Abstract
In pea (Pisum sativum), normal fruit growth requires the presence of the seeds. The coordination of growth between the seed and ovary tissues involves phytohormones; however, the specific mechanisms remain speculative. This study further explores the roles of the gibberellin (GA) biosynthesis and catabolism genes during pollination and fruit development and in seed and auxin regulation of pericarp growth. Pollination and fertilization events not only increase pericarp PsGA3ox1 message levels (codes for GA 3-oxidase that converts GA20 to bioactive GA1) but also reduce pericarp PsGA2ox1 mRNA levels (codes for GA 2-oxidase that mainly catabolizes GA20 to GA29), suggesting a concerted regulation to increase levels of bioactive GA1 following these events. 4-Chloroindole-3-acetic acid (4-Cl-IAA) was found to mimic the seeds in the stimulation of PsGA3ox1 and the repression of PsGA2ox1 mRNA levels as well as the stimulation of PsGA2ox2 mRNA levels (codes for GA 2-oxidase that mainly catabolizes GA1 to GA8) in pericarp at 2 to 3 d after anthesis, while the other endogenous pea auxin, IAA, did not. This GA gene expression profile suggests that both seeds and 4-Cl-IAA can stimulate the production, as well as modulate the half-life, of bioactive GA1, leading to initial fruit set and subsequent growth and development of the ovary. Consistent with these gene expression profiles, deseeded pericarps converted [14C]GA12 to [14C]GA1 only if treated with 4-Cl-IAA. These data further support the hypothesis that 4-Cl-IAA produced in the seeds is transported to the pericarp, where it differentially regulates the expression of pericarp GA biosynthesis and catabolism genes to modulate the level of bioactive GA1 required for initial fruit set and growth.
In pea (Pisum sativum), normal pericarp growth requires the presence of seeds. Removal or destruction of the seeds at 2 to 3 d after anthesis (DAA) results in the slowing of pericarp growth and subsequently abscission (Eeuwens and Schwabe, 1975; Ozga et al., 1992). Signaling molecules originating from the seeds may be responsible for continued fruit development by maintaining hormone levels in the surrounding tissue (Eeuwens and Schwabe, 1975; Sponsel, 1982). Developing pea seeds and pericarps contain gibberellins (GAs; Garcia-Martinez et al., 1991; Rodrigo et al., 1997) and auxins (4-chloroindole-3-acetic acid [4-Cl-IAA] and IAA; Marumo et al., 1968; Magnus et al., 1997). During early pericarp growth (2 DAA), application of the naturally occurring hormones 4-Cl-IAA (Reinecke et al., 1995) and GA (Eeuwens and Schwabe, 1975; Ozga and Reinecke, 1999) to deseeded pericarps can substitute for seeds and stimulate pericarp growth. However, the other naturally occurring auxin in pea fruit, IAA, was ineffective at promoting growth (Reinecke et al., 1995).
Studies comparing the growth-promoting properties of 4-, 5-, 6- and 7-Cl-IAAs and the corresponding F-IAA analogs (Reinecke et al., 1995) and the physicochemical properties of 4-Cl-IAA and 4-substituted analogs (Reinecke et al., 1999) found that the 4-position of the indole ring and the 4-substituents's size and lipophilicity were required for significant biological activity in pea pericarp growth. Pea pericarps respond in a qualitatively different fashion to two naturally occurring auxins (IAA and 4-Cl-IAA), which, in a variety of other auxin bioassays, exhibited only quantitative differences in activity (Reinecke, 1999). These data suggest unique ways of auxin action based on alternative molecular recognition mechanisms in this tissue.
Pea plants metabolize GAs by the early 13-hydroxylation pathway: GA12 → GA53 → GA44 → GA19 → GA20 → GA1 (Sponsel, 1995). Previous studies using the pea split-pericarp assay (test compounds are applied to the inner walls of split and deseeded 2-DAA pericarps) have shown that the presence of seeds or the application of 4-Cl-IAA to deseeded pea pericarps stimulated pericarp GA biosynthesis gene expression, specifically PsGA20ox1 (codes for enzyme that converts GA53 to GA20; van Huizen et al., 1997) and PsGA3ox1 (codes for enzyme that converts GA20 to GA1; Ozga et al., 2003). IAA was ineffective at stimulating pericarp PsGA20ox1 (Ngo et al., 2002) and PsGA3ox1 expression (Ozga et al., 2003) and pericarp growth (Reinecke et al., 1995). Furthermore, elongating pollinated pericarps (3 DAA) were capable of converting [14C]GA12 and [14C]GA19 to [14C]GA20; however, conversion to [14C]GA1 was not detected after a 24-h incubation period (Ozga et al., 1992; van Huizen et al., 1995). Maki and Brenner (1991) reported metabolism of [2H]GA53 to [2H]GA1 in pollinated pericarp tissue after a 48-h incubation period; however, Rodrigo et al. (1997), using 5-DAA pollinated pericarps and [14C]GA12, obtained results similar to those of Ozga et al. (1992). Therefore, the ability of young pea pericarps to metabolize GA20 to GA1 remains unclear.
In order to understand further the developmental and hormonal regulation of the GA biosynthesis and catabolism pathways during early pea fruit development, this study explored the developmental-, pollination-, seed-, and auxin-specific regulation of PsCPS1 (codes for ent-copalyl diphosphate synthase [CPS; Ait-Ali et al., 1997], a key step early in the GA biosynthesis pathway [Hedden and Phillips, 2000]), PsGA20ox1 and PsGA20ox2 (code for GA 20-oxidases that convert GA53 to GA20; Martin et al., 1996; Ait-Ali et al., 1997), PsGA3ox1 and PsGA3ox2 (code for GA 3-oxidases that convert GA20 to bioactive GA1; Lester et al., 1997; Martin et al., 1997; Weston et al., 2008), and PsGA2ox1 and PsGA2ox2 (code for GA 2-oxidases that catabolize GA20 to GA29 and GA1 to GA8; Lester et al., 1999) gene expression in pericarps and seeds, and the effects of seeds and auxin (4-Cl-IAA) on the metabolism of GA12 to GA1 in young pea fruits (pericarps). In addition, the role PsGA2ox1 in GA metabolism in the pericarp was examined in the slender (sln) pea mutant, which contains a null mutation of PsGA2ox1.
RESULTS AND DISCUSSION
Pericarp and Seed Growth
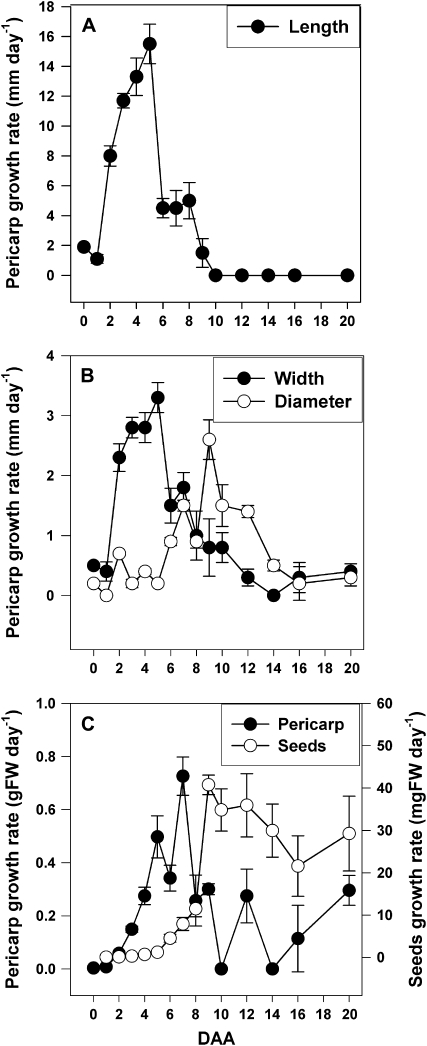
Pericarp growth rate in length and width was rapid from 2 to 5 DAA (Fig. 1, A and B). Subsequently, the growth rate of the pericarp in diameter increased rapidly from 6 to 9 DAA to accommodate the developing seeds (Fig. 1B). Pericarp fresh weight increased rapidly from 3 to 7 DAA and increased to a lesser extent to 20 DAA (Fig. 1C). Following pollination, seed fresh weight increased rapidly from 9 to 20 DAA (Fig. 1C). Pericarps from flowers emasculated at −2 DAA and harvested at the equivalent to −1, 0, 1, 2, and 3 DAA (pericarps from unpollinated ovaries) ranged from 7 to 10 mm in length.
Figure 1.
Pea pericarp and seed development from 0 to 20 DAA. Growth rate of pericarps in length (A), width (B), and diameter (B) and growth rate of pericarps and seeds in fresh weight (FW; C) are shown. Data are means ± se (n = 4–12 for pericarps; n = 2–4 for seed fresh weight).
Pollination and Fertilization Events Modify GA Biosynthesis in the Ovary
In order to determine if GA biosynthesis in the ovary is modified by pollination and fertilization events, the expression of GA biosynthesis and catabolism genes was monitored in the pericarp prior to and following these events and in pericarps of ovaries that were emasculated prior to pollination. Prior to pollination (−2 DAA), the relative mRNA levels of pericarp GA biosynthesis genes PsCPS1, PsGA20ox1, and PsGA20ox2 were elevated (Table I; Fig. 2, A and B), suggesting that unpollinated pericarps are capable of GA biosynthesis from ent-copalyl diphosphate through to GA20. However, at −2 DAA, pericarp mRNA levels of PsGA3ox1 and PsGA3ox2 (code for GA 3-oxidases that convert GA20 to biologically active GA1) were minimal (Table I; Fig. 2C; Ozga et al., 2003) and transcript levels of PsGA2ox2 (codes for the GA 2-oxidase that preferentially converts GA1 to biologically inactive GA8) were elevated (Table I; Fig. 2D), suggesting minimal presence of bioactive GA1 in unpollinated pericarps. Consistent with these gene expression data, emasculated (unpollinated) ovaries (at 0 DAA) were found to contain GA20 (approximately 5 ng g−1 fresh weight; cv Alaska) but minimal to no detectable levels of GA1 (Garcia-Martinez et al., 1991).
Table I.
Effects of pollination on PsCPS1, PsGA20ox1, PsGA20ox2, PsGA2ox1, PsGA2ox2, PsGA3ox1, and PsGA3ox2 mRNA levels in pea pericarps
Quantitation for PsCPS1, PsGA20ox, and PsGA2ox genes was performed using an Applied Biosystems 7700 sequence detector, and that for the PsGA3ox genes was performed using an Applied Biosystems StepOnePlus sequence detector with 200 ng of total RNA per sample. Due to different PCR efficiencies between the two models, the abundance of PsCPS1, PsGA20ox, and PsGA2ox genes cannot be directly compared with that of the PsGA3ox genes. Data are means ± se (n = 2–3 for pollinated pericarps; n = 2 for unpollinated pericarps).
| Pollination Statusa | −2 DAA
|
−1 DAA
|
0 DAA
|
1 DAA
|
2 DAA
|
3 DAA
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| − | + | − | + | − | + | − | + | − | + | − | |
| PsCPS1 | 28,135 ± 734 | –b | 35,432 ± 7,261 | 15,777 ± 846 | 13,927 ± 1,691 | 9,554 ± 516 | 12,618 ± 196 | 21,166 ± 2,064 | 9,259 ± 1,091 | 17,561 ± 2,197 | 12,647 ± 2,345 |
| PsGA20ox1 | 191,230 ± 12,324 | – | 246,859 ± 20,923 | 166,630 ± 12,106 | 218,289 ± 27,713 | 124,093 ± 1,272 | 266,480 ± 58,598 | 71,596 ± 7,185 | 246,111 ± 13,546 | 72,797 ± 83 | 87,527 ± 48,690 |
| PsGA20ox2 | 5,495 ± 1,942 | – | 4,239 ± 467 | 951 ± 373 | 1,997 ± 712 | 12 ± 5 | 410 ± 24 | 3 ± 1 | 869 ± 62 | 4.8 ± 1 | 143 ± 82 |
| PsGA2ox1 | 1,016 ± 462 | – | 60,522 ± 7,091 | 14,990 ± 2,878 | 51,045 ± 18,378 | 4,899 ± 438 | 14,164 ± 666 | 1,803 ± 559 | 13,092 ± 2,701 | 887 ± 116 | 21,058 ± 5,898 |
| PsGA2ox2 | 330 ± 105 | – | 347 ± 134 | 39 ± 3 | 126 ± 44 | 41 ± 8 | 79 ± 7 | 254 ± 15 | 214 ± 138 | 449 ± 197 | 644 ± 94 |
| PsGA3ox1 | 28 ± 6 | – | 270 ± 50 | 2,542 ± 850 | 878 ± 21 | 402 ± 36 | 704 ± 142 | 156 ± 30 | 914 ± 199 | 465 ± 25 | 3,884 ± 177 |
| PsGA3ox2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
Pollination status: +, pericarps from pollinated ovaries; −, unpollinated pericarps emasculated at −2 DAA, which were green and turgid through the equivalent to 3 DAA but did not exceed 10 mm in length.
–, Not determined.
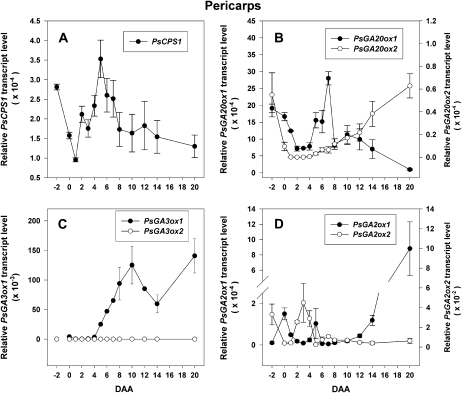
Figure 2.
Developmental regulation of PsCPS1 (A), PsGA20ox1 (B), PsGA20ox2 (B), PsGA3ox1 (C), PsGA3ox2 (C), PsGA2ox1 (D), and PsGA2ox2 (D) mRNA levels in pea pericarps from −2 to 20 DAA. Data are means ± se (n = 2–3).
Following pollination and fertilization (complete by 0 DAA), the pericarp is the major nutrient sink tissue in developing pea fruit until approximately 8 to 12 DAA, when the seeds become the terminal sink (cv Alaska; Johnstone, 2004). The transcription profiles of the GA biosynthesis and catabolism genes dramatically changed in the pericarp following the pollination and fertilization events. From −2 (unpollinated) to 0 DAA (pollinated), transcript abundance of pericarp PsCPS1, PsGA20ox1, and PsGA20ox2 that code for enzymes that synthesize precursors of bioactive GA1 declined (Table I; Fig. 2A and B), while mRNA levels of PsGA3ox1 that codes for the enzyme that synthesizes GA1 increased (Table I; Ozga et al., 2003) and PsGA2ox2 that codes for an enzyme that catabolizes GA1 decreased (Table I; Fig. 2D). Pollination and fertilization also lessened the increase in transcript abundance of pericarp PsGA2ox1 (also codes for an enzyme that catabolizes GA1 to GA8) from 50-fold to 15-fold (pollinated versus unpollinated pericarps at 0 DAA; Table I). In pollinated pericarps from 0 to 1 DAA, the abundance of PsGA3ox1 mRNA decreased (Table I; Ozga et al., 2003), along with further declines in the mRNA abundance of the GA biosynthesis genes PsCPS1, PsGA20ox1, and PsGA20ox2. The GA catabolic gene PsGA2ox1 also decreased from 0 to 1 DAA (Table I; Fig. 2). PsGA3ox2 mRNA transcripts were not detected in the pericarp tissue during this developmental phase (Table I). This coordination of GA biosynthesis and catabolism gene transcript levels in the pericarp suggests that regulation at the level of transcript production or stability is involved in a transitory increase in bioactive GA1 in the pericarp following pollination and fertilization to stimulate initial growth and fruit set.
Although steady-state GA1 levels were reported to be minimal to undetectable in pollinated and unpollinated ovaries at 0 DAA, GA8 levels were two times higher in pollinated than unpollinated ovaries at this time (Garcia-Martinez et al., 1991). Since GA8 is the immediate biologically inactive product of GA1 (as a result of 2β-hydroxylation), these data suggest that GA1 was synthesized to a greater extent in pollinated than unpollinated pericarps and/or ovules by 0 DAA.
In the absence of pollination and fertilization, pericarp PsGA3ox1 mRNA abundance did not peak at 0 DAA (as in pollinated pericarps) but peaked at 3 DAA, likely the result of feedback regulation due to minimal levels (or the absence) of bioactive GA1 (Table I; Ozga et al., 2003). Interestingly, PsGA20ox1 mRNA levels were higher in unpollinated pericarps than those observed in pollinated pericarps (Table I; Garcia-Martinez et al., 1997; van Huizen et al., 1997); however, the pool of GA20 was reduced by half in the unpollinated pericarps by 2 DAA (Garcia-Martinez et al., 1991). This decline in GA20 in unpollinated pericarps is likely a result of catabolism to GA29 by GA 2-oxidase, as PsGA2ox1 mRNA levels increased approximately 50-fold from −2 to 0 DAA in unpollinated pericarps (a smaller increase of 15-fold was observed in pollinated pericarps at this time; Table I). Consistent with the elevated mRNA abundance of PsGA2ox1, a 3.3-fold higher GA29-to-GA20 ratio was found in unpollinated ovaries compared with pericarps from pollinated ovaries (Santes and Garcia-Martinez, 1995). By 4 DAA, the unpollinated ovaries degenerated (data not shown), likely in part due to minimal GA1 levels in the tissue, as treatment with bioactive GA can rescue unpollinated ovaries from degeneration (Rodrigo et al., 1997).
Seeds Required for Pericarp Growth and Bioactive GA1 Production
As the pericarps from pollinated ovaries continued to develop (2–5 DAA), an increased potential for flux through the GA biosynthesis pathway occurs, as observed by increasing mRNA levels of PsCPS1 (Fig. 2A), PsGA20ox1 (Fig. 2B), and PsGA3ox1 (Fig. 2C; Ozga et al., 2003) and lower levels of PsGA2ox1 (Fig. 2D), concomitant with high pericarp growth rates (length and fresh weight; Fig. 1). In parallel with these pericarp GA biosynthesis and catabolism gene expression and growth profiles, bioactive GA1 levels were approximately 0.4 to 1.2 ng g−1 fresh weight from 2 to 5 DAA (Table II, intact and SP; Ozga et al., 1992; Rodrigo et al., 1997) in pericarps from pollinated ovaries.
Table II.
A profile of GAs and [14C]GA12 metabolites in pericarps with (intact and SP) or without (SPNS) seeds and in pericarps without seeds treated with 4-Cl-IAA
Values shown are ng g−1 fresh weight quantitated by GC-MS-SIM.
| Treatment | GA19
|
GA20
|
GA1
|
GA29
|
GA8
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| [12C] | [14C] | [12C] | [14C] | [12C] | [14C] | [12C] | [14C] | [12C] | [14C] | |
| Intact 3 DAA | 2.45 | –a | 2.31 | – | 1.13 | – | 20.74 | – | 2.57 | – |
| SP 4 DAA | 0.72 | – | 2.05 | – | 1.19 | – | 16.38 | – | 1.04 | – |
| SPNS 4 DAA | 0.83 | – | 0.24 | – | 0.27 | – | 11.78 | – | Lb | – |
| SPc 5 DAA | 1.01 | 0.31 | 1.04 | 0.04 | 0.42 | ndd | 5.02 | 0.22 | 1.55 | nd |
| SPNSc 5 DAA | 1.34 | 0.76 | nd | nd | nd | nd | 7.25 | 0.35 | nd | nd |
| SPNS + 4-Cl-IAAc 5 DAA | 0.43 | 0.24 | 0.76 | 0.17 | 0.11 | 0.21 | 4.65 | 0.47 | L | L |
| KRIe STD/sample | 2,652/2,652 | 2,546/2,544 | 2,711/2,711 | 2,715/2,714 | 2,866/2,863 | |||||
–, Not determined.
L, Sample was lost at the GC-MS step.
Pericarps treated with [14C]GA12 24 h prior to harvest.
nd, Not detected.
KRI, Kovats retention index.
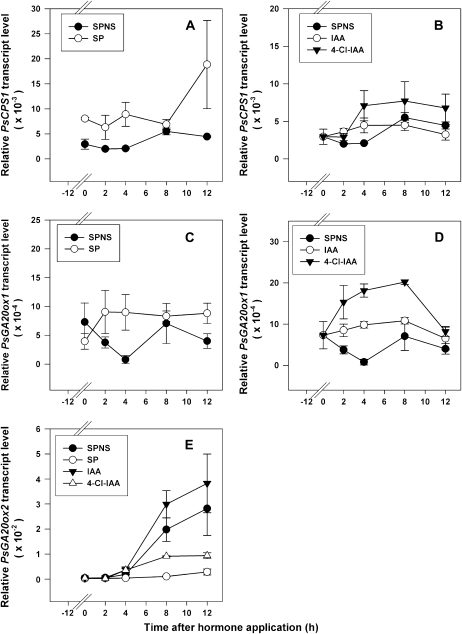
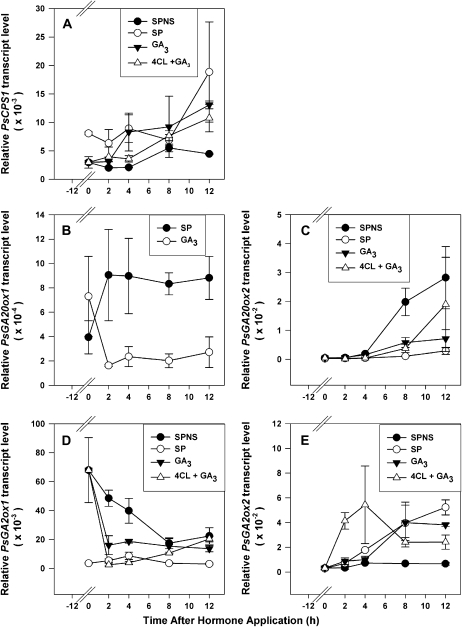
The presence of developing seeds in the ovary is required for continued pericarp growth. Pea pericarps continued to grow after splitting of the pericarp at 2 or 3 DAA without disturbing the seeds (SP); however, removal of the seeds at 2 or 3 DAA results in slowing of pericarp growth and subsequent abscission (Ozga et al., 1992). The presence of developing seeds in the ovary is also required to modulate the abundance of pea pericarp GA biosynthesis and catabolic genes (Table I, pericarps from pollinated versus unpollinated ovaries; Ozga et al., 2003) and for maintenance of pericarp GA1 levels (Table II, SP versus split pod no seeds [SPNS] treatments; Ozga et al., 1992). In order to further understand the effect of seeds on pericarp GA biosynthesis, GA biosynthesis and catabolism gene transcript profiles were monitored in pollinated pericarps with and without seeds. Transcript levels were first monitored 12 h after deseeding (0 h in Figs. 3 and 4) to allow sufficient time for the pericarps to become depleted of seed-produced factors that might affect gene expression. Over a 12-h experimental time course, seed removal (SPNS) decreased the transcript abundance of the pericarp GA biosynthesis genes PsCPS1 (Fig. 3A), PsGA20ox1 (Fig. 3C), and PsGA3ox1 (Ozga et al., 2003) and increased the pericarp transcript abundance of the GA catabolic gene PsGA2ox1 (Fig. 4A) when compared with pericarps with seeds (SP). PsGA3ox2 transcript abundance was minimal to not detectable in pericarps with or without seeds over the time course (data not shown). Seed removal increased PsGA20ox2 message levels (10-fold by the 12-h treatment period [24 h after seed removal]; Fig. 3E); however, this change in PsGA20ox2 transcript levels will likely have only minimal effects on the levels of PsGA20ox mRNA in this tissue, since PsGA20ox2 mRNA levels are only a minor contributor to the PsGA20ox mRNA pool in the pericarp compared with those of PsGA20ox1 (200-fold greater than PsGA20ox2 at the 12-h time point), unless PsGA20ox2 mRNA is localized in pericarp cells or tissues distinct from those of PsGA20ox1. Regulation of PsGA20ox2 mRNA levels, in general, differs from that of PsGA20ox1. In addition to the difference in seed regulation of pericarp mRNA levels above, PsGA20ox2 mRNA levels were not feedback regulated by bioactive GA, as is the case for PsGA20ox1 (Ayele et al., 2006b). Furthermore, the tissue-specific expression of PsGA20ox1 varies from that of PsGA20ox2. PsGA20ox1 transcript abundance was higher in young actively growing pericarps and seeds (Figs. 2B and 5B), and PsGA20ox2 transcripts were more abundant in mature tissues (pericarps [Fig. 2B] and roots [Ayele et al., 2006a]) and in seeds when the embryo was actively accumulating storage reserves (Fig. 5B). Seed removal also inhibited the transitory increase in gene expression of the pericarp GA catabolism gene PsGA2ox2 that was observed in pericarps with seeds (Figs. 2D and 4C) during this developmental time window.
Figure 3.
Effects of seeds, IAA, and 4-Cl-IAA on PsCPS1 (A and B), PsGA20ox1 (C and D), and PsGA20ox2 (E) mRNA levels in pea pericarps. Two DAA pericarps were split (SP) or split and deseeded and treated with 50 μm (30 μL) 4-Cl-IAA, IAA, or 0.1% aqueous Tween 80 (SPNS control) at 12 h after splitting or splitting and deseeding (0 h on the x axis). mRNA levels were monitored at 2, 4, 8, and 12 h after hormone application. Data are means ± se (n = 2–3).
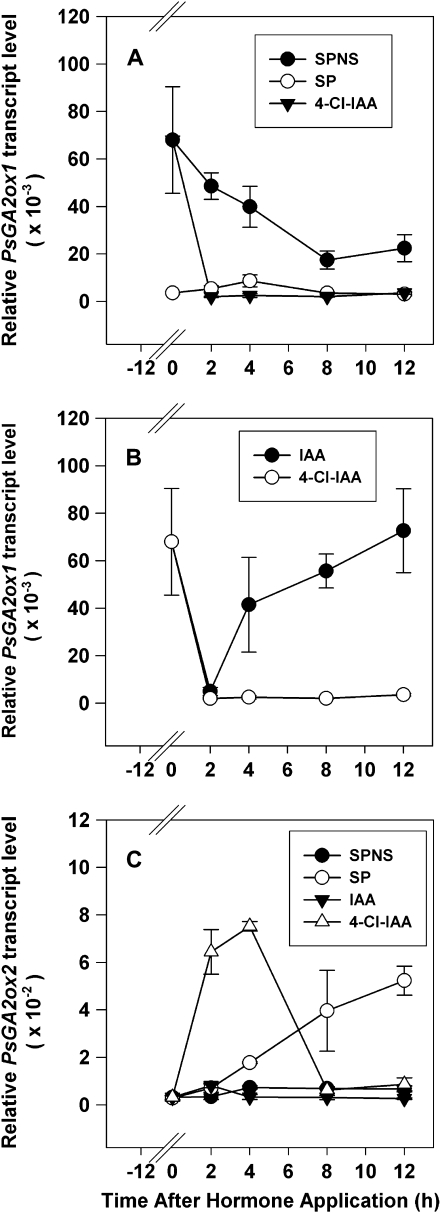
Figure 4.
Effects of seeds, IAA, and 4-Cl-IAA on PsGA2ox1 (A and B) and PsGA2ox2 (C) mRNA levels in pea pericarps. Two DAA pericarps were split (SP) or split and deseeded and treated with 50 μm (30 μL) 4-Cl-IAA, IAA, or 0.1% aqueous Tween 80 (SPNS control) at 12 h after splitting or splitting and deseeding (0 h on the x axis). mRNA levels were monitored at 2, 4, 8, and 12 h after hormone application. Data are means ± se (n = 2–3).
Figure 5.
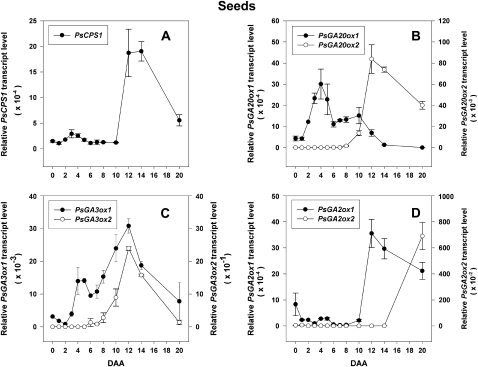
Developmental regulation of PsCPS1 (A), PsGA20ox1 (B), PsGA20ox2 (B), PsGA3ox1(C), PsGA3ox2 (C), PsGA2ox1 (D), and PsGA2ox2 (D) mRNA levels in pea seeds from 0 to 20 DAA. Data are means ± se (n = 2–3).
Consistent with the reduction in mRNA levels of the GA biosynthesis genes and the increase in the message levels of the GA catabolism gene PsGA2ox1, seed removal from pollinated pericarps resulted in a marked depletion of pericarp GA20 and GA1 levels 24 h after removal (4 DAA SP versus SPNS; Table II), and by 48 h neither GA20 nor GA1 could be detected (5 DAA SP versus SPNS; Table II; Ozga et al., 1992). These results suggest that pericarp GA biosynthesis and catabolism gene expression and production of bioactive GA are regulated by the seeds in young fruits.
During rapid pericarp expansion (4–7 DAA), mesocarp cells continue to expand and the only increase in cell number occurs in cell layers of the endocarp middle zone layer (pericarp wall thickness; Ozga et al., 2002). Consistent with the hypothesis that seeds at least partially regulate pericarp GA biosynthesis during this developmental period, the mRNA levels of pericarp GA biosynthesis genes PsCPS1 (Fig. 2A), PsGA20ox1 (Fig. 2B), and PsGA3ox1 (Fig. 2C; Ozga et al., 2003) sequentially peak from approximately 4 to 10 DAA, coincident with rapid pericarp diameter expansion (6–12 DAA; Fig. 1) to accommodate the growing seeds. The reduction in mRNA abundance of PsGA2ox2 by 5 DAA along with lower levels of PsGA2ox1 mRNA through 12 DAA (Fig. 2D) suggest seed repression of the expression of these GA catabolic genes during this phase of pericarp expansion (5–12 DAA).
Auxin (4-Cl-IAA) Regulates GA Biosynthesis and Catabolism in Young Pericarps
To further explore if auxin can mimic the seeds in regulation of the GA biosynthesis and catabolic pathways in the surrounding pericarp tissue, auxin regulation of these pathways in pea pericarp was investigated over a 12-h period. Hormones were applied to the pericarps 12 h after deseeding (0 h in Figs. 3 and 4) to allow sufficient time for the pericarps to become depleted of seed-produced factors that might affect pericarp growth. In deseeded pericarps, neither IAA nor 4-Cl-IAA affected the transcript abundance of PsCPS1 (Fig. 3B), which codes for a key GA biosynthesis enzyme that occurs early in the pathway (appears to be a single-copy gene responsible for ent-copalyl diphosphate synthesis in pea; Ait-Ali et al., 1997). Bioactive GA levels also appear not to feedback regulate PsCPS1 mRNA levels in deseeded pea pericarps (Fig. 6A), roots, or shoots (Davidson et al., 2005; Ayele et al., 2006b). These data confirm the results of a number of studies that hormonal regulation of GA biosynthesis occurs mainly later in the pathway (Hedden and Phillips, 2000; Yamaguchi, 2008).
Figure 6.
Effects of seeds, GA3, and 4-Cl-IAA + GA3 on PsCPS1 (A), PsGA20ox1 (B), PsGA20ox2 (C), PsGA2ox1 (D), and PsGA2ox2 (E) mRNA levels in pea pericarps. Pericarps at 2 DAA were split (SP) or split and deseeded and treated with 50 μm (30 μL) GA3, a mixture of 4-Cl-IAA + GA3 (50 μm each in 30 μL total), or 0.1% aqueous Tween 80 (SPNS control) at 12 h after splitting or splitting and deseeding (0 h on the x axis). mRNA levels were monitored at 2, 4, 8, and 12 h after hormone application. Data are means ± se (n = 2–3).
4-Cl-IAA, the biologically active auxin in pea pericarp growth, increased the mRNA abundance of PsGA20ox1 (Fig. 3D; van Huizen et al., 1997; Ngo et al., 2002) and PsGA3ox1 (Ozga et al., 2003) in deseeded pericarps within 2 h of hormone application. IAA did not stimulate PsGA20ox1 (no significant change in PsGA20ox1 transcript abundance in IAA-treated pericarps from the 0 h control; Fig. 3D; Ngo et al., 2002) or PsGA3ox1 (Ozga et al., 2003) mRNA levels or pericarp growth (Reinecke et al., 1995). These data suggest that 4-Cl-IAA-induced pericarp growth occurs in part by coordinated regulation of PsGA20ox1 and PsGA3ox1 transcription in the GA biosynthesis pathway.
PsGA20ox2 mRNA levels were regulated in a different manner by IAA and 4-Cl-IAA than PsGA20ox1 mRNA levels in the pericarp (Fig. 3, D and E). 4-Cl-IAA did not increase PsGA20ox2 mRNA levels in deseeded pericarps. Instead, PsGA20ox2 transcript abundance increased in nongrowing tissues, including deseeded pericarps (SPNS), IAA-treated pericarps (Fig. 4E), and nonpollinated pericarps (Table I). Also, in contrast to PsGA3ox1, the mRNA abundance of PsGA3ox2 was minimal to not detectable in the pericarp tissue (Fig. 2C), and transcript abundance was not increased by application of 4-Cl-IAA to deseeded pericarps (data not shown). PsGA3ox2 transcript abundance also remained minimal to not detectable in deseeded pericarps (SPNS) and pericarps treated with IAA or GA3 (data not shown). Indeed, as with PsGA20ox2, the highest transcript abundance for PsGA3ox2 was observed in the seeds (Fig. 5C).
Treatment with both IAA and 4-Cl-IAA markedly decreased PsGA2ox1 transcript abundance 2 h after application to deseeded pericarps (Fig. 4, A and B). However, by 4 h after application, PsGA2ox1 mRNA abundance in the IAA-treated deseeded pericarps sharply increased and remained elevated, while PsGA2ox1 mRNA abundance in the 4-Cl-IAA-treated pericarps remained at low levels throughout the 12-h treatment period (Fig. 4B). These data suggest that the initial auxin signaling events for down-regulating PsGA2ox1 transcript abundance are similar for both IAA and 4-Cl-IAA; however, longer term down-regulation and pericarp growth are only maintained by 4-Cl-IAA, suggesting subsequent divergent signaling pathways for these two naturally occurring auxins in pea. An alternative explanation is that 4-Cl-IAA is more stable in the tissue than IAA, leading to a longer term down-regulation of PsGA2ox1 transcript abundance. Application of GA3 to deseeded pericarps also decreased PsGA2ox1 transcript abundance within 2 h (Fig. 6D), although transcript levels were always significantly higher in the GA3-treated deseeded pericarps compared with 4-Cl-IAA-treated deseeded pericarps (Fig. 4A) or pericarps with seeds (Fig. 6E). These data suggest that the transcript levels of PsGA2ox1 in the pericarp early in development are regulated by bioactive auxin as well as other factors present in growth induction conditions.
Auxin regulation of the transcript abundance of the GA catabolic gene PsGA2ox2 is dramatically different than that of PsGA2ox1 in pericarp tissue. 4-Cl-IAA substantially increased PsGA2ox2 mRNA abundance within 2 h of application to the deseeded pericarp (Fig. 4C). The elevated levels of PsGA2ox2 mRNA were transitory, and within 8 h of 4-Cl-IAA application, PsGA2ox2 mRNA levels were similar to that of the deseeded control (SPNS). Furthermore, during this developmental period, the presence of the seeds also stimulated an increase in pericarp PsGA2ox2 mRNA levels (SP; Fig. 4C). This seed-induced increase in PsGA2ox2 mRNA levels was also transitory, as PsGA2ox2 message levels decreased in the pericarp by 5 DAA (Fig. 2C). IAA did not affect the abundance of PsGA2ox2 transcripts throughout the 12-h treatment period (Fig. 4C). The 4-Cl-IAA-induced transitory increase in PsGA2ox2 mRNA levels suggests that bioactive auxin can also modulate GA gene expression to keep bioactive GA1 levels within limits that are appropriate for specific developmental stages during pea fruit development.
Similarly, application of IAA to decapitated pea plants reduced the level of the GA catabolic gene PsGA2ox1 while increasing the transcript levels of the biosynthetic GA gene PsGA3ox1 as well as the catabolic gene PsGA2ox2 in the subtending elongating internode (O'Neill and Ross, 2002). O'Neill and Ross (2002) suggested that PsGA2ox2 is not directly regulated by IAA and that the IAA-induced up-regulation of this gene is attributable to a feed-forward mechanism whereby increased levels of bioactive GA up-regulate genes encoding GA deactivation enzymes (Thomas et al., 1999). Bioactive GA3 applied to deseeded pericarps decreased the transcript levels of PsGA20ox1 (Fig. 6B) and PsGA3ox1 (Ozga et al., 2003) in a feedback regulation manner, as observed in pea shoots and roots (Ayele et al., 2006b). However, the marked increase in PsGA2ox2 transcript levels at 2 h after 4-Cl-IAA application to deseeded pericarps (Fig. 5C) did not occur with GA3 application (Fig. 6E). PsGA2ox2 message levels did increase at 8 h after GA3 application. These data show that the 4-Cl-IAA-induced up-regulation of pericarp PsGA2ox2 is not directly attributable to a bioactive GA feed-forward mechanism but is likely a direct effect of this bioactive auxin on PsGA2ox2 message levels to regulate the half-life of GA1 in this tissue. The delayed increase in PsGA2ox2 levels (first observed at 8 h after hormone treatment) by GA3 may indicate that a feed-forward mechanism is present, but it is distinct from the early 4-Cl-IAA-induced response on PsGA2ox2 gene expression. When GA3 was applied in combination with 4-Cl-IAA, the bioactive auxin response of stimulation of PsGA2ox2 expression was again observed within 2 h after hormone application, followed by the GA3 response of a delayed stimulation of PsGA2ox2 transcript levels (8–12 h after hormone application; Fig. 6E).
Overall, the GA biosynthesis and catabolic transcription profiles suggest that 4-Cl-IAA can stimulate the production of bioactive GA1 in the pericarp as well as modulate the half-life of GA1 by regulating the mRNA levels of the catabolic gene PsGA2ox2. Interestingly, conversion of labeled GAs to GA1 in pericarps has been difficult to obtain using conventional methods of detection (Ozga et al., 1992; van Huizen et al., 1995; Rodrigo et al., 1997). In the current study, when [14C]GA12 was applied to pericarps with seeds, endogenous and 14C-labeled GA19, GA20, and GA29 were detected along with endogenous GA1 and GA8, but [14C]GA1 was not detected (SP 5 DAA; Table II). In deseeded pericarps treated with [14C]GA12, only endogenous and 14C-labeled GA19 and GA29 were detected (SPNS 5 DAA; Table II). These results suggest that seeds are required for GA biosynthesis in the pericarp and are consistent with our previous findings (Ozga et al., 1992; van Huizen et al., 1995). However, when deseeded pericarps were treated with 4-Cl-IAA, endogenous and 14C-labeled GA1, along with endogenous and 14C-labeled GA19, GA20, and GA29, were detected (SPNS + 4-Cl-IAA 5 DAA; Table II). The radiolabeling of GA1 is most likely due to the ability of 4-Cl-IAA to stimulate the transcript levels of the GA biosynthesis genes PsGA20ox1 and PsGA3ox1 and decrease the mRNA levels of the catabolic gene PsGA2ox1 in pericarp tissue. In addition, a labeled substrate with higher specific radioactivity and one farther up the pathway ([14C]GA12 used in this study compared with [14C]GA19 used by van Huizen et al. [1995]) are likely both important to increase sensitivity and avoid increased metabolism of the labeled substrate into the inactive GA pool (GA29 and other inactive GA metabolites).
It also has been proposed that the sensitivity of fruit to bioactive GA may be substantially greater than that of the stem internodes in pea. Comparison of near-isogenic lines of pea that contain either the wild-type PsGA3ox1 gene (LELE) or a 1-bp point mutation of LE that greatly increases the Km of the encoded GA 3-oxidase (lele) demonstrated that the mutation (lele genotype) reduced GA1 content and growth of internodes (Ross et al., 1992). The content of GA1 was also lower (seven to 10 times) in young lele pericarps compared with those of LELE, but only minor effects on fruit growth were observed (Santes et al., 1993; MacKenzie-Hose et al., 1998). In order to determine the minimum amount of bioactive GA necessary for pea fruit set and growth, the size of the pericarp was plotted against the GA1 concentration in nonpollinated fruit growing after application of GA1 or GA3 to the leaf subtending the fruit (Rodrigo et al., 1997). A linear relationship of GA concentration with pericarp growth was found from about 0.1 (the minimum amount necessary for fruit set and growth) to 2 ng g−1 fresh weight. Higher concentrations of GA1 in the pericarp (20 ng g−1 fresh weight) did not result in substantial further growth. Therefore, the concentration of GA1 in young lele pods (0.1 ng g−1 fresh weight at 6 DAA; Santes et al., 1993) may be sufficient, if not optimal, to stimulate fruit set and growth in this tissue.
Effect of a PsGA2ox1 Null Mutation on Seed and Auxin-Induced Pericarp Growth and GA Metabolism
To further understand the role of GA 2-oxidase in seed and auxin regulation of GA biosynthesis in the fruit, we monitored pericarp growth and metabolism of [14C]GA19 in the pea sln mutant (a null mutation in PsGA2ox1; Lester et al., 1999; Martin et al., 1999) and its associated wild type. In both SLN and sln genotypes, pericarps with seeds (SP) continued to grow, while deseeding at 2 DAA (SPNS) inhibited pericarp growth (Table III) and the deseeded pericarps subsequently senesced. Application of 4-Cl-IAA stimulated the growth of deseeded pericarps of SLN and sln (Table III). Consistent with the mutation in the PsGA2ox1 gene, more [14C]GA20 accumulated and less [14C]GA20 was converted to [14C]GA29 in the pericarp with seeds (SP) of sln compared with that in SLN plants (Table III). Minimal production of [14C]GA20 and [14C]GA29 occurred in deseeded pericarps (SPNS) of both genotypes. 4-Cl-IAA application to deseeded pericarps stimulated a 3-fold increase in [14C]GA20 in SLN and a 9-fold increase in sln plants compared with the SPNS controls, mimicking the presence of the seeds (SP treatment; Table III). The amount of [14C]GA29 produced in sln deseeded pericarps treated with 4-Cl-IAA was also approximately 3-fold lower than in the SLN pericarps. These data demonstrate that substantial reduction in pericarp GA 2-oxidase activity neither stimulated pericarp growth nor maintained the pool of pericarp GA20 required as a substrate for conversion to bioactive GA1 in the absence of seeds. 4-Cl-IAA also mimicked the effect of seeds in stimulating pericarp growth and metabolism of [14C]GA19 to [14C]GA20 and [14C]GA29 in both the SLN and sln pericarps.
Table III.
Accumulation of [14C]GA20, [14C]GA29, and [14C]GA29-catabolite in wild-type SLN and sln mutants after 24 h of incubation with [14C]GA19
Values shown are percentages calculated as (dpm 14C-metabolite after 24 h of incubation)/(dpm [14C]GA19 added to tissue) × 100. Values in parentheses are 14C-metabolite in dpm × 10−3 ± se (n = 3 for wild-type SLN and n = 2 for sln). Amount of [14C]GA19 added per three pods (three pods per extraction) was as follows: all wild-type SLN, 162,525 dpm; SP (sln), 155,954 dpm; 4-Cl-IAA (sln), 111,054 dpm; SPNS (sln), 111,054 dpm.
| Treatmenta | Pericarp Growthb |
14C-Metabolite
|
||
|---|---|---|---|---|
| [14C]GA20 | [14C]GA29 | [14C]GA29-Catabolite | ||
| mm | ||||
| SP (wild-type SLN) | 18.6 ± 2.3 | 7.7 ± 1.1 (12.57 ± 1.83) | 3.3 ± 1.20 (5.36 ± 1.87) | 1.2 ± 0.32 (1.97 ± 0.52) |
| SPNS (wild-type SLN) | 2.0 ± 0.3 | 1.91 ± 0.14 (3.10 ± 0.31) | 0.94 ± 0.23 (1.52 ± 0.36) | 1.5 ± 0.59 (2.51 ± 0.96) |
| 4-Cl-IAA (wild-type SLN) | 8.0 ± 0.5 | 5.9 ± 0.76 (11.36 ± 4.23) | 14.8 ± 2.50 (24.00 ± 4.05) | 4.5 ± 0.43 (7.40 ± 0.70) |
| SP (sln) | 13.6 ± 4.2 | 13.1 ± 3.9 (16.62 ± 2.21) | 1.0 ± 1.00 (1.67 ± 1.67) | 0.16 ± 0.16 (0.24 ± 0.24) |
| SPNS (sln) | 0 ± 0 | 1.19 ± 1.19 (1.32 ± 1.32) | 0.30 ± 0.30 (0.33 ± 0.27) | ndc |
| 4-Cl-IAA (sln) | 11.7 ± 2.0 | 10.8 ± 2.5 (15.0 ± 5.75) | 4.4 ± 1.30 (6.12 ± 2.71) | nd |
Seeds were either left intact (SP) or removed (SPNS) from the pericarp, or seeds were removed and 4-Cl-IAA was applied to the pericarp at 12 h after deseeding.
Pericarp growth in length at 48 h after initial pericarp splitting or splitting and seed removal (±se; n = 6–9).
nd, Not detected.
Additionally, regardless of treatment, the sln pericarps produced little to no detectable GA29-catabolite compared with the SLN pericarps. MacKenzie-Hose et al. (1998) found that the steady-state levels of GA20 were higher (2-fold) and those of GA29 were lower (3.6-fold) in 4- to 7-DAA sln pericarps compared with the SLN wild type. However, they reported that the pericarp GA29-catabolite levels did not differ between these genotypes. Our [14C]GA19 metabolism data support that both the conversion of GA20 to GA29 and GA29 to GA29-catabolite are reduced by the sln mutation in the pericarp tissue (similar to that shown for pea seed coat tissue at 20 DAA; Ross et al., 1995) and that 4-Cl-IAA stimulation of pericarp PsGA20ox1 leads to higher accumulation of GA20 in the sln pericarp compared with SLN due to the block in catabolism of GA20 to both GA29 and GA29-catabolite. Furthermore, although higher endogenous GA1 levels were observed in the pericarps of sln (5.3 ng g−1 fresh weight) than those of SLN (1.7 ng g−1 fresh weight; MacKenzie-Hose et al., 1998), the [14C]GA19 metabolism method used was not sensitive enough to monitor the synthesis of [14C]GA1 in the pericarps of SLN or sln plants after the 24-h incubation period. Using similar extraction and detection methods, [14C]GA1 was also not detected in pericarp [14C]GA19 metabolism experiments in cv Alaska plants (I3; van Huizen et al., 1995).
Comparisons with Other Species
In tomato (Solanum lycopersicum) fruit, data from semiquantitative reverse transcription (RT)-PCR gene expression analysis suggest that only GA 20-oxidase mRNA levels are regulated (increased) by pollination and fertilization events (Serrani et al., 2007). These results are substantially different from those in pea, where pollination and fertilization events increase GA 3-oxidase (PsGA3ox1) and decrease GA 2-oxidase (PsGA2ox1 and PsGA2ox2) and GA 20-oxidase (PsGA20ox1 and PsGA20ox2) mRNA levels. However, auxin regulation of GA biosynthesis appears to be similar in the fruit of these species. Data from quantitative RT-PCR expression and GA quantitation studies (Serrani et al., 2008) suggest that the synthetic auxin 2,4-dichlorophenoxyacetic acid induced parthenocarpic tomato fruit growth in part by increasing SlGA20ox and SlGA3ox1 and decreasing SlGA2ox2 message levels, similar to the effects of the endogenous auxin 4-Cl-IAA on GA biosynthesis and catabolism genes in pea pericarps.
In Arabidopsis (Arabidopsis thaliana), the synthetic auxin 1-naphthalene acetic acid stimulated message levels of specific AtGA20ox genes, as well as AtGA2ox genes, but not AtGA3ox genes when applied to light-grown Arabidopsis seedlings (Frigerio et al., 2006). It is apparent that auxin regulation of GA biosynthesis and catabolism in plants is a mechanism whereby specific bioactive auxins can developmentally, temporally, and spatially regulate levels of another class of hormones (GAs) at the transcript level to coordinate growth and development. Auxin responses mediated through the GA biosynthesis pathway and those mediated directly through auxin likely involve Aux/IAA and ARF signaling elements (Frigerio et al., 2006; Goetz et al., 2006; Serrani et al., 2008).
Fertilization Changes GA Biosynthesis Transcript Profiles in the Ovule
Following pollination of the ovary and fertilization of the ovule (0 DAA), a temporary increase in PsGA3ox1 mRNA levels (19-fold) in the ovules occurred followed by a substantial decline by 1 DAA (Ozga et al., 2003; Fig. 5C), with no marked changes in the transcript abundance of PsCPS1, PsGA20ox1, and PsGA20ox2 from 0 to 1 DAA (Fig. 5, A and B). The GA2ox catabolic genes were elevated in the ovules following fertilization (PsGA2ox1 at 0 DAA; PsGA2ox2 relative mRNA level of 493 ± 145 at 0 DAA and of 702 ± 252 at 1 DAA; Fig. 5D) and then decreased by 1 DAA (PsGA2ox1; Fig. 5D) or 2 DAA (PsGA2ox2; 104 ± 6). This flux in the transcript abundance of the GA biosynthesis and catabolic genes suggests that an increase in bioactive GA1 is triggered by pollination and fertilization, followed by an immediate reduction in GA1 levels in the fertilized ovules. Consistent with this hypothesis, minimal to no GA1 or GA8 was detected at 2 DAA in the developing seeds (Rodrigo et al., 1997). It is possible that bioactive GA levels are minimized during this developmental window (approximately 1–2 DAA) in the fertilized ovule to allow for the formation or development of the proembryo mass (Pharis and King, 1985).
Young Developing Seeds Are Active Sites of GA Biosynthesis
From 2 to 6 DAA, a sequential increase in mRNA abundance of GA biosynthesis genes occurred throughout the pathway in the developing seed, including PsCPS1 (Fig. 5A), PsGA20ox1 (Fig. 5B), and PsGA3ox1 (Fig. 5C; Ozga et al., 2003). The transcript abundance of the GA2ox catabolic genes, PsGA2ox1 and PsGA2ox2, was minimally affected during this developmental stage (Fig. 5D). Correspondingly, approximately 45 ng g −1 fresh weight GA1 was detected in the developing seeds by 4 DAA, with a peak in GA1 levels in the seeds at 6 DAA (about 90 ng g−1 fresh weight; Rodrigo et al., 1997). At 4 DAA, 86% of the GA1 observed in the seeds occurred in the testa, with the remainder in the endosperm (no GA1 was detected in the embryo; Rodrigo et al., 1997). A subsequent increase in seed PsGA3ox1 and PsGA3ox2 transcript abundance was observed from 8 to 12 DAA (Fig. 5C), coincident with rapid seed coat and embryo growth (Fig. 1C) and maximum endosperm volume (reached at 12 DAA; Eeuwens and Schwabe, 1975). However, GA1 levels decreased to 20 ng g−1 fresh weight coincident with a peak in GA8 production (approximately 70 ng g−1 fresh weight) at 8 DAA followed by minimally detectable GA1 levels and moderately high levels of GA8 (approximately 40 ng g−1 fresh weight at 12 DAA; Rodrigo et al., 1997).
The marked increase in seed PsGA2ox1 transcript abundance from 8 to 12 DAA suggests that the GA 2-oxidase encoded by this gene is responsible for decreasing the half-life of GA1 in the seed tissues, leading to lower steady-state levels of this bioactive GA. It must also be noted that during this developmental window, seeds consist of three major distinct tissues (seed coat, endosperm, and embryo) that have different functions and developmental patterns. Therefore, it is highly likely that tissue-specific regulation of GA biosynthesis occurs for unique developmental outcomes within each tissue of the seed that is not reflected in whole seed analysis. Furthermore, studies that compare GA biosynthesis and catabolism gene expression patterns and GA levels within each major tissue would clarify the tissue-specific nature of GA biosynthesis within the seed at this developmental stage.
Seed Maturation Is Accompanied by Increased Seed GA Catabolic Gene Expression
By 12 DAA, accumulation of cotyledonary storage reserves begins in the rapidly developing pea embryo (Pate and Flinn, 1977). The mRNA profiles of the GA biosynthesis and catabolism genes in the seeds from 12 to 20 DAA are likely primarily reflective of the developing embryo as it grows to fill the seed coat by 20 DAA (contact point; Ozga et al., 2003). Seed PsGA20ox1 mRNA abundance decreased after 10 DAA (Fig. 5B), and PsGA3ox1 and PsGA3ox2 mRNA levels decreased after 12 DAA and continued to decrease until 20 DAA (Fig. 5C). However, from 10 to 12 DAA, the transcript abundance of PsCPS1, PsGA20ox2, and PsGA2ox1 dramatically increased and remained elevated through 20 DAA (Fig. 5, A, B, and D). The high levels of PsCPS1 and PsGA20ox2 transcripts are likely mainly localized in the embryo of the seed, as high levels of PsCPS1 and PsGA20ox2 were found in the embryos compared with the testa of 20-DAA pea seeds (cv Torsdag; Ait-Ali et al., 1997). In contrast, high levels of PsGA2ox1 mRNA were found in the testa compared with the cotyledons of 26-DAA seeds of pea (cv Progress No. 9; Martin et al., 1999). This change in the GA biosynthesis and catabolism gene mRNA abundance profile from 12 to 20 DAA reflects the change in the GA profile in the developing seed at the contact point (20 DAA), with high levels of GA20 (446 ng g−1 fresh weight) and GA29 (189 ng g−1 fresh weight) and no detectable levels of GA1 and GA8 in the seeds (cv Alaska [I3]; Ayele et al., 2006a).
SUMMARY
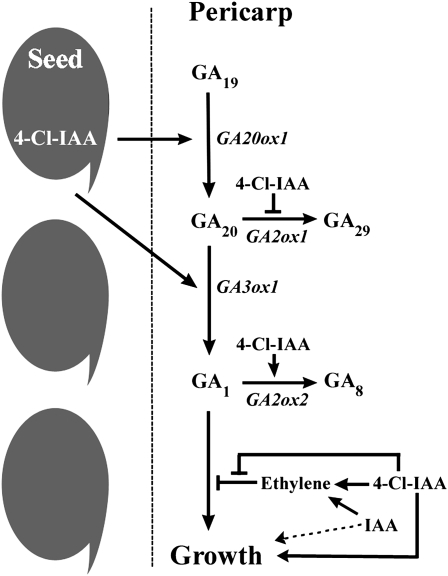
We propose the following working model for hormonally directed fruit set and seed and pericarp coordinated development. Pollination and fertilization events stimulate a pulse of bioactive GA1 synthesis in the pericarp and the ovules (via an increase in PsGA3ox1 and a decrease in PsGA2ox1 and PsGA2ox2 mRNA levels in the pericarp and an increase in PsGA3ox1 in the ovules) to promote initial seed and fruit set. Subsequently, seeds maintain pericarp growth (both in length [2–8 DAA] and in width [6–12 DAA] to accommodate the developing seeds) at least in part by stimulating pericarp GA biosynthesis (increasing PsGA20ox1 and PsGA3ox1 and decreasing PsGA2ox1 message levels), thereby maintaining a critical level of GA1 for pericarp growth. Furthermore, we hypothesize that auxin (4-Cl-IAA in pea) is one of the seed-derived signals that is involved in stimulation of GA biosynthesis in the pericarp at an early developmental stage (2–5 DAA) to promote growth (Fig. 7). 4-Cl-IAA is present in both pea seed and pericarp tissues at levels that suggest that transport from the seed to the pericarp is possible (Magnus et al., 1997). 4-Cl-IAA can stimulate deseeded pericarp growth and the production of bioactive GA1 in the pericarp (via an increase in PsGA20ox1 and PsGA3ox1 and a decrease in PsGA2ox1 mRNA levels) as well as modulate the half-life of GA1 (by regulating the mRNA levels of the catabolic gene PsGA2ox2). Additionally, 4-Cl-IAA (but not IAA) can potentiate a higher response to the bioactive GA produced in the pericarp through inhibition of ethylene action (Johnstone et al., 2005). 4-Cl-IAA can also affect fruit growth directly through auxin-mediated responses (van Huizen et al., 1996).
Figure 7.
Model of seed-derived auxin stimulation of pericarp growth. Seed-derived 4-Cl-IAA stimulates pericarp growth and the production of bioactive GA1 in the pericarp (via an increase in PsGA20ox1 and PsGA3ox1 and a decrease in PsGA2ox1 mRNA levels) as well as modulates the half-life of GA1 (by regulating the mRNA levels of the catabolic gene PsGA2ox2). Both 4-Cl-IAA and IAA stimulate ethylene production in the pericarp. However, only 4-Cl-IAA, through inhibition of ethylene action, can potentiate a higher response to the bioactive GA produced in the pericarp.
In developing seeds, bioactive GA1 synthesis is triggered by pollination and fertilization events, followed by an immediate reduction in GA1 levels in the fertilized ovules, possibly to allow the formation or development of the proembryo mass. From 2 to 6 DAA, a sequential increase in mRNA abundance of GA biosynthesis genes occurs to promote the production of GA1 to drive seed growth (mainly testa tissue). From 8 to 12 DAA, a transition in the seed GA biosynthesis and catabolism pathways occurs to produce sufficient bioactive GA for continued seed tissue growth and development, with a shift to the production of GA20 and minimal bioactive GA in the embryo as the seed enters into its maturation phase.
MATERIALS AND METHODS
Labeled GAs
[14C]GA12 was biosynthesized from R,S-[4,5-14C]mevalonic acid (110 μCi μmol−1) using a cell-free system of pumpkin (Cucurbita maxima) endosperm as described by Birnberg et al. (1986) and modified by Ozga et al. (1992). The specific radioactivity of [14C]GA12 was determined from its mass spectra to be 180.5 μCi μmol−1 using the method described by Bowen et al. (1972). Protio- and deutero-GA19, GA20, GA1, GA8, and [14C]GA19 were purchased from or provided by Dr. L.N. Mander. [13C-3H]GA29 was a gift from Dr. B.O. Phinney.
Plant Material and Treatments
Plants of pea (Pisum sativum ‘Alaska’ [I3]) were grown under a 16-/8-h light/dark photoperiod (19°C/17°C) with an average photon flux density of 402 μE m−2 s−1 (van Huizen et al., 1995). Pericarps (−2, 0, 1, 2, 3, 4, 5, 6, 7, 8, 10, 12, 14, and 20 DAA) and ovules (−2 DAA) or seeds (0, 1, 2, 3, 4, 5, 6, 7, 8, 10, 12, 14, and 20 DAA) were collected from the first to the fifth flowering nodes for experiments monitoring GA gene expression during fruit development. For nonpollinated pericarps, flowers (first to fifth flowering nodes) were emasculated at −2 DAA, and the pericarps were harvested from the plant at equivalent to −1, 0, 1, 2, and 3 DAA (ovules removed at harvest). For hormone-treated pericarps, one fruit between the third and fifth flowering nodes was used per plant; subsequent flowers and lateral buds were removed as they developed. Terminal apical meristems of plants were intact, and pericarps remained attached to the plant throughout the experiments.
Pericarps were treated with hormones using a split-pod technique (Ozga et al., 1992). Fruits at 2 DAA measuring 15 to 20 mm in length were split down the dorsal suture 1 h prior to the 8-h dark period, and seeds were either left intact (SP) or removed (SPNS). Surgical manipulation of the pea fruit was completed 12 h prior to all hormone applications. Deseeded pericarps were treated with IAA, 4-Cl-IAA, or GA3 at 50 μm in 0.1% (v/v) aqueous Tween 80 (30 μL total) or 4-Cl-IAA plus GA3 (50 μm each in 0.1% [v/v] aqueous Tween 80; 30 μL total). All solutions were applied directly to the inside surface of the pericarp wall (endocarp). The SP and SPNS controls were treated with 30 μL of 0.1% (v/v) aqueous Tween 80. Treated pericarps were covered with plastic bags to maintain high humidity. Pericarps were harvested at 0, 2, 4, 8, and 12 h after the hormone treatment. Seeds, if present, were removed from the pericarp at harvest. All tissues were harvested into liquid N2 and subsequently stored at −80°C until extraction.
For the [14C]GA12 metabolism experiments, pea plants (cv Alaska [I3]) were grown as described by Maki and Brenner (1991). Pericarps at 3 DAA were split (SP) or split and deseeded (SPNS) and treated with a 10-s dip of either 4-Cl-IAA (50 μm) in 0.1% Tween 80 or 0.1% Tween 80 (control) immediately and 20 h after surgical treatment. Splitting of the pericarp and seed removal were completed 24 h prior to [14C]GA12 application to the inside surface (endocarp) of the pericarp. Pericarps were harvested onto dry ice 24 h after [14C]GA12 application and stored at −80°C until extraction. Intact pericarps (3 DAA) and 4-DAA SP and SPNS pericarps (harvested 24 h after surgical treatment at 3 DAA) were also harvested for endogenous GA determination. For all treatments, the seeds were removed from the pericarps prior to harvest of tissue if present, and the pericarps remained attached to the plant throughout the experiment.
For the [14C]GA19 metabolism experiments, plants of genotype SLN (cv Torsdag wild type; JI992) and sln (JI3011) from the John Innes Pisum collection were grown as described above. SLN and sln pericarps were deseeded and treated with 4-Cl-IAA as described above (30 μL, 50 μm in 0.1% [v/v] aqueous Tween 80), and SP and SPNS controls were treated with 0.1% (v/v) aqueous Tween 80. 17-[14C]GA19 (specific radioactivity of 54 mCi mmol−1) was applied (60,000 dpm per pod in 5 μL of 50% [v/v] aqueous methanol) at 12 h after hormone application (24 h after deseeding) to the inside surface of the pericarp wall (endocarp). Treated pericarps (seeds were removed if present) were harvested onto dry ice at 24 h after [14C]GA19 application and stored at −80°C until extraction.
RNA Isolation
Two whole or half pericarps and seeds were ground to a fine powder in liquid N2, and 100 to 500 mg fresh weight of pericarp or 10 to 500 mg fresh weight of seed tissue subsamples was used for RNA extraction. Total RNA was extracted using a modified TRIzol (Invitrogen) procedure. In brief, after initial extraction with the TRIzol reagent and centrifugation, the supernatant was cleaned by chloroform partitioning (0.2 mL mL−1 TRIzol reagent). Subsequently, for further purification, the following steps were carried out in order: high-salt precipitation (1.2 m sodium citrate, 0.8 m NaCl) to remove polysaccharides and proteoglycans, 4 m LiCl precipitation, and precipitation with a 1:20 (v/v) solution of 3 m sodium acetate (pH 5):100% ethanol. The total RNA samples were then treated with DNase (Ambion DNA-free kit) and stored at −80°C prior to real-time RT-PCR analysis.
Gene Expression Analysis
Primers and Probes
The target gene quantifying amplicons CPS1-92 (used for PsCPS1 quantification), GA20ox1-104 (used for PsGA20ox1 quantification), GA20ox2-88 (used for PsGA20ox2 quantification), GA2ox1-73 (used for PsGA2ox1 quantification), and GA2ox2-83 (used for PsGA2ox2 quantification) were designed by Ayele et al. (2006b). Primers and probes used to quantify PsGA3ox1 (PsGA3ox1-87) and the reference gene amplicon 18S-62 (used for pea 18S rRNA quantification) were designed by Ozga et al. (2003). For PsGA3ox2 quantitation, the following primers and probe were used to produce a 104-bp amplicon (PsGA3ox2-104) that spans nucleotides 476 to 579 of DQ864759: forward primer, 5′-ATCATGGGGTCACCGTCTAA-3′; reverse primer, 5′-GCTAGTGTCTTCATTTGCTTTTGA-3′; probe, 5′-CCTAATGACTACGAATATT-3′. The primers for PsGA3ox2 produced a single product of correct length, and sequencing of the product confirmed the specificity (data not shown). TaqMan probes were used for all gene expression studies. All probes for the GA genes were labeled at the 5′ end with the fluorescent reporter dye 6-carboxyfluorescein and at the 3′ end with a minor-groove-binding nonfluorescent quencher (Applied Biosystems). The 18S-62 reference gene probe was labeled at the 5′ end with the VIC fluorescent reporter dye and at the 3′ end with the TAMRA quencher (Applied Biosystems).
Real-Time RT-PCR Assay
Real-time RT-PCR assays were performed on an Applied Biosystems StepOnePlus sequence detector (PsGA3ox1 and PsGA3ox2) or an Applied Biosystems model 7700 sequence detector (all other genes) using the TaqMan One-Step RT-PCR Master Mix Reagent Kit (Applied Biosystems). For each 25-μL reaction, 200 ng of total RNA was mixed with 1× Master Mix (final concentration; containing AmpliTaq Gold DNA polymerase), 2.5× MultiScribe (final concentration; reverse transcriptase and RNase inhibitor mix), 300 nm (final concentration) each forward and reverse primer, 100 nm probe (final concentration), and diethyl pyrocarbonate-treated water (to bring the reaction volume to 25 μL). Samples were subjected to thermal cycling conditions as described previously (Ozga et al., 2003), and the average of the two subsamples was used to calculate the sample transcript abundance. Total RNA from one sample was run on each plate and used as a control to correct for plate-to-plate amplification differences (Ayele et al., 2006a). The relative transcript abundance of the target genes in the individual plant samples was determined by the 2−ΔCt method (Livak and Schmittgen, 2001), where ΔCt is the difference between the target sample Ct and the average Ct of the reference sample. For PsCPS1, PsGA20ox1, PsGA20ox2, PsGA2ox1, and PsGA2ox2, transcript levels were compared across genes, developmental stages, and tissues using the lowest sample average Ct value (Ct = 37.975) obtained in the study for these genes as the reference sample. For PsGA3ox1 and PsGA3ox2, transcript levels were compared across genes, developmental stages, and tissues using the lowest sample average Ct value of 40 obtained for these genes as the reference sample. At least two, and often three, replicate plant samples were assayed.
The pea 18S small subunit nuclear rRNA gene was used as a loading control to estimate the variation in total RNA input of the samples. For PsCPS1, PsGA20ox1, PsGA20ox2, PsGA2ox1, and PsGA2ox2 genes, 10 pg of DNase-treated total RNA was used for 18S rRNA quantitation using the same reaction and thermocycling conditions described above on the Applied Biosystems model 7700 sequence detector. The coefficient of variation of the 18S rRNA amplicon Ct values among the samples was between 1% and 1.3%; therefore, the target amplicon mRNA values were not normalized to the 18S signal (Livak and Schmittgen, 2001).
For PsGA3ox1 and PsGA3ox2 genes, a competitive primer approach was used to quantify 18S rRNA transcript levels. 18S rRNA was quantified on 3 ng of DNase-treated total RNA generated from a single dilution of the original 25 ng μL−1 stocks (final concentration of 120 pg μL−1) using the same reaction and thermocycling conditions described above on the Applied Biosystems StepOnePlus sequence detector. The addition of competitive primers [primers with the same sequence as the 18S primers but lacking the 3′ hydroxyl group necessary for DNA polymerase elongation, in this case (CH2)6NH2] along with primers containing a 3′ hydroxyl group to the PCR mixture allows a larger amount of template RNA to be used while still maintaining an acceptable reaction profile, decreasing the variation inherent in serial dilutions of RNA samples. An optimal 1:9 ratio of primers containing 3′ hydroxyl and 3′ (CH2)6NH2 chain terminators was used as determined through experiments varying input RNA and the ratio of 3′ hydroxyl to 3′ (CH2)6NH2 primers. The coefficient of variation of the 18S rRNA amplicon Ct values among these samples was again low (2.4%–2.9%); therefore, the target amplicon mRNA values were not normalized to the 18S signal (Livak and Schmittgen, 2001).
[14C]GA12 Metabolism Experiments
Pericarps (39–41; 4–25 g fresh weight) were homogenized in cold 80% aqueous methanol (4 mL g−1 tissue) containing 10 mg L−1 butylated hydroxytoluene using a Polytron homogenizer. To each of these extracts, [2H]GA19, [2H]GA20, [2H]GA1, [2H]GA8, and [13C-3H]GA29 were added at homogenization as internal standards for gas chromatography-mass spectrometry-selected ion monitoring (GC-MS-SIM). The tissue homogenates were extracted for 12 h by shaking and gently stirring in darkness at 4°C. The tissue was allowed to settle, the supernatant was decanted, and the tissue was reextracted with the same volume of solvent for an additional 12 h as described above. After decanting the supernatant, the tissue pellet was washed three times with 2 mL of extraction solvent, and both supernatants and the solvent wash were pooled and evaporated to dryness under vacuum in silylated glass vials.
The extracts were resuspended in 20 mm imidazole buffer (pH 7.0) and applied to a conditioned DEAE-Sephacel (Pharmacia) column (DEAE-Sephacel [1.2 mL g−1 fresh weight tissue] conditioned with the following solvents sequentially [ratio of solvent to bed volume]: hexane [2×], acetonitrile [2×], water [2×], 0.2 m imidazole buffer [pH 7.0; 2×], and water [10× to remove excess buffer]). The columns were washed sequentially with the following solvents (volume of solvent 2× bed volume): hexane, ethyl acetate, acetonitrile, methanol, and methanol with 2% acetic acid. The methanol fraction containing the GAs was collected, dried under vacuum, and then brought up in 1 mL of methanol:ethyl acetate (1:1) followed by two drops of water. This extract was applied to a hydrated silica column (1 g of hydrated SiO2 at 22% water [w/w] per 1 g fresh weight of tissue) packed in 95:5 hexane:ethyl acetate (formate saturated). The hydrated silica column was then washed with 10 times its bed volume with 5:95 hexane:ethyl acetate (v/v; formate saturated), and this fraction, which contained the free-GAs, was dried under vacuum prior to further purification using HPLC.
HPLC
The partially purified extracts were resuspended in 400 μL of 20% aqueous methanol and passed through 0.45-μm nylon filters prior to injection onto a 4.5- × 250-mm Spherisorb C18 column (5 μm; Beckman). The samples were eluted at a flow rate of 1.0 mL min−1 using a linear gradient of 0.01% aqueous trifluoroacetic acid (solvent A) and 100% methanol (solvent B). Conditions of the linear gradient were 20% solvent B for 1 min, gradient to 100% solvent B in 45 min, and isocratic 100% solvent B for 5 min. Radioactivity in the effluent was monitored using a flow-through radiochemical detector (Beckman 171). Radioactive fractions eluting near standard retention times of GA8 (9.2 min), GA29 (12.6 min), GA1 (17.8 min), GA20 (26.4 min), and GA19 (29.2 min) were collected and dried down.
[14C]GAs were converted to their methyl esters using diazomethane. The [14C]GA methyl esters were subsequently converted to trimethylsilyl ether derivatives (Gaskin and MacMillan, 1991) for identification and quantitation by GC-MS-SIM. Mass spectral analyses of derivatized samples were performed using a Hewlett-Packard model 5890 Series II Gas Chromatograph interfaced to a Hewlett-Packard model 5972A Mass Selective Detector equipped with a HP-5 MS column (30 m × 0.25 mm × 0.25 m film thickness). Helium was used as the carrier gas at a flow rate of 1 mL min−1. The samples were injected on-column with the initial column temperature at 50°C for 2 min, followed by temperature programming at 10°C min−1 to 150°C and then at 3°C min−1 to 300°C. Selective ion monitoring of three prominent ions for each GA of interest and Kovats retention index data were used for confirmation of [14C]GA identity. For quantitation, the protio- and deutero-ions monitored were corrected for donation of natural isotopes to the peak area. A separate calibration curve of peak area ratio versus molar ratio of [2H]GA/protio-GA was constructed for GA19, GA20, GA1, and GA8 (Gaskin and MacMillan, 1991). Using the corrected peak area and the calibration curve, the total amounts of the protio-GAs were calculated. For GA29 and the 14C-labeled GAs (protio-ion + 16 atomic mass units; 8 14C-molecules per GA molecule), the total amounts were calculated by reference to the stable isotope-labeled internal standard using equations for isotope dilution analysis (Bandurski and Schulze, 1977). For calculation of endogenous GAs, the most prominent ion measured (usually M+) was used for quantitation, and the calculated value was checked for reproducibility using the second most prominent ion.
[14C]GA19 Metabolism Experiments
Using a Polytron homogenizer, radiolabeled pericarps (three per sample) were homogenized in silylated 30-mL Corex tubes with 10 mL of cold 80% (v/v) methanol containing 10 mg L−1 butylated hydroxytoluene. An external standard, 10,000 dpm of 17-[14C]GA7, was added at the time of homogenization for determination of radioactive metabolite recovery. After homogenization, samples were gently shaken in darkness at 4°C for 12 to 16 h and then centrifuged at 10,000g for 30 min. The supernatant was removed, and the residue was resuspended in 10 mL of the homogenization solvent and gently shaken at 4°C in darkness for at least 4 h. The residue extracts were centrifuged at 10,000g for 30 min, and the combined supernatants were reduced to the aqueous phase using a vacuum concentrator (Savant). The pH of the aqueous extracts was adjusted to 8.0 with NH4OH (0.1 n) and the extracts partitioned against n-hexane (5 mL) four times in silylated 20-mL glass scintillation vials. The aqueous fraction was then adjusted to pH 3.0 with 0.1 n HCl and partitioned against ethyl acetate (5 mL) five times. The combined ethyl acetate extracts were reduced to approximately 3 mL using a vacuum concentrator and partitioned against 5% (w/v) aqueous NaHCO3 (5 mL) four times. The combined NaHCO3 extracts were transferred to 30-mL silylated Pyrex tubes, the pH was adjusted to 3.0 with concentrated HCl on ice, and the extracts partitioned against ethyl acetate (5 mL) five times. The ethyl acetate extracts were combined and evaporated to near dryness, transferred to 7-mL silylated scintillation vials, and dried down under vacuum.
The ethyl acetate extracts were subject to the same HPLC procedure as described above. Radioactive fractions eluting near standard retention times of GA8 (9.2 min), GA29 (12.6 min), GA3 (16.3 min), GA1 (17.8 min), GA5 (25.0 min), GA20 (26.4 min), GA19 (29.2 min), and GA7 (31.0 min) were collected and dried down. Collected [14C]GAs were methylated using diazomethane and rechromatographed as their methyl esters by C18-HPLC using the same solvent system.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AF001219, AF056935, AF100954, DQ864759, U58830, U63652, and U70471.
This work was supported by the Natural Sciences and Engineering Research Council of Canada.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Jocelyn A. Ozga (jocelyn.ozga@ualberta.ca).
Open Access articles can be viewed online without a subscription.
References
- Ait-Ali T, Swain SM, Reid JB, Sun TP, Kamiya Y (1997) The LS locus of pea encodes the gibberellin biosynthesis enzyme ent-kaurene synthase A. Plant J 11 443–454 [DOI] [PubMed] [Google Scholar]
- Ayele BT, Ozga JA, Kurepin LV, Reinecke DM (2006. a) Developmental and embryo axis regulation of gibberellin biosynthesis during germination and young seedling growth of pea. Plant Physiol 142 1267–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayele BT, Ozga JA, Reinecke DM (2006. b) Regulation of GA biosynthesis genes during germination and young seedling growth of pea (Pisum sativum L.). J Plant Growth Regul 25 219–232 [Google Scholar]
- Bandurski RS, Schulze A (1977) Concentration of indole-3-acetic acid and its derivatives in plants. Plant Physiol 60 211–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnberg PR, Maki SL, Brenner ML, Davis GC, Carnes MG (1986) An improved enzymatic synthesis of labeled gibberellin A12-aldehyde and gibberellin A12. Anal Biochem 153 1–8 [DOI] [PubMed] [Google Scholar]
- Bowen DH, MacMillan J, Graebe JE (1972) Determination of specific radioactivity of [14C]-compounds by mass spectroscopy. Phytochemistry 11 2253–2257 [Google Scholar]
- Davidson SE, Swain SM, Reid JB (2005) Regulation of the early GA biosynthesis pathway in pea. Planta 222 1010–1019 [DOI] [PubMed] [Google Scholar]
- Eeuwens CJ, Schwabe WW (1975) Seed and pod wall development in Pisum sativum L. in relation to extracted and applied hormones. J Exp Bot 26 1–14 [Google Scholar]
- Frigerio M, Alabadi D, Perez-Gomez J, Garcia-Carcel L, Phillips AL, Hedden P, Blazquez MA (2006) Transcriptional regulation of gibberellin metabolism genes by auxin signaling in Arabidopsis. Plant Physiol 142 553–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Martinez JL, Lopez-Diaz I, Sanchez-Beltran MJ, Phillips AL, Ward DA, Gaskin P, Hedden P (1997) Isolation and transcript analysis of gibberellin 20-oxidase genes in pea and bean in relation to fruit development. Plant Mol Biol 33 1073–1084 [DOI] [PubMed] [Google Scholar]
- Garcia-Martinez JL, Santes C, Croker SJ, Hedden P (1991) Identification, quantitation and distribution of gibberellins in fruits of Pisum sativum L. cv. Alaska during pod development. Planta 184 53–60 [DOI] [PubMed] [Google Scholar]
- Gaskin P, MacMillan J (1991) GC-MS of the Gibberellins and Related Compounds: Methodology and a Library of Spectra. University of Bristol (Cantock's Enterprises), Bristol, UK
- Goetz M, Vivian-Smith A, Johnson SD, Koltunow AM (2006) AUXIN RESPONSE FACTOR8 is a negative regulator of fruit initiation in Arabidopsis. Plant Cell 18 1873–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P, Phillips AL (2000) Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci 5 523–530 [DOI] [PubMed] [Google Scholar]
- Johnstone MMG (2004) Phytohormones in early pea fruit growth. MSc thesis. University of Alberta, Edmonton, Alberta, Canada
- Johnstone MMG, Reinecke DM, Ozga JA (2005) The auxins IAA and 4-Cl-IAA differentially modify gibberellin action via ethylene response in developing pea fruit. J Plant Growth Regul 24 214–225 [Google Scholar]
- Lester DR, Ross JJ, Davies PJ, Reid JB (1997) Mendel's stem length gene (Le) encodes a gibberellin 3β-hydroxylase. Plant Cell 9 1435–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester DR, Ross JJ, Smith JJ, Elliott RC, Reid JB (1999) Gibberellin 2-oxidation and the SLN gene of Pisum sativum. Plant J 19 65–73 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25 402–408 [DOI] [PubMed] [Google Scholar]
- MacKenzie-Hose AK, Ross JJ, Davies NW, Swain SM (1998) Expression of gibberellin mutations in fruits of Pisum sativum L. Planta 204 397–403 [Google Scholar]
- Magnus V, Ozga JA, Reinecke DM, Pierson GL, Larue TA, Cohen JD, Brenner ML (1997) 4-Chloroindole-3-acetic acid and indole-3-acetic acid in Pisum sativum. Phytochemistry 46 675–681 [Google Scholar]
- Maki SL, Brenner ML (1991) [14C]GA12-aldehyde, [14C]GA12, and [2H]- and [14C]GA53 metabolism by elongating pea pericarp. Plant Physiol 97 1359–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DN, Proebsting WM, Hedden P (1997) Mendel's dwarfing gene: cDNAs from the Le alleles and function of the expressed proteins. Proc Natl Acad Sci USA 94 8907–8911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DN, Proebsting WM, Hedden P (1999) The SLENDER gene of pea encodes a gibberellin 2-oxidase. Plant Physiol 121 775–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DN, Proebsting WM, Parks TD, Dougherty WG, Lange T, Lewis MJ, Gaskin P, Hedden P (1996) Feed-back regulation of gibberellin biosynthesis and gene expression in Pisum sativum L. Planta 200 159–166 [DOI] [PubMed] [Google Scholar]
- Marumo S, Hattori H, Abe H, Munakata K (1968) Isolation of 4-chloroindolyl-3-acetic acid from immature seeds of Pisum sativum. Nature 219 959–960 [DOI] [PubMed] [Google Scholar]
- Ngo P, Ozga JA, Reinecke DM (2002) Specificity of auxin regulation of gibberellin 20-oxidase gene expression in pea pericarp. Plant Mol Biol 49 439–448 [DOI] [PubMed] [Google Scholar]
- O'Neill DP, Ross JJ (2002) Auxin regulation of the gibberellin pathway in pea. Plant Physiol 130 1974–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozga JA, Brenner ML, Reinecke DM (1992) Seed effects on gibberellin metabolism in pea pericarp. Plant Physiol 100 88–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozga JA, Reinecke DM (1999) Interaction of 4-chloroindole-3-acetic acid and gibberellins in early pea fruit development. Plant Growth Regul 27 33–38 [Google Scholar]
- Ozga JA, van Huizen R, Reinecke DM (2002) Hormone and seed-specific regulation of pea fruit growth. Plant Physiol 128 1379–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozga JA, Yu J, Reinecke DM (2003) Pollination-, development-, and auxin-specific regulation of gibberellin 3β-hydroxylase gene expression in pea fruit and seeds. Plant Physiol 131 1137–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pate JS, Flinn AM (1977) Fruit and seed development. In JF Sutcliffe, JS Pate, eds, The Physiology of the Garden Pea. Academic Press, New York, pp 431–468
- Pharis RP, King RW (1985) Gibberellins and reproductive development in seed plants. Annu Rev Plant Physiol 36 517–568 [Google Scholar]
- Reinecke DM (1999) 4-Chloroindole-3 acetic acid and plant growth. Plant Growth Regul 27 3–13 [Google Scholar]
- Reinecke DM, Ozga JA, Ilic N, Magnus V, Kojic-Prodic B (1999) Molecular properties of 4-substituted indole-3-acetic acids affecting pea pericarp elongation. Plant Growth Regul 27 39–48 [Google Scholar]
- Reinecke DM, Ozga JA, Magnus V (1995) Effect of halogen substitution of indole-3-acetic acid on biological activity in pea fruit. Phytochemistry 40 1361–1366 [Google Scholar]
- Rodrigo MJ, Garcia-Martinez JL, Santes CM, Gaskin P, Hedden P (1997) The role of gibberellins A1 and A3 in fruit growth of Pisum sativum L. and the identification of gibberellins A4 and A7 in young seeds. Planta 201 446–455 [Google Scholar]
- Ross JJ, Reid JB, Dungey HS (1992) Ontogenetic variation in levels of gibberellin A1 in Pisum: implications for the control of stem elongation. Planta 186 166–171 [DOI] [PubMed] [Google Scholar]
- Ross JJ, Reid JB, Swain SM, Hasan O, Poole AT, Hedden P, Willis CL (1995) Genetic regulation of gibberellin deactivation in Pisum. Plant J 7 513–523 [Google Scholar]
- Santes CM, Garcia-Martinez JL (1995) Effect of the growth retardant 3,5-dioxo-4-butyryl-cyclohexane carboxylic acid ethyl ester, an acylcyclohexanedione compound, on fruit growth and gibberellin content of pollinated and unpollinated ovaries in pea. Plant Physiol 108 517–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santes CM, Hedden P, Sponsel VM, Reid JM, Garcia-Martinez JL (1993) Expression of the le mutation in young ovaries of Pisum sativum and its effect on fruit development. Plant Physiol 101 759–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrani JC, Ruiz-Rivero O, Fos M, Garcia-Martinez JL (2008) Auxin-induced fruit-set in tomato is mediated in part by gibberellins. Plant J 56 922–934 [DOI] [PubMed] [Google Scholar]
- Serrani JC, Sanjuan R, Ruiz-Rivero O, Fos M, Garcia-Martinez JL (2007) Gibberellin regulation of fruit set and growth in tomato. Plant Physiol 145 246–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sponsel VM (1982) Effects of applied gibberellins and naphthylacetic acid on pod development in fruits of Pisum sativum L. cv. Progress No. 9. J Plant Growth Regul 1 147–152 [Google Scholar]
- Sponsel VM (1995) The biosynthesis and metabolism of gibberellins in higher plants. In PJ Davies, ed, Plant Hormones: Physiology, Biochemistry and Molecular Biology, Ed 2. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 66–97
- Thomas SG, Phillips AL, Hedden P (1999) Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc Natl Acad Sci USA 96 4698–4703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Huizen R, Ozga JA, Reinecke DM (1996) Influence of auxin and gibberellin on in vivo protein synthesis during early pea fruit growth. Plant Physiol 112 53–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Huizen R, Ozga JA, Reinecke DM (1997) Seed and hormonal regulation of gibberellin 20-oxidase expression in pea pericarp. Plant Physiol 115 123–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Huizen R, Ozga JA, Reinecke DM, Twitchin B, Mander LN (1995) Seed and 4-chloroindole-3-acetic acid regulation of gibberellin metabolism in pea pericarp. Plant Physiol 109 1213–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston DE, Elliott RC, Lester DR, Rameau C, Reid JB, Murfet IC, Ross JJ (2008) The pea DELLA proteins LA and CRY are important regulators of gibberellin synthesis and root growth. Plant Physiol 147 199–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S (2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59 225–251 [DOI] [PubMed] [Google Scholar]