endosperm is rapidly expanding, the pericarp starch has been largely remobilized (Duffus and Cochrane, 1993; Sreenivasulu et al., 2004). It is unknown whether the identical or specific sets of genes, coding for starch-synthesizing enzymes, are activated in pericarp and endosperm of the developing grain. The developing barley caryopsis also contains AMY, BAM, PHO, and α-glucosidase enzyme activities (Duffus, 1987) associated with starch degradation. The α-amylolytic activity in the developing barley caryopsis was detected by Duffus (1969) with a later reappraisal (Allison et al., 1974; MacGregor and Dushnicky, 1989a, 1989b), and it was shown to be mostly associated with the pericarp (Allison et al., 1974). The disappearance of starch granules in this tissue strongly coincides with AMY enzyme activity (MacGregor and Dushnicky, 1989a). AMY enzyme activity was also detected in the embryo-surrounding region of the endosperm (MacGregor and Dushnicky, 1989b). BAM enzyme activity was measured in the barley endosperm at later developmental stages (Beck and Ziegler, 1989). However, data on genes coding for the corresponding starch degradation enzymes in the developing grain are missing. Furthermore, the physiological role of transient starch accumulation in certain grain tissues remains unexplored. Measurements of enzyme activities, transcript levels, and metabolite contents have been routinely either averaged over the whole caryopsis or restricted to the starchy endosperm.
In this investigation, we have comprehensively analyzed the expression patterns of gene families involved in both starch synthesis and degradation in distinct tissues of the developing barley caryopsis via macroarray and/or northern-blot hybridizations. Localizations in situ of key gene transcripts and measurements of key metabolites and enzyme activities have allowed us to establish the genes involved in starch biosynthesis and degradation of the respective caryopsis tissues and to propose a role for starch remobilization in the developing barley grain.
RESULTS
Localization of Starch in the Developing Barley Grain
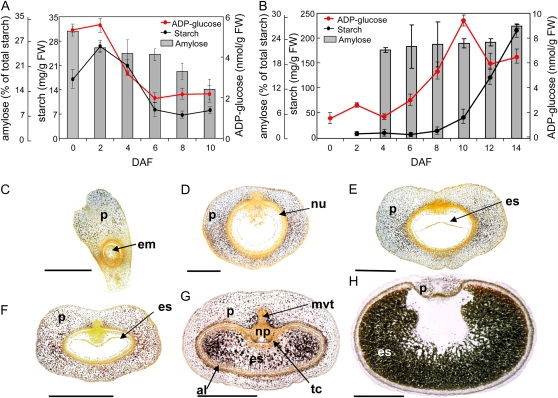
Starchy endosperm is the main storage tissue of the barley grain, accumulating starch during middle and late stages of caryopsis development. However, starch is also detected in the pericarp between anthesis and 4 d after flowering (DAF; Fig. 1, A and B). The pattern of starch accumulation is dynamic, both temporally and spatially. The pericarp cells of the gynoecium have been filled with starch granules already at anthesis (Fig. 1C). With the onset of seed development, starch begins to accumulate around the lateral vascular bundles (Fig. 1, D and E), eventually filling the whole pericarp (Fig. 1F). The pericarp cells flanking the main vascular bundle accumulate starch at the latest (Fig. 1G). A few days after anthesis, the pericarp cells begin to disintegrate (Sreenivasulu et al., 2006) and, simultaneously, their starch granules disappear, starting from the distal part of the pericarp and ending in its ventral region. By 14 DAF, starch is barely detectable in the pericarp (Fig. 1H). Starch accumulates in the starchy endosperm starting around 6 DAF and continuing throughout the main grain filling period (Fig. 1, B, G, and H). The aleurone cells and the transfer cell layer are largely free of starch (Fig. 1, G and H). Amylose makes up around 20% of starch at the beginning of storage starch accumulation, and this proportion is increased (Fig. 1B) to around 30% in the mature grain.
Figure 1.
Starch accumulation in the developing barley grains. A and B, Starch, ADP-Glc, and amylose contents in pericarp (A) and endosperm (B). FW, Fresh weight. C to H, Spatial distribution of starch granules (visualized in black) in the developing grains in a longitudinal section (C) and in median transverse sections of developing grains at anthesis (C), 2 DAF (D), 4 DAF (E), 5 DAF (F), 8 DAF (G), and 14 DAF (H). al, Aleurone; em, embryo sac; es, endosperm; mvt, main vascular tissue; np, nucellar projection; nu, nucellus; p, pericarp; tc, transfer cells. Bars = 500 μm in C to E and 1 mm in F to H.
Since ADP-Glc is the key metabolite for starch synthesis, its level was measured in both the pericarp and endosperm fractions of the developing caryopsis (Fig. 1, A and B). In the pericarp, the highest level of ADP-Glc is present between anthesis and 2 DAF and falls thereafter, consistent with the accumulation of starch in this tissue (Fig. 1A). Highest levels of ADP-Glc in the endosperm were observed from 4 DAF onward, peaking around 10 DAF (Fig. 1B).
Enzymes for Starch Biosynthesis and Degradation Are Encoded by Small Gene Families
cDNA sequences of the relevant barley genes involved in the synthesis and degradation of starch were selected from EST databases (Zhang et al., 2004; http://compbio.dfci.harvard.edu) using already described sequences as references (Table I). The small subunit of AGPase in barley is encoded by Hv.AGP.S.1 and Hv.AGP.S.2 genes (Rösti et al., 2006), abbreviated here as AGP-S1 and AGP-S2. AGP-S1 encodes both cytosolic and plastidial isoforms (AGP-S1a and AGP-S1b) produced by alternative splicing (Thorbjørnsen et al., 1996; Rösti et al., 2006). The AGP-S1b and AGP-S2 translation products are predicted to be plastid targeted, while the AGP-S1a subunit was shown to be cytosolic (Rösti et al., 2006). Only two AGP-L cDNAs, AGP-L1 and AGP-L2, were identified, despite evidence for four genes in the rice genome (Ohdan et al., 2005). Both gene products are predicted as plastid localized. With respect to GBSS, we identified only already known GBSS1a and GBSS1b sequences (Patron et al., 2002). A full-length cDNA for SSI (Gubler et al., 2000) and a partial cDNA for SSIIa (Li et al., 2003) were recovered. The EST database screen also identified SSIIb (sharing 71.6% identity with barley SSIIa), SSIIIa (91.8% nucleotide identity with wheat SSIII; Li et al., 2000) and SSIIIb (79.1% identity with wheat SSIII and 91.0% identity with rice SSIII; Dian et al., 2005), and a partial SSIV sequence sharing 97.4% identity with wheat SSIV (GenBank accession no. AAK97773). Because SSs are large and rather poorly expressed proteins, only partial sequences of the corresponding cDNAs were recovered (Table I). Three SBE cDNAs were identified, each almost identical with the cloned SBEI (Mutisya et al., 2003) and SBEIIa and SBEIIb (Sun et al., 1998) transcripts. Three different ISA cDNA sequences, one full and two partial, were identified, while one partial sequence was homologous with PUL cDNA (Burton et al., 1999, 2002a). We did not find another PUL gene (Kristensen et al., 1999) among the seed-specific ESTs, possibly because of its preferential expression during seed germination.
Table I.
List of genes involved in starch biosynthesis
cTP, Chloroplast transit peptide; n.d., signal peptide was not possible to determine because of the partial length of the corresponding nucleotide sequence available.
| Enzyme | Gene | Reference EST (Full Length or Partial) | Length of the Signal Peptide, Amino Acids (Probability) | Highest BLAST Hit (Percentage Identity) | Reference |
|---|---|---|---|---|---|
| ADP-Glc pyrophosphorylase (EC 2.7.7.27) | |||||
| Small subunit | AGP-S1a | HB16K13 (full) | No | CAA88449 (100%) | Thorbjørnsen et al. (1996) |
| AGP-S1b | HI02B12 (full) | 64 cTP (0.944) | CAA88450 (99.4%) | Thorbjørnsen et al. (1996) | |
| AGP-S2 | HW03I05 (partial) | 32 cTP (0.866) | AAO16183 (99.2%) | Johnson et al. (2003) | |
| Large subunit | AGP-L1 | HY04M18 (full) | 43 cTP (0.854) | CAA47626 (100%) | Villand et al. (1992) |
| AGP-L2 | HX03I15 (full) | 91 cTP (0.307) | AAC49729 (98.8%) | Eimert et al. (1997) | |
| Starch synthase (EC 2.4.1.21) | SSI | HV04E12 (partial) | 31 cTP (0.864) | AAF37876 (100%) | Gubler et al. (2000) |
| SSIIa | HB02N20 (partial) | n.d. | AAN28307 (100%) | Li et al. (2003) | |
| SSIIb | HT03C10 (partial) | n.d. | AAN28307 (71.6%) | Morell et al. (2003) | |
| SSIIIa | HB14E10 (partial) | n.d. | AAF87999 (91.8%) | Li et al. (2000) | |
| SSIIIb | HB14B08 (partial) | n.d. | AAL40942 (91.0%) | Dian et al. (2005) | |
| SSIV | HF05C15 (partial) | n.d. | AAK97773 (97.4%) | ||
| Granule-bound starch synthase (EC 2.4.1.21) | GBSS1a | HY10D16 (full) | 69 cTP (0.841) | AAM74051 (99.8%) | Patron et al. (2002) |
| GBSS1b | HI06O16 (partial) | n.d. | AAM74054 (98.1%) | Patron et al. (2002) | |
| Starch-branching enzyme (EC 2.4.1.18) | SBE1 | HB02A08 (partial) | n.d. | AAP72268 (96.8%) | Mutisya et al. (2003) |
| SBE2a | HZ64I18 (full) | 7 cTP (0.599) | AAC69753 (99.7%) | Sun et al. (1998) | |
| SBE2b | HB03H24 (partial) | n.d. | AAC69754 (100%) | Sun et al. (1998) | |
| Starch-debranching enzyme | |||||
| Isoamylase (EC 3.2.1.68) | ISA1 | HB05N18 (partial) | n.d. | AAM46866 (99.2%) | Burton et al. (2002a) |
| ISA2 | HI05I24 (full) | 49 cTP (0.888) | BAD08581 (100%) | ||
| ISA3 | HM13A11 (partial) | n.d. | BAD89532 (100%) | ||
| Limit dextrinase (EC 3.2.1.41) | PUL | HB26F24 (partial) | n.d. | AAD34348 (100%) | Burton et al. (1999) |
A set of genes encoding enzymes of starch degradation has also been identified (Table II). Four full-length AMY cDNAs were found, including the newly described AMY4. The latter shares only limited homology with the three known barley AMY sequences but is highly homologous to a putative rice AMY (GenBank accession no. Os04g0403300) and to a large number of dicot sequences, including those from Plantago major (71.2% identity; Pommerrenig et al., 2006), Malus domestica (70.0% identity; Wegrzyn et al., 2000), and Arabidopsis (68.0% identity; GenBank accession no. At1g76130). AMY1 to AMY3 are predicted to be secretory proteins (Table II). Two already known and five new cDNAs encoding β-amylases were identified. BAM1 and BAM2 are full-length sequences and predicted to belong to the secretory pathway. The localization of the others has not been possible to estimate because only partial sequences are available. However, barley BAM3, BAM4, BAM5, and BAM6 predicted proteins share the highest similarities to Arabidopsis AtBAM1, AtBAM2, and AtBAM3 (Table II; Supplemental Fig. S1). All of these Arabidopsis BAMs were proven to be active β-amylases targeted to the chloroplasts (Fulton et al., 2008). Therefore, we assume that barley BAM3, BAM4, BAM5, and BAM6 are also plastid localized. Two putative GWD cDNA sequences and one PWD cDNA sequence were also identified, as well as one putatively plastidial isoform (DPE1) and one cytosolic isoform (DPE2) of the disproportionating enzyme. All of them are only partial sequences, but they share high levels of similarity to the corresponding proteins of Arabidopsis (Table II).
Table II.
List of starch-degrading genes of barley
n.d., Signal peptide was not possible to determine because of the partial length of the corresponding nucleotide sequence; SP, signal peptide.
| Enzyme | Gene | Reference EST (Full Length or Partial) | Length of the Signal Peptide (Probability) | Highest BLAST Hit (Percentage Identity) | Reference |
|---|---|---|---|---|---|
| α-Amylase (EC 3.2.1.1) | AMY1 | HZ37L10 (full) | 24 SP (0.995) | P00693 (99.8%) | Rogers and Milliman (1983) |
| AMY2 | HV02D09 (full) | 24 SP (0.994) | AAA98615 (98.8%) | Khursheed and Rogers (1988) | |
| AMY3 | HV14L21 (full) | 24 SP (0.994) | P04063 (99.8%) | Rahmatullah et al. (1989) | |
| AMY4 | HO12I21 (full) | No | Os04g33040 (87.6%) | ||
| β-Amylase (EC 3.2.1.2) | BAM1 | HY09N12 (full) | 22 SP (0.548) | AAO67355 (100%) | Clark et al. (2003) |
| BAM2 | HZ50O15 (full) | 22 SP (0.742) | AAC64904 (96.0%) | Jung et al. (2001) | |
| BAM3 | HS08F24 (partial) | n.d. | At3g23920 (68.3%) | Fulton et al. (2008) | |
| BAM4 | HI09E01 (partial) | n.d. | At3g23920 (63.9%) | Fulton et al. (2008) | |
| BAM5 | HI06F14 (partial) | n.d. | At4g17090 (66.3%) | Fulton et al. (2008) | |
| BAM6 | HDP09L01 (partial) | n.d. | At4g00490 (75.2%) | Fulton et al. (2008) | |
| BAM7 | AV833737 (partial) | n.d. | At5g45300 (66.2%) | Fulton et al. (2008) | |
| α-Glucan phosphorylase (EC 2.4.1.1) | |||||
| Plastidic | PHO1 | HB21H16 (partial) | n.d. | AAQ73180 (98.1%) | Schupp and Ziegler (2004) |
| Cytosolic | PHO2 | HO06A20 (partial) | n.d. | AAF82787 (96.7%) | Schupp and Ziegler (2004) |
| Glucan, water dikinase (EC 2.7.9.4) | GWD1 | HO15I19 (partial) | n.d. | At1g10760 (75.0%) | Baunsgaard et al. (2005) |
| GWD2 | HDP27A11 (partial) | n.d. | ACG69788 (82.5%) | Nashilevitz et al. (2009) | |
| Phosphoglucan, water dikinase (EC 2.7.9.5) | PWD | BI951013 (partial) | n.d. | At5g26570 (66.3%) | Kötting et al. (2005) |
| Disproportionating enzyme (EC 2.4.1.25) | DPE1 | HB03F13 (partial) | n.d. | AAZ23612 (88.0%) | Bresolin et al. (2006) |
| DPE2 | HV04E07 (partial) | n.d. | At2g40840 (72.9%) | Lu and Sharkey (2004) |
Distinct Genes Are Involved in Starch Biosynthesis in Pericarp and Endosperm
Patterns of gene expression were determined by transcript profiling of developing barley caryopses between anthesis and desiccation (24 DAF) using a cDNA macroarray filter carrying about 12,000 seed-expressed sequences (Sreenivasulu et al., 2006). The caryopses were separated into pericarp and endosperm fractions. To analyze the expression of genes not present on the filter, northern-blot hybridizations were performed using gene-specific sequences.
Transcripts of AGP-L1, AGP-L2, AGP-S1b, and AGP-S2 were detected in both pericarp and endosperm tissues, while cytosol-specific AGP-S1a transcripts were evident only in the endosperm between 6 and 18 DAF (Fig. 2A). In the pericarp, peak expression of AGP-S1b and AGP-L2 is attained around 2 DAF. In the endosperm, all AGPase transcripts except AGP-S2 are strongly up-regulated during seed filling. AGP-S2 mRNA is present in both pericarp and endosperm at low but almost constant levels (Fig. 2A). During the late stages of endosperm development (after 20 DAF), AGP-L1 and AGP-S2 transcripts remain detectable, suggesting that starch synthesis continues at this time. In situ localization revealed that the AGP-S1a mRNAs are restricted to the starchy endosperm at 14 DAF (Fig. 3, A and B). In contrast, AGP-S1b transcripts are present around the pericarp lateral vascular bundles at 4 DAF (Fig. 3C), coinciding spatially with starch accumulation (Fig. 1E). At 6 DAF, a strong AGP-S1b signal is associated with the transfer cells of the endosperm as well as with the pericarp (Fig. 3D). Similar to the AGP-S1a mRNA, AGP-S2 transcripts are also present in the starchy endosperm (Fig. 3E). While AGP-S1a mRNA is abundant in the center of the endosperm, AGP-S2 expression is concentrated more peripherally (Fig. 3, B and E).
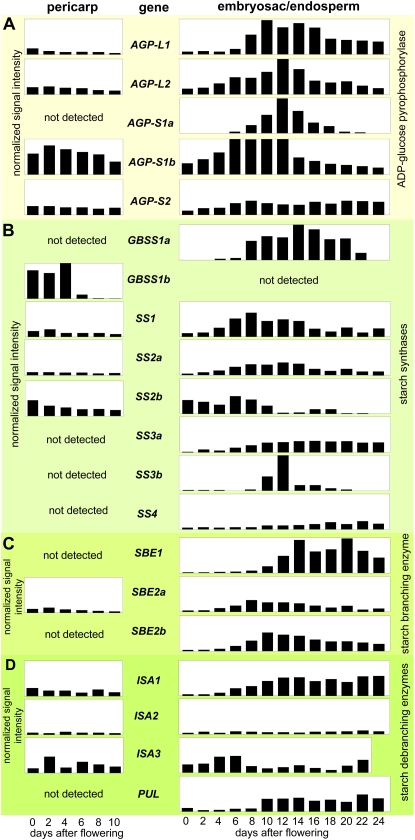
Figure 2.
Transcript profiles of genes involved in starch synthesis in pericarp (left) and endosperm (right) fractions of developing barley grains. Transcript levels were determined by cDNA macroarray or northern-blot analyses for AGPase (A), starch synthases (B), starch-branching enzymes (C), and starch-debranching enzymes (D). The relative values were calculated as means of at least two independent experiments and are shown in normalized signal intensities.
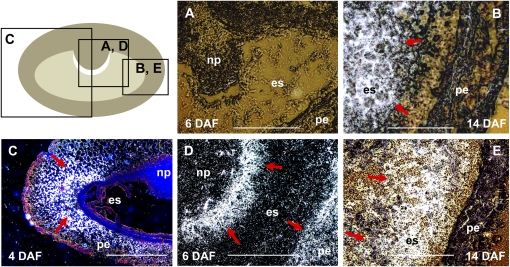
Figure 3.
In situ localization of transcripts of small subunits of AGPase in developing barley caryopses. The top left panel shows a median transverse section of a developing caryopsis with the regions shown in A to E. Hybridization sites in A to E are visualized as white signals (indicated by red arrows). A, AGP-S1a transcripts were not detected in the developing caryopsis at 4 DAF. B, AGP-S1a mRNA is localized exclusively in endosperm at 14 DAF. C and D, Expression patterns of the AGP-S1b gene at 4 DAF (C) and 6 DAF (D). E, Endosperm-specific localization of AGP-S2 mRNA at 14 DAF. es, Endosperm; np, nucellar projection; pe, pericarp. Bars = 500 μm.
The two GBSS genes are differentially expressed (Fig. 2B). GBSS1b transcripts were highly abundant between anthesis and 4 DAF in the pericarp, diminishing thereafter, while they were not detected in endosperm. In contrast, GBSS1a gene expression was restricted to the endosperm, mainly between 8 and 22 DAF. The SS transcript profiles were highly variable, but their expression levels were low (Fig. 2B). Only SS1, SS2a, and SS2b mRNAs were detected in the pericarp, with a moderate accumulation peak between anthesis and 4 DAF. The SS2b transcript levels were high in the early endosperm fraction (anthesis to 8 DAF) and diminished gradually after the beginning of seed filling. SS1 and SS2a shared almost identical temporal expression patterns, showing a rapid increase with the beginning of starch accumulation, remaining high during seed filling, and finally decreasing as the endosperm matured. SS3a and SS4 transcript accumulation slowly increased from 2 DAF until around 10 DAF and remained thereafter at almost constant levels until maturation. SS3b was transiently but very strongly up-regulated between 10 and 16 DAF, with highest expression at 12 DAF (Fig. 2B).
Only SBE2a but not SBE1 or SBE2b was expressed in the pericarp, and although the level was low, a peak was detectable at 2 DAF (Fig. 2C). In the endosperm, a moderate level of SBE2a expression peaked at 8 DAF and diminished gradually until late development. Accumulation of SBE2b transcripts began from 4 DAF at a higher level, peaking at 10 DAF, and then diminishing gradually until 24 DAF. SBE1 expression was found from 10 DAF onward, increasing to a high level at 14 DAF, and stayed almost constant at this level until desiccation (Fig. 2C).
Of the three ISA genes, ISA1 and ISA3 were expressed in the pericarp at low levels with similar temporal profiles (Fig. 2D). However, their expression patterns were different in the endosperm. While ISA3 transcripts were more abundant during early endosperm development, with an expression maximum around 4 to 6 DAF, ISA1 transcripts began to accumulate from 6 DAF and remained abundant until desiccation (Fig. 2D). ISA2 was barely expressed both in pericarp and endosperm during caryopsis development. The PUL gene was not expressed in the pericarp but up-regulated in the endosperm between 10 and 24 DAF (Fig. 2D).
Genes Coding for Starch Breakdown Enzymes Are Also Expressed during Seed Development
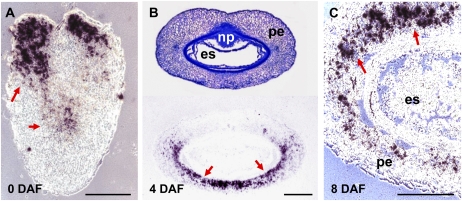
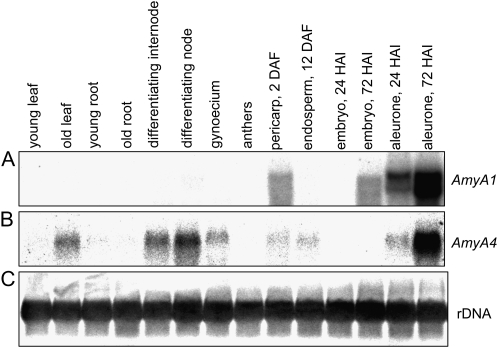
Of the four AMY genes, only AMY1 and AMY4 were expressed in the developing caryopsis (Fig. 4A). AMY1 was specifically and highly up-regulated in the pericarp, with a pronounced peak of expression around 2 DAF, but its expression was not detected in the endosperm. AMY4 transcripts were highly abundant in the pericarp between anthesis and 10 DAF as well as in the early endosperm fraction, where its expression was gradually down-regulated from 6 DAF onward, although remaining detectable until maturity. AMY1 and AMY4 mRNA was localized by in situ hybridization and the pattern was compared with starch distribution (Figs. 5 and 6). At anthesis, AMY1 transcripts were strongly accumulated in the style region and central parts of the pericarp (Fig. 5A). By 4 DAF, the transcripts were detectable within the dorsal part of the pericarp (Fig. 5B), and by 8 DAF, they were detectable in its ventral region (Fig. 5C). No accumulation of AMY1 transcripts was detected in endosperm (Fig. 5). AMY4 transcripts were localized at anthesis to the nucellar tissue of the gynoecium and to a region in pericarp presumably representing the developing vascular bundle (Fig. 6C). These tissues contain small starch granules (arrows in Fig. 6B). During development, the nucellus disintegrates to be replaced by growing endosperm (Radchuk et al., 2006). Differentiation of tracheary elements of vascular tissue is also involved in programmed cell death (Gunawardena, 2008). Later during grain development, AMY4 transcripts were detectable only in the nucellar projection (Fig. 6F), the tissue where continuous cell death also occurs (Radchuk et al., 2006; Thiel et al., 2008). Hence, spatial distribution of the AMY1 and AMY4 transcripts was limited to maternal tissues of the developing barley grain.
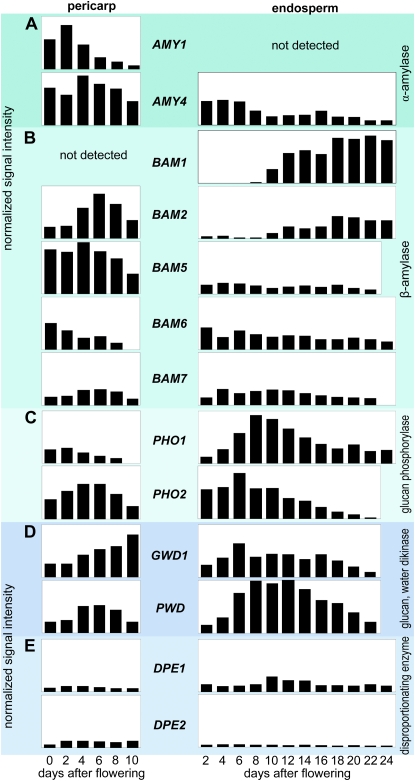
Figure 4.
Transcript profiles of genes involved in starch degradation in pericarp (left) and endosperm (right) fractions of developing barley grains. Transcript levels were determined by macroarray or northern-blot analyses for α-amylases (A), β-amylases (B), α-glucan phosphorylases (C), glucan, water dikinases (D), and disproportionating enzyme (E). The bars represent relative values calculated from normalized signal intensities derived from at least two independent experiments.
Figure 5.
In situ localization of α-amylase AMY1 transcripts (visualized in black and indicated by red arrows) in the pericarp of developing barley grains at anthesis (A), 4 DAF (B), and 8 DAF (C). es, Endosperm; np, nucellar projection; pe, pericarp. Bars = 250 μm.
Figure 6.
Distribution of starch granules and in situ localization of α-amylase AMY4 transcripts in developing barley grains. A and D, Transverse sections of barley grains at 2 DAF (A) and 14 DAF (D), toluidine blue staining. B and E, Distribution of starch granules in nucellus (B) and nucellar projection (E; visualized in black and shown by red arrows). C, Localization of AMY4 transcripts in outer cell rows of the nucellus and developing vascular tissue at 2 DAF (indicated by red arrows), bright-field visualization. F, Localization of AMY4 transcripts in the nucellar projection at 14 DAF, dark field visualization. es, Endosperm; np, nucellar projection; nu, nucellus; pe, pericarp; tc, transfer cells. Bars = 120 μm in A, C, D, and F and 250 μm in B and E.
The expression of AMY1 and AMY4 genes in other barley tissues was studied by northern-blot analyses using 3′ untranslated region (UTR) fragments of the corresponding cDNAs as probes. Besides being present in the developing pericarp, AMY1 transcripts were detected also in embryo and aleurone of germinating grains (Fig. 7A). The AMY4 gene was expressed in senescing leaves, young differentiating internodes, and nodes and in aleurone during seed germination (Fig. 7B).
Figure 7.
Expression patterns of AMY1 and AMY4 α-amylase genes in different barley tissues as determined by northern-blot hybridization. Total RNA (10 μg per line) isolated from different barley tissues was subsequently hybridized with 3′ UTRs of AMY1 (A) and AMY4 (B) cDNA. Hybridization with a 25S rDNA probe (C) was performed as a loading control. HAI, Hours after imbibition.
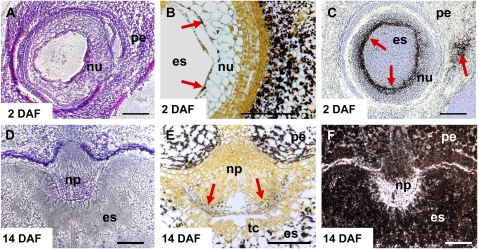
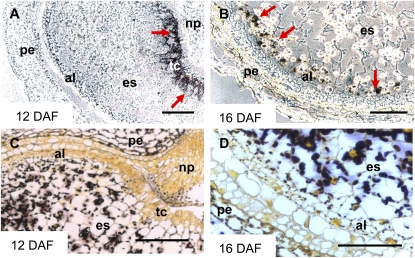
Five of seven BAM genes were expressed during caryopsis development (Fig. 4B). The other genes, BAM3 and BAM4, could be germination specific, as concluded from an EST in silico analysis (data not shown). Transcripts of both β-amylases with predicted signal peptides (BAM1 and BAM2) showed similar patterns of accumulation in endosperm during middle and late development. However, their expression in pericarp was different. While BAM1 transcripts were not detected, the BAM2 gene was highly expressed in pericarp, peaking at 6 DAF (Fig. 4B). In situ localization of the BAM1 transcripts revealed their accumulation in the transfer cell layer (Fig. 8A) and in the aleurone/subaleurone (Fig. 8B) during the middle stages of endosperm development. These tissues do not accumulate storage starch (Duffus and Cochrane, 1993). However, some small starch granules were visible in aleurone at 16 DAF (Fig. 8D).
Figure 8.
In situ localization of β-amylase BAM1 transcripts (A and B) and distribution of starch granules (C and D) in developing caryopses at 12 DAF (A and C) and 16 DAF (B and D). The BAM1 mRNA was detected (indicated by red arrows) in transfer cell layer (A) and in aleurone/subaleurone (A and B). These tissues contain only small or no starch granules (C and D). al, Aleurone; es, endosperm; np, nucellar projection; pe, pericarp; tc, transfer cells. Bars = 250 μm.
Transcripts of three β-amylases (BAM5, BAM6, and BAM7), which had been predicted as being plastid localized (Supplemental Fig. S1), were accumulated in both pericarp and endosperm during seed development (Fig. 4B). Their mRNAs were detected throughout endosperm development, but at rather low levels. In pericarp, BAM5 was highly expressed between anthesis and 6 DAF, slowly decreasing afterward. BAM6 mRNA was abundant at anthesis but gradually decreased during pericarp development to undetectable levels at 10 DAF. The rather moderate expression of BAM7 in pericarp peaked at 6 DAF (Fig. 4B).
The plastidial PHO1 was transcriptionally up-regulated in endosperm at the beginning of the filling phase and peaked around 8 DAF (Fig. 4C). Highest PHO1 mRNA accumulation in pericarp was found at 2 DAF. The expression pattern of PHO1 was similar to the expression of the AGP-L1, AGP-L2, and AGP-S1b subunits of AGPase in both pericarp and endosperm (Fig. 2A). The expression maximum of cytosolic PHO2 in pericarp was detected at 4 to 6 DAF. In the endosperm fraction, PHO2 mRNA was more abundant during early development, peaking at 4 DAF and rapidly decreasing afterward (Fig. 4C).
GWD1 and PWD genes were active in pericarp as well as in endosperm (Fig. 4D), while GWD2 was not expressed. GWD1 expression increased with the aging of the pericarp. Its expression in the endosperm fraction was moderate but almost constant during early development and the seed filling stages, and it decreased during seed maturation (after 18 DAF). Both DPE1 and DPE2 genes of disproportionating enzyme were active in pericarp and endosperm fractions, although at rather low and almost constant levels (Fig. 4D).
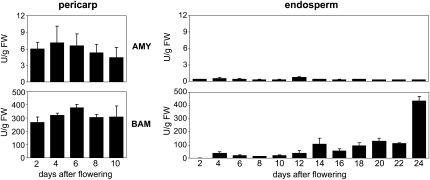
Patterns of AMY and BAM Enzyme Activities Largely Coincide with Transcript Profiles of the Corresponding Genes
Relatively weak α-amylase enzyme activity was measured in pericarp with a maximum around 4 DAF (Fig. 9), or 2 d later as a maximum of AMY transcripts in this tissue (Fig. 4). No or very little AMY enzyme activity was detected in the developing endosperm (Fig. 9). Activity of β-amylase enzyme was very strong at all studied stages in pericarp, rather moderate in endosperm during early and middle stages of development, but strongly increased at very late stages (Fig. 9).
Figure 9.
Activities (in units per gram fresh weight [FW]) of α-amylase (AMY) and β-amylase (BAM) enzymes in pericarp (left) and endosperm (right) fractions of developing barley grains.
DISCUSSION
We have analyzed in detail the expression patterns of the genes involved in starch accumulation and breakdown in the pericarp and endosperm fractions of the developing barley caryopsis between anthesis and maturation. Transcript profiling in conjunction with mRNA in situ localization and enzyme activity measurements has revealed different roles for starch accumulation and mobilization in maternal and filial parts of the developing seed.
Pericarp Is the Main Starch Storage Tissue during Early Caryopsis Development
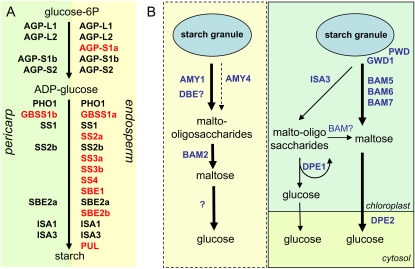
Pericarp is the first site of short-term starch storage, with an accumulation maximum already at 2 DAF (Fig. 1). The profile of starch accumulation coincides with the expression patterns of genes coding for starch biosynthetic enzymes (Fig. 2), involving, however, a smaller number of active genes compared with endosperm (Fig. 10A). AGP-S1b transcripts are highly abundant in the pericarp (Fig. 2A) and spatially colocalize with starch accumulation (Figs. 1D and 3C), implying a role of AGP-S1b in the accumulation of pericarp starch. This subunit is also a major subunit in leaves (Rösti et al., 2006). GBSS1b gene expression is found in both pericarp (Fig. 2B) and embryo (Sreenivasulu et al., 2006) but not in endosperm (Fig. 2B; noted also by Patron et al., 2002). Similarly, the ortholog GBSSII in wheat is expressed in pericarp and embryo as well as in vegetative organs (Vrinten and Nakamura, 2000). Based on the expression patterns in wheat, we assume that GBSS1b is also expressed in barley leaves, contributing there to amylose biosynthesis, while GBSS1a is a major enzyme involved in amylose synthesis in endosperm. Of the six barley SS genes involved in amylopectin synthesis, only SS2b and SS1 are expressed in the pericarp (Fig. 2B), indicating that pericarp starch may differ in structure from endosperm storage starch. Different phenotypes of mutants affected in SS expression suggest that each isoform tends to contribute to the extension of a specific subset of the available nonreducing ends within amylopectin (Gao et al., 1998; Li et al., 2000). Distinct physicochemical properties of endosperm and pericarp starch have also been noted in maize (Zea mays; Li et al., 2007). Out of three SBE genes, only SBE2a transcripts were detected in pericarp. In maize, SBE2a plays a specific role in leaves, which is necessary for normal plant development (Blauth et al., 2001), while the SBE2b gene is not expressed (Gao et al., 1996). Based on the expression of leaf-specific AGP-S1b, GBSS1b, and SBE2a genes in pericarp, we speculate that starch biosynthesis in pericarp is performed by the same isoforms as in leaves. The coordinated expression of specific isoforms may be important for proper functioning of starch biosynthesis, since starch biosynthetic enzymes in maize and wheat have recently been shown to be associated in multisubunit complexes (Hennen-Bierwagen et al., 2008, 2009; Tetlow et al., 2008).
Figure 10.
Schemes illustrating the involvement of differing suites of isoforms of starch biosynthetic enzymes in pericarp and endosperm (A), and proposed starch degradation pathways in the pericarp of developing barley grains (B). A, Distinct genes encoding enzymes of starch synthesis are expressed in maternal and filial tissues of developing barley grains. Genes in red are those that are exclusively expressed in the respective tissue. AGP-L, Large subunit of AGPase; AGP-S, small subunit of AGPase; GBSS, granule-bound starch synthase; ISA, isoamylase; PHO, glucan phosphorylase; PUL, pullanase (limit dextrinase); SBE, starch-branching enzyme; SS, starch synthase. B, The proposed pathways of starch breakdown in different cells of the pericarp. In the cells undergoing programmed cell death, starch degradation possibly occurs via AMY1 and AMY4 α-amylases. Linear maltooligosaccharides released by the action of α-amylases and an unidentified DBE may provide substrates for the BAM2 β-amylase. The question mark indicates an unidentified enzyme converting maltose in Glc. In living cells of the pericarp, GWD1 and PWD may phosphorylate the surface of the starch granule, making it accessible for β-amylase action. BAM5, BAM6, and BAM7 β-amylases may be involved in maltose production, acting either at the granule surface or on linear maltooligosaccharides. The action of ISA3 on the granule may release soluble maltooligosaccharides. Short oligosaccharides can be metabolized by DPE1, liberating Glc and larger maltooligosaccharides for continued degradation. After transport in cytosol, maltose may be further converted to Glc by DPE2.
The fixation of sugars as starch in the young pericarp determines Suc influx into the caryopsis and is likely to be responsible for the establishment of an assimilate sink at early developmental stages, when the endosperm is still not differentiated. Beginning with endosperm cellularization around 4 DAF, the role of the pericarp as a sink tissue diminishes, accumulated starch is progressively degraded, and the pericarp cells disintegrate through programmed cell death (Sreenivasulu et al., 2006).
Gene Expression Patterns Suggest Two Different Pathways for Starch Degradation in the Pericarp
In pericarp, growing and degenerating processes are interconnected. Growth prevails in early pericarp (0–4 DAF), while degeneration predominates from 6 DAF onward (Sreenivasulu et al., 2006). However, the chlorenchyma cell layer of the pericarp remains photosynthetically active until the end of seed filling (Rolletschek et al., 2004).
Starch decrease in pericarp coincides with the expression of genes encoding starch-degrading enzymes (Fig. 4). Two α-amylase genes, AMY1 and AMY4, are expressed in pericarp (Fig. 4A), and none encodes a plastid-localized protein (Table II). AMY1 contains a signal peptide and is one of the main α-amylase isoforms in germinating barley grains (Bak-Jensen et al., 2007), produced and secreted by aleurone and, possibly, scutellum (Beck and Ziegler, 1989). AMY1 expression in the developing pericarp coincides spatially with the regions where disintegration of pericarp takes place. The presence of a signal peptide suggests that AMY1 is secreted into regions where programmed cell death has already occurred to mobilize starch granules. Further breakdown of linear maltooligosaccharides may proceed by putatively secreted BAM2 that is also up-regulated in pericarp (Fig. 4).
The newly described AMY4 does not possess any predicted transit peptide (Table II). Northern blotting revealed expression of the gene in pericarp and endosperm fractions (Fig. 4). However, its mRNA was in situ localized to differentiating vascular tissue (isolated as part of the pericarp fraction), nucellus, and, after nucellus disintegration, to the remaining nucellar projection (both isolated as parts of the endosperm fraction). AMY4 transcripts were also detected in senescing leaves and in differentiating internodes and nodes besides being present in germinating seeds (Fig. 7). All of these tissues undergo programmed cell death (Radchuk et al., 2006; Gunawardena, 2008; Thiel et al., 2008). After fertilization, the nucellus degenerates in a few days, providing space and nutrients for the developing endosperm (Radchuk et al., 2006). Later on, the nucellar projection establishes as a maternal part of the nutrient transport route, releasing nutrients into the apoplastic endosperm cavity. This process is accompanied by programmed cell death (Radchuk et al., 2006; Thiel et al., 2008). Differentiation of the tracheary elements as part of the vascular system and the senescence process also involves programmed cell death (Gunawardena, 2008). During senescence, plastids degrade initially followed by disruption of the nucleus and vacuole (Gunawardena, 2008). AMY4 may participate in starch breakdown in those tissues that undergo programmed cell death with the initial disruption of plastids, making starch granules accessible for α-amylase action.
The homologous genes of all currently known enzymes involved in transient starch breakdown in Arabidopsis leaves (Zeeman et al., 2007) were up-regulated in barley pericarp, including transcripts of probably chloroplast-targeted BAM5, BAM6, BAM7 (homologous to plastidial BAMs of Arabidopsis; Fulton et al., 2008), GWD1, PWD, DPE1, DPE2, and ISA3 enzymes (Fig. 4). A possible site of transient starch metabolism is the chlorenchyma, which is able to produce photosynthetic starch (Rolletschek et al., 2004). Relatively weak but almost constant mRNA levels of BAM5, BAM6, BAM7, DPE1, and DPE2 in the endosperm fraction could be explained by their expression in the chlorenchyma, which is coisolated with the endosperm fraction from 4 DAF onward (Sreenivasulu et al., 2002, 2004). Up-regulation of almost all genes involved in transient starch degradation in the pericarp fraction supposes that transient starch turnover occurs also in plastids of nongreen pericarp cells in order to avoid carbon starvation and optimize the supply of carbon to the developing endosperm, similar to transient starch functions in leaves (Smith and Stitt, 2007).
Transcript profiling and in situ localization coupled with starch and enzyme measurements led us to propose two different pathways for starch breakdown in maternal tissues (Fig. 10B). Amylolytic starch degradation could be connected with tissues undergoing programmed cell death, while transient starch breakdown may occur in living cells of pericarp and/or chlorenchyma.
In the Developing Endosperm, Starch Breakdown Occurs in the Transfer Cells and the Aleurone Layer
The starchy endosperm is the major tissue for starch synthesis and storage, while the aleurone, the transfer cell layer, and the embryo-surrounding region contain little or no starch at maturity (Duffus and Cochrane, 1993; Olsen, 2001). The relative amount of starch in the developing barley caryopsis increases steadily between 6 DAF and approximately 20 DAF (Weschke et al., 2000), thereafter staying at a constant level and reaching 60% to 70% of the dry weight of the mature caryopsis (Duffus and Cochrane, 1993). The genetic machinery required for starch synthesis in the developing endosperm is much more complex than that in the pericarp (Fig. 10A). A very strong and exclusively endosperm-specific expression of the cytosolic AGP-S1a subunit corresponds to the finding that most of the AGPase activity in the endosperm of barley is cytosolic (Thorbjørnsen et al., 1996; Johnson et al., 2003). AGP-S1a and AGP-S2 subunits are simultaneously expressed in distinct regions of the endosperm (Fig. 3), with the former more abundant in central regions and the latter at the periphery at 14 DAF. Amylose accumulation in endosperm lags behind that of amylopectin and continues after amylopectin synthesis ceases (Fig. 1B; Tomlinson and Denyer, 2003). This could be explained, at least in part, by the expression patterns of endosperm-specific GBSS1a and SS genes (Fig. 2B). While GBSS1a expression begins relatively late in the developing endosperm and continues until maturity, the accumulation of SS2b transcripts begins very early and is followed by an increase in the levels of SS1, SS2a, and SS3b transcripts during the middle stage. SS3a and SS4 expression also continues during maturity/desiccation. The supposition that some starch synthesis may occur after the main filling phase is also supported by the expression of the AGP-S2 and AGP-L1 genes, which are transcriptionally active over the whole period of caryopsis development, including maturation (Fig. 2A). Expression of ISA1 and PUL genes is consistent with starch accumulation in endosperm, assuming a role of debranching enzymes in starch biosynthesis. A mandatory role of both isoamylase and pullanase types of debranching enzyme for amylopectin biosynthesis was clearly shown in maize endosperm and Arabidopsis leaves (Dinges et al., 2003; Streb et al., 2008). Mutation of the plastidial α-glucan phosphorylase gene affects the synthesis and fine structure of amylopectin in rice endosperm (Satoh et al., 2008). Therefore, the strong expression of PHO1 in barley may also be connected with starch synthesis. Expression of PHO1 in rice (Satoh et al., 2008), wheat (Schupp and Ziegler, 2004), and barley (Fig. 4C) is higher during the early stages of endosperm development, indicating that PHO1 may be essential during early steps of starch synthesis. It is notable that PHO1 transcript abundance in pericarp also coincides with the starch accumulation profile as well as AGP-L2 and AGP-S1b expression patterns (Figs. 1A and 2A).
The rate of starch accumulation is believed to be very similar to that of starch synthesis (Tomlinson and Denyer, 2003), ensuring that there is no significant turnover of starch in the starchy endosperm during the filling period. Thus, the expression of starch-degrading enzymes during endosperm development (Fig. 4) is unexpected. Transcript in situ localization helped to reveal that the breakdown of starch in the endosperm during caryopsis development occurs at least in transfer cells and aleurone.
Transfer cells facilitate the transport of nutrients into the endosperm (Olsen, 2001); therefore, they are enriched by diverse transport proteins, including hexose and Suc transporters. Cell wall invertases, vacuolar invertases, and Suc synthases were also localized to this tissue (Weschke et al., 2003). AGP-S1b transcripts are detected in the transfer cells as well. Since transfer cells do not accumulate starch, starch synthesis occurring here is transitory and might serve as an energy and carbon source for intracellular processes, including the maintenance of cell wall biosynthesis and nutrient transfer.
A few starch granules are present in cells of developing aleurone, but only protein bodies and lipids are stored in the mature aleurone (Duffus and Cochrane, 1993; Neuberger et al., 2008). Consistent with these histological observations, BAM1 transcripts begin to accumulate from 10 DAF onward (Fig. 4B) in the aleurone/subaleurone layer (Fig. 8), indicating that starch breakdown occurs in the tissue.
BAM1 and BAM2 transcripts also accumulate at very high levels in the endosperm fraction during maturation/desiccation (Fig. 4) and persist in the dry caryopses (Sreenivasulu et al., 2008), coinciding with high β-amylase enzyme activity at these stages (Fig. 9). In dry barley grains, β-amylase is found in two forms: a free, active form and a bound (less active or latent) form (Beck and Ziegler, 1989). Immunochemical (Lauriere et al., 1986) and biochemical (Hara-Nishimura et al., 1986) studies have demonstrated that β-amylase is deposited on starch granules in the subaleurone layer during caryopsis desiccation. The active form appears earlier in caryopsis development, converting to a bound one during desiccation (Hara-Nishimura et al., 1986). Obviously, BAM enzyme activity during late seed development results from BAM1 and BAM2 gene expression. However, its physiological role is unclear, because starch degradation has not been reported at seed maturation. These enzymes might be presynthesized during maturation, so that they can become rapidly involved in starch remobilization upon imbibition.
In summary, we have established that starch mobilization in the pericarp directly follows its accumulation. This ensures the maintenance of sink strength in the young caryopsis and probably fulfills a regulatory role for carbon transport and availability to endosperm and embryo. Starch synthesis in the pericarp is largely performed by the same genes as in leaves. In contrast, the genetic machinery involved in storage starch synthesis in endosperm is more complex. Starch metabolism in the endosperm transfer cells may contribute to sugar transport and reallocation. Temporally accumulated aleurone starch is degraded and replaced by storage proteins and lipids. The allocation of starch breakdown to specific caryopsis tissues clarifies the function of starch degradation during grain development, when storage starch synthesis is the predominant physiological event.
MATERIALS AND METHODS
Plant Material and Tissue Preparation
Barley plants (Hordeum vulgare ‘Barke’) were cultivated under standard greenhouse conditions at 18°C with 16 h of light and a relative air humidity of 60%. Determination of developmental stages for developing barley seeds and tissue isolations were performed as described (Weschke et al., 2000). The developing seeds were harvested from the middle region of the ear at 2-d intervals starting from anthesis until 24 DAF. Pericarp and endosperm tissue fractions were separated by hand dissection with a light microscope (Sreenivasulu et al., 2002).
Sequence Isolation and Identification of Genes
To identify genes encoding enzymes of starch synthesis and degradation, sequence similarity searches were performed with publicly available sequences from monocot plants by BLASTN, and ESTs with high percentages of identity were selected. To define gene family members, Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa) genome sequence annotations were used. The longest cDNA clones were sequenced using primers generated out of the flanking plasmid sequences as well as gene-specific primers (Metabion). Sequence data were processed using the Lasergene software (DNAstar). The TargetP tool (Emanuelsson et al., 2000) was used for the prediction of the localization of putative proteins.
Evaluation of Expression Profiles from Macroarray
Upon updating annotation information, mRNA expression profiles of corresponding starch-synthesizing and starch-degrading genes were extracted from the previously described seed transcriptome data set generated from a macroarray encompassing 12,000 cDNA sequences from developing barley grains (Sreenivasulu et al., 2006). The full macroarray data set is available at the IPK home page (http://pgrc.ipk-gatersleben.de/seeds/12k_EST_Brenda_seed_development_gene_expression_data.php). The median-centered, normalized, log2-transformed expression values collected from pericarp (0–10 DAF) and endosperm (0–24 DAF) fractions, covering seed development at 2-d intervals, were considered for evaluation of expression profiles. Furthermore, quality checks were performed to consider transcriptome data collected from independently grown biological material to remove low-quality and nonreproduced data (Sreenivasulu et al., 2006), and differentially expressed starch-synthesizing and starch-degrading genes were listed (Supplemental Table S1). All experiments were replicated once with independently grown material as described (Sreenivasulu et al., 2006). The normalized data are represented in relative units in the figures to better correlate with northern-blot data.
Northern-Blot Analyses
Northern blot analyses were performed to study the expression of those genes whose cDNAs were not present on the macroarray. Total RNA was extracted from tissue samples using the Gentra RNA isolation kit (Biozym). Isolated RNA (10 μg lane−1) was fractionated on a 1% agarose/formaldehyde gel, transferred to a nylon filter, and hybridized as described (Radchuk et al., 2006), applying gene-specific DNA fragments as probes.
For alternatively spliced AGP-S1a and AGP-S1b transcripts, the specific cDNA fragments were used. The primers for the amplification of the 5′ UTR with the first exon of AGP-S1a cDNA (EST HI02B12) were 5′-GTCGCTCTCCCTTACCCCTCG-3′ and 5′-GTGCTTGCGTCGGGGTCGAG-3′ (fragment size, 314 nucleotides); for amplification of the alternative 5′ UTR and the first exon of the AGP-S1a cDNA (EST HB16K13), the primers were 5′-CACGAGGCCTCGTGCCGAATTC-3′ and 5′-GAACACTATCAATAGCATGGGGATTG-3′ (fragment size, 201 nucleotides).
As a probe for AGP-L2, an 850-nucleotide DNA fragment was isolated from the EST HX03I15 (http://pgrc.ipk-gatersleben.de/est/index.php) by DraI/SacI endonuclease digestion. An EcoRV/XhoI fragment (513 nucleotides) from the EST HT08K10 was used as a probe for ISA3. GBSS1a was probed with a PstI fragment (946 nucleotides in length) of the EST HY10D16, and GBSS1a was probed with the HincII fragment (487 nucleotides in length) of the EST HZ60A01. The primers 5′-CTACATTGAATTGTATCATGCCGCTC-3′ and 5′-GCTTCCACATGATCAGAGAGCCTAAC-3′ were used for amplification of a 419-nucleotide 3′ UTR of SS3b (EST HB14E10). The 3′ UTR fragment of the SS2b gene (350 nucleotides in length) was amplified from the EST HT03C10 with the primers 5′-GTCCTCGTCAAGGCCAAGTACCAG-3′ and 5′-CAACTCCCTATTTGATCGCCTAC-3′. A StuI/NcoI fragment (751 nucleotides) of EST HI06F14 was used as a probe for the BAM5, and a PstI fragment (750 nucleotides) of EST HO06A20 was taken to probe PHO2 mRNA expression. A probe for BAM6 was PCR amplified using the following primers: 5′-CGTCATGGTCGACTGCTGGTGG-3′ and 5′-CCACCAGTGTGTACCTGACACCTTGAC-3′; and a probe for BAM7 was directly amplified by reverse transcription-PCR from seed mRNA using the primers 5′-CACGAGGGTTATTGTTGACTGTTGGTG-3′ and 5′-CGTGTGACCTTGAATTATAGTAGCCAGC-3′.
After PCR amplification or enzyme digestion, all fragments were separated on 0.8% agarose gels, extracted from agarose using the Nucleospin Extract kit (Macherey-Nagel), and labeled with [32P]dCTP using the Rediprime kit (Amersham Biosciences). For quantification, filters were hybridized with a 26S rDNA fragment for estimation of RNA loading. The hybridization signals were quantified as described by Weschke et al. (2000) and are given in relative units to correlate their expression profiles with macroarray data. All northern-blot hybridizations were performed at least twice with independently grown plant material for each probe used. The representative northern-blot hybridizations are summarized in Supplemental Figure S2.
Histochemical Techniques and in Situ Hybridization
Caryopses were fixed in 2.5% (v/v) glutaraldehyde, 50 mm sodium cacodylate (pH 7.0) or in 4% (w/v) paraformaldehyde, 50 mm potassium phosphate buffer (pH 7.0) under slight vacuum for 4 h at room temperature, rinsed in cacodylate buffer, dehydrated, and embedded in Paraplast Plus (Sherwood Medical). Sections were cut at 10 to 15 μm on a microtome, transferred on poly-l-Lys-treated slides (Sigma Diagnostics), and dried overnight at 45°C. As a general stain, toluidine blue was used according to Gerlach (1977) and Carson (1990). In situ hybridization was performed according to Borisjuk et al. (1995). The same cDNA fragments as for blot hybridizations were used as probes after labeling with [33P]dCTP.
Biochemical Measurements
Starch and the relative amounts of amylose and amylopectin in starch were analyzed as described by Hovenkamp-Hermelink et al. (1988). ADP-Glc was extracted and measured by mass spectrometry (Rolletschek et al., 2005). All measurements were performed in duplicate from independently grown plant material with three technical repetitions each.
Enzyme Assays
Activity of α-amylase was assayed using the Ceralpha assay kit (Megazyme International) as described by McCleary and Sheehan (1987) with modifications to use small sample volumes. For extraction, ground and frozen material was suspended in 1 m sodium malate, 1 m sodium chloride, 40 mm calcium chloride, and 0.1% (w/v) sodium azide (pH 5.2) and incubated at 40°C for 20 min under constant stirring. After centrifugation at 1,000g for 10 min, extracts were diluted in extraction buffer. Activity of α-amylase was determined by mixing the extract with amylase HR reagent (blocked p-nitrophenyl maltoheptaoside, glucoamylase, and thermostable α-glucosidase) in equal parts. After incubation for exactly 20 min at 40°C, the reaction was stopped by adding trisodium phosphate solution (2%, w/v). The absorbance was measured at 400 nm, and the activity was calculated according to the supplier's formula. One unit of enzyme activity is defined as the amount of enzyme required to release 1 μmol of p-nitrophenol from blocked p-nitrophenyl maltoheptaoside per minute under the specified assay conditions.
Activity of β-amylase was determined according to McCleary and Codd (1989) using the Betamyl assay kit (Megazyme International) with modifications to use small sample volumes. Ground and frozen material was suspended in extraction buffer (50 mm Tris-HCl, 1 mm EDTA, and 100 mm Cys-HCl, pH 8.0), and the enzyme was extracted for 1 h at room temperature. After centrifugation at 1,000g for 10 min, the supernatant was diluted in 0.1 m maleate buffer, 1 mm EDTA, 1.0 mg mL−1 bovine serum albumin, and 0.02% (w/v) sodium azide at pH 6.2 to get a reaction absorbance below 0.8. Activity was determined by mixing enzyme extract with Betamyl substrate solution (p-nitrophenyl-α-d-maltopentaoside and α-glucosidase). After incubation for exactly 10 min at 40°C, the reaction was stopped by adding 1% (w/v) Trizma solution. The absorbance was measured at 400 nm, and the activity was calculated according to the supplier's formula. One unit of enzyme activity is defined as the amount of enzyme required, in the presence of excess α-glucosidase, to release 1 μmol of p-nitrophenol from p-nitrophenyl-α-d-maltopentaoside in 1 min under the defined assay conditions.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers FN179369 to FN179406.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phylogenetic tree of β-amylases.
Supplemental Figure S2. Results of northern-blot hybridizations.
Supplemental Table S1. Complete list of starch biosynthesis-related and starch degradation-related genes together with annotation and normalized macroarray expression values obtained from two experimental series based on independently grown plant material.
Supplementary Material
Acknowledgments
We thank Angela Stegmann, Gabriele Einert, Elsa Fessel, Uta Siebert, and Katrin Blaschek for excellent technical assistance.
This work was supported by the German Ministry of Education and Research within the German Plant Genome Initiative (grant nos. GABI–SEEDII, FKZ 0313115 and GABI–sysSEED, FKZ 0315044A).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Volodymyr V. Radchuk (radchukv@ipk-gatersleben.de).
The online version of this article contains Web-only data.
References
- Allison MJ, Ellis RP, Swanston JS (1974) Tissue distribution of α-amylase and phosphorylase in developing barley grains. J Inst Brew 80 488–491 [Google Scholar]
- Asatsuma S, Sawada C, Itoh K, Okito M, Kitajima A, Mitsui T (2005) Involvement of α-amylase I-1 in starch degradation in rice chloroplasts. Plant Cell Physiol 46 858–869 [DOI] [PubMed] [Google Scholar]
- Bak-Jensen KS, Laugesen S, Ostergaard O, Finnie C, Roepstorff P, Svensson B (2007) Spatio-temporal profiling and degradation of α-amylase isozymes during barley seed germination. FEBS J 274 2552–2565 [DOI] [PubMed] [Google Scholar]
- Baunsgaard L, Lutken H, Mikkelsen R, Glaring MA, Pham TT, Blennow A (2005) A novel isoform of glucan, water dikinase phosphorylates pre-phosphorylated α-glucans and is involved in starch degradation in Arabidopsis. Plant J 41 595–605 [DOI] [PubMed] [Google Scholar]
- Beck E, Ziegler P (1989) Biosynthesis and degradation of starch in higher plants. Annu Rev Plant Physiol Plant Mol Biol 40 95–117 [Google Scholar]
- Blauth SL, Yao Y, Klucinec JD, Shannon JC, Thompson DB, Guilitinan MJ (2001) Identification of mutator insertional mutants of starch-branching enzyme 2a in corn. Plant Physiol 125 1396–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisjuk L, Weber H, Panitz R, Manteuffel R, Wobus U (1995) Embryogenesis of Vicia faba L.: histodifferentiation in relation to starch and storage protein synthesis. J Plant Physiol 147 203–218 [Google Scholar]
- Bresolin NS, Li Z, Kosar-Hashemi B, Tetlow IJ, Chatterjee M, Rahman S, Morell MK, Howitt CA (2006) Characterisation of disproportionating enzyme from wheat endosperm. Planta 224 20–31 [DOI] [PubMed] [Google Scholar]
- Burton RA, Jenner H, Carrangis L, Fahy B, Fincher GB, Hylton C, Laurie DA, Parker M, Waite D, van Wegen S, et al (2002. a) Starch granule initiation and growth are altered in barley mutants that lack isoamylase activity. Plant J 31 97–112 [DOI] [PubMed] [Google Scholar]
- Burton RA, Johnson PE, Beckles DM, Fincher GB, Jenner HL, Naldrett MJ, Denyer K (2002. b) Characterization of the genes encoding the cytosolic and plastidial forms of ADP-glucose pyrophosphorylase in wheat endosperm. Plant Physiol 130 1464–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RA, Zhang XQ, Hrmova M, Fincher GB (1999) A single limit dextrinase gene is expressed both in the developing endosperm and in germinated grains of barley. Plant Physiol 119 859–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson FL (1990) Histotechnology: A Self-Instructional Text. Chicago, ASCP Press
- Clark SE, Hayes PM, Henson CA (2003) Effects of single nucleotide polymorphisms in β-amylase 1 alleles from barley on functional properties of the enzymes. Plant Physiol Biochem 41 798–804 [Google Scholar]
- Critchley JH, Zeeman SC, Takaha T, Smith AM, Smith SM (2001) A critical role for disproportionating enzyme in starch breakdown is revealed by a knock-out mutation in Arabidopsis. Plant J 26 89–100 [DOI] [PubMed] [Google Scholar]
- Delatte T, Umhang M, Trevisan M, Eicke S, Thorneycroft D, Smith SM, Zeeman SC (2006) Evidence for distinct mechanisms of starch granule breakdown in plants. J Biol Chem 281 12050–12059 [DOI] [PubMed] [Google Scholar]
- Dian W, Jiang H, Wu P (2005) Evolution and expression analysis of starch synthase III and IV in rice. J Exp Bot 56 623–632 [DOI] [PubMed] [Google Scholar]
- Dinges JR, Colleoni C, James MG, Myers AM (2003) Mutational analysis of the pullulanase-type debranching enzyme of maize indicates multiple functions in starch metabolism. Plant Cell 15 666–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffus CM (1969) α-Amylase activity in the developing barley grain and its dependence on gibberellic acid. Phytochemistry 8 1205–1209 [Google Scholar]
- Duffus CM (1987) Physiological aspects of enzymes during grain development and germination. In JE Kruger, D Lineback, CE Stauffer, eds, Enzymes and Their Role in Cereal Technology. American Association of Cereal Chemists, St Paul, pp 83–111
- Duffus CM, Cochrane MP (1993) Formation of the barley grain: morphology, physiology and biochemistry. In AW MacGregor, RS Bhatty, eds, Barley: Chemistry and Technology. American Association of Cereal Chemists, St Paul, pp 31–72
- Eimert K, Luo C, Dejardin A, Villand P, Thorbjornsen T, Kleczkowski LA (1997) Molecular cloning and expression of the large subunit of ADP-glucose pyrophosphorylase from barley (Hordeum vulgare) leaves. Gene 189 79–82 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300 1005–1016 [DOI] [PubMed] [Google Scholar]
- Emes MJ, Bowsher CG, Hedley C, Burrell MM, Scrase-Field ES, Tetlow IJ (2003) Starch synthesis and carbon partitioning in developing endosperm. J Exp Bot 54 569–575 [DOI] [PubMed] [Google Scholar]
- Fulton DC, Stettler M, Mettler T, Vaughan CK, Li J, Francisco P, Gil M, Reinhold H, Eicke S, Messerli G, et al (2008) β-AMYLASE4, a noncatalytic protein required for starch breakdown, acts upstream of three active β-amylases in Arabidopsis chloroplasts. Plant Cell 20 1040–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Fisher DK, Kim KN, Shannon JC, Guiltinan MJ (1996) Evolutionary conservation and expression patterns of maize starch branching enzyme I and IIb genes suggest isoform specialization. Plant Mol Biol 30 1223–1232 [DOI] [PubMed] [Google Scholar]
- Gao M, Wanat J, Stinard PS, James MG, Myers AM (1998) Characterization of dull1, a maize gene coding for a novel starch synthase. Plant Cell 10 399–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach D (1977) Botanische Mikrotechnik, Vol 2. Thieme Verlag, Stuttgart, Germany
- Gubler F, Li Z, Fieg S, Jacobsen JV, Morell MK (2000) Cloning and characterization of a starch synthase I gene (accession no. AF234163) from barley. Plant Physiol 122 1457–145910759541 [Google Scholar]
- Gunawardena AH (2008) Programmed cell death and tissue remodelling in plants. J Exp Bot 59 445–451 [DOI] [PubMed] [Google Scholar]
- Hara-Nishimura I, Nishimura M, Daussant J (1986) Conversion of free β-amylase to bound β-amylase on starch granules in the barley endosperm during desiccation phase of seed development. Protoplasma 134 149–153 [Google Scholar]
- Hennen-Bierwagen TA, Lin Q, Grimaud F, Planchot V, Keeling PL, James MG, Myers AM (2009) Proteins from multiple metabolic pathways associate with starch biosynthetic enzymes in high molecular weight complexes: a model for regulation of carbon allocation in maize amyloplasts. Plant Physiol 149: 1541–1559 [DOI] [PMC free article] [PubMed]
- Hennen-Bierwagen TA, Liu F, Marsh RS, Kim S, Gan Q, Tetlow IJ, Emes MJ, James MG, Myers AM (2008) Starch biosynthetic enzymes from developing Zea mays endosperm associate in multisubunit complexes. Plant Physiol 146 1892–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovenkamp-Hermelink JH, de Vries JN, Adamse P, Jacobsen E, Witholt B, Feenstra WJ (1988) Rapid estimation of the amylose/amylopectin ratio in small amounts of tuber and leaf tissue of the potato. Potato Res 31 241–246 [Google Scholar]
- James MG, Denyer K, Myers AM (2003) Starch synthesis in the cereal endosperm. Curr Opin Plant Biol 6 215–222 [DOI] [PubMed] [Google Scholar]
- Johnson PE, Patron NJ, Bottrill AR, Dinges JR, Fahy BF, Parker ML, Waite DN, Denyer K (2003) A low-starch barley mutant, Riso16, lacking the cytosolic small subunit of ADP-glucose pyrophosphorylase, reveals the importance of the cytosolic isoform and the identity of the plastidial small subunit. Plant Physiol 131 684–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung W, Skadsen RW, Peterson DM (2001) Characterization of a novel barley β-amylase gene expressed only during early grain development. Seed Sci Res 11 325–334 [Google Scholar]
- Khursheed B, Rogers JC (1988) Barley α-amylase genes: quantitative comparison of steady-state mRNA levels from individual members of the two different families expressed in aleurone cells. J Biol Chem 263 18953–18960 [PubMed] [Google Scholar]
- Kötting O, Pusch K, Tiessen A, Geigenberger P, Steup M, Ritte G (2005) Identification of a novel enzyme required for starch metabolism in Arabidopsis leaves: the phosphoglucan, water dikinase. Plant Physiol 137 242–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen M, Lok F, Planchot V, Svendsen I, Leah R, Svensson B (1999) Isolation and characterization of the gene encoding the starch debranching enzyme limit dextrinase from germinating barley. Biochim Biophys Acta 1431 538–546 [DOI] [PubMed] [Google Scholar]
- Lauriere C, Lauriere M, Daussant J (1986) Immunohistochemical localization of β-amylase in resting barley seeds. Physiol Plant 67 383–388 [Google Scholar]
- Li L, Blanco M, Jane J (2007) Physicochemical properties of endosperm and pericarp starches during maize development. Carbohydr Polymers 67 630–639 [Google Scholar]
- Li Z, Mouille G, Kosar-Hashemi B, Rahman S, Clarke B, Gale KR, Appels R, Morell MK (2000) The structure and expression of the wheat starch synthase III gene: motifs in the expressed gene define the lineage of the starch synthase III gene family. Plant Physiol 123 613–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Sun F, Xu S, Chu X, Mukai Y, Yamamoto M, Ali S, Rampling L, Kosar-Harshemi B, Rahman S, et al (2003) The structural organisation of the gene encoding class II starch synthase of wheat and barley and the evolution of the genes encoding starch synthases in plants. Funct Integr Genomics 3 76–85 [DOI] [PubMed] [Google Scholar]
- Lloyd JR, Kossmann J, Ritte G (2005) Leaf starch degradation comes out of the shadows. Trends Plant Sci 10 130–137 [DOI] [PubMed] [Google Scholar]
- Lu Y, Sharkey TD (2004) The role of amylomaltase in maltose metabolism in the cytosol of photosynthetic cells. Planta 218 466–473 [DOI] [PubMed] [Google Scholar]
- MacGregor AW, Dushnicky L (1989. a) α-Amylase in developing barley kernel: a reappraisal. J Inst Brew 95 29–33 [Google Scholar]
- MacGregor AW, Dushnicky L (1989. b) Starch degradation in endosperms of developing barley kernels. J Inst Brew 95 321–325 [Google Scholar]
- McCleary BV, Codd RJ (1989) Measurement of β-amylase in cereal flours and commercial enzyme preparations. J Cereal Sci 9 17–33 [Google Scholar]
- McCleary BV, Sheehan HJ (1987) Measurement of cereal α-amylase: a new assay procedure. J Cereal Sci 6 237–251 [Google Scholar]
- Morell MK, Kosar-Hashemi B, Cmiel M, Samuel MS, Chandler P, Rahman S, Buleon A, Batey IL, Li Z (2003) Barley sex6 mutants lack starch synthase IIa activity and contain a starch with novel properties. Plant J 34 173–185 [DOI] [PubMed] [Google Scholar]
- Morell MK, Myers AM (2005) Towards the rational design of cereal starches. Curr Opin Plant Biol 8 204–210 [DOI] [PubMed] [Google Scholar]
- Mutisya J, Sathish P, Sun C, Andersson L, Ahlandsberg S, Baguma Y, Palmqvist S, Odhiambo B, Aman P, Jansson C (2003) Starch branching enzymes in sorghum (Sorghum bicolor) and barley (Hordeum vulgare): comparative analyses of enzyme structure and gene expression. J Plant Physiol 160 921–930 [DOI] [PubMed] [Google Scholar]
- Nashilevitz S, Melamed-Bessudo C, Aharoni A, Kossmann J, Wolf S, Levy AA (2009) The legwd mutant uncovers the role of starch phosphorylation in pollen development and germination in tomato. Plant J 57 1–13 [DOI] [PubMed] [Google Scholar]
- Neuberger T, Sreenivasulu N, Rokitta M, Rolletschek H, Göbel C, Rutten T, Radchuk V, Feussner I, Wobus U, Jakob P, et al (2008) Quantitative imaging of oil storage in developing crop seeds. Plant Biotechnol J 6 31–45 [DOI] [PubMed] [Google Scholar]
- Ohdan T, Francisco PB, Sawada T, Hirose T, Terao T, Satoh H, Nakamura Y (2005) Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J Exp Bot 56 3229–3244 [DOI] [PubMed] [Google Scholar]
- Olsen OA (2001) Endosperm development: cellularization and cell fate specification. Annu Rev Plant Physiol Plant Mol Biol 52 233–267 [DOI] [PubMed] [Google Scholar]
- Patron NJ, Smith AM, Fahy BF, Hylton CM, Naldrett MJ, Rossnagel BG, Denyer K (2002) The altered pattern of amylose accumulation in the endosperm of low-amylose barley cultivars is attributable to a single mutant allele of granule-bound starch synthase I with a deletion in the 5'-non-coding region. Plant Physiol 130 190–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommerrenig B, Barth I, Niedermeier M, Kopp S, Schmid J, Dwyer RA, McNair RJ, Klebl F, Sauer N (2006) Common plantain: a collection of expressed sequence tags from vascular tissue and a simple and efficient transformation method. Plant Physiol 142 1427–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radchuk V, Borisjuk L, Radchuk R, Steinbiss HH, Rolletschek H, Broeders S, Wobus U (2006) Jekyll encodes a novel protein involved in the sexual reproduction of barley. Plant Cell 18 1652–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmatullah RJ, Huang JK, Clark KL, Reeck GR, Chandra GR, Muthukrishnan S (1989) Nucleotide and predicted amino acid sequences of two different genes for high-pI α-amylases from barley. Plant Mol Biol 12 119–121 [DOI] [PubMed] [Google Scholar]
- Ritte G, Lloyd JR, Eckermann N, Rottmann A, Kossmann J, Steup M (2002) The starch-related R1 protein is an α-glucan, water dikinase. Proc Natl Acad Sci USA 99 7166–7171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers JC, Milliman C (1983) Isolation and sequence analysis of a barley α-amylase cDNA clone. J Biol Chem 258 8169–8174 [PubMed] [Google Scholar]
- Rolletschek H, Koch K, Wobus U, Borisjuk L (2005) Positional cues for the starch/lipid balance in maize kernels and resource partitioning to the embryo. Plant J 42 69–83 [DOI] [PubMed] [Google Scholar]
- Rolletschek H, Weschke W, Weber H, Wobus U, Borisjuk L (2004) Energy state and its control on seed development: starch accumulation is associated with high ATP and steep oxygen gradients within barley grains. J Exp Bot 55 1351–1359 [DOI] [PubMed] [Google Scholar]
- Rösti S, Rudi H, Rudi K, Opsahl-Sorteberg HG, Fahy B, Denyer K (2006) The gene encoding the cytosolic small subunit of ADP-glucose pyrophosphorylase in barley endosperm also encodes the major plastidial small subunit in the leaves. J Exp Bot 57 3619–3626 [DOI] [PubMed] [Google Scholar]
- Satoh H, Shibahara K, Tokunaga T, Nishi A, Tasaki M, Hwang SK, Okita TW, Kaneko N, Fujita N, Yoshida M, et al (2008) Mutation of the plastidial α-glucan phosphorylase gene in rice affects the synthesis and structure of starch in the endosperm. Plant Cell 20 1833–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupp N, Ziegler P (2004) The relation of starch phosphorylases to starch metabolism in wheat. Plant Cell Physiol 45 1471–1484 [DOI] [PubMed] [Google Scholar]
- Smith AM (1999) Making starch. Curr Opin Plant Biol 2 223–229 [DOI] [PubMed] [Google Scholar]
- Smith AM, Stitt M (2007) Coordination of carbon supply and plant growth. Plant Cell Environ 30 1126–1149 [DOI] [PubMed] [Google Scholar]
- Smith AM, Zeeman SC, Smith SM (2005) Starch degradation. Annu Rev Plant Biol 56 73–98 [DOI] [PubMed] [Google Scholar]
- Sreenivasulu N, Altschmied L, Panitz R, Hahnel U, Michaliek W, Weschke W, Wobus U (2002) Identification of genes specifically expressed in maternal and filial tissues of barley caryopses: a cDNA array analysis. Mol Genet Genomics 266 758–767 [DOI] [PubMed] [Google Scholar]
- Sreenivasulu N, Altschmied L, Radchuk V, Gubatz S, Wobus U, Weschke W (2004) Transcript profiles and deduced changes of metabolic pathways in maternal and filial tissues of developing barley grains. Plant J 37 539–553 [DOI] [PubMed] [Google Scholar]
- Sreenivasulu N, Radchuk V, Strickert M, Miersch O, Weschke W, Wobus U (2006) Gene expression patterns reveal tissue-specific signaling networks controlling programmed cell death and ABA-regulated maturation in developing barley seeds. Plant J 47 310–327 [DOI] [PubMed] [Google Scholar]
- Sreenivasulu N, Usadel B, Winter A, Radchuk V, Scholz U, Stein N, Weschke W, Strickert M, Close TJ, Stitt M, et al (2008) Barley grain maturation and germination: metabolic pathway and regulatory network commonalities and differences highlighted by new MapMan/PageMan profiling tools. Plant Physiol 146 1738–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streb S, Delatte T, Umhang M, Eicke S, Schorderet M, Reinhardt D, Zeeman SC (2008) Starch granule biosynthesis in Arabidopsis is abolished by removal of all debranching enzymes but restored by the subsequent removal of an endoamylase. Plant Cell 20 3448–3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Sathish P, Ahlandsberg S, Jansson C (1998) The two genes encoding starch-branching enzymes IIa and IIb are differentially expressed in barley. Plant Physiol 118 37–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetlow IJ, Beisel KG, Cameron S, Makhmoudova A, Liu F, Bresolin NS, Wait R, Morell MK, Emes MJ (2008) Analysis of protein complexes in wheat amyloplasts reveals functional interactions among starch biosynthetic enzymes. Plant Physiol 146 1878–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel J, Weier D, Sreenivasulu N, Strickert M, Weichert N, Melzer M, Czauderna T, Wobus U, Weber H, Weschke W (2008) Different hormonal regulation of cellular differentiation and function in nucellar projection and endosperm transfer cells: a microdissection-based transcriptome study of young barley grains. Plant Physiol 148 1436–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorbjørnsen T, Villand P, Kleczkowski LA, Olsen OA (1996) A single gene encodes two different transcripts for the ADP-glucose pyrophosphorylase small subunit from barley (Hordeum vulgare). Biochem J 313 149–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson K, Denyer K (2003) Starch synthesis in cereal grains. Adv Bot Res 40 1–61 [Google Scholar]
- Villand P, Olsen OA, Kilian A, Kleczkowski LA (1992) ADP glucose pyrophosphorylase large subunit from barley endosperm. Plant Physiol 100 1617–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrinten PL, Nakamura T (2000) Wheat granule-bound starch synthase I and II are encoded by separate genes that are expressed in different tissues. Plant Physiol 122 255–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattebled F, Dong Y, Dumez S, Delvallé D, Planchot V, Berbezy P, Vyas D, Colonna P, Chatterjee M, Ball S, et al (2005) Mutants of Arabidopsis lacking a chloroplastic isoamylase accumulate phytoglycogen and an abnormal form of amylopectin. Plant Physiol 138 184–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegrzyn T, Reilly K, Cipriani G, Murphy P, Newcomb R, Gardner R, MacRae E (2000) A novel α-amylase gene is transiently upregulated during low temperature exposure in apple fruit. Eur J Biochem 267 1313–1322 [DOI] [PubMed] [Google Scholar]
- Weschke W, Panitz R, Gubatz S, Wang Q, Radchuk R, Weber H, Wobus U (2003) The role of invertases and hexose transporters in controlling sugar ratios in maternal and filial tissues of barley caryopses during early development. Plant J 33 395–411 [DOI] [PubMed] [Google Scholar]
- Weschke W, Panitz R, Sauer N, Wang Q, Neubohn B, Weber H, Wobus U (2000) Sucrose transport into barley seeds: molecular characterization of two transporters and implications for seed development and starch accumulation. Plant J 21 455–467 [DOI] [PubMed] [Google Scholar]
- Yu TS, Zeeman SC, Thorneycroft D, Fulton DC, Dunstan H, Lue WL, Hegemann B, Tung SY, Umemoto T, Chapple A, et al (2005) α-Amylase is not required for breakdown of transitory starch in Arabidopsis leaves. J Biol Chem 280 9773–9779 [DOI] [PubMed] [Google Scholar]
- Zeeman SC, Delatte T, Messerli G, Umhang M, Stettler M, Mettler T, Streb S, Reinhold H, Kotting O (2007) Starch breakdown: recent discoveries suggest distinct pathways and novel mechanisms. Funct Plant Biol 34 465–473 [DOI] [PubMed] [Google Scholar]
- Zhang H, Sreenivasulu N, Weschke W, Stein N, Rudd S, Radchuk V, Potokina E, Scholz U, Schweizer P, Zierold U, et al (2004) Large-scale analysis of the barley transcriptome based on expressed sequence tags. Plant J 40 276–290 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.