Abstract
The phytohormone abscisic acid (ABA) is well known for its regulatory roles in integrating environmental constraints with the developmental programs of plants. Here, we characterize the biological function of the Arabidopsis (Arabidopsis thaliana) RING-H2 protein RHA2a in ABA signaling. The rha2a mutant is less sensitive to ABA than the wild type during seed germination and early seedling development, whereas transgenic plants overexpressing RHA2a are hypersensitive, indicating that RHA2a positively regulates ABA-mediated control of seed germination and early seedling development. Double mutant analyses of rha2a with several known ABA-insensitive mutants suggest that the action of RHA2a in ABA signaling is independent of that of the transcription factors ABI3, ABI4, and ABI5. We provide evidence showing that RHA2a also positively regulates plant responses to salt and osmotic stresses during seed germination and early seedling development. RHA2a is a functional E3 ubiquitin ligase, and its conserved RING domain is likely important for the biological function of RHA2a in ABA signaling. Together, these results suggest that the E3 ligase RHA2a is an important regulator of ABA signaling during seed germination and early seedling development.
The phytohormone abscisic acid (ABA) is well known for its regulatory roles in integrating environmental constraints with the developmental programs of plants (for review, see Leung and Giraudat, 1998; Finkelstein et al., 2002; Zhu, 2002; Assmann, 2003; Himmelbach et al., 2003; Nambara and Marion-Poll, 2003; Chow and McCourt, 2004; Christmann et al., 2006; Yamaguchi-Shinozaki and Shinozaki, 2006). ABA affects a broad range of physiological processes during different developmental stages. For example, ABA helps maintain seed dormancy to ensure that seeds germinate under favorable conditions. Immediately after germination, ABA may inhibit the establishment and subsequent development of young seedlings, with this postgerminative arrest representing an early developmental checkpoint to slow seedling growth until better conditions arise (Lopez-Molina et al., 2001; Finkelstein et al., 2002; Nambara and Marion-Poll, 2003). During more advanced developmental stages, ABA also regulates plant responses to various abiotic stresses, largely by directing guard cell functioning (Finkelstein et al., 2002; Assmann, 2003; Christmann et al., 2006; Yamaguchi-Shinozaki and Shinozaki, 2006). Therefore, ABA-regulated processes are generally divided into two broad and overlapping categories: ABA signaling in seeds (maintenance of seed dormancy and control of early seedling development) and ABA signaling in guard cells of more mature plants (Pandey et al., 2006).
Molecular genetics studies have significantly advanced our understanding on the molecular basis of ABA signaling in seeds and seedlings. Notably, through characterization of a series of ABA-insensitive mutants, which are resistant to ABA-mediated inhibition of germination and/or postgerminative growth, several components regulating ABA signaling in seeds and/or guard cells have been identified in Arabidopsis (Arabidopsis thaliana; Finkelstein et al., 2002; Assmann, 2003; Hegedus et al., 2003; Nambara and Marion-Poll, 2003). Among them, ABI1 (Leung et al., 1994) and ABI2 (Rodriguez et al., 1998) are protein phosphatases that negatively regulate ABA signaling during seed dormancy and germination. These phosphatases were later shown to be involved in ABA-mediated guard cell signaling as well (Allen et al., 1999). In contrast, the ABI (for ABA-insensitive) transcription factors, including ABI3, ABI4, and ABI5, act positively to regulate ABA signaling in seeds. ABI3 is a B3-domain transcription factor (Giraudat et al., 1992) that is essential for embryogenesis, and null mutations of this transcription factor lead to severe phenotypes in both seed development and ABA sensitivity (Finkelstein et al., 2002). Compared with ABI3, the AP2-type transcription factor ABI4 (Finkelstein et al., 1998) and the basic leucine zipper (bZIP) transcription factor ABI5 (Finkelstein and Lynch, 2000) mainly act in ABA-mediated control of seed germination and early seedling development (Finkelstein et al., 2002).
The ubiquitin/26S proteasome pathway, which is conserved in all eukaryotic cells, is proposed to be the dominant selective protein turnover system in plants (Moon et al., 2004; Schwechheimer and Schwager, 2004; Smalle and Vierstra, 2004; Dreher and Callis, 2007). Covalent attachment of ubiquitin molecules to a substrate protein is a prerequisite for its degradation by the 26S proteasome (Hershko and Ciechanover, 1998; Callis and Vierstra, 2000). The ubiquitin attachment process, called ubiquitination, is achieved through the sequential action of ubiquitin-activating enzyme E1, ubiquitin-conjugating enzyme E2, and ubiquitin ligase E3. The specificity of ubiquitination is largely determined by E3, which recruits appropriate substrate(s) (Smalle and Vierstra, 2004). In the Arabidopsis genome, more than 1,300 genes are predicted to encode different classes of E3 ligases (Smalle and Vierstra, 2004), implying that the ubiquitin-dependent proteolysis is extensively involved in different cellular processes.
Among the more than 1,300 predicted potential E3 ligases in Arabidopsis, more than 450 belong to the RING (for Really Interesting New Gene) finger class (Stone et al., 2005). The RING finger is defined by eight conserved Cys and His residues that together coordinate two zinc ions in a cross-braced fashion (Fang and Weissman, 2004). Initial functional evaluation has indicated that a large number of them are capable of mediating protein ubiquitination (Stone et al., 2005). Recently, some of these RING class E3 enzymes have been implicated in specific plant signaling pathways, including ABA signaling (Callis and Vierstra, 2000; Hellmann and Estelle, 2002; Devoto et al., 2003; Moon et al., 2004; Schwechheimer and Schwager, 2004; Smalle and Vierstra, 2004; Hoecker, 2005; Huq, 2006; Dreher and Callis, 2007). For example, the ABI3-interacting protein AIP2, which is a RING domain-containing E3 ligase, serves as a negative regulator of ABA signaling by targeting ABI3 for degradation (Zhang et al., 2005). Previous studies have indicated that the bZIP transcription factor ABI5 is possibly regulated by the ubiquitin-dependent proteolysis (Lopez-Molina and Chua, 2000; Lopez-Molina et al., 2001, 2003; Smalle et al., 2003). It has recently been shown that the novel RING E3 ligase KEG probably targets ABI5 for degradation (Stone et al., 2006). In addition, SDIR1 (for SALT- AND DROUGHT-INDUCED RING FINGER1), another RING finger E3 ligase, acts upstream of ABI3 and ABI5 in ABA signaling and regulates plant responses to drought and salt stresses (Zhang et al., 2007).
Here, we report that the previously described Arabidopsis RING-H2 protein RHA2a (Jensen et al., 1998; Greve et al., 2003) regulates ABA-mediated control of seed germination and early seedling development. Genetic analyses indicate that the action of RHA2a in ABA signaling is independent of that of the ABI transcription factors ABI3, ABI4, and ABI5. We provide evidence showing that RHA2a is a functional E3 ubiquitin ligase and that its conserved RING domain is likely important for the biological function of RHA2a in ABA signaling.
RESULTS
Expression of RHA2a
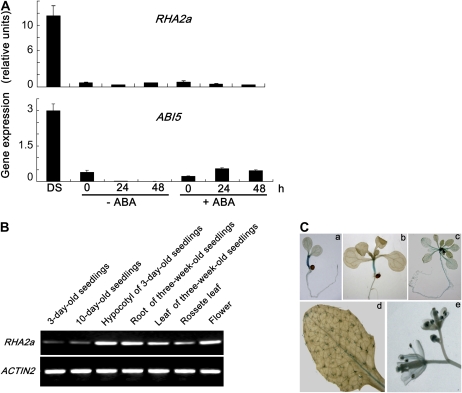
From publicly available microarray data, we noticed that the expression level of RHA2a (At1g15100), a gene that encodes a C3H2C3-type RING finger protein, is relatively high in dry seeds and undergoes significant reduction after imbibition (Nakabayashi et al., 2005; Tatematsu et al., 2008). We examined the expression pattern of RHA2a following stratification as well as during the early stages of seed germination with or without ABA. In parallel, the expression of ABI5, an important regulator of ABA response during seed germination (Lopez-Molina et al., 2001, 2002), was also examined. Our quantitative real-time reverse transcription-PCR (qRT-PCR) assays indicated that transcripts of RHA2a were abundant in dry seeds but were dramatically reduced to a barely detectable level when seeds were stratified at 4°C for 72 h. When stratified seeds were transferred to ABA-free medium for germination, the expression of RHA2a remained at a low level (Fig. 1A). This expression pattern shows high similarity to that of ABI5 (Fig. 1A). We then transferred the stratified seeds to ABA-containing medium for germination and found that the expression of RHA2a was not obviously induced by ABA at the time points investigated. However, consistent with several previous reports (Finkelstein and Lynch, 2000; Lopez-Molina et al., 2001, 2002), the expression of ABI5 was significantly induced by ABA (Fig. 1A).
Figure 1.
Expression of RHA2a. A, RHA2a expression in dry seeds (DS) and during seed imbibition analyzed by qRT-PCR. Col-0 seeds were kept in darkness at 4°C for 72 h and then transferred to medium with or without ABA (5 μm) in constant light at 22°C for germination. Total RNA was extracted at the indicated times (0 indicates the time immediately following transfer). Transcript levels of RHA2a and ABI5 were quantified by qRT-PCR against ACTIN2. Each value is the mean ± sd of three independent biological determinations. B, RT-PCR analysis of RHA2a expression in different organs. ACTIN2 primers were used in PCR as an internal control. C, GUS staining of RHA2apro:GUS plants from different growth stages. a, Three-day-old seedling; b, 10-d-old seedling; c, 3-week-old seedling; d, rosette leaf of a 5-week-old plant; e, flowers from 7-week-old plants. [See online article for color version of this figure.]
RT-PCR analysis indicated that, in addition to seeds, RHA2a expression was also detected in young seedlings and in multiple organs of more mature plants (Fig. 1B). For a more detailed analysis of the RHA2a expression pattern, a 2,360-bp promoter sequence of RHA2a was fused with the GUS gene to generate transgenic plants. Representative transgenic plants named RHA2apro:GUS were used to follow RHA2a expression in different developmental stages. Strong GUS staining was observed in hypocotyls of 3-d-old and 10-d-old seedlings (Fig. 1C, a and b). In 3-week-old plants, GUS staining was detected in shoot apical meristems and relatively weak in petioles and roots (Fig. 1C, c). In rosette leaves, GUS staining was clearly exhibited in trichomes (Fig. 1C, d). In flowers, GUS staining was observed in the anthers (Fig. 1C, e).
ABA Response of RHA2a Knockdown and RHA2a Overexpression Plants during Seed Germination
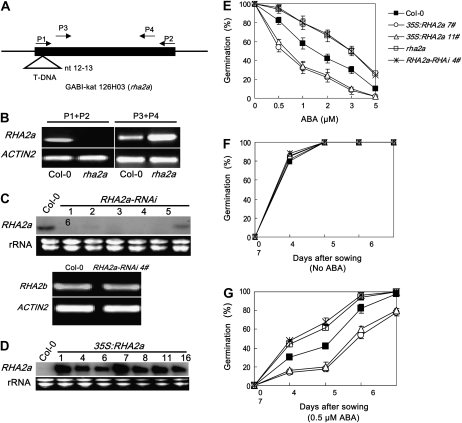
To study the physiological function of the RHA2a gene using a genetic approach, we obtained mutant and transgenic plants showing reduced and elevated expression of RHA2a, respectively. A T-DNA insertion line named rha2a was obtained from the Nottingham Arabidopsis Stock Centre (GABI-kat 126H03, N378380). Gene expression analyses revealed that the T-DNA in rha2a disrupted the expression of RHA2a (Fig. 2, A and B). Furthermore, genomic DNA of RHA2a driven by its native promoter rescued the ABA-related phenotype of rha2a (Supplemental Fig. S1). In addition, we generated six homozygous T3 RNA interference (RNAi) lines of RHA2a (Fig. 2C). RNAi lines with significantly reduced expression levels of RHA2a showed a similar ABA response phenotype to that of the rha2a mutant. Results of one of these RNAi lines named RHA2a-RNAi 4#, together with those of rha2a, are presented in this paper.
Figure 2.
ABA response of rha2a, RHA2a-RNAi, and 35S:RHA2a plants in seed germination. A, Diagram of the RHA2a gene illustrating a T-DNA inserted between nucleotides 12 and 13 in the rha2a mutant. The positions of the primer pairs P1/P2 and P3/P4 are indicated. B, Expression of the RHA2a gene in the rha2a mutant analyzed by RT-PCR using the primer pairs P1/P2 and P3/P4. As shown in A, the primer pair P1/P2 spans the T-DNA insertion, whereas the primer pair P3/P4 locates downstream of the T-DNA insertion site. ACTIN2 primers were used in PCR as an internal control. C, Expression of RHA2a and RHA2b in RHA2a-RNAi lines. Top, RNA gel blot analysis showing reduced expression of RHA2a in different RHA2a-RNAi lines. Lines 1, 3, and 4 showed similar ABA-insensitive phenotypes; results from line 4 (labeled as RHA2a-RNAi 4#) are shown in this report. Two-week-old plants were collected for RNA extraction. Thirty micrograms of RNA was loaded per lane. A duplicated gel stained with ethidium bromide was used as a loading control. Bottom, RT-PCR analysis showing expression of the RHA2b gene in RHA2a-RNAi 4#. ACTIN2 primers were used in PCR as an internal control. D, RNA gel blot analysis showing elevated expression of RHA2a in different 35S:RHA2a lines. All lines showed similar ABA-hypersensitive phenotypes; results from lines 7 and 11 (labeled as 35S:RHA2a 7# and 35S:RHA2a 11#) are shown in this report. Two-week-old plants were collected for RNA extraction. Ten micrograms of RNA was loaded per lane. A duplicated gel stained with ethidium bromide was used as a loading control. E, Quantification of seed germination. Seed germination percentage of the indicated genotypes grown on different concentrations of ABA was recorded at 3 d after the end of stratification. Data show the mean ± sd of three replicates. At least 100 seeds per genotype were measured in each replicate. F and G, Seed germination time course of the four genotypes grown on medium without ABA (F) or containing 0.5 μm ABA (G). Plates were kept at 4°C in the dark for 3 d and then transferred to a growth chamber. Germination was recorded starting 24 h after transfer. Data show the mean ± sd of three replicates. At least 100 seeds per genotype were measured in each replicate. For E to G, at least three independent experiments were conducted and similar results were obtained.
At the same time, we generated RHA2a overexpression plants by expressing the RHA2a cDNA under the control of the cauliflower mosaic virus (CaMV) 35S promoter. Homozygous transgenic lines containing the 35S:RHA2a transgene were identified based on their resistance to the antibiotic kanamycin. The selected transgenic lines were then examined for increased levels of RHA2a expression by RNA gel-blot analysis (Fig. 2D). All of the RHA2a overexpression lines showed a similar ABA-related phenotype; results obtained with two representative lines named 35S:RHA2a 7# and 35S:RHA2a 11# are shown below.
The rha2a mutant, RHA2a-RNAi 4#, and 35S:RHA2a plants were compared with wild-type plants for their response to the inhibitory effect of ABA in seed germination. In an ABA dose-response assay, seeds were germinated on ABA-free or ABA-containing medium and seed germination (obvious radicle emergence) percentage was scored at 3 d after the end of stratification. In the absence of ABA, the seed germination percentages of different genotypes were similar (Fig. 2E). In the presence of different concentrations of ABA, rha2a and RHA2a-RNAi 4# plants showed higher seed germination percentages than wild-type plants, whereas the two 35S:RHA2a lines exhibited significantly reduced seed germination percentages (Fig. 2E). In the absence of ABA, the four genotypes showed similar seed germination kinetics (Fig. 2F). When germinated on medium containing 0.5 μm ABA, the seed germination of rha2a and RHA2a-RNAi 4# plants occurred earlier than in wild-type plants, whereas this concentration of ABA significantly inhibited the seed germination of 35S:RHA2a plants (Fig. 2G). These results demonstrated that, while rha2a and RHA2a-RNAi 4# plants are less sensitive to ABA than wild-type plants, 35S:RHA2a plants are more sensitive to this hormone, suggesting that the RHA2a gene acts as a positive regulator of ABA signaling during seed germination.
It has been shown that fluridone, an inhibitor of ABA biosynthesis, can effectively reduce endogenous ABA levels (Ullah et al., 2002; Dekkers et al., 2004; Chen et al., 2006; Pandey et al., 2006). To challenge the possibility that RHA2a may regulate ABA response by affecting ABA biosynthesis, we examined the effect of fluridone on ABA sensitivity of RHA2a overexpression lines, the rha2a mutant, and the wild type. Consistent with previous reports (Ullah et al., 2002; Chen et al., 2006), pretreatment with fluridone generally reduced the seed ABA sensitivity of different genotypes (Supplemental Fig. S2). However, in the presence of fluridone pretreatment, seed germination of 35S:RHA2a plants still showed increased sensitivity to ABA, whereas seed germination of the rha2a mutant was less sensitive to ABA (Supplemental Fig. S2). The finding that fluridone treatment did not diminish the difference in ABA-mediated inhibition of seed germination among genotypes suggested that RHA2a affects ABA signaling rather than ABA biosynthesis.
ABA Response of RHA2a Knockdown and RHA2a Overexpression Plants during Early Seedling Development
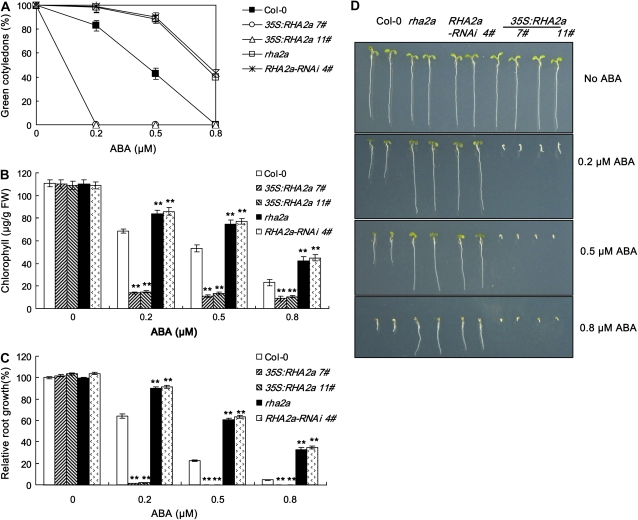
The RHA2a knockdown and RHA2a overexpression plants were also assessed for their response to ABA during early seedling development. For these experiments, seeds germinated on ABA-free medium were transferred to medium containing different concentrations of ABA and cotyledon-greening percentage was scored (see “Materials and Methods”). In the absence of ABA, the cotyledon-greening percentages of the different genotypes were similar (Fig. 3A). In the presence of different concentrations of ABA, rha2a and RHA2a-RNAi 4# plants showed higher cotyledon-greening percentages than wild-type plants, whereas the two lines of 35S:RHA2a plants exhibited significantly reduced cotyledon-greening percentages (Fig. 3A). To quantify the effect of ABA on cotyledon greening, we measured chlorophyll contents of the seedlings. In the absence of ABA, the chlorophyll contents were largely similar among genotypes (Fig. 3B). In line with the cotyledon-greening data, ABA treatment exerted differential effects on chlorophyll accumulation of the RHA2a knockdown and RHA2a overexpression plants: while rha2a and RHA2a-RNAi 4# seedlings showed higher levels of chlorophyll accumulation than wild-type seedlings, 35S:RHA2a 7# and 35S:RHA2a 11# seedlings showed significantly reduced levels of chlorophyll accumulation than wild-type seedlings (Fig. 3B), again indicating that rha2a and RHA2a-RNAi 4# plants are less sensitive to ABA in cotyledon greening but 35S:RHA2a plants are hypersensitive. The action of RHA2a in ABA signaling was also assessed by investigating the ABA-mediated retardation of seedling root growth. In the presence of different concentrations of ABA, root growth of rha2a and RHA2a-RNAi 4# seedlings was significantly better than that of wild-type seedlings, whereas root growth of the 35S:RHA2a plants was more severely inhibited (Fig. 3, C and D). Taken together, the contrasting ABA sensitivities displayed by the RHA2a knockdown plants (T-DNA mutant and RNAi lines) and RHA2a overexpression plants suggested that RHA2a acts as a positive regulator of ABA signaling during early seedling development.
Figure 3.
ABA response of rha2a, RHA2a-RNAi, and 35S:RHA2a plants during early seedling development. A, Quantification of cotyledon greening. Cotyledon-greening percentage of the indicated genotypes grown on medium containing different concentrations of ABA was recorded at 5 d after the end of stratification (see “Materials and Methods”). Data show the mean ± sd of three replicates. At least 100 seeds per genotype were measured in each replicate. B, Quantification of chlorophyll content. Seedlings described in A were collected for chlorophyll a/b extraction and measurement. Values represent the mean ± sd of three replicates. FW, Fresh weight of whole seedlings. C, Root growth measurements. Seedling root length of the indicated genotypes grown on medium containing different concentrations of ABA was measured at 5 d after the end of stratification (see “Materials and Methods”). Relative root growth compared with that on ABA-free medium is indicated. Data show the mean ± sd of three replicates. At least 30 seedlings per genotype were measured in each replicate. D, Photographs of seedlings at 5 d after the end of stratification. Twenty-four hours after stratification, germinated seeds were transferred from ABA-free medium to medium containing 0.5 μm ABA. Asterisks in B and C indicate the significance of the difference from the corresponding wild-type values determined by Student's t test (** P < 0.01). At least three independent experiments were conducted and similar results were obtained. [See online article for color version of this figure.]
Under our experimental conditions, the above-described distinct effects of ABA on RHA2a knockdown plants and RHA2a overexpression plants could be easily observed if the seedlings were transferred to ABA-containing medium less than 48 h after stratification, but the differential effects were less apparent when the transfer took place more than 48 h after stratification, suggesting that the action of RHA2a in ABA signaling may specifically be seen during germination and early stages of seedling development.
Consistently, gene expression analyses using 2-week-old seedlings indicated that the ABA-induced expression levels of several marker genes, including RD29A, RAB18, KIN1, and NCED3 (Shinozaki and Yamaguchi-Shinozaki, 1997), did not show significant differences among 35S:RHA2a, rha2a, and wild-type plants (data not shown). To test whether RHA2a affects ABA signaling in stomata of more mature plants, we compared leaf water loss of 35S:RHA2a, rha2a, and wild-type plants. The leaf water loss did not show significant differences among the three genotypes (Supplemental Fig. S3), suggesting that RHA2a has little effect on ABA signaling in more mature plants.
Genetic Analysis of rha2a Double Mutants
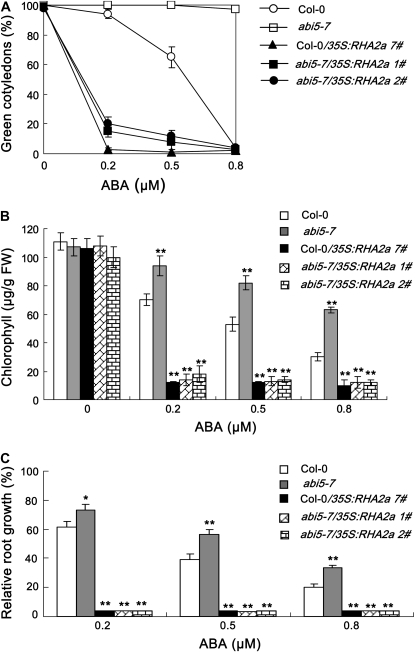
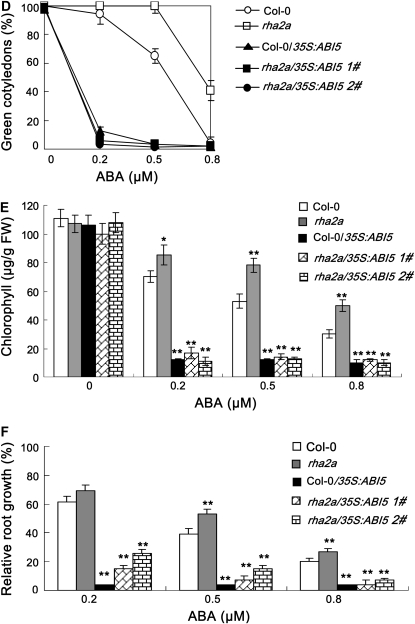
The opposite responses of RHA2a knockdown and RHA2a overexpression plants to ABA show high similarity to those of the abi5 mutants and ABI5 overexpression plants (Finkelstein et al., 2002). To test whether the RHA2a gene acts in the same or a different pathway to ABI5, we generated 35S:RHA2a plants in the genetic background of the ABA-insensitive abi5-7 mutant (abi5-7/35S:RHA2a). In our ABA response assays, abi5-7/35S:RHA2a plants were hypersensitive to ABA (Fig. 4, A–C), suggesting that RHA2a might act downstream of ABI5 in ABA signaling. Alternatively, RHA2a might act in a pathway independent of ABI5 to control ABA-mediated regulation of seed germination and early seedling development. To distinguish these two possibilities, we further generated 35S:ABI5 plants in the genetic background of the rha2a mutant (rha2a/35S:ABI5). In our ABA response assays, rha2a/35S:ABI5 plants also showed ABA-hypersensitive phenotype (Fig. 4, D–F), suggesting that RHA2a might act upstream or independently of ABI5 in ABA-mediated regulation of seed germination and early seedling development. Together, these results support the idea that RHA2a functions in parallel with ABI5 in ABA signaling.
Figure 4.
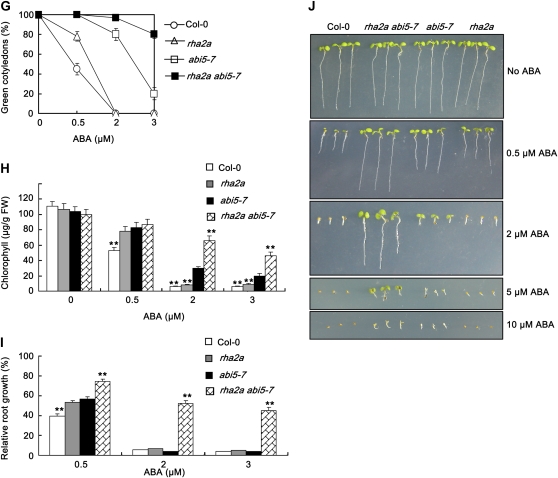
Genetic relationship between RHA2a and ABI5. A to C, Overexpression of RHA2a in abi5-7 leads to ABA hypersensitivity. A, Quantification of cotyledon greening. Cotyledon-greening percentage of Col-0, abi5-7, and RHA2a overexpression plants in the genetic background of Col-0 (35S:RHA2a 7#) and two lines of RHA2a overexpression plants in the genetic background of abi5-7 (abi5-7/35S:RHA2a 1# and abi5-7/35S:RHA2a 2#) grown on medium containing different concentrations of ABA was recorded at 5 d after the end of stratification. Data show the mean ± sd of three replicates. At least 100 seeds per genotype were measured in each replicate. B, Quantification of chlorophyll content. Seedlings described in A were collected for chlorophyll a/b extraction and measurement. Values represent the mean ± sd of three replicates. FW, Fresh weight of whole seedlings. C, Root growth measurements. Seedling root length of the indicated genotypes grown on medium containing different concentrations of ABA was measured at 5 d after the end of stratification (see “Materials and Methods”). Relative root growth compared with that on ABA-free medium is indicated. Data show the mean ± sd of three replicates. At least 30 seedlings per genotype were measured in each replicate. Asterisks indicate the significance of the difference from the corresponding wild-type values determined by Student's t test (* 0.01 ≤ P < 0.05, ** P < 0.01). At least three independent experiments were conducted and similar results were obtained. D to F, Overexpression of ABI5 in rha2a leads to ABA hypersensitivity. D, Quantification of cotyledon greening. Cotyledon-greening percentage of Col-0, rha2a, and ABI5 overexpression plants in the genetic background of Col-0 (Col-0/35S:ABI5) and two lines of RHA2a overexpression plants in the genetic background of rha2a (rha2a/35S:ABI5 1# and rha2a/35S:ABI5 2#) grown on medium containing different concentrations of ABA was recorded at 5 d after the end of stratification. Data show the mean ± sd of three replicates. At least 100 seeds per genotype were measured in each replicate. E, Quantification of chlorophyll content. Seedlings described in D were collected for chlorophyll a/b extraction and measurement. Values represent the mean ± sd of three replicates. F, Root growth measurements. Seedling root length of the indicated genotypes grown on medium containing different concentrations of ABA was measured at 5 d after the end of stratification (see “Materials and Methods”). Relative root growth compared with that on ABA-free medium is indicated. Data show the mean ± sd of three replicates. At least 30 seedlings per genotype were measured in each replicate. Asterisks indicate the significance of the difference from the corresponding wild-type values determined by Student's t test (* 0.01 ≤ P < 0.05, ** P < 0.01). At least three independent experiments were conducted and similar results were obtained. G to J, Double mutant analysis between rha2a and abi5-7. G, Quantification of cotyledon greening. Cotyledon-greening percentage of the indicated genotypes grown on medium containing different concentrations of ABA was recorded at 5 d after the end of stratification. Data show the mean ± sd of three replicates. At least 100 seeds per genotype were measured in each replicate. H, Quantification of chlorophyll content. Seedlings described in G were collected for chlorophyll a/b extraction and measurement. Values represent the mean ± sd of three replicates. I, Root growth measurements. Seedling root length of the indicated genotypes grown on different concentrations of ABA was measured at 5 d after the end of stratification. Relative root growth compared with that on ABA-free medium is indicated. Data show the mean ± sd of three replicates. At least 30 seedlings per genotype were measured in each replicate. J, Photographs of seedlings at 5 d after the end of stratification. Twenty-four hours after stratification, germinated seeds were transferred from ABA-free medium to medium containing different concentrations of ABA. Asterisks indicate the significance of the difference from the corresponding abi5-7 values determined by Student's t test (* 0.01 ≤ P < 0.05, ** P <0.01). At least three independent experiments were conducted and similar results were obtained. [See online article for color version of this figure.]
To confirm the above scenario, we further generated the rha2a abi5-7 double mutant and compared its response to ABA with that of the two single mutants. Given that both rha2a and abi5-7 are less sensitive to ABA, we reasoned that if RHA2a would indeed act in parallel with ABI5, the double mutant should be more insensitive to ABA than the single mutants alone. As expected, our ABA response assays, including cotyledon greening (Fig. 4G), chlorophyll accumulation (Fig. 4H), and root growth inhibition (Fig. 4I), all indicated that the double mutant was significantly more insensitive to ABA than the single mutants. For example, under our experimental conditions, when the ABA concentration was raised to 2 μm, both rha2a and abi5-7 were severely inhibited in cotyledon greening and root growth, but the double mutant still exhibited an insensitive phenotype (Fig. 4, G–J). The double mutant seedlings showed obvious growth retardation only when the ABA concentration was raised to 5 μm or higher (Fig. 4J). These data suggested that RHA2a functions in parallel with ABI5 in ABA signaling.
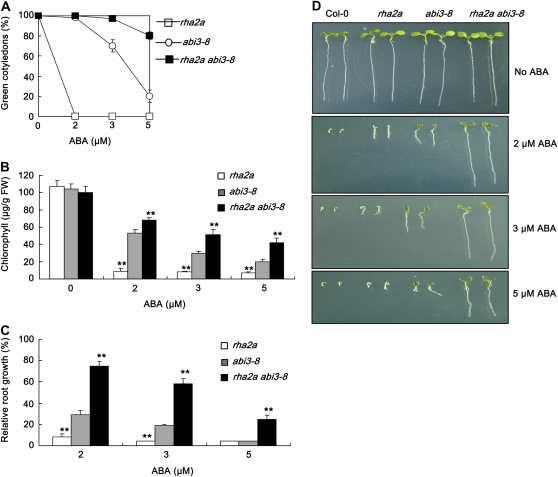
The ABI5 gene has been shown to act downstream of ABI3 in the same pathway to execute ABA-mediated regulation of seed germination and early seedling development (Lopez-Molina et al., 2002). ABA response assays indicated that overexpression of RHA2a in the abi3-8 mutant background led to ABA hypersensitivity (Supplemental Fig. S4). Furthermore, the rha2a abi3-8 double mutant was significantly more insensitive to ABA than the rha2a and abi3-8 single mutants (Fig. 5). These data suggested that the RHA2a gene may act in parallel with the ABI3-ABI5 pathway in ABA signaling.
Figure 5.
Double mutant analysis between rha2a and abi3-8. A, Quantification of cotyledon greening. Cotyledon-greening percentage of the indicated genotypes grown on medium containing different concentrations of ABA was recorded at 5 d after the end of stratification. Data show the mean ± sd of three replicates. At least 100 seeds per genotype were measured in each replicate. B, Quantification of chlorophyll content. Seedlings described in A were collected for chlorophyll a/b extraction and measurement. Values represent the mean ± sd of three replicates. FW, Fresh weight of whole seedlings. C, Root growth measurements. Seedling root length of the indicated genotypes grown on different concentrations of ABA was measured at 5 d after the end of stratification. Relative root growth compared with that on ABA-free medium is indicated. Data show the mean ± sd of three replicates. At least 30 seedlings per genotype were measured in each replicate. D, Photographs of seedlings at 5 d after the end of stratification. Twenty-four hours after stratification, germinated seeds were transferred from ABA-free medium to medium containing different concentrations of ABA. Asterisks in B and C indicate the significance of the difference from the corresponding abi3-8 values determined by Student's t test (** P < 0.01). Three independent experiments were conducted and similar results were obtained. [See online article for color version of this figure.]
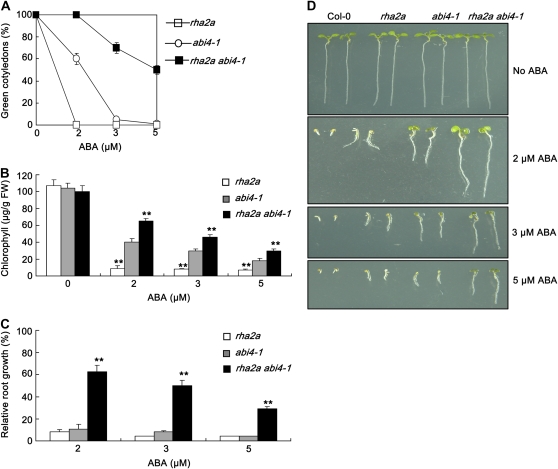
Two lines of evidence suggested that the function of RHA2a in ABA signaling is independent of that of ABI4. First, overexpression of RHA2a in the abi4-1 background led to an ABA-hypersensitive phenotype (Supplemental Fig. S5). Second, the rha2a abi4-1 double mutant was significantly more insensitive to ABA than either abi4-1 or rha2a (Fig. 6). Taken together, our genetic data support a hypothesis that the action of RHA2a in ABA signaling is independent of that of the ABI transcription factor genes, including ABI3, ABI4, and ABI5.
Figure 6.
Double mutant analysis between rha2a and abi4-1. A, Quantification of cotyledon greening. Cotyledon-greening percentage of the indicated genotypes grown on medium containing different concentrations of ABA was recorded at 5 d after the end of stratification. Data show the mean ± sd of three replicates. At least 100 seeds per genotype were measured in each replicate. B, Quantification of chlorophyll content. Seedlings described in A were collected for chlorophyll a/b extraction and measurement. Values represent the mean ± sd of three replicates. FW, Fresh weight of whole seedlings. C, Root growth measurements. Seedling root length of the indicated genotypes grown on different concentrations of ABA was measured at 5 d after the end of stratification. Relative root growth compared with that on ABA-free medium is indicated. Data show the mean ± sd of three replicates. At least 30 seedlings per genotype were measured in each replicate. D, Photographs of seedlings at 5 d after the end of stratification. Twenty-four hours after stratification, germinated seeds were transferred from ABA-free medium to medium containing different concentrations of ABA. Asterisks in B and C indicate the significance of the difference from the corresponding abi4-1 values determined by Student's t test (** P < 0.01). Three independent experiments were conducted and similar results were obtained. [See online article for color version of this figure.]
Salt and Osmotic Responses of rha2a and 35S:RHA2a Plants during Seed Germination
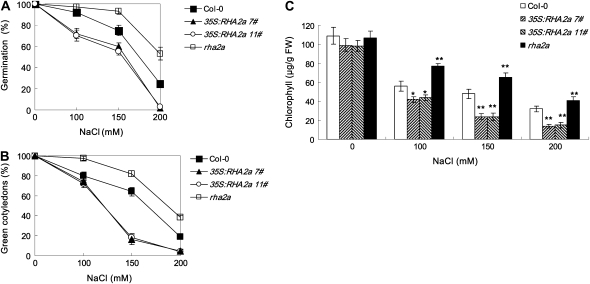
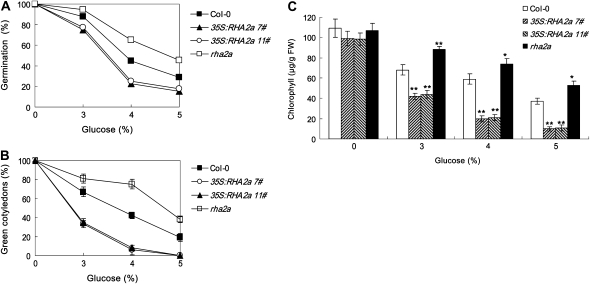
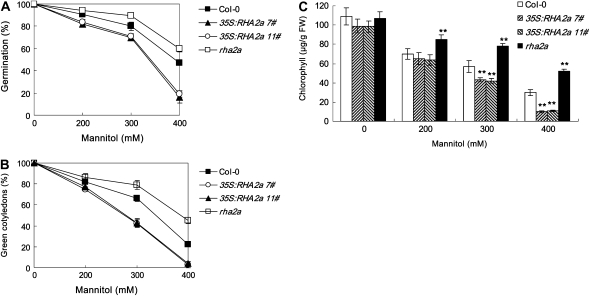
Salts inhibit seed germination and postgerminative growth in an ABA-dependent or ABA-independent manner (Zhu, 2002). Since the RHA2a gene regulates ABA signaling during seed germination and early seedling development, we tested whether RHA2a affects the plant response to salt stress during these processes. Seeds of rha2a, 35S:RHA2a, and the wild type were sown on medium containing different concentrations of NaCl and germination percentages were scored. In medium supplemented with 100 to 200 mm NaCl, germination percentages of rha2a were significantly higher than those of the wild type (Fig. 7A). On the contrary, germination percentages of the two 35S:RHA2a lines were significantly lower than those of the wild type (Fig. 7A). To distinguish whether RHA2a is involved in salt-specific or general osmotic responses, we compared the responses of different genotypes to the osmotic reagents Glc and mannitol. Seed germination of rha2a plants showed insensitivity to the inhibition effects of both Glc (Fig. 8A) and mannitol (Fig. 9A), whereas seed germination of 35S:RHA2a plants was more sensitive to these reagents (Figs. 8A and 9A), suggesting that RHA2a positively regulates osmotic stress responses during seed germination.
Figure 7.
Response of rha2a and 35S:RHA2a plants to NaCl. A, Quantification of seed germination. Seed germination percentage of the indicated genotypes grown on different concentrations of NaCl was recorded at 3 d after the end of stratification. Data show the mean ± sd of three replicates. At least 100 seeds per genotype were measured in each replicate. B, Quantification of cotyledon greening. Cotyledon-greening percentage of the indicated genotypes grown on medium containing different concentrations of NaCl was recorded at 5 d after the end of stratification. Data show the mean ± sd of three replicates. At least 100 seeds per genotype were measured in each replicate. C, Quantification of chlorophyll content of seedlings described in B. Values represent the mean ± sd of three replicates. FW, Fresh weight of whole seedlings. Asterisks in C indicate the significance of the difference from the corresponding wild-type values determined by Student's t test (* 0.01 ≤ P < 0.05, ** P < 0.01). Three independent experiments were conducted and similar results were obtained.
Figure 8.
Response of rha2a and 35S:RHA2a plants to Glc. A, Quantification of seed germination. Seed germination percentage of the indicated genotypes grown on different concentrations of Glc was recorded at 3 d after the end of stratification. Data show the mean ± sd of three replicates. At least 100 seeds per genotype were measured in each replicate. B, Quantification of cotyledon greening. Cotyledon-greening percentage of the indicated genotypes on medium containing different concentrations of Glc was recorded at 5 d after the end of stratification. Data show the mean ± sd of three replicates. At least 100 seeds per genotype were measured in each replicate. C, Quantification of chlorophyll content of seedlings described in B. Values represent the mean ± sd of three replicates. FW, Fresh weight of whole seedlings. Asterisks in C indicate the significance of the difference from the corresponding wild-type values determined by Student's t test (* 0.01 ≤ P < 0.05, ** P < 0.01). Three independent experiments were conducted and similar results were obtained.
Figure 9.
Response of rha2a and 35S:RHA2a plants to mannitol. A, Quantification of seed germination. Seed germination percentage of the indicated genotypes grown on medium containing different concentrations of mannitol was recorded at 3 d after the end of stratification. Data show the mean ± sd of three replicates. At least 100 seeds per genotype were measured in each replicate. B, Quantification of cotyledon greening. Cotyledon-greening percentage of the indicated genotypes on medium containing different concentrations of mannitol was recorded at 5 d after the end of stratification. Data show the mean ± sd of three replicates. At least 100 seeds per genotype were measured in each replicate. C, Quantification of chlorophyll content of seedlings described in B. Values represent the mean ± sd of three replicates. FW, Fresh weight of whole seedlings. Asterisks in C indicate the significance of the difference from the corresponding wild-type values determined by Student's t test (** P < 0.01). Three independent experiments were conducted and similar results were obtained.
Salt and Osmotic Responses of rha2a and 35S:RHA2a Plants during Early Seedling Development
rha2a, 35S:RHA2a, and wild-type plants were also investigated for their responses to NaCl, Glc, and mannitol during early seedling development. For these experiments, germinated seeds were transferred 24 h after stratification from control medium to osmotic stress-containing medium (see “Materials and Methods”). In our cotyledon-greening assays, rha2a seedlings were less sensitive than wild-type seedlings to NaCl (Fig. 7B), Glc (Fig. 8B), and mannitol (Fig. 9B). On the contrary, seedlings of the 35S:RHA2a lines were more sensitive than wild-type seedlings to NaCl (Fig. 7B), Glc (Fig. 8B), and mannitol (Fig. 9B). Similar results were obtained when measuring chlorophyll accumulation (Figs. 7C, 8C, and 9C). These results suggested that RHA2a also positively regulates osmotic stress responses during early seedling development.
The RHA2a Protein Is a Functional E3 Ligase
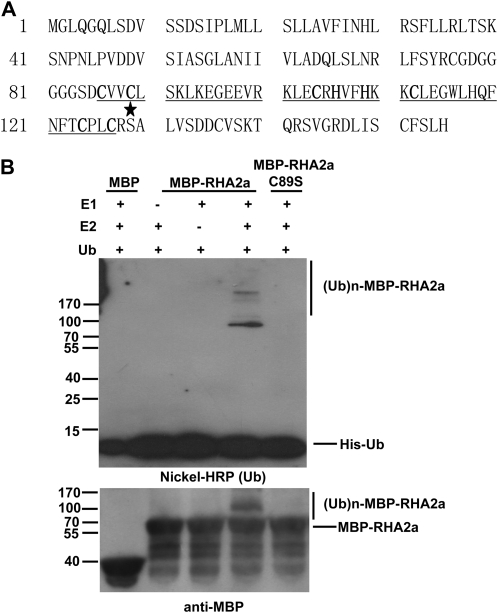
The C terminus of RHA2a contains a C3H2C3-type RING domain with conserved Cys and His residues (Fig. 10A). An increasing body of evidence indicates that RING motif-containing proteins can act as an active E3 ligase and function in a broad range of biological processes (Xie et al., 2002; Seo et al., 2003; Zhang et al., 2005, 2007; Dong et al., 2006; Stone et al., 2006; Qin et al., 2008). To examine whether RHA2a has E3 activity, we conducted in vitro autoubiquitination assays. RHA2a was expressed in Escherichia coli as a fusion protein with maltose binding protein (MBP) and affinity purified from the soluble fraction. In the presence of wheat (Triticum aestivum) E1 and a human E2 (UbcH5B) enzyme, purified MBP-RHA2a was able to execute autoubiquitination, as evidenced by the formation of large molecular mass proteins detected by immunoblot analyses using anti-His (Fig. 10B, top) or anti-MBP (Fig. 10B, bottom) antibodies. As negative controls, when E1 or E2 was omitted from the reaction, no polyubiquitination of MBP-RHA2a was detected (Fig. 10B, lanes 2 and 3 from the left). These results indicated that RHA2a has E3 ligase activity. To test whether an intact RING finger domain is required for the E3 ligase activity of RHA2a, a single amino acid substitution allele named RHA2aC89S, in which Cys-89 mutated to Ser-89, was produced (Fig. 10A), as this mutation might disrupt the RING domain (Xie et al., 2002; Stone et al., 2006). In vitro ubiquitination assays indicated that the E3 ligase activity was completely abolished in RHA2aC89S (Fig. 10B, lane 5 from the left), demonstrating that an intact RING domain is required for the E3 ligase activity of RHA2a.
Figure 10.
E3 ubiquitin ligase activity of RHA2a. A, Amino acid sequence of RHA2a. The RING finger domain is underlined, the conserved Cys and His residues are shown in boldface, and the star indicates Cys-89 changed to Ser-89 in RHA2aC89S, a mutant version of RHA2a. B, MBP-RHA2a and its mutant form MBP-RHA2aC89S fusion proteins were assayed for E3 activity in the presence of E1 (from wheat), E2 (UbcH5B), and His-tagged ubiquitin (Ub). MBP itself was used as a negative control (left lane). RHA2aC89S lost E3 ligase activity (right lane). The numbers on the left denote the molecular masses of marker proteins in kilodaltons. The nickel-nitrilotriacetic acid conjugated to horseradish peroxidase (HRP; KPL) was used to detect His-tagged ubiquitin (top), and the anti-MBP antibody was used to detect maltose fusion proteins (bottom).
Furthermore, we generated transgenic lines overexpressing RHA2aC89S directed by the 35S promoter. As shown in Supplemental Figure S6A, the expression level of RHA2aC89S in lines 35S:RHA2aC89S 1# and 35S:RHA2aC89S 2# was comparable to the expression level of RHA2a in the above-described 35S:RHA2a 7# plants. ABA response assays indicated that, in contrast to 35S:RHA2a 7# plants, which were hypersensitive to ABA, the response of 35S:RHA2aC89S plants to ABA was essentially similar to that of the wild type (Supplemental Fig. S6, B–D). These results suggested that the RING domain is important for the biological function of RHA2a in ABA signaling.
RHA2a Physically Interacts with the NAC Transcription Factors ANAC019 and ANAC055
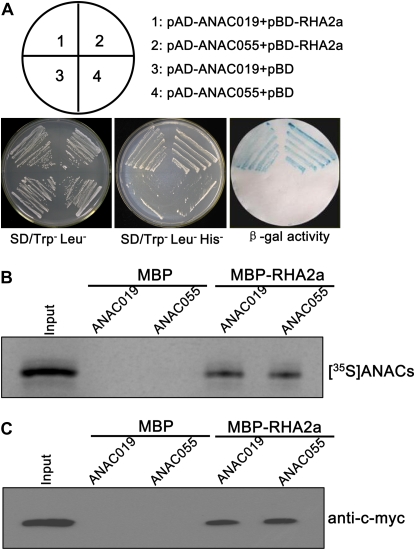
A previous yeast two-hybrid screen identified a NAC family protein (At1g52890) named ANAC (for ABA-inducible NAC) as an interacting protein of RHA2a (Greve et al., 2003). Based on the nomenclature established for the NAC family proteins in Arabidopsis (Ooka et al., 2003; Olsen et al., 2005), ANAC was renamed as ANAC019. Our yeast two-hybrid assays indicated that, in addition to ANAC019, RHA2a was also able to interact with ANAC055 (At3g15500; Fig. 11A). ANAC019 and ANAC055 represent the most closely related members among the NAC family proteins in Arabidopsis (Tran et al., 2004).
Figure 11.
RHA2a interacts with ANAC019 and ANAC055. A, RHA2a interacts with ANAC019 and ANAC055 in yeast. Full-length ANAC019 or ANAC055 was cloned into pGADT7 to generate pAD-ANAC019 or pAD-ANAC055. Full-length RHA2a was cloned into pGBKT7 to generate pBD-RHA2a. Yeast cells were cotransformed with a combination of the indicated plasmids and streaked on plates lacking Leu and Trp or Leu, Trp, and His. To corroborate the interactions between the two ANACs and RHA2a, β-galactosidase (β-gal) activity was assayed on replica filters. SD, Synthetic dextrose. B, In vitro pull-down assay. Purified MBP and the MBP-RHA2a fusion proteins were incubated with in vitro-translated [35S]ANAC019 and [35S]ANAC055. Bound proteins were pulled down with amylose resin, separated by 10% SDS-PAGE, and analyzed by autoradiography. Input proteins [35S]ANAC019 and [35S]ANAC055 exhibited the same size, and only [35S]ANAC019 is shown. C, In vitro pull-down assay. Purified MBP and MBP-RHA2a fusion proteins were incubated with the 6myc-ANAC019 and 6myc-ANAC055 expressed in N. benthamiana leaves. Bound proteins were pulled down with amylose resin, separated by 10% SDS-PAGE, and probed with anti-c-myc antibody. Input proteins 6myc-ANAC019 and 6myc-ANAC055 exhibited the same size, and only 6myc-ANAC019 is shown. [See online article for color version of this figure.]
The interaction of RHA2a with ANAC019 and ANAC055 was further confirmed by in vitro pull-down assays. MBP and the MBP-RHA2a fusion proteins were produced in E. coli and purified using amylose resin. Purified MBP-RHA2a or MBP was incubated with the in vitro-translated [35S]ANAC019 and [35S]ANAC055 polypeptides. As shown in Figure 11B, the MBP-RHA2a fusion protein, but not the MBP control, was able to pull down [35S]ANAC019 and [35S]ANAC055. In addition, purified MBP-RHA2a was also incubated with the 6myc-ANAC019 and 6myc-ANAC055 fusion proteins expressed in Nicotiana benthamiana leaves. Again, MBP-RHA2a, but not MBP alone, was able to pull down the 6myc-ANAC019 or the 6myc-ANAC055 fusion protein in the presence of the proteasome-specific inhibitor MG132 (Fig. 11C). These results provided further evidence that RHA2a physically interacts with the two NAC proteins.
DISCUSSION
The RHA2a Gene Product Plays an Important Role in ABA Signaling during Seed Germination and Early Seedling Development
We demonstrate here that the RHA2a gene, which encodes a deduced C3H2C3-type RING finger protein, plays an important role in ABA signaling. ABA response assays indicated that while the rha2a mutant and RHA2a-RNAi plants were less sensitive to ABA, 35S:RHA2a plants were more sensitive, suggesting that RHA2a positively regulates ABA signaling during seed germination and early seedling development. Our findings that down-regulation or overexpression of RHA2a did not significantly affect leaf water loss or ABA-induced marker gene expression support the idea that RHA2a may be an ABA signaling component specifically effective during seed germination and early stages of seedling development.
Among the known positive regulators of ABA signaling, ABI5 encodes a bZIP transcription factor whose accumulation inhibits seed germination and early seedling establishment (Finkelstein and Lynch, 2000; Lopez-Molina et al., 2001, 2002). Three lines of evidence suggested that the action of the RHA2a gene in ABA signaling may be in parallel with that of ABI5. First, overexpression of RHA2a in the ABA-insensitive abi5-7 mutant led to an ABA-hypersensitive phenotype (Fig. 4, A–C). Second, overexpression of ABI5 in the genetic background of the rha2a mutant also led to an ABA-hypersensitive phenotype (Fig. 4, D–F). Finally, the rha2a abi5-7 double mutant was significantly more insensitive to ABA than the rha2a and abi5-7 single mutants (Fig. 4, G–I).
The ABI3 protein had been shown to interact with ABI5 in a yeast two-hybrid assay (Nakamura et al., 2001). Previous genetic analyses revealed that ABI5 acts downstream of ABI3 in the same pathway to execute ABA-mediated regulation of seed germination and early seedling development (Lopez-Molina et al., 2002). In this context, our finding that RHA2a acts in parallel with ABI5 (Fig. 4) suggests that the function of RHA2a may also be in parallel with ABI3. Indeed, our ABA response assays indicated that the rha2a abi3-8 double mutant is also significantly more insensitive to ABA than the single mutants (Fig. 5). Therefore, our data suggest that RHA2a acts in parallel with the ABI3-ABI5 pathway in ABA signaling.
In addition to ABI3 and ABI5, the ABI4 gene, which encodes an AP2-type transcription factor, also plays an important role in ABA-regulated seed germination (Finkelstein et al., 1998). However, relatively less is known regarding the physical and genetic interactions of ABI4 with ABI3 and ABI5 (Finkelstein et al., 2002). Nevertheless, our ABA response assays showing that the rha2a abi4-1 double mutant was significantly more insensitive than the single mutants to ABA suggest that the action of RHA2a in ABA signaling is also in parallel with that of ABI4 (Fig. 6). Together, these data support the idea that RHA2a is an important positive regulator of ABA signaling during seed germination and early seedling development and that the action of RHA2a in ABA signaling is independent of that of the ABI transcription factor genes, including ABI3, ABI4, and ABI5.
RHA2a Is a Functional E3 Ligase
Prior to this work, several studies have postulated that RHA2a may encode a C3H2C3-type RING finger E3 ligase (Greve et al., 2003; Stone et al., 2005). Our analyses here demonstrate that the RHA2a protein is indeed an active E3 ligase based on the occurrence of autoubiquitination of the MBP-RHA2a fusion protein in the presence of the E1 and E2 enzymes (Fig. 10B). Thus, RHA2a joins SDIR1 as positive regulators of ABA signaling as active C3H2C3-type E3 ligases (Zhang et al., 2007; this work). However, RHA2a and SDIR1 differ from each other in their functional relationship with ABI5. While the former acts in parallel with ABI5 (this work), the latter functions upstream of ABI5 (Zhang et al., 2007). The XERICO gene, which is predicted to encode a small protein (162 amino acids) containing a RING-H2 zinc finger motif, has been genetically characterized to be a positive regulator of ABA signaling for drought tolerance in Arabidopsis (Ko et al., 2006). However, an E3 ligase activity remains to be demonstrated for the protein product of XERICO (Ko et al., 2006). In contrast to RHA2a, SDIR1, and XERICO, two novel E3 ligases (AIP2 and KEG) have recently been found to be negative regulators in several aspects of ABA signaling (Zhang et al., 2005; Stone et al., 2006). AIP2 contains a RING motif and targets ABI3 for posttranslational degradation (Zhang et al., 2005; Stone et al., 2006). KEG has a complex structure composed of a RING-HCa motif, a kinase domain, ankyrin repeats, and HERC2-like repeats and is required for keeping the ABI5 protein at a low level under normal growth conditions (Stone et al., 2006).
Based on our results, we hypothesize that RHA2a functions as an E3 ligase that mediates the degradation of its substrate(s) (yet to be identified) through the ubiquitin-proteasome machinery (Zhang et al., 2005; Stone et al., 2006). Given that RHA2a itself positively regulates ABA signaling, we propose that the degraded proteins by RHA2a are negative regulators of ABA signaling and that removing these molecules has the effect of activating ABA signaling. In a previous study, RHA2a was shown to interact with the NAC family protein ANAC019 in yeast two-hybrid assays (Greve et al., 2003). Our yeast two-hybrid assays indicated that in addition to ANAC019, RHA2a also interacts with ANAC055, which shows high sequence similarity to ANAC019 (Fig. 11A). The physical interaction of RHA2a with ANAC019 and ANAC055 was further confirmed by in vitro pull-down assays. Previous work has elegantly demonstrated that ANAC019 and ANAC055 are functional transcription activators for ABA- and dehydration-responsive gene expression (Tran et al., 2004). Given that RHA2a is a functional E3 ligase, it will be interesting to test weather ANAC019 and ANAC055 are substrates of RHA2a in ABA signaling. Further studies are required to examine the physiological relevance of the interaction between RHA2a and the two NAC proteins in ABA signaling.
Alternatively, RHA2a could activate positive regulators by monoubiquitination and stabilize some key regulators of the ABA signaling pathway. Could these positive regulators be ABI3, ABI4, or ABI5? If RHA2a modifies ABI5, we would expect that the ABI5 locus is epistatic to the RHA2a locus in ABA signaling. However, our genetic analyses indicated that overexpression of RHA2a in the ABA-insensitive abi5-7 mutant leads to an ABA-hypersensitive phenotype (Fig. 4, A–C) and that the rha2a abi5-7 double mutant is significantly more insensitive to ABA than the single mutants (Fig. 4, G–J). Similarly, our data indicated that the rha2a abi3-8 double mutant is significantly more insensitive to ABA than the single mutants (Fig. 5) and that the rha2a abi4-1 double mutant is significantly more insensitive to ABA than the single mutants (Fig. 6). These data do not favor the hypothesis that RHA2a activates ABA signaling through modification of ABI5, ABI3, or ABI4. Thus, further functional studies of RHA2a, its target protein(s), and their interplay are necessary for complete understanding of the ABA signaling networks in plants.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana ecotype Columbia [Col-0]) was used as the wild type in this study. Seeds of each genotype were surface sterilized with 10% bleach and 0.001% Triton X-100 for 10 min and washed three times with sterile water. Sterilized seeds were then suspended in 0.2% agarose and plated on Murashige and Skoog (MS) medium. Plants were stratified at 4°C in darkness for 3 d and then transferred to a Phytotron set at 22°C with a 16-h-light/8-h-dark photoperiod (light intensity of 120 μmol m−2 s−1). After 2 to 3 weeks, seedlings were also potted in soil and placed in a growth room at 22°C with a 16-h-light/8-h-dark cycle (light intensity of 120 μmol m−2 s−1).
Mutant Identification of RHA2a, ABI5, ABI3, and ABI4
A putative homozygous T-DNA insertion mutant line (GABI-kat 126H03, N378380) of RHA2a (At1g15100) was obtained from the Nottingham Arabidopsis Stock Centre at the University of Nottingham. N378380 seedlings show 100% resistance to sulfadiazine. Diagnostic PCR analyses with gene-specific primers 5′-GGAAACCAAAGCAGATCTACAAAG-3′ and 5′-ACACTGAAAGTGAAAACTAACGGG-3′ in combination with the T-DNA primer 5′-ATATTGACCATCATACTCATTGC-3′ indicated that line N378380 contains a T-DNA inserted between nucleotides 12 and 13 of RHA2a (Fig. 2A). Homozygous N378380 was named rha2a in this study. RT-PCR analysis using primer pair P3 (5′-TCTCTCTCCTCGCCGTCTTC-3′) and P4 (5′-TCCAGCTTCCTCACCTCTTCA-3′) indicated that the T-DNA insertion led to increased levels of RHA2a mRNA fragment (Fig. 2B). However, no RHA2a mRNA was detected in the rha2a mutant when the primers P1 (5′-ACTGGATCCAAGATGGGGCTACAAGGTCAG-3′) and P2 (5′-ACTGAGCTCTACTCAGTGGAGAGAGAAAC-3′) that span the T-DNA insertion were used (Fig. 2B). A possible explanation is that the T-DNA insertion leads to transcription activation of a nonfunctional RHA2a fragment. The pAC161 used to generate GABI-Kat lines contains the 35S CaMV promoter, which could act as an activation tagging element after T-DNA integration (Rosso et al., 2003). Genomic DNA of RHA2a driven by its native promoter rescues the ABA-insensitive phenotype of rha2a (Supplemental Fig. S1).
The ABA-insensitive mutants abi5-7 (Nambara et al., 2002; Tamura et al., 2006), abi4-1 (Finkelstein et al., 1998), and abi3-8 (Nambara et al., 2002; Tamura et al., 2006) were used to generate double mutants with rha2a. Homozygous rha2a and abi5-7 were crossed, and putative double mutant plants were screened from the resulting F2 progeny by their insensitivity to ABA (2 μm) during seed germination and early seedling development. A rha2a abi5-7 double mutant line was identified by PCR-based genotyping of the RHA2a locus and sequence confirmation of the ABI5 locus (Nambara et al., 2002; Tamura et al., 2006).
Similarly, we generated rha2a abi3-8 and rha2a abi4-1 double mutant lines. Identification of the abi3-8 mutation (Nambara et al., 2002; Tamura et al., 2006) and the abi4-1 mutation (Finkelstein et al., 1998) was performed as described previously.
Generation of Transgenic Plants
To get RNAi plants of RHA2a, a 319-bp fragment of the RHA2a cDNA was PCR amplified with the primers 5′-ACTGGATCCAAGATGGGGCTACAAGGTCAG-3′ and 5′-CGTGTCGACATTCCAGCTTCCTCACC-3′. This fragment was sequentially cloned into the BamHI/SalI and BglII/XhoI sites of the pUC-RNAi vector (Luo et al., 2006) to target the gene in both the sense and antisense orientations. Finally, the resulting construct was cloned into the pCAMBIA2300-35S vector for plant transformation. Homozygous RHA2a-RNAi plants were identified and verified for reduced endogenous RHA2a expression by RNA gel blot analysis (Fig. 2C). RHA2a-RNAi 4# was used for the ABA response assay. RT-PCR analysis indicated that the expression of RHA2b, a RING finger gene most closely related to RHA2a (Stone et al., 2005), was not affected in RHA2a-RNAi 4# (Fig. 2C).
To complement the rha2a mutant, a 3,110-bp genomic DNA fragment containing the entire RHA2a coding region, a 1,765-bp upstream sequence, and an 874-bp downstream sequence was PCR amplified with the primers 5′-ACTAAGCTTTCTCTCCCGGTAATGGATGC-3′ and 5′-AGAGAATTCCTCGTCATTCGGAGCCAA-3′ using Col-0 genomic DNA as a template. The resulting fragments were cloned into the HindIII and EcoRI sites of the binary vector pCAMBIA1300 for transformation of the rha2a mutant plants.
For overexpression of RHA2a, the RHA2a coding sequence was PCR amplified with the primers 5′-ACTGGATCCAAGATGGGGCTACAAGGTCAG-3′ and 5′-ACTGAGCTCTACTCAGTGGAGAGAGAAAC-3′ using Col-0 genomic DNA as a template. The resulting PCR product was cloned into the BamHI and SacI sites of the binary vector pBI121 under the control of the CaMV 35S promoter. The resulting 35S:RHA2a construct was introduced into Col-0 and the ABA-insensitive mutants abi3-8, abi4-1, and abi5-7. Similarly, RHA2aC89S, a mutant version of RHA2a (see below), was also cloned into the BamHI and SacI sites of the binary vector pBI121 under the control of the 35S promoter.
For overexpression of ABI5, the coding region of ABI5 was fused with the myc tag in the N terminus and cloned into the pCAMBIA1300-221 vector under the control of the 35S promoter (Zhang et al., 2007). The resulting 35S:myc-ABI5 construct was transformed into Col-0 and the rha2a mutant to get Col-0/35S:ABI5 or rha2a/35S:ABI5 transgenic plants.
For RHA2a promoter:GUS fusion, DNA fragments covering 2,360 bp upstream of the translational start site of RHA2a were amplified by PCR with the primers 5′-GGAAGCTTTCCAACTGTCATATATAACCCC-3′ and 5′-GGGGATCCTCTTCTTCTTCTCTCTCTGTTTTC-3′ using genomic DNA as a template. This fragment was cloned into the HindIII and BamHI sites of the binary vector pBI121 containing a GUS reporter. GUS staining assay was done essentially as described previously (Zheng et al., 2006).
Transformation of Arabidopsis plants was performed by the floral dip infiltration method (Bechtold and Pelletier, 1998) using Agrobacterium tumefaciens strain GV3101 (pMP90). T2 seeds from each of the selected transgenic plants were plated on germination medium containing 50 μg mL−1 kanamycin (for pBI121 and pCAMBIA2300-35S) or 20 μg mL−1 hygromycin (for pCAMBIA1300) as selection antibiotics, and the homozygous lines were selected. Homozygous T3 progeny were then examined for expression levels of the target genes by RNA gel blot analysis. Progeny from the homozygous T3 plants were used for ABA response assays.
ABA-Mediated Inhibition of Seed Germination and Early Seedling Development Assays
Seed Germination
Plants of different genotypes were grown in the same conditions, and seeds were collected at the same time. For each comparison, seeds were planted on the same plate containing MS medium (0.5× MS salts, 1% Suc, and 0.8% agar) without or with different concentrations of ABA or other osmotic stresses as indicated. Plates were chilled at 4°C in the dark for 3 d (stratified) and moved to 22°C with a 16-h-light/8-h-dark cycle. The percentage of seed germination was scored at the indicated times. Germination was defined as an obvious emergence of the radicle through the seed coat.
Cotyledon Greening
To study the effect of ABA on cotyledon greening, seeds were sown on ABA-free medium as described above. Twenty-four hours after stratification, germinated seeds were transferred to medium containing different concentrations of ABA or other osmotic agents (NaCl, Glc, and mannitol) as indicated. The percentage of cotyledon greening was recorded at 5 d after the end of stratification. Cotyledon greening is defined as obvious cotyledon expansion and turning green.
The effect of ABA or other osmotic agents on cotyledon greening was also quantified by measurement of chlorophyll content of the seedlings as follows: after recording cotyledon-greening percentage, the seedlings were harvested, weighed, and ground into fine powder in liquid nitrogen for chlorophyll extraction (Lichtenthaler, 1987). Chlorophyll a/b contents were determined according to a described method (Lichtenthaler, 1987).
Seedling Root Growth
To study the effect of ABA on seedling root growth, seeds were sown on ABA-free medium as described above. Twenty-four hours after stratification, germinated seeds were transferred to medium containing different concentrations of ABA. Plates were placed vertically in a growth chamber, and root growth was measured at 5 d after the end of stratification.
Leaf Water Loss Assays
For leaf water loss measurements, fully expanded leaves were removed from 4-week-old plants and incubated under the same conditions used for seedling growth, and each sample (consisting of four individual leaves) was weighed at the indicated times.
Gene Expression Analyses
For RNA gel blot analysis, total RNA from vegetative tissues was prepared by a guanidine thiocyanate extraction method (Zheng et al., 2006). Total RNA was separated on an agarose gel containing 10% formaldehyde, blotted onto a Hybond N+ membrane (Amersham), and probed with the PCR-amplified DNA fragments using the gene-specific primers. Probes were labeled with [α-32P]dCTP using random primer labeling reagents (Pharmacia).
For RT-PCR analysis, total RNA from 14-d-old seedlings was extracted by the TRIzol method. Reverse transcription was performed using 5 μg of total RNA and M-MLV reverse transcriptase (Promega). The cDNA was then used for PCR amplification. For RHA2a, the above-described primers P1/P2 and P3/P4 were used. For RHA2b, the primers were 5′-GGTCTAGAATGGGACTACAAGGTCAGCTC-3′ and 5′-GGGGTACCATGAGATGATGCAGTAGAGGT-3′. For ACTIN2, the primers were 5′-TTGACTACGAGCAGGAGATGG-3′ and 5′-ACAAACGAGGGCTGGAACAAG-3′.
For qRT-PCR analysis, total RNA was isolated from seeds during germination at the indicated times as described previously (Soderman et al., 2000). Poly(dT) cDNA was prepared from 5 μg of total RNA with SuperScript III reverse transcriptase (Invitrogen) and quantified with a cycler apparatus (Bio-Rad) with the qPCR core kit for SYBR Green I (Tiangen). PCR was performed on 96-well optical reaction plates heated for 5 min to 95°C to activate hot-start Taq DNA polymerase, followed by 40 cycles of denaturation for 60 s at 95°C and annealing/extension for 60 s at 59°C. Targets were quantified with gene-specific primer pairs designed with the Beacon Designer 4.0 (Premier Biosoft International). Expression levels were normalized to that of ACTIN2. For RHA2a, the primers were 5′-TCTCTCTCCTCGCCGTCTTC-3′ and 5′-TCCAGCTTCCTCACCTCTTCA-3′. For ABI5, the primers were 5′-TGGTAACTAGAGAAACGAAG-3′ and 5′-TTACTACTACTACTACGTCC-3′. For RD29A (At5g52310), the primers were 5′-ATCACTTGGCTCCACTGTTGTTC-3′ and 5′-ACAAAACACACATAAACATCCAAAGT-3′. For NCED3 (At3g14440), the primers were 5′-TGGCTTCTTTCACGGCAAC-3′ and 5′-CAATGGCGGGAGAGTTTGA-3′. For RAB18 (At5g66400), the primers were 5′-CAGCAGCAGTATGACGAGTA-3′ and 5′-CAGTTCCAAAGCCTTCAGTC-3′. For KIN1 (At5g15960), the primers were 5′-ACCAACAAGAATGCCTTCCA-3′ and 5′-CCGCATCCGATACACTCTTT-3′. For ACTIN2, the primers were 5′-TTGACTACGAGCAGGAGATGG-3′ and 5′-ACAAACGAGGGCTGGAACAAG-3′. All qRT-PCR experiments were done in triplicate.
Expression of MBP-RHA2a Fusion Protein and Ubiquitination Assays
To generate MBP-RHA2a, the coding sequence of RHA2a was amplified with the primers 5′-ACTGAATTCAAGATGGGGCTACAAGGTCAG-3′ and 5′-ACTGGATCCGTGGAGAGAGAAACACGAGA-3′ and cloned into the EcoRI and BamHI sites of pMal-c2 (New England Biolabs). MBP-RHA2a fusion proteins were prepared following the manufacturer's instructions.
Recombinant proteins of wheat (Triticum aestivum) E1 (GI: 136632) and human E2 (UbcH5B) were prepared based on a published method (Xie et al., 2002). The Arabidopsis UBQ14 gene (At4g02890) contains tandem repeats of 228 bp that encode a ubiquitin monomer. The 228-bp ubiquitin monomer cDNA was cloned into the His tag-containing vector pET-28a (Novagen) to generate the His-ubiquitin monomer construct. Crude extracts containing recombinant wheat E1, human UbcH5B (E2; approximately 40 ng), purified MBP-RHA2a (E3; approximately 1 μg), and purified UBQ14-His (approximately 2 μg) were used for E3 ubiquitin ligase activity assay as described (Xie et al., 2002; Zhang et al., 2007). RHA2aC89S, a mutant version of RHA2a, in which Cys-89 was mutated to Ser-89, was generated using the Quick-Change site-directed mutagenesis kit (Stratagene). After reaction, proteins were separated by SDS-PAGE, blotted, and probed by the His Detector nickel-nitrilotriacetic acid conjugated to horseradish peroxidase (KPL) for detection of His-tagged ubiquitin; the blot was also probed by MBP antibody (New England Biolabs) and visualized using chemiluminescence as instructed by the manufacturer (Millipore).
In Vitro Translation of the NAC Proteins
The coding sequences of ANAC019 and ANAC055 were cloned into the BamHI and SacI sites of the Luciferase SP6 Control DNA (Promega) to generate the SP6-ANAC019 and SP6-ANAC055 constructs for in vitro translation. The primers used to amplify ANAC019 were 5′-CGAGGATCCATGGGTATCCAAGAAACTGAC-3′ and 5′-GATGAGCTCTCACATAAACCCAAACCCACC-3′, and the primers used to amplify ANAC055 were 5′-TACGGATCCATGGGTCTCCAAGAGCTTGAC-3′ and 5′-TGTGAGCTCTCAAATAAACCCGAACCCACT-3′. [35S]Met-labeled ANAC019 and ANAC055 were generated by in vitro translation with Rabbit Reticulocyte Lysate using a SP6-coupled TnT kit (Promega).
A. tumefaciens-Mediated Protein Transient Expression of the NAC Proteins
6myc-ANAC019 and 6myc-ANAC055 fusion proteins were transiently expressed in Nicotiana benthamiana leaves by agroinfiltration. The coding sequence of ANAC019 was amplified with the primers 5′-CGAGGATCCATGGGTATCCAAGAAACTGAC-3′ and 5′-GATGAGCTCTCACATAAACCCAAACCCACC-3′, whereas the coding sequence of ANAC055 was amplified with the primers 5′-TACGGATCCATGGGTCTCCAAGAGCTTGAC-3′ and 5′-TGTGAGCTCTCAAATAAACCCGAACCCACT-3′. These DNA fragments were cloned into the BamHI and SacI sites of the myc-pBA vector, respectively. A. tumefaciens strain GV3101 (pMP90) carrying 35S:6myc-ANAC019 or 35S:6myc-ANAC055 was infiltrated into leaves of N. benthamiana using described methods (Day et al., 2006).
In Vitro Pull-Down Assay
MBP-RHA2a fusions on amylose resin beads (approximately 1 μg) were incubated with 10 μL of in vitro translation mix of the NAC proteins in 500 μL of immunoprecipitation (IP) buffer at 4°C for 2 h. After incubation, the beads were washed five times with 1 mL of IP buffer. MBP-RHA2a fusions on amylose resin beads (approximately 1 μg) were also incubated with 1 mL of supernatant of the transiently expressed 6myc-ANAC protein lysates for 2 h at 4°C. After incubation, the beads were washed five times with 1 mL of IP buffer. The bound proteins were then resuspended in 50 μL of 1× SDS loading buffer, separated by 10% SDS-PAGE, and detected by immunoblot analysis using anti-c-myc antibody (Santa Cruz Biotechnology). The purified MBP was used as a negative control.
Yeast Assays
The full-length coding sequence of RHA2a was PCR amplified with a forward EcoRI linker primer (5′-ACTGAATTCAAGATGGGGCTACAAGGTCAG-3′) and a reverse BamHI linker primer (5′-ACTGGATCCGTGGAGAGAGAAACACGAGA-3′) and cloned into the Gal4 DNA-binding domain in the pGBKT7 vector. Full-length ANAC019 was PCR amplified with the primers 5′-CGAGGATCCATATGGGTATCCAAGAAACTG-3′ and 5′-GATGAGCTCTCACATAAACCCAAACCCACC-3′ and cloned into the BamHI and SacI sites of the pGADT7 vector to generate pAD-ANAC019. Full-length ANAC055 was PCR amplified with the primers 5′-TACGAATTCATGGGTCTCCAAGAGCTTGAC-3′ and 5′-TACGGATCCAATAAACCCGAACCCACTAGA-3′ and cloned into the EcoRI and BamHI sites of the pGADT7 vector to generate pAD-ANAC055. Yeast two-hybrid assay was performed using the yeast strain HF7c as described (Xie et al., 2002).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: RHA2a, At1g15100; ABI5, At2g36270; ABI3, At3g24650; ABI4, At2g40220.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Complementation of the rha2a mutant.
Supplemental Figure S2. Effects of fluridone on the ABA responses of different genotypes.
Supplemental Figure S3. Leaf water loss assay.
Supplemental Figure S4. ABA response analysis of RHA2a overexpression in abi3-8.
Supplemental Figure S5. ABA response analysis of RHA2a overexpression in abi4-1.
Supplemental Figure S6. Transgenic plants overexpressing RHA2C89S (35S:RHA2aC89S) do not show the ABA-hypersensitive phenotype.
Supplementary Material
Acknowledgments
We thank Professor Yongbiao Xue (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing) for critical reading of the manuscript. We thank the Arabidopsis Biological Resource Center and the Nottingham Arabidopsis Stock Centre for providing T-DNA insertion mutants. We thank Dr. Eiji Nambara (RIKEN Plant Science Center, Japan) for providing the abi5-7 and abi3-8 mutants.
This work was supported by the Knowledge Innovation Program of the Chinese Academy of Sciences (grant no. KSCX2–YW–N–045), the Ministry of Science and Technology of China (grant no. 2006CB910604), and the National Natural Science Foundation of China (grant nos. 30530440 and 90717007).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Chuanyou Li (cyli@genetics.ac.cn).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Allen GJ, Kuchitsu K, Chu SP, Murata Y, Schroeder JI (1999) Arabidopsis abi1-1 and abi2-1 phosphatase mutations reduce abscisic acid-induced cytoplasmic calcium rises in guard cells. Plant Cell 11 1785–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assmann SM (2003) OPEN STOMATA1 opens the door to ABA signaling in Arabidopsis guard cells. Trends Plant Sci 8 151–153 [DOI] [PubMed] [Google Scholar]
- Bechtold N, Pelletier G (1998) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol 82 259–266 [DOI] [PubMed] [Google Scholar]
- Callis J, Vierstra RD (2000) Protein degradation in signaling. Curr Opin Plant Biol 3 381–386 [DOI] [PubMed] [Google Scholar]
- Chen Y, Ji FF, Xie H, Liang JS, Zhang JH (2006) The regulator of G-protein signaling proteins involved in sugar and abscisic acid signaling in Arabidopsis seed germination. Plant Physiol 140 302–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow B, McCourt P (2004) Hormone signalling from a developmental context. J Exp Bot 55 247–251 [DOI] [PubMed] [Google Scholar]
- Christmann A, Moes D, Himmelbach A, Yang Y, Tang Y, Grill E (2006) Integration of abscisic acid signalling into plant responses. Plant Biol (Stuttg) 8 314–325 [DOI] [PubMed] [Google Scholar]
- Day B, Dahlbeck D, Staskawicz BJ (2006) NDR1 interaction with RIN4 mediates the differential activation of multiple disease resistance pathways in Arabidopsis. Plant Cell 18 2782–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers BJ, Schuurmans JA, Smeekens SC (2004) Glucose delays seed germination in Arabidopsis thaliana. Planta 218 579–588 [DOI] [PubMed] [Google Scholar]
- Devoto A, Muskett PR, Shirasu K (2003) Role of ubiquitination in the regulation of plant defence against pathogens. Curr Opin Plant Biol 6 307–311 [DOI] [PubMed] [Google Scholar]
- Dong CH, Agarwal M, Zhang Y, Xie Q, Zhu JK (2006) The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc Natl Acad Sci USA 103 8281–8286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher K, Callis J (2007) Ubiquitin, hormones and biotic stress in plants. Ann Bot (Lond) 99 787–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S, Weissman AM (2004) A field guide to ubiquitylation. Cell Mol Life Sci 61 1546–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell (Suppl) 14 S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Wang ML, Lynch TJ, Rao S, Goodman HM (1998) The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell 10 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM (1992) Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greve K, La Cour T, Jensen MK, Poulsen FM, Skriver K (2003) Interactions between plant RING-H2 and plant-specific NAC (NAM/ATAF1/2/CUC2) proteins: RING-H2 molecular specificity and cellular localization. Biochem J 371 97–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegedus D, Yu M, Baldwin D, Gruber M, Sharpe A, Parkin I, Whitwill S, Lydiate D (2003) Molecular characterization of Brassica napus NAC domain transcriptional activators induced in response to biotic and abiotic stress. Plant Mol Biol 53 383–397 [DOI] [PubMed] [Google Scholar]
- Hellmann H, Estelle M (2002) Plant development: regulation by protein degradation. Science 297 793–797 [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67 425–479 [DOI] [PubMed] [Google Scholar]
- Himmelbach A, Yang Y, Grill E (2003) Relay and control of abscisic acid signaling. Curr Opin Plant Biol 6 470–479 [DOI] [PubMed] [Google Scholar]
- Hoecker U (2005) Regulated proteolysis in light signaling. Curr Opin Plant Biol 8 469–476 [DOI] [PubMed] [Google Scholar]
- Huq E (2006) Degradation of negative regulators: a common theme in hormone and light signaling networks? Trends Plant Sci 11 4–7 [DOI] [PubMed] [Google Scholar]
- Jensen RB, Jensen KL, Jespersen HM, Skriver K (1998) Widespread occurrence of a highly conserved RING-H2 zinc finger motif in the model plant Arabidopsis thaliana. FEBS Lett 436 283–287 [DOI] [PubMed] [Google Scholar]
- Ko JH, Yang SH, Han KH (2006) Upregulation of an Arabidopsis RING-H2 gene, XERICO, confers drought tolerance through increased abscisic acid biosynthesis. Plant J 47 343–355 [DOI] [PubMed] [Google Scholar]
- Leung J, Bouvier-Durand M, Morris PC, Guerrier D, Chefdor F, Giraudat J (1994) Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science 264 1448–1452 [DOI] [PubMed] [Google Scholar]
- Leung J, Giraudat J (1998) Abscisic acid signal transduction. Annu Rev Plant Physiol Plant Mol Biol 49 199–222 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler H (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148 350–382 [Google Scholar]
- Lopez-Molina L, Chua NH (2000) A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol 41 541–547 [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Chua NH (2001) A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci USA 98 4782–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Kinoshita N, Chua NH (2003) AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev 17 410–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua NH (2002) ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J 32 317–328 [DOI] [PubMed] [Google Scholar]
- Luo A, Qian Q, Yin H, Liu X, Yin C, Lan Y, Tang J, Tang Z, Cao S, Wang X, et al (2006) EUI1, encoding a putative cytochrome P450 monooxygenase, regulates internode elongation by modulating gibberellin responses in rice. Plant Cell Physiol 47 181–191 [DOI] [PubMed] [Google Scholar]
- Moon J, Parry G, Estelle M (2004) The ubiquitin-proteasome pathway and plant development. Plant Cell 16 3181–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E (2005) Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J 41 697–709 [DOI] [PubMed] [Google Scholar]
- Nakamura S, Lynch TJ, Finkelstein RR (2001) Physical interactions between ABA response loci of Arabidopsis. Plant J 26 627–635 [DOI] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A (2003) ABA action and interactions in seeds. Trends Plant Sci 8 213–217 [DOI] [PubMed] [Google Scholar]
- Nambara E, Suzuki M, Abrams S, McCarty DR, Kamiya Y, McCourt P (2002) A screen for genes that function in abscisic acid signaling in Arabidopsis thaliana. Genetics 161 1247–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen AN, Ernst HA, Leggio LL, Skriver K (2005) NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 10 79–87 [DOI] [PubMed] [Google Scholar]
- Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, et al (2003) Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res 10 239–247 [DOI] [PubMed] [Google Scholar]
- Pandey S, Chen JG, Jones AM, Assmann SM (2006) G-protein complex mutants are hypersensitive to abscisic acid regulation of germination and postgermination development. Plant Physiol 141 243–256 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Qin F, Sakuma Y, Tran LS, Maruyama K, Kidokoro S, Fujita Y, Fujita M, Umezawa T, Sawano Y, Miyazono K, et al (2008) Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 20 1693–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez PL, Benning G, Grill E (1998) ABI2, a second protein phosphatase 2C involved in abscisic acid signal transduction in Arabidopsis. FEBS Lett 421 185–190 [DOI] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53 247–259 [DOI] [PubMed] [Google Scholar]
- Schwechheimer C, Schwager K (2004) Regulated proteolysis and plant development. Plant Cell Rep 23 353–364 [DOI] [PubMed] [Google Scholar]
- Seo HS, Yang JY, Ishikawa M, Bolle C, Ballesteros ML, Chua NH (2003) LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423 995–999 [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K (1997) Gene expression and signal transduction in water-stress response. Plant Physiol 115 327–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Kurepa J, Yang P, Emborg TJ, Babiychuk E, Kushnir S, Vierstra RD (2003) The pleiotropic role of the 26S proteasome subunit RPN10 in Arabidopsis growth and development supports a substrate-specific function in abscisic acid signaling. Plant Cell 15 965–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55 555–590 [DOI] [PubMed] [Google Scholar]
- Soderman EM, Brocard IM, Lynch TJ, Finkelstein RR (2000) Regulation and function of the Arabidopsis ABA-insensitive4 gene in seed and abscisic acid response signaling networks. Plant Physiol 124 1752–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Hauksdottir H, Troy A, Herschleb J, Kraft E, Callis J (2005) Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol 137 13–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Williams LA, Farmer LM, Vierstra RD, Callis J (2006) KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18 3415–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura N, Yoshida T, Tanaka A, Sasaki R, Bando A, Toh S, Lepiniec L, Kawakami N (2006) Isolation and characterization of high temperature-resistant germination mutants of Arabidopsis thaliana. Plant Cell Physiol 47 1081–1094 [DOI] [PubMed] [Google Scholar]
- Tatematsu K, Nakabayashi K, Kamiya Y, Nambara E (2008) Transcription factor AtTCP14 regulates embryonic growth potential during seed germination in Arabidopsis thaliana. Plant J 53 42–52 [DOI] [PubMed] [Google Scholar]
- Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early response to dehydration stress 1 promoter. Plant Cell 16 2481–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah H, Chen JG, Wang S, Jones AM (2002) Role of a heterotrimeric G protein in regulation of Arabidopsis seed germination. Plant Physiol 129 897–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Guo HS, Dallman G, Fang S, Weissman AM, Chua NH (2002) SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419 167–170 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57 781–803 [DOI] [PubMed] [Google Scholar]
- Zhang X, Garreton V, Chua NH (2005) The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev 19 1532–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yang C, Li Y, Zheng N, Chen H, Zhao Q, Gao T, Guo H, Xie Q (2007) SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell 19 1912–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Zhai Q, Sun J, Li CB, Zhang L, Li H, Zhang X, Li S, Xu Y, Jiang H, et al (2006) Bestatin, an inhibitor of aminopeptidases, provides a chemical genetics approach to dissect jasmonate signaling in Arabidopsis. Plant Physiol 141 1400–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.