Abstract
Receptor-Like Kinase (RLK)/Pelle genes play roles ranging from growth regulation to defense response, and the dramatic expansion of this family has been postulated to be crucial for plant-specific adaptations. Despite this, little is known about the history of or the factors that contributed to the dramatic expansion of this gene family. In this study, we show that expansion coincided with the establishment of land plants and that RLK/Pelle subfamilies were established early in land plant evolution. The RLK/Pelle family expanded at a significantly higher rate than other kinases, due in large part to expansion of a few subfamilies by tandem duplication. Interestingly, these subfamilies tend to have members with known roles in defense response, suggesting that their rapid expansion was likely a consequence of adaptation to fast-evolving pathogens. Arabidopsis (Arabidopsis thaliana) expression data support the importance of RLK/Pelles in biotic stress response. We found that hundreds of RLK/Pelles are up-regulated by biotic stress. Furthermore, stress responsiveness is correlated with the degree of tandem duplication in RLK/Pelle subfamilies. Our findings suggest a link between stress response and tandem duplication and provide an explanation for why a large proportion of the RLK/Pelle gene family is found in tandem repeats. In addition, our findings provide a useful framework for potentially predicting RLK/Pelle stress functions based on knowledge of expansion pattern and duplication mechanism. Finally, we propose that the detection of highly variable molecular patterns associated with specific pathogens/parasites is the main reason for the up-regulation of hundreds of RLK/Pelles under biotic stress.
The Receptor-Like Kinase (RLK)/Pelle protein kinase family is the largest gene family in Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa), with more than 600 and 1,100 family members, respectively (Shiu and Bleecker, 2001a, 2001b, 2003; Shiu et al., 2004). In contrast to plants, animals have a much smaller number of RLK/Pelles (Shiu and Bleecker, 2001b). The Pelle kinase is the sole member of the RLK/Pelle family in Drosophila melanogaster and is involved in dorsal-ventral axis determination (Hecht and Anderson, 1993; Shelton and Wasserman, 1993) as well as in innate immunity (Belvin and Anderson, 1996). The human RLK/Pelle family includes four interleukin-1 receptor-associated kinases that are involved in innate and adaptive immunity (Janssens and Beyaert, 2003). No RLK/Pelle genes have been identified in fungi (Shiu and Bleecker, 2003). These cursory surveys of various eukaryotic genomes indicate that dramatic expansion of the RLK/Pelle family appears to be limited to plants. In addition to the large family size, another important feature of plant members of the RLK/Pelle family is that they can have two main configurations: (1) Receptor-Like Cytoplasmic Kinases (RLCKs) lack an extracellular domain (ECD), and (2) RLKs contain an intracellular kinase domain, a transmembrane domain, and an ECD with one or more of a diverse array of protein domains (Shiu and Bleecker, 2001a). ECDs likely allow RLKs to respond to a variety of extracellular signals. These signals include those derived from the plant itself, such as the small protein ligand, CLAVATA3, which binds to the RLK, CLAVATA1, to restrict meristem proliferation (Ogawa et al., 2008), as well as those derived from microbes, including flagellin (Felix et al., 1999) and cell wall components such as chitin and peptidoglycans (Madsen et al., 2003; Radutoiu et al., 2003; Miya et al., 2007; Wan et al., 2008). Fusions between diverse ECDs and kinase domains provide a source of innovation in signaling networks by linking novel inputs to existing response networks. It is likely, therefore, that the evolutionary success of this gene family is tied to the need for a diverse array of receptors to perceive highly variable, and in the case of biotic agents, rapidly evolving stimuli (Shiu et al., 2004).
As suggested by the diversity and size of the RLK/Pelle family, its members are involved in many different processes in plants. RLK/Pelle members function in a wide range of developmental processes, such as the regulation of meristem proliferation (Clark et al., 1997; DeYoung et al., 2006), organ specification (Torii et al., 1996; Muto et al., 2004; Shpak et al., 2004; Chevalier et al., 2005; Kwak et al., 2005; Eyuboglu et al., 2007), reproduction (Albrecht et al., 2005; Colcombet et al., 2005; Escobar-Restrepo et al., 2007), and hormone signal transduction (Li and Chory, 1997; Li et al., 2002; Nam and Li, 2002; Zhou et al., 2004). Other RLK/Pelle members function in signaling networks involving environmental stimuli, both abiotic (Sivaguru et al., 2003; Hou et al., 2005) and biotic (Song et al., 1995; Gomez-Gomez and Boller, 2000; Endre et al., 2002; Stracke et al., 2002; Chen et al., 2003; Fontes et al., 2004; Sun et al., 2004; Diener and Ausubel, 2005; Florentino et al., 2006; Zipfel et al., 2006; Acharya et al., 2007; Miya et al., 2007). In some cases, RLK/Pelle genes exhibit multifunctionality, playing roles in both development and defense (for review, see Bent and Mackey, 2007).
A great deal of research has focused on the roles of RLK/Pelle genes in defense response. RLK/Pelle genes have been shown to play roles in basal immunity, where components such as flagellin and chitin that are common to both pathogenic and nonpathogenic microbes (microbe-associated molecular patterns [MAMPs]) are perceived and lead to activation of defense signal transduction networks. Other RLK/Pelles function in resistance gene (R)-mediated defense, where pathogen-specific effectors are recognized (Bent and Mackey, 2007). For example, FLS2 and EFR are MAMP receptors (Gomez-Gomez and Boller, 2000; Zipfel et al., 2006), and the R gene Xa21 mediates resistance to the rice blast pathogen Xanthomonas oryzae (Song et al., 1995). It is thought that many more RLK/Pelle members may function as either MAMP receptors or R genes and that they might be identified based on characteristics such as type and/or structure of ECD, subfamily classification, mode of gene duplication, expression pattern after treatment with pathogens, or kinase sequence signature (Shiu et al., 2004; Dardick and Ronald, 2006; Thilmony et al., 2006; Afzal et al., 2008).
The ability to predict RLK/Pelle functions would be extremely useful because the vast majority of them still do not have known functions. This is particularly true for genes in RLK/Pelle subfamilies that have undergone dramatic expansion and those that are derived from tandem duplication. Therefore, we set out to determine the relationship between the diversity of RLK/Pelle genes, the degree of RLK/Pelle family expansion, duplication mechanism, and plant stress responses. We first conducted a computational analysis of the RLK/Pelle family using genomic information from four land plants and two green algae to determine: (1) when the receptor configuration arose, (2) how often new RLK/Pelle subfamilies have been created, and (3) how subfamilies differ in their patterns of expansion and loss in different plant lineages. To better understand the properties of RLK/Pelle genes that are involved in stress response, we used publicly available Arabidopsis microarray data to identify RLK/Pelle genes that are responsive to abiotic and biotic stresses. We then asked if stress responsiveness of RLK/Pelle members is correlated with patterns of lineage-specific expansion and duplication mechanism and how well characteristics of RLK/Pelle genes can predict stress responsiveness. Our findings indicate a significant positive correlation between RLK/Pelle subfamily expansion, the degree of tandem duplication, and stress responsiveness as well as complex interactions between tandem duplication, expansion, and receptor configuration. Based on these results, we discuss why there are hundreds of stress-responsive RLK/Pelle genes.
RESULTS AND DISCUSSION
The Evolutionary History of RLK/Pelle Family Expansion in Viridiplantae
Our earlier studies established that the RLK/Pelle family has expanded dramatically in rice and Arabidopsis (Shiu and Bleecker, 2001b, 2003, Shiu et al., 2004). However, it is not clear when the RLK/Pelle family expanded and why there are such large differences in family size between Arabidopsis and rice. To address these questions, we first identified Ser/Thr/Tyr protein kinase domain-containing protein sequences (referred to as “kinase”) from two algae, Ostreococcus tauri (Derelle et al., 2006) and Chlamydomonas reinhardtii (Merchant et al., 2007), and four land plants, a moss (Physcomitrella patens; Rensing et al., 2008), rice (International Rice Genome Sequencing Project, 2005), poplar (Populus trichocarpa; Tuskan et al., 2006), and Arabidopsis (Arabidopsis Genome Initiative, 2000). Kinase domain sequences from all six species were aligned and used to construct a phylogenetic tree. Protein kinases were then classified based on grouping with known kinase families from Arabidopsis and rice (see “Materials and Methods”).
The availability of sequences from these six genomes allowed us to examine the evolutionary trajectories of protein kinases throughout the evolution of the land plants, from the transition to land to the divergence of nonvascular and vascular plants to the divergence of monocots from dicots (Fig. 1). First, we examined the kinase superfamily in two green algae (O. tauri and C. reinhardtii) to determine if RLK/Pelle members were present and if the receptor configuration (i.e. ECD + kinase) had been established before the green algae diverged from the land plants approximately 109 years ago (Table I; Yoon et al., 2004). Of the 426 kinases in C. reinhardtii, only two are RLK/Pelles, and the predicted proteins do not have recognizable ECDs. O. tauri, the smallest known free-living eukaryote (Courties et al., 1994), has 93 kinases but lacks any recognizable RLK/Pelle. RLK/Pelles have also been isolated from the multicellular alga Nitella axillaris and the unicellular alga Closterium ehrenbergii (Sasaki et al., 2007). Most interestingly, transmembrane domains and/or domains in the ECDs of land plant RLKs are found in C. ehrenbergii and N. axillaris RLK/Pelle genes. For example, the C. ehrenbergii RLK/Pelle gene, CeRLK8, has leucine-rich repeats (LRRs) in its ECD. Based on the parsimony assumption, the receptor configuration was likely established before the divergence of land plants from charophytes (including C. ehrenbergii and N. axillaris) but before the divergence of charophytes from chlorophytes (including C. reinhardtii and O. tauri; Bhattacharya and Medlin, 1998).
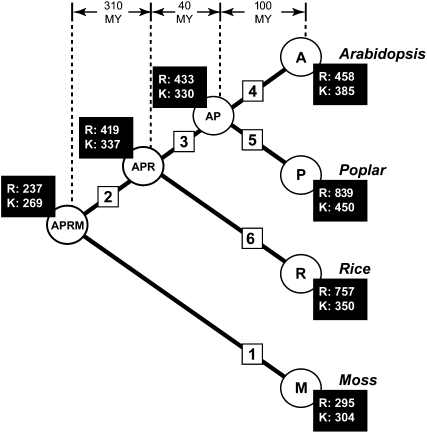
Figure 1.
Expansion of RLK/Pelle and other kinase families during land plant evolution. The phylogeny indicates the relationships between the four land plant species analyzed (A, Arabidopsis; P, poplar; R, rice; M, moss). The internal nodes are labeled indicating the common ancestors of the four plants (AP, APR, and APRM). The branches are labeled 1 to 6 to facilitate comparisons. The numbers of RLK/Pelle kinases (R) and other kinases (K), calculated based on subfamilies shared between all four species, at each node are shown in black boxes. Ancestral gene numbers were estimated by reconciling the ML gene tree of each kinase subfamily with the four-plant species tree (see “Materials and Methods”).
Table I.
Representation of the kinase superfamily and the RLK/Pelle family in selected eukaryotic genomes
| Species | No. of Kinases (K)a | No. of RLK/Pelles (R)b
|
No. of Protein-Coding Genes (G)c | Percentage (K/G)d | Percentage (R/G)e | |
|---|---|---|---|---|---|---|
| RLK | RLCK | |||||
| Saccharomyces cerevisiae | 113 | 0 | 0 | 6,449 | 1.75 | 0 |
| Candida albicans | 95 | 0 | 0 | 6,146 | 1.55 | 0 |
| Drosophila melanogaster | 262 | 0 | 1 | 13,808 | 1.90 | 0.007 |
| Homo sapiens | 472 | 0 | 4 | 22,980 | 2.05 | 0.017 |
| Ostreococcus tauri | 93 | 0 | 0 | 8,166 | 1.14 | 0 |
| Chlamydomonas reinhardtii | 426 | 0 | 2 | 15,143 | 2.81 | 0.013 |
| Physcomitrella patens (moss) | 685 | 136 | 193 | 39,796 | 1.72 | 0.83 |
| Oryza sativa subsp. japonica (rice) | 1,446 | 786 | 284 | 42,653 | 3.39 | 2.48 |
| Populus trichocarpa (poplar) | 1,643 | 726 | 466 | 45,555 | 3.61 | 2.40 |
| Arabidopsis thaliana (Arabidopsis) | 998 | 443 | 167 | 26,751 | 3.73 | 2.28 |
Number of protein kinase domain-containing sequences (including RLK/Pelle family members).
Number of RLK/Pelle sequences.
Number of predicted protein-coding genes (S. cerevisiae, C. albicans, D. melanogaster, and H. sapiens [Shiu and Bleecker, 2003], O. tauri [Derelle et al., 2006], C. reinhardtii [Merchant et al., 2007], moss [Rensing et al., 2008], rice [Yuan et al., 2005], poplar [Tuskan et al., 2006], and Arabidopsis [TAIR version 6]).
Percentage of protein-coding genes that are protein kinases.
Percentage of protein-coding genes that are RLK/Pelle family members.
Based on cDNA evidence, at least 29 RLK/Pelle genes are present in liverwort (Marchantia polymorpha; Sasaki et al., 2007), indicating that the RLK/Pelle family began to expand early in the land plant lineage. In P. patens (referred to as “moss”), a relatively large number (329) of RLK/Pelles are found, including 136 RLKs and 193 RLCKs (Table I). It is clear that the RLK/Pelle family continued to expand in the vascular plant lineage after the divergence from the moss lineage, since there are 1.9, 3.3, and 3.6 times as many members in Arabidopsis, rice, and poplar, respectively, compared with moss. Based on estimated numbers of ancestral RLK/Pelle genes, the most dramatic expansion occurred immediately following the divergence of the vascular plants and independently in the rice and poplar lineages (Fig. 1; Supplemental Fig. S1). Interestingly, the number of all other non-RLK/Pelle kinases is approximately 400 in C. reinhardtii and all the land plants, indicating that variation in the RLK/Pelle family is the major contributor to variation in the kinase superfamily size among plant species (Table I). In fact, in all lineages except for the Arabidopsis lineage, RLK/Pelles expanded at a higher rate than other kinases (Supplemental Fig. S1).
Note that the RLK/Pelle family can be subdivided into multiple subfamilies based on phylogenetic relationships between members and that RLK/Pelle genes with related kinase sequences tend to have similar ECDs (Shiu and Bleecker, 2003; Shiu et al., 2004). Variation in the number of RLK/Pelle family members among land plants may reflect substantial differences in subfamily composition among species. Therefore, to understand how the RLK/Pelle subfamilies diversified over land plant evolution, we next analyzed the RLK/Pelle subfamily composition in each species and expansion rates for each subfamily.
Rate of RLK Diversification via Domain Fusion
The diversity of ECDs makes the RLK/Pelle family one of the most versatile plant gene families; members are capable of recognizing a wide range of ligands. In addition, some subfamily members have different predicted protein domains within the ECD or lack ECDs completely compared with other members of that subfamily (Shiu and Bleecker, 2003; Shiu et al., 2004). New receptors may be able to broaden a plant's ability to sense extracellular signals and to link those signals to existing signaling networks. There are two outstanding questions regarding the domain content evolution of RLK/Pelle members. The first is when the different RLK/Pelle subfamilies, defined based on kinase sequence similarity, were established. The second is how frequently novel RLKs with distinct ECDs arose.
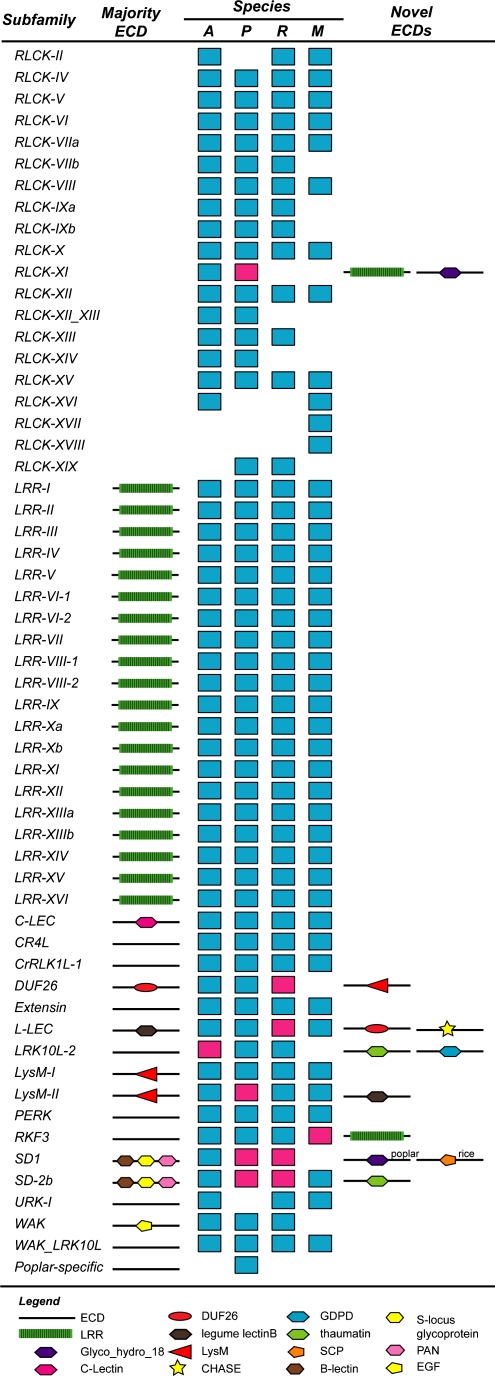
To evaluate how many different RLK/Pelle subfamilies were established in the Arabidopsis-poplar-rice-moss (APRM) common ancestor, we first determined which subfamilies were present in each of the four plant species (Fig. 2). Strikingly, most RLK/Pelle subfamilies (44 of 57 or 77%) are found in moss. Moreover, there are only a few species-specific subfamilies, including two moss-specific RLCK families and one poplar-specific family. Note that previously we identified rice-specific subfamilies, including RLCK-OS1-4 and WAKL-OS (Shiu et al., 2004); however, in this study, these rice genes did not clearly resolve into separate subfamilies after adding poplar and moss sequences. We found that most subfamilies with receptor kinase members are conserved (31 of 37), usually with most subfamily members having the same ECDs (Fig. 2; Supplemental Table S1). In addition, 16 RLK/Pelle subfamilies are found in liverwort (Sasaki et al., 2007). Therefore, many events that resulted in the fusion between ECDs and kinase domains occurred early in land plant evolution, at least prior to the divergence between liverwort and the rest of the land plant lineages. Notable exceptions to the early establishment of receptor kinases are the vascular plant-specific DUF26, LRK10L-2, SD1, and WAK subfamilies, raising the question of how frequently novel receptors have arisen during land plant evolution.
Figure 2.
RLK/Pelle subfamily representation among four plant species and innovation in receptor kinase configuration. The first column shows subfamily names. The second column indicates the presence of an ECD found in most members of that subfamily. ECDs with known protein domains are indicated by symbols, with legends shown at the bottom. In the third column (species), the colored rectangles indicate the presence of subfamily member(s) in Arabidopsis (A), poplar (P), rice (R), and moss (M). A red rectangle indicates the gain of a protein domain. The identities of the novel domains are shown to the right. For the SD1 subfamily, poplar and rice RLK/Pelle genes gained novel ECDs independently, and these are labeled accordingly.
Using the presence of a protein domain different from the majority of subfamily members as an indication of potential domain gain, we found that innovation in receptor configuration has occurred in all land plant lineages analyzed (Fig. 2, red rectangles; Supplemental Table S2). In most cases, the domains gained, such as thaumatin, LysM, LRR, and DUF26, are not novel in the sense that they are already found in other RLK subfamilies. However, these “old” domains were paired with kinases from different RLK subfamilies with potentially different downstream components. In addition, several RLCKs have predicted signal sequences and transmembrane regions that resemble ECDs. However, these putative ECDs have no known protein domains, and it remains to be seen if they are truly cell surface receptors (Supplemental Table S1). One common feature among these newly acquired protein domains in the ECDs (Fig. 2; Supplemental Table S2) is that they have been found in genes implicated in defense response, and several have been shown to bind pathogen components or have the potential to do so (Perrakis et al., 1994; Song et al., 1995; Gomez-Gomez and Boller, 2000; Santelli et al., 2004; Kaku et al., 2006; Miya et al., 2007; Wan et al., 2008).
Taken together, the potential for many of these newly acquired domains to recognize microbial components underscores the importance of the RLK/Pelle family in recognition of plant pathogen components and suggests that repeated innovation through domain acquisition was likely selected for. Although RLK domain content is rather dynamic, we should emphasize that receptor configuration has remained largely the same since the divergence of the vascular plants from moss. Only 12 clear examples of domain-gain events in the flowering plant lineage have occurred in the past 150 million years (Supplemental Table S2; Chaw et al., 2004). Overall, it appears that dramatically changing the configuration of RLKs is not the most common way in which novel RLKs were established. Instead, the expansion of existing RLK/Pelle kinase subfamilies seems to have played the major role given the large size of the RLK/Pelle family in land plants (Fig. 1; Table I).
RLK/Pelle Expansion at the Subfamily Level
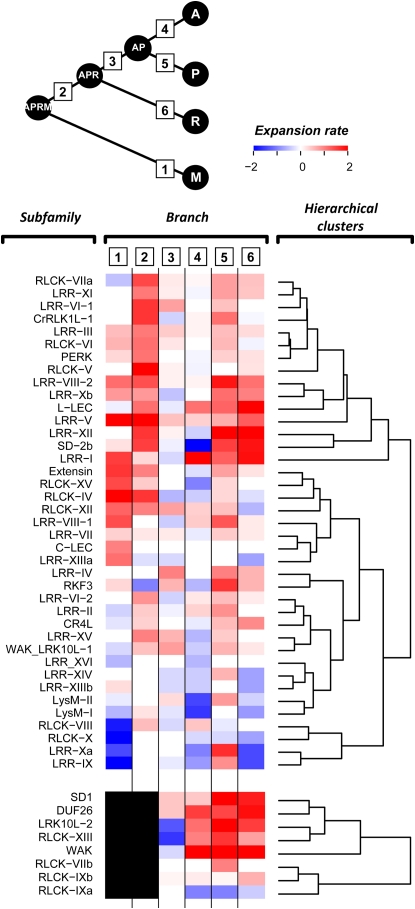
To obtain a more detailed picture of RLK/Pelle expansion, we next looked at the expansion patterns of individual RLK/Pelle subfamilies among different branches in the four-species tree. A heat map of expansion rates clearly shows that subfamilies have expanded at different rates during land plant evolution (Fig. 3). One of the most striking features is the expansion of the majority of subfamilies in the APRM-APR (branch 2) and the poplar (branch 5) lineages. Although loss (negative expansion rate) has occurred in many subfamilies in the rice (branch 6) and Arabidopsis (branch 4) lineages, many subfamilies have expanded in parallel with poplar. Interestingly, some members in most of the extensively expanded subfamilies (Fig. 3), including DUF26, LRK10L-2, LRR-I, LRR-XII, SD1, SD-2b, and WAK, have been implicated in biotic stress responses in Arabidopsis and/or other plants (Feuillet et al., 1997; He et al., 1998; Gomez-Gomez and Boller, 2000; Ohtake et al., 2000; Endre et al., 2002; Robatzek and Somssich, 2002; Stracke et al., 2002; Chen et al., 2003, 2006; Diener and Ausubel, 2005; Zipfel et al., 2006; Acharya et al., 2007). Consistent with the acquisition of ECDs implicated in defense response, substantial expansion of RLK/Pelle subfamilies with members involved in biotic interactions may have been selected for and retained.
Figure 3.
Heat map of subfamily expansion rates in different lineages. Relative expansion rate (defined as the log ratio of the number of genes at the nodes flanking each branch with the more recent node as the numerator) is shown for each subfamily at each branch. Shades of blue indicate the degree of loss, white indicates no net gain or loss, and shades of red indicate the degree of gain. The black box indicates the absence of a subfamily in the APRM-M (branch 1) and APRM-APR (branch 2) lineages. Branches are numbered as shown in the tree diagram.
It is intuitive that diversification of RLKs, particularly of their ECDs, was selected for to allow continued detection of rapidly evolving biotic signals. It is less clear why RLCK subfamilies, which mostly lack members with ECDs, have also undergone expansion, particularly immediately following the divergence from moss (Fig. 3). Because RLCKs lack an ECD, they may not be directly involved in the perception of extracellular signals, particularly those derived from pathogens, For example, the RLCK PTI helps mediate resistance to Pseudomonas syringae but does not appear to interact with effector proteins directly (Zhou et al., 1995). However, there are a couple of examples where RLCKs (PTO and PBS1) interact directly with P. syringae effector proteins (Scofield et al., 1996; Tang et al., 1996; Shao et al., 2003). Therefore, it is possible that diversification of RLCKs may be important for the perception of intracellular biotic signals. It can also be argued that the differential expansion of RLCK (as well as RLK) subfamilies is due to random genomic drift, such as has been postulated for animal chemosensory receptor gene families (Nozawa et al., 2007; Nei et al., 2008). In this view, the large differences in gene family size are largely the result of random gene duplication and loss and may not always reflect an adaptive advantage. In some cases, a larger gene family size generated by random forces provides a selective advantage and allows a group of individuals to occupy a new niche (Nozawa et al., 2007). Further amplification of the gene family by random genomic drift may then occur (Nei et al., 2008). However, the random drift hypothesis does not fully explain why the RLK/Pelle families have expanded independently in multiple plant lineages. Given the functions of known RLK/Pelles, their ability to perceive signals and regulate the propagation of signals, and the finding that expansion tends to involve genes responsive to stress (see next section), we argue that some form of adaptive evolution must have occurred in this family in addition to genomic drift.
Stress Responsiveness of RLK/Pelle Members in Arabidopsis
We have shown that lineage-specific expansion occurred in RLK/Pelle subfamilies with members that have been implicated in biotic stress response. In addition, the novel extracellular protein domains that have been acquired tend to be those involved in biotic interactions. Although these findings suggest that many members of this gene family are likely involved in plant defense responses, functional data for RLK/Pelles remain scarce. Our findings also suggest that expansion of this gene family is likely a consequence of the selection pressures imposed by pathogens. However, this interpretation necessarily requires the establishment of a relationship between expansion of this gene family and stress responsiveness. Using the publicly available AtGenExpress microarray data (Kilian et al., 2007; see “Materials and Methods”), we asked whether RLK/Pelle genes are more likely to be involved in stress response than other Arabidopsis genes and whether stress-responsive RLK/Pelle subfamily members tend be those involved in lineage-specific expansion.
Because several RLK/Pelles have roles in plant-pathogen interactions (Song et al., 1995; Gomez-Gomez and Boller, 2000; Sun et al., 2004; Zipfel et al., 2006) and abiotic stress (Sivaguru et al., 2003; Osakabe et al., 2005), we first asked whether RLK/Pelles are more likely to be up- or down-regulated by abiotic or biotic stress conditions than other Arabidopsis genes. We found that the number of up-regulated RLK/Pelles is significantly overrepresented for several of the stress conditions tested (Table II). In particular, RLK/Pelle genes are significantly enriched in up-regulated genes for seven of the eight biotic stress conditions we examined (χ2 test, α = 5%). Interestingly, RLK/Pelle genes are also enriched in down-regulated genes for nine conditions. This may reflect the fact that while many RLK/Pelle genes are involved in stress response, many others have roles in development and may be down-regulated because of the need for resource allocation upon stress treatment.
Table II.
Expression of RLK/Pelle genes under abiotic and biotic stress conditions
| Response: | Up-Regulation
|
Down-Regulation
|
||||||
|---|---|---|---|---|---|---|---|---|
| Statistical Test: | RLK/Pellea
|
Tandem/Nontandemb
|
RLK/Pellea
|
Tandem/Nontandemb
|
||||
| P | O/Uc | P | O/Uc | P | O/Uc | P | O/Uc | |
| Abiotic stress conditions | ||||||||
| UV-B | 9.36E-25 | O (28)d | 0.002 | O (37)e | 4.58E-05 | O (17)d | 5.64E-04 | U (6)e |
| Wounding | 1.61E-06 | O (10) | 0.237 | (12) | 8.42E-06 | O (5) | 0.062 | (2) |
| Drought | 0.033f | U (0.5) | 0.244 | (0) | 0.158 | (0.2) | 0.139 | (0.5) |
| Cold (4°C) | 0.718 | (13) | 0.690 | (12) | 2.84E-07 | O (25) | 0.064 | (19) |
| Heat | 1.00E-04 | U (10) | 0.699 | (9) | 0.377 | (21) | 0.174 | (17) |
| Salt | 0.771 | (8) | 0.141 | (5) | 0.561 | (9) | 0.479 | (10) |
| Osmotic | 5.15E-04 | O (19) | 0.601 | (19) | 1.49E-06 | O (24) | 5.76E-04 | U (14) |
| Biotic stress conditions | ||||||||
| Flg22 | 1.37E-47 | O (18) | 0.010f | O (23) | 0.279 | (5) | 0.008f | U (1) |
| GST-NPP1 | 2.51E-28 | O (13) | 0.012f | O (17) | 0.195 | (3) | 0.108 | (1) |
| HrcC− | 1.31E-48 | O (19) | 6.62E-05 | O (28) | 0.002 | O (6) | 0.104 | (3) |
| P. infestans | 2.52E-17 | O (18) | 0.076 | (22) | 0.008f | O (12) | 0.011f | U (6) |
| P. syringae pv phaseolicola | 1.57E-44 | O (25) | 0.012f | O (31) | 0.029f | O (10) | 0.009f | U (4) |
| HrpZ | 4.02E-43 | O (23) | 0.013f | O (28) | 0.555 | (7) | 0.008f | U (2) |
| AvrRpm1 | 3.18E-04 | O (14) | 0.676 | (12) | 0.024f | O (12) | 0.207 | (9) |
| DC3000 | 0.393 | (12) | 0.710 | (12) | 6.61E-07 | O (20) | 0.514 | (17) |
χ2 tests were conducted to determine whether RLK/Pelle genes were overrepresented among genes up- or down-regulated by stress conditions compared with all Arabidopsis genes.
χ2 tests were conducted to determine whether tandem RLK/Pelle genes were over-represented among genes up- or down-regulated by stress conditions compared with nontandem RLK/Pelles.
O and U indicate whether significant differences are due to overrepresentation or underrepresentation, respectively.
Numbers in parentheses indicate the percentage of RLK/Pelle genes that are up- or down-regulated.
Numbers in parentheses indicate the percentage of tandem RLK/Pelle genes that are up- or down-regulated.
Not significant after adjusting for multiple testing with sequential Bonferroni adjustment.
The only biotic condition where the RLK/Pelle family is not significantly enriched in up-regulated genes is treatment with P. syringae pv tomato DC3000 (Table II), a virulent bacterial strain capable of infecting Arabidopsis (Dong et al., 1991; Whalen et al., 1991). The other P. syringae pv tomato strains in this data set, avrRpm1 and HrcC−, are avirulent due to recognition of avrRpm1 by the Arabidopsis protein RPM1 and a mutation in HrcC that prevents the formation of a functional type III secretion system (Grant et al., 1995; Deng et al., 1998). Importantly, a significant number of RLK/Pelles are up-regulated when treated with P. syringae pv phaseolicola, a bacterial pathogen of bean (Phaseolus vulgaris) that does not cause disease in Arabidopsis but still activates defense networks (Lindgren et al., 1986; Ham et al., 2007). Several genes including RLK/Pelles are up-regulated specifically by avirulent P. syringae but are targeted for down-regulation by DC3000 (Thilmony et al., 2006). Therefore, the fact that RLK/Pelles as a whole tend to be up-regulated specifically by avirulent P. syringae is consistent with the hypothesis that RLK/Pelle genes are involved in basal defense networks suppressed by DC3000.
Divergence in Stress Responsiveness at the Subfamily Level
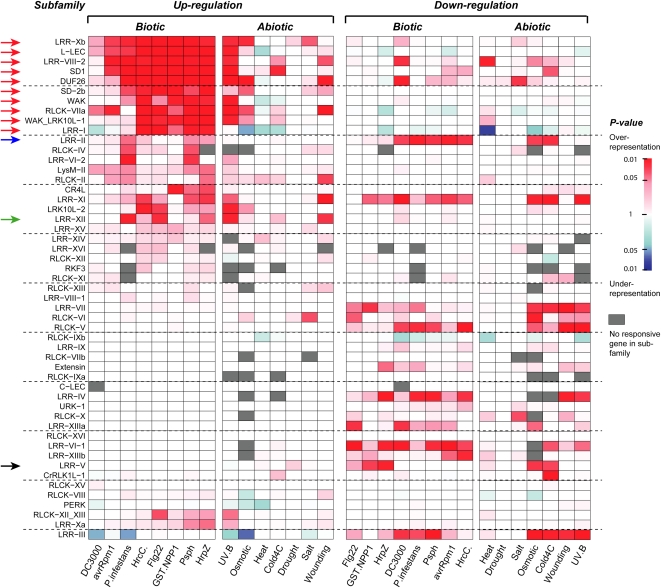
We showed that RLK/Pelle family members are significantly overrepresented among stress response genes, particularly under biotic stress conditions (Table II). To determine the contribution of different subfamilies to this overrepresentation, we looked for enrichment in genes up- and down-regulated by each stress condition in each subfamily. Interestingly, several subfamilies are broadly overrepresented in genes up-regulated by most biotic stresses, including DUF26, L-LEC, LRR-I, LRR-VIII-2, LRR-Xb, RLCK-VIIa, SD1, SD-2b, WAK, and WAK_LRK10L-1 (Fig. 4, red arrows). In addition, members of subfamilies do not appear to respond significantly to a particular biotic agent. For example, the LRR-XII subfamily (Fig. 4, green arrow), of which EFR and FLS2 are members, is enriched for genes up-regulated by the bacterial flagellin elicitor flg22 as well as the oomycetous fungal pathogen Phytophthora infestans. Therefore, signaling networks leading to RLK/Pelle up-regulation seem to overlap upon treatment with different pathogens.
Figure 4.
RLK/Pelle subfamilies enriched in up- and down-regulated genes under abiotic and biotic stress conditions. Enrichment of stress-responsive members in each subfamily was determined by Fisher's exact test, with red shading indicating overrepresentation and blue shading indicating underrepresentation. A gray box indicates that no gene in that subfamily was up- or down-regulated. Red arrows indicate subfamilies with responsiveness to a broad range of biotic signals. The black arrow indicates the LRR-V subfamily whose members have functions in development, and the blue arrow indicates the LRR-II subfamily whose members function in both development and disease resistance. The green arrow indicates the LRR-XII subfamily. Two members in this family are MAMP receptors.
There is also an overlap between subfamilies with members induced by biotic and abiotic stresses. RLK/Pelle subfamilies tend to be enriched in genes up-regulated mainly by UV-B, wounding, and osmotic stress (Fig. 4), and these subfamilies tend to be those that are enriched in genes up-regulated under biotic stress conditions as well. The overlap between RLK/Pelle genes up-regulated by UV-B and biotic stress is particularly striking; for example, 80% of RLK genes up-regulated by P. syringae pv phaseolicola are also up-regulated by UV-B. Previous studies have also revealed an overlap between genes induced by herbivory and UV-B (Izaguirre et al., 2003), systemin binding and UV-B (Yalamanchili and Stratmann, 2002; Holley et al., 2003; Ulm and Nagy, 2005), as well as wounding and other abiotic and biotic stresses (Walley et al., 2007), indicating that some abiotic and biotic signals lead to the induction of similar gene sets.
Taken together, we found that 284 of the 577 (49%) Arabidopsis RLK/Pelle genes for which microarray data are available are up-regulated by one or more stress conditions, supporting the idea that the RLK/Pelle family plays an important role in stress response, particularly under biotic stress conditions. In general, the expression patterns reflect what is known about the functions of RLK/Pelle genes (Fig. 4), indicating that it is appropriate to use expression as a proxy for RLK/Pelle function. Most importantly, we show that some subfamilies contain members that are consistently up-regulated under biotic stress conditions. Interestingly, most of these subfamilies have experienced substantial expansion (Fig. 3).
Relationship between Degree of Expansion, Duplication Mechanism, and Stress Responsiveness
Based on the functions of a limited number of RLK/Pelles, we previously hypothesized that RLK/Pelle genes located in tandem clusters tend to have roles in stress/defense response (Shiu et al., 2004). Several studies have shown that membrane-bound proteins and proteins containing kinase domains are overrepresented in tandem repeats compared with other proteins (Shiu et al., 2004; Rizzon et al., 2006; Tuskan et al., 2006; Hanada et al., 2008). In addition, we have shown that tandem duplicates created by lineage-specific expansion tend to be involved in stress response in plants (Hanada et al., 2008). Therefore, we next asked if duplication mechanism (i.e. tandem versus nontandem) is a predictor of RLK/Pelle function in stress response and if expansion of RLK/Pelle subfamilies is correlated with the degree of tandem duplication.
We found that 50%, 39%, and 30% of RLK/Pelle family members are found in tandem repeats in rice, poplar, and Arabidopsis, respectively. For example, in Arabidopsis, this represents a significant enrichment of RLK/Pelles in tandem repeats compared with other genes (P < 1.0e-20). The percentage of subfamilies in tandem repeats and the expansion rate for each subfamily is significantly and positively correlated (Supplemental Fig. S2), indicating that much of the lineage-specific expansion of RLK/Pelle subfamilies was due to tandem duplication. In addition, RLK/Pelles in tandem repeats are more likely than nontandem RLKs to be up-regulated by biotic stress conditions and UV-B (Table II). In contrast, down-regulated RLK/Pelles are more likely to be nontandem duplicates (Table II). Our findings suggest that by knowing the mechanism of gene duplication we can predict which RLK/Pelles are likely to be up-regulated by biotic stress.
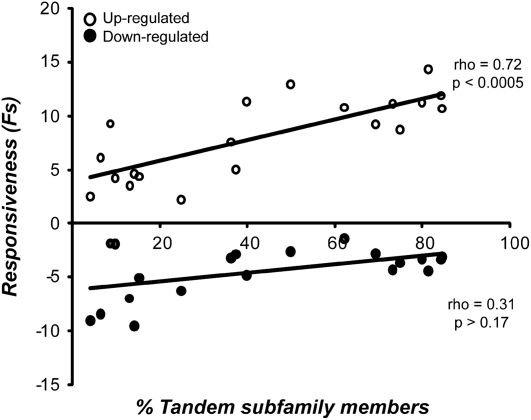
To further test the relationship between stress responsiveness and tandem duplication, we devised a measure of stress responsiveness. We first determined fold change (FC) in expression upon stress treatment for each condition using the maximum or minimum fold change among the time points when considering up-regulation or down-regulation, respectively. FC values of an RLK/Pelle were then summed across conditions (both biotic and abiotic) and averaged across genes in a subfamily to generate a subfamily-wide “responsiveness” measure, FS (sum of fold changes). Responsiveness for each subfamily with four or more members with two or more of them tandem was plotted against the percentage of tandem duplicates found in that subfamily. Up- and down-regulation were evaluated separately. Among tandem RLKs, there is a significant positive correlation between FS and the percentage of tandem subfamily members for up-regulation but not down-regulation (Fig. 5). Interestingly, when conditions were tested individually, a significant correlation was observed for most biotic stress conditions and for three abiotic conditions (Supplemental Table S3). This result is consistent with the hypothesis that tandemly arrayed RLK/Pelles tend to be expressed under stress conditions, particularly biotic, and presumably have a role in stress response. Furthermore, the more tandem members a subfamily has, the more “stress responsive” that family is. This does not mean that only tandem RLK/Pelles are stress responsive, however. For example, the SD-2b subfamily has no tandem duplicate subfamily members but has a higher responsiveness (FS) than DUF26, which has a high proportion (81%) of tandem duplicates.
Figure 5.
Relationship between tandem duplication and stress responsiveness. Average subfamily stress responsiveness (FS), as defined in the text, was plotted against the percentage of subfamily members found in tandem repeats for subfamilies with two or more tandem members. Responsiveness for up-regulated and down-regulated genes is shown by white and black circles, respectively. The best-fit lines, Spearman's ρ, and P values for up- and down-regulation are shown.
Because the proportion of tandem RLK/Pelle subfamily members is positively correlated with expansion rates (Supplemental Fig. S2), we also expected and found a positive relationship between expansion rate of subfamilies and stress responsiveness of subfamily members (Supplemental Fig. S3). Previously, we speculated that there is a correlation between stress functions of RLK/Pelles, tandem duplicates, and genes derived from lineage-specific expansion (Shiu et al., 2004). Our findings are consistent with this observation, but we should emphasize that the positive correlations between responsiveness and the percentage of subfamily members in tandem repeats as well as between responsiveness and expansion rate are significant only when looking at subfamilies that have tandem members or have an expansion rate greater than zero, respectively. This is due to the fact that nonexpanded and nontandem subfamilies are also stress responsive.
Factors Affecting RLK/Pelle Stress Responsiveness
Our findings so far highlight several important characteristics of RLK/Pelles that are responsive to biotic stress. We reasoned that it might be possible to predict stress responsiveness of RLK/Pelles based on knowledge of duplication mechanism, expansion, subfamily identity, and other characteristics that we and others have established to be important predictors of biotic stress responsiveness. We employed multiple regression to test the combined effects of (1) tandem duplication (tandem versus nontandem [T]), (2) receptor configuration (RLK versus RLCK [R]), (3) lineage-specific expansion (member of an expanded or nonexpanded subfamily [E]), and (4) kinase signature sequence (non-RD [no conserved Arg-Asp motif within the active site; Dardick and Ronald, 2006] versus other [K]) on biotic stress responsiveness (see “Materials and Methods”).
The adjusted r2 value for the model is significant (P < 2.8e-08) but small (0.077), explaining only 7.7% of the variance in biotic stress responsiveness. There are clearly additional unidentified factors that affect stress responsiveness among RLK/Pelle genes. It is also possible that the measure for responsiveness was not entirely satisfactory, since we only considered the averaged effects of genes over multiple conditions. In addition, the timing of up-regulation may be crucial. This simplified model, however, does reveal several interesting features of the RLK/Pelle family, particularly interactions between factors (Table III). Surprisingly, RLKs have a lower biotic stress responsiveness compared with RLCKs (Table III), and no significant effect of tandemness (T) alone was observed. However, the significant T*R term indicates that the effect of T is dependent upon receptor configuration (R). Tandem RLKs tend to have a higher stress responsiveness than nontandem RLKs, whereas there is no significant difference between tandem and nontandem RLCKs. Instead, RLCKs belonging to expanded subfamilies have a higher stress responsiveness than RLCKs belonging to nonexpanded subfamilies (see interaction plots in Supplemental Fig. S4). Taken together, these results suggest that tandem duplication is a predictor of RLK stress responsiveness, while expansion may be a predictor of stress responsiveness for RLCKs. In addition to these factors, we expected to see a significant effect of kinase signature sequence (modeled as the parameter K) because of the correlation between non-RD kinase sequence signature and the role in MAMP perception (Dardick and Ronald, 2006). A significant effect of K alone was not observed; however, due to the relatively small number of non-RD kinases, we may have lacked statistical power to detect significant effects.
Table III.
Effects of tandem duplication, expansion, receptor configuration, and kinase signature sequence on stress responsiveness
The highest fold change during the time course was summed across biotic stress conditions and box-cox transformed to improve normality (see “Materials and Methods”).
| Variable | Regression Coefficenta | se | Pb |
|---|---|---|---|
| Intercept | 1.678 | 0.021 | 2.00e-16 |
| T (nontandem)c | 0.004 | 0.048 | 0.94 |
| E (expanded) | −0.094 | 0.047 | 0.047d |
| R (RLCK) | −0.084 | 0.027 | 0.002 |
| K (other) | −0.017 | 0.089 | 0.85 |
| T*E | −0.110 | 0.082 | 0.18 |
| T*R | 0.158 | 0.053 | 0.003 |
| T*K | −0.102 | 0.112 | 0.36 |
| E*R | 0.114 | 0.053 | 0.031d |
| E*K | 0.181 | 0.095 | 0.056e |
| R*K | 0.000 | 0.087 | 0.999 |
Regression coefficients are on the box-cox-transformed scale (λ = −0.315).
P value, determined by t test, indicates whether the effect of a factor is significantly greater than zero. Significant P values (α = 0.05) are shown in boldface.
Dummy variables are assigned values of 1 or 0 (with reference shown in parentheses).
Not significant after sequential Bonferroni adjustment.
A significant effect of E*K was observed for analysis including two potential outliers (see “Materials and Methods”).
We note that the regression coefficients reported in Table III must be interpreted with caution because of potential multicollinearity among the predictors (see “Materials and Methods”). However, this model does reveal important trends. In particular, the fact that tandem RLKs but not RLCKs have a higher median stress responsiveness was not evident from looking at the effect of tandem duplication alone. This interaction is consistent with a whole genome study of gene ontology categories and stress responsiveness that showed that tandemly duplicated membrane-bound proteins were more likely to be stress responsive than nontandem proteins but that the same trend was not apparent for intracellular signal transducers (Hanada et al., 2008). The identification of other factors, such as cis elements, and the use of better measures of stress responsiveness will help improve this model and our ability to predict the functions of RLK/Pelle genes.
CONCLUSION
Based on the analysis of the RLK/Pelle gene family in algae and land plants, we found that the receptor kinase configuration was likely established prior to the divergence between algae and land plants. In addition, most receptor kinase subfamilies were established before the divergence of vascular plants from moss. One of the common themes in RLK/Pelle evolution appears to be a history of selection for innovation, either by domain acquisition or expansion, in biotic response signaling. Supporting the importance of RLK/Pelle genes in stress response, our analysis of microarray expression data revealed that RLK/Pelles are more responsive to stress, particularly biotic stress, than Arabidopsis genes in general. Among stress-responsive RLK/Pelle genes, significantly more tandem genes are responsive to stress than nontandem genes. Previous studies showed that genes involved in biotic stress response tend to be located in tandem repeats (Rizzon et al., 2006; Tuskan et al., 2006; Hanada et al., 2008), and our findings indicate that the RLK/Pelle family is a major contributor to the overrepresentation of tandem genes among stress-responsive genes.
Why are RLKs that have functions in biotic stress more likely to be found in tandem repeats? Tandem duplication occurs on a much shorter time scale than other duplication mechanisms such as whole genome duplication and is positively correlated with recombination rates (Zhang and Gaut, 2003). It has been demonstrated that recombination rates increase when plants are under stress (Molinier et al., 2006), raising the intriguing possibility that stress might result in increased tandem duplication and therefore duplication of the genes found in tandem repeats. Duplication of RLK/Pelle genes involved in biotic stress via tandem duplication might allow selection for the detection of diverse pathogens. We speculate that the rapid lineage-specific expansion of RLK/Pelle genes with stress functions via tandem duplication led to an increased ability to perceive signals from pathogens and was a major factor contributing to the rapid parallel expansion of the RLK/Pelle family in land plants. We also showed that, although expansion and degree of tandem duplication are significantly correlated with the stress responsiveness of RLK/Pelle subfamilies, these factors can only explain a small portion of the variance in stress responsiveness. In addition to alternative ways to define stress responsiveness, it is also necessary to identify other properties of RLK/Pelles that can be important determinants of how they respond to stress. One potential parameter is the level of sequence variation of a given RLK/Pelle in natural populations. This is because RLK/Pelles involved in biotic stress responses are likely subjected to strong positive selection due to the intense selection pressure imposed by biotic agents.
If RLK/Pelle genes are mostly involved in detecting molecular components of pathogens, intuitively, most of these detectors should be expressed constitutively. The underlying assumption is that, to recognize the pathogen components efficiently, the signal perception mechanism should be present at all times. But why are hundreds of RLK/Pelle genes induced by pathogen treatment? In yeast, it has been shown that the function of some genes induced by a stress condition is not to survive that stress but to prepare the cell to survive future stresses (Berry and Gasch, 2008). Similarly, the induction of RLK/Pelles might be required to prepare the plant to respond to additional threats. Supporting this idea, Navarro et al. (2004) found that treatment with flg22, a bacterial MAMP, induces the transcription of genes that may respond to other signals from the same bacterium and may even mediate resistance to other nonbacterial pathogens such as fungi, oomycetes, and viruses. This led them to propose that the up-regulation of RLKs and R genes after flg22 treatment indicated an interaction between MAMP signaling pathways and an increased ability to recognize pathogens. We propose a model that expands upon this idea to explain why so many RLK/Pelle genes are up-regulated by stress conditions. This two-stage model, the “receptor swarm hypothesis” (Supplemental Fig. S5), involves (1) perception and signaling of MAMPs by RLK/Pelles that are constitutively expressed and (2) induction of a large number of RLK/Pelles for detecting specific pathogens/parasites. Since MAMP receptors, such as FLS2, are activated by molecular patterns from both pathogens and nonpathogens, the plant must then make a decision about whether to mount a defense response after MAMP perception. We hypothesize that this decision is made by presenting a large number of potential receptors, including products of the RLK/Pelle genes, and a strong defense response is mounted if any molecular patterns result in the activation of up-regulated receptors. There are multiple direct and indirect pieces of evidence that RLK/Pelle genes are central to biotic defense responses, judging from the fact that loss of function of these RLK/Pelles abolish or diminish plant defense (Song et al., 1995; Gomez-Gomez and Boller, 2000; Sun et al., 2004; Diener and Ausubel, 2005; Chen et al., 2006; Wan et al., 2008). In addition, stress-induced gene expression is a regulated phenomenon that likely has important biological consequences. Therefore, the assumption in the model that RLK/Pelles are capable of eliciting defense response upon activation is likely correct. Because these induced RLK/Pelles may be directly involved in sensing the presence of pathogens/parasites, this model also implies that these RLK/Pelles must be fast evolving due to strong selection on pathogens/parasites to evade detection. This is consistent with the high expansion rate reported here and the significantly higher sequence variation of this gene family among Arabidopsis populations (Clark et al., 2007).
Although this model may explain why many RLK/Pelles are induced by biotic stress, there are several outstanding questions. First, how are these stress-responsive RLK/Pelle transcripts regulated at the translational and posttranslational levels? Second, what molecular patterns are perceived by these RLK/Pelles? Are they simply derived from pathogens/parasites, or are they molecular complexes of pathogen/parasite and host components? Given the size of the RLK/Pelle family, can a single molecular pattern be detected by multiple RLK/Pelles or vice versa? Third, how is the defense response elicited? Does it involve one or a few active RLK/Pelles that provide most of the signals for triggering the defense response? Or is there integration of signal inputs from tens or even hundreds of RLK/Pelles? Finally, it has been shown that disease resistance genes reduce fitness significantly in the absence of any pathogen (Tian et al., 2003). This not only provides a good reason for not turning on these potential resistance genes but also raises the question of what is the fitness cost in up-regulating hundreds of RLK/Pelles that are potential agents of cell death. Answers to these questions will provide not only more detailed understanding of RLK/Pelle functions but also of the selection pressures that contribute to the expansion of this gene family and its rapid evolution.
MATERIALS AND METHODS
Sequence Retrieval and Alignment
An HMM search of the predicted protein sequences of Arabidopsis (Arabidopsis thaliana; The Arabidopsis Information Resource [TAIR], version 6), poplar (Populus trichocarpa; Joint Genome Institute [JGI], version 1.1), rice (Oryza sativa subsp. japonica; International Rice Genome Sequencing Project, version 4), moss (Physcomitrella patens; JGI, version 1.1), Chlamydomonas reinhardtii (JGI, version 3), and Ostreococcus tauri (JGI, version 4) was done to identify sequences containing Ser/Thr/Tyr kinase domains. The “trusted cutoff” of the kinase domain HMM established by Pfam was used as the threshold for detecting kinase domains. Kinase domain protein sequences from all six species were aligned by profile alignment with seed sequences from animal and plant Ser/Thr/Tyr kinases (Shiu and Bleecker, 2001b) in ClustalW (Supplemental Fig. S6; Higgins et al., 1996) with the Gonnet matrix, gap-opening penalty of 10, and extension penalty of 0.2. Some proteins had two kinase domains, and these were concatenated before alignment.
Phylogenetic Analysis, Subfamily Classification, and Orthology Inference
Based on the alignments of kinase domain sequences, first a phylogenetic tree of the kinase domain-containing sequences for all six species was generated with the neighbor-joining method (Saitou and Nei, 1987) with Poisson correction (Supplemental Fig. S7). Kinase families were then defined based on clustering with classified kinase sequences from Arabidopsis and rice (Shiu et al., 2004) in the phylogenetic tree (Supplemental Table S4). RLK/Pelle kinases were further classified into subfamilies based on the classification of Shiu et al. (2004). Among 5,290 RLK/Pelle sequences, 865 could not be confidently assigned to known subfamilies. To classify these sequences, the kinase domain sequences of these unclassified RLK/Pelles were used in a BLAST (Altschul et al., 1997) search against the kinase domains of the classified sequences. The unclassified sequences were then tentatively assigned to the subfamily of their top match. To further refine the classification, phylogenetic trees were generated for each subfamily. First, full-length sequences for each subfamily were aligned using ClustalW (Thompson et al., 1997) with a Blosum 65 matrix, 5.0 gap-opening penalty, and 10.0 gap-extension penalty. Protein distances among subfamily members were then estimated with PHYLIP (Felsenstein, 2005) using the JTT matrix and the gamma correction with a coefficient of variation of 0.3126. Based on the JTT model with gamma correction, a maximum likelihood (ML) tree for each subfamily was generated with RaxML (Stamatakis et al., 2005) and rooted using Raf kinase At1g18160 and Aurora kinase At2g25880 as outgroups, since these two kinases belong to families other than RLK/Pelle. In some cases, putative RLK/Pelle family members that failed to cluster with the assigned subfamily were assigned to new subfamilies. Those that failed to be in the RLK/Pelle clade in the ML trees were excluded from further RLK/Pelle family analysis. Inference of orthologous groups and ancestral gene number was done by reconciling the ML subfamily trees with the species tree of four plants using Notung (Chen et al., 2000).
Determination of Expansion Rate
Each orthologous group indicates the presence of one ancestral gene in the common ancestor of species included in the orthologous group. Therefore, the total number of orthologous groups is used as an estimate for ancestral gene number. Depending on whether subfamilies contained genes from all species making up the orthologous groups, we estimated ancestral gene numbers two different ways. In the first approach, the ancestral number of genes at each internal node shown in Figure 1 was calculated only when at least one subfamily member was present in all species being compared. Alternatively, we also took into account cases where a subfamily was absent in one or more species by assuming that at least one ancestral gene was present in the common ancestor and that the absence of subfamily members in some species was due to gene loss. Therefore, two sets of expansion rates were generated: “shared,” where only subfamilies present in all species compared are included in the calculation of expansion rate, and “all,” where all subfamilies are included and the number at the ancestral node for species lacking those subfamilies is assumed to be one. The results were very similar, so here we only present the results for shared subfamilies.
Domain Family Designation and Tandem Duplicate Definition
To define domain configuration, full-length RLK/Pelle sequences were queried against the SMART (Schultz et al., 2000; Letunic et al., 2006) and PFAM (Finn et al., 2008) databases to annotate ECDs. RLK/Pelles that did not have a receptor configuration (defined by the lack of a signal sequence or transmembrane domain, or a predicted ECD of less than 30 amino acids) were defined as RLCKs. Subfamily names, however, were determined based on clustering of a gene within an existing subfamily; therefore, an RLCK may belong to a subfamily named after protein domains found in the extracellular regions or vice versa. Tandem RLK/Pelle genes in Arabidopsis, poplar, rice, and moss were defined as those genes that (1) are less than or equal to 10 genes apart, (2) belong to the same subfamily, and (3) are within 100 kb (Arabidopsis and moss) or 350 kb (poplar and rice). Because shotgun sequencing was used to sequence the poplar and moss genomes, many scaffolds remain unassigned to linkage groups and the number of tandem duplicates may be underestimated.
Processing of Microarray Data
AtGenExpress stress microarray data were obtained from TAIR (Swarbreck et al., 2008). LIMMA was used to identify genes that were significantly up- or down-regulated under each abiotic and biotic stress condition compared with the mock-treated ecotype Columbia control. Data are available for several time points for each condition; therefore, a gene was considered up- or down-regulated by a stress condition if significant up- or down-regulation was observed at any time point. The genotoxic treatment was not included in the analysis because no RLK/Pelle members were up- or down-regulated by these conditions. Significant up- or down-regulation was determined using the moderated t test in LIMMA at adjusted P ≤ 0.05. The raw test P values were adjusted based on false discovery rate (Storey and Tibshirani, 2003).
Linear Model of Stress Responsiveness
Multiple regression analysis was done to test the importance of several factors on stress responsiveness (S), defined as the highest fold change during the time course summed across eight biotic stress conditions from the AtGenExpress stress microarray data set. A box-cox transformation was done on S to improve the normality required for multiple regression analysis. S included negative values; therefore, 8.12 was added to each data point to bring the minimum value of S to 1. The value, λ = −0.315, that maximized normality was found using the box.cox.powers function in R (version 2.4.1; http://www.r-project.org/), and the data were transformed according to the formula Y′ = (Y λ − 1)/λ (Sokal and Rohlf, 1995). The lm function in the R environment was used to fit the model:
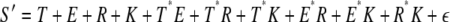 |
where S′ is box-cox-transformed data, T is tandem or nontandem, E is expanded or nonexpanded, R is RLK or RLCK, K is kinase signature sequence non-RD or other, and ε is error. Dummy variables were assigned a value of 1 or 0 with nontandem, expanded, RLCK, and other kinase sequence signatures serving as the reference levels (assigned a value of 0). Regression coefficients, therefore, reflect the effect of the levels assigned a value of 1 (tandem, nonexpanded, RLK, and non-RD). A q plot of residuals revealed two potential outliers, the LRR-XI subfamily members At3g49670 and At5g56040. The model was refitted after deleting these two data points. Deleting the potential outliers slightly reduced the predicted effect sizes of most factors but changed the significance of only one term (E*K). Here, we present the results obtained excluding these potential outliers. There is a potential for multicollinearity among factors, especially T and E, to affect the interpretation of the effects of each factor. Using the vif function in the R environment, we calculated the variance inflation factor (VIF), a measure of how much of the variance is elevated due to correlation between factors. VIF values were 6.08, 5.52, 1.83, and 6.51 for T, E, R, and S′, respectively. The interaction terms had VIF values ranging from 2.8 to 6.8. This indicates that multicollinearity is a potential problem and that the magnitude of regression coefficients should be interpreted with caution.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Kinase expansion rates during land plant evolution.
Supplemental Figure S2. Correlation between the percentage of tandem duplicates per subfamily and subfamily expansion rate in Arabidopsis, poplar, and rice.
Supplemental Figure S3. Relationship between subfamily stress responsiveness and subfamily expansion rate.
Supplemental Figure S4. Plots illustrating interaction effects determined by multiple regression.
Supplemental Figure S5. The receptor swarm hypothesis.
Supplemental Figure S6. Kinase amino acid sequence alignments.
Supplemental Figure S7. Phylogenetic tree of kinases in Arabidopsis, poplar, rice, moss, C. reinhardtii, and O. tauri.
Supplemental Table S1. Predicted signal sequences, transmembrane domains, and ECDs of RLK/Pelle genes.
Supplemental Table S2. RLK/Pelle genes that have gained new ECDs.
Supplemental Table S3. Summary statistics for regression of subfamily stress responsiveness (up-regulation) against the percentage of tandem subfamily members
Supplemental Table S4. List of kinases identified in Arabidopsis, poplar, rice, moss, C. reinhardtii, and O. tauri and their subfamily classifications.
Supplementary Material
Acknowledgments
We thank Lori Imboden for reading and discussing the manuscript. We also thank the Arabidopsis Functional Genomics Network for making the Arabidopsis stress expression data sets available.
This work was supported by grants from the Rackham Foundation and the National Science Foundation (grant nos. DBI 0638591 and MCB 0749634).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Shin-Han Shiu (shius@msu.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Acharya BR, Raina S, Maqbool SB, Jagadeeswaran G, Mosher SL, Appel HM, Schultz JC, Klessig DF, Raina R (2007) Overexpression of CRK13, an Arabidopsis cysteine-rich receptor-like kinase, results in enhanced resistance to Pseudomonas syringae. Plant J 50 488–499 [DOI] [PubMed] [Google Scholar]
- Afzal AJ, Wood AJ, Lightfoot DA (2008) Plant receptor-like serine threonine kinases: roles in signaling and plant defense. Mol Plant Microbe Interact 21 507–517 [DOI] [PubMed] [Google Scholar]
- Albrecht C, Russinova E, Hecht V, Baaijens E, de Vries S (2005) The Arabidopsis thaliana SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASES1 and 2 control male sporogenesis. Plant Cell 17 3337–3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815 [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, Medlin L (1998) Algal phylogeny and the origin of land plants. Plant Physiol 116 9–15 [Google Scholar]
- Belvin MP, Anderson KV (1996) A conserved signaling pathway: the Drosophila Toll-Dorsal pathway. Annu Rev Cell Dev Biol 12 393–416 [DOI] [PubMed] [Google Scholar]
- Bent AF, Mackey D (2007) Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu Rev Phytopathol 45 399–436 [DOI] [PubMed] [Google Scholar]
- Berry DB, Gasch AP (2008) Stress-activated genomic expression changes serve a preparative role for impending stress in yeast. Mol Biol Cell 19 4580–4587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaw SM, Chang CC, Chen HL, Li WH (2004) Dating the monocot-dicot divergence and the origin of core eudicots using whole chloroplast genomes. J Mol Evol 58 424–441 [DOI] [PubMed] [Google Scholar]
- Chen K, Du L, Chen Z (2003) Sensitization of defense responses and activation of programmed cell death by a pathogen-induced receptor-like protein kinase in Arabidopsis. Plant Mol Biol 53 61–74 [DOI] [PubMed] [Google Scholar]
- Chen K, Durand D, Farach-Colton M (2000) NOTUNG: a program for dating gene duplications and optimizing gene family trees. J Comput Biol 7 429–447 [DOI] [PubMed] [Google Scholar]
- Chen X, Shang J, Chen D, Lei C, Zou Y, Zhai W, Liu G, Xu J, Ling Z, Cao G, et al (2006) A B-lectin receptor kinase gene conferring rice blast resistance. Plant J 46 794–804 [DOI] [PubMed] [Google Scholar]
- Chevalier D, Batoux M, Fulton L, Pfister K, Yadav RK, Schellenberg M, Schneitz K (2005) STRUBBELIG defines a receptor kinase-mediated signaling pathway regulating organ development in Arabidopsis. Proc Natl Acad Sci USA 102 9074–9079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RM, Schweikert G, Toomajian C, Ossowski S, Zeller G, Shinn P, Warthmann N, Hu TT, Fu G, Hinds DA, et al (2007) Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science 317 338–342 [DOI] [PubMed] [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM (1997) The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89 575–585 [DOI] [PubMed] [Google Scholar]
- Colcombet J, Boisson-Dernier A, Ros-Palau R, Vera CE, Schroeder JI (2005) Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASES1 and 2 are essential for tapetum development and microspore maturation. Plant Cell 17 3350–3361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courties C, Vaquer A, Troussellier M, Laurtier J (1994) Smallest eukaryotic organism. Nature 370 255 [Google Scholar]
- Dardick C, Ronald P (2006) Plant and animal pathogen recognition receptors signal through non-RD kinases. PLoS Pathog 2 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng WL, Preston G, Collmer A, Chang CJ, Huang HC (1998) Characterization of the hrpC and hrpRS operons of Pseudomonas syringae pathovars syringae, tomato, and glycinea and analysis of the ability of hrpF, hrpG, hrcC, hrpT, and hrpV mutants to elicit the hypersensitive response and disease in plants. J Bacteriol 180 4523–4531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derelle E, Ferraz C, Rombauts S, Rouzé P, Worden AZ, Robbens S, Partensky F, Degroeve S, Echeynié S, Cooke R, et al (2006) Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc Natl Acad Sci USA 103 11647–11652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeYoung BJ, Bickle KL, Schrage KJ, Muskett P, Patel K, Clark SE (2006) The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. Plant J 45 1–16 [DOI] [PubMed] [Google Scholar]
- Diener AC, Ausubel FM (2005) RESISTANCE TO FUSARIUM OXYSPORUM 1, a dominant Arabidopsis disease-resistance gene, is not race specific. Genetics 171 305–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Mindrinos M, Davis KR, Ausubel FM (1991) Induction of Arabidopsis defense genes by virulent and avirulent Pseudomonas syringae strains and by a cloned avirulence gene. Plant Cell 3 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endre G, Kereszt A, Kevei Z, Mihacea S, Kalo P, Kiss GB (2002) A receptor kinase gene regulating symbiotic nodule development. Nature 417 962–966 [DOI] [PubMed] [Google Scholar]
- Escobar-Restrepo JM, Huck N, Kessler S, Gagliardini V, Gheyselinck J, Yang WC, Grossniklaus U (2007) The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science 317 656–660 [DOI] [PubMed] [Google Scholar]
- Eyuboglu B, Pfister K, Haberer G, Chevalier D, Fuchs A, Mayer KF, Schneitz K (2007) Molecular characterisation of the STRUBBELIG-RECEPTOR FAMILY of genes encoding putative leucine-rich repeat receptor-like kinases in Arabidopsis thaliana. BMC Plant Biol 7 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix G, Duran JD, Volko S, Boller T (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 18 265–276 [DOI] [PubMed] [Google Scholar]
- Felsenstein J (2005) PHYLIP (Phylogeny Inference Package) Version 3.6. Department of Genome Sciences, University of Washington, Seattle
- Feuillet C, Schachermayr G, Keller B (1997) Molecular cloning of a new receptor-like kinase gene encoded at the Lr10 disease resistance locus of wheat. Plant J 11 45–52 [DOI] [PubMed] [Google Scholar]
- Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, et al (2008) The Pfam protein families database. Nucleic Acids Res 36 D281–D288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florentino LH, Santos AA, Fontenelle MR, Pinheiro GL, Zerbini FM, Baracat-Pereira MC, Fontes EP (2006) A PERK-like receptor kinase interacts with the geminivirus nuclear shuttle protein and potentiates viral infection. J Virol 80 6648–6656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontes EP, Santos AA, Luz DF, Waclawovsky AJ, Chory J (2004) The geminivirus nuclear shuttle protein is a virulence factor that suppresses transmembrane receptor kinase activity. Genes Dev 18 2545–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gomez L, Boller T (2000) FLS2: an LRR receptor-like kinase involved in the perception of bacterial elicitor flagellin in Arabidopsis. Mol Cell 5 1003–1011 [DOI] [PubMed] [Google Scholar]
- Grant MR, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes RW, Dangl JL (1995) Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 269 843–846 [DOI] [PubMed] [Google Scholar]
- Ham JH, Kim MG, Lee SY, Mackey D (2007) Layered basal defenses underlie non-host resistance of Arabidopsis to Pseudomonas syringae pv. phaseolicola. Plant J 51 604–616 [DOI] [PubMed] [Google Scholar]
- Hanada K, Zou C, Lehti-Shiu MD, Shinozaki K, Shiu SH (2008) Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol 148 993–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z-H, He D, Kohorn BD (1998) Requirement for the induced expression of a cell wall associated receptor kinase for survival during the pathogen response. Plant J 14 55–63 [DOI] [PubMed] [Google Scholar]
- Hecht PM, Anderson KV (1993) Genetic characterization of tube and pelle, genes required for signaling between toll and dorsal in the specification of the dorsal-ventral pattern of the Drosophila embryo. Genetics 135 405–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins DG, Thompson JD, Gibson TJ (1996) Using CLUSTAL for multiple sequence alignments. Methods Enzymol 266 383–402 [DOI] [PubMed] [Google Scholar]
- Holley SR, Yalamanchili RD, Moura DS, Ryan CA, Stratmann JW (2003) Convergence of signaling pathways induced by systemin, oligosaccharide elicitors, and ultraviolet-B radiation at the level of mitogen-activated protein kinases in Lycopersicon peruvianum suspension-cultured cells. Plant Physiol 132 1728–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Tong H, Selby J, Dewitt J, Peng X, He ZH (2005) Involvement of a cell wall-associated kinase, WAKL4, in Arabidopsis mineral responses. Plant Physiol 139 1704–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436 793–800 [DOI] [PubMed] [Google Scholar]
- Izaguirre MM, Scopel AL, Baldwin IT, Ballare CL (2003) Convergent responses to stress: solar ultraviolet-B radiation and Manduca sexta herbivory elicit overlapping transcriptional responses in field-grown plants of Nicotiana longiflora. Plant Physiol 132 1755–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens S, Beyaert R (2003) Functional diversity and regulation of different interleukin-1 receptor-associated kinase (IRAK) family members. Mol Cell 11 293–302 [DOI] [PubMed] [Google Scholar]
- Kaku H, Nishizawa Y, Ishii-Minami N, Akimoto-Tomiyama C, Dohmae N, Takio K, Minami E, Shibuya N (2006) Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc Natl Acad Sci USA 103 11086–11091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D'Angelo C, Bornberg-Bauer E, Kudla J, Harter K (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50 347–363 [DOI] [PubMed] [Google Scholar]
- Kwak SH, Shen R, Schiefelbein J (2005) Positional signaling mediated by a receptor-like kinase in Arabidopsis. Science 307 1111–1113 [DOI] [PubMed] [Google Scholar]
- Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P (2006) SMART 5: domains in the context of genomes and networks. Nucleic Acids Res 34 D257–D260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chory J (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90 929–938 [DOI] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110 213–222 [DOI] [PubMed] [Google Scholar]
- Lindgren PB, Peet RC, Panopoulos NJ (1986) Gene cluster of Pseudomonas syringae pv. “phaseolicola” controls pathogenicity of bean plants and hypersensitivity of nonhost plants. J Bacteriol 168 512–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, Sato S, Kaneko T, Tabata S, Sandal N, et al (2003) A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425 637–640 [DOI] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, et al (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K, Narusaka Y, Kawakami N, Kaku H, Shibuya N (2007) CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci USA 104 19613–19618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinier J, Ries G, Zipfel C, Hohn B (2006) Transgeneration memory of stress in plants. Nature 442 1046–1049 [DOI] [PubMed] [Google Scholar]
- Muto H, Yabe N, Asami T, Hasunuma K, Yamamoto KT (2004) Overexpression of constitutive differential growth 1 gene, which encodes a RLCKVII-subfamily protein kinase, causes abnormal differential and elongation growth after organ differentiation in Arabidopsis. Plant Physiol 136 3124–3133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam KH, Li J (2002) BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110 203–212 [DOI] [PubMed] [Google Scholar]
- Navarro L, Zipfel C, Rowland O, Keller I, Robatzek S, Boller T, Jones JD (2004) The transcriptional innate immune response to flg22: interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol 135 1113–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Niimura Y, Nozawa M (2008) The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nat Rev Genet 9 951–963 [DOI] [PubMed] [Google Scholar]
- Nozawa M, Kawahara Y, Nei M (2007) Genomic drift and copy number variation of sensory receptor genes in humans. Proc Natl Acad Sci USA 104 20421–20426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Shinohara H, Sakagami Y, Matsubayashi Y (2008) Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319 294. [DOI] [PubMed] [Google Scholar]
- Ohtake Y, Takahashi T, Komeda Y (2000) Salicylic acid induces the expression of a number of receptor-like kinase genes in Arabidopsis thaliana. Plant Cell Physiol 41 1038–1044 [DOI] [PubMed] [Google Scholar]
- Osakabe Y, Maruyama K, Seki M, Satou M, Shinozaki K, Yamaguchi-Shinozaki K (2005) Leucine-rich repeat receptor-like kinase1 is a key membrane-bound regulator of abscisic acid early signaling in Arabidopsis. Plant Cell 17 1105–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrakis A, Tews I, Dauter Z, Oppenheim AB, Chet I, Wilson KS, Vorgias CE (1994) Crystal structure of a bacterial chitinase at 2.3 A resolution. Structure 2 1169–1180 [DOI] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Grønlund M, Sato S, Nakamura Y, Tabata S, Sandal N, et al (2003) Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425 585–592 [DOI] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud PF, Lindquist EA, Kamisugi Y, et al (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319 64–69 [DOI] [PubMed] [Google Scholar]
- Rizzon C, Ponger L, Gaut BS (2006) Striking similarities in the genomic distribution of tandemly arrayed genes in Arabidopsis and rice. PLOS Comput Biol 2 e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S, Somssich IE (2002) Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev 16 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4 406–425 [DOI] [PubMed] [Google Scholar]
- Santelli E, Schwarzenbacher R, McMullan D, Biorac T, Brinen LS, Canaves JM, Cambell J, Dai X, Deacon AM, Elsliger MA, et al (2004) Crystal structure of a glycerophosphodiester phosphodiesterase (GDPD) from Thermotoga maritima (TM1621) at 1.60 A resolution. Proteins 56 167–170 [DOI] [PubMed] [Google Scholar]
- Sasaki G, Katoh K, Hirose N, Suga H, Kuma K, Miyata T, Su ZH (2007) Multiple receptor-like kinase cDNAs from liverwort Marchantia polymorpha and two charophycean green algae, Closterium ehrenbergii and Nitella axillaris: extensive gene duplications and gene shufflings in the early evolution of streptophytes. Gene 401 135–144 [DOI] [PubMed] [Google Scholar]
- Schultz J, Copley RR, Doerks T, Ponting CP, Bork P (2000) SMART: a Web-based tool for the study of genetically mobile domains. Nucleic Acids Res 28 231–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield SR, Tobias CM, Rathjen JP, Chang JH, Lavelle DT, Michelmore RW, Staskawicz BJ (1996) Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato. Science 274 2063–2065 [DOI] [PubMed] [Google Scholar]
- Shao F, Golstein C, Ade J, Stoutemyer M, Dixon JE, Innes RW (2003) Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science 301 1230–1233 [DOI] [PubMed] [Google Scholar]
- Shelton CA, Wasserman SA (1993) Pelle encodes a protein kinase required to establish dorsoventral polarity in the Drosophila embryo. Cell 72 515–525 [DOI] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB (2001. a) Plant receptor-like kinase gene family: diversity, function, and signaling. Sci STKE 2001 RE22. [DOI] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB (2001. b) Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA 98 10763–10768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB (2003) Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol 132 530–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu SH, Karlowski WM, Pan R, Tzeng YH, Mayer KF, Li WH (2004) Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16 1220–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak ED, Berthiaume CT, Hill EJ, Torii KU (2004) Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation. Development 131 1491–1501 [DOI] [PubMed] [Google Scholar]
- Sivaguru M, Ezaki B, He ZH, Tong H, Osawa H, Baluska F, Volkmann D, Matsumoto H (2003) Aluminum-induced gene expression and protein localization of a cell wall-associated receptor kinase in Arabidopsis. Plant Physiol 132 2256–2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal R, Rohlf F (1995) Biometry: The Principles and Practice of Statistics in Biological Research, Ed 3. W.H. Freeman, New York
- Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Holsten T, Gardner J, Wang B, Zhai WX, Zhu LH, et al (1995) A receptor kinase-like protein encoded by the rice disease resistance gene Xa21. Science 270 1804–1806 [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Ludwig T, Meier H (2005) RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 21 456–463 [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke S, Kistner C, Yoshida S, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Szczyglowski K, et al (2002) A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417 959–962 [DOI] [PubMed] [Google Scholar]
- Sun X, Cao Y, Yang Z, Xu C, Li X, Wang S, Zhang Q (2004) Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J 37 517–527 [DOI] [PubMed] [Google Scholar]
- Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L, et al (2008) The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res 36 D1009–D1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Frederick RD, Zhou J, Halterman DA, Jia Y, Martin GB (1996) Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science 274 2060–2063 [DOI] [PubMed] [Google Scholar]
- Thilmony R, Underwood W, He SY (2006) Genome-wide transcriptional analysis of the Arabidopsis thaliana interaction with the plant pathogen Pseudomonas syringae pv. tomato DC3000 and the human pathogen Escherichia coli O157:H7. Plant J 46 34–53 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D, Traw MB, Chen JQ, Kreitman M, Bergelson J (2003) Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 423 74–77 [DOI] [PubMed] [Google Scholar]
- Torii KU, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier RF, Komeda Y (1996) The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8 735–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313 1596–1604 [DOI] [PubMed] [Google Scholar]
- Ulm R, Nagy F (2005) Signalling and gene regulation in response to ultraviolet light. Curr Opin Plant Biol 8 477–482 [DOI] [PubMed] [Google Scholar]
- Walley JW, Coughlan S, Hudson ME, Covington MF, Kaspi R, Banu G, Harmer SL, Dehesh K (2007) Mechanical stress induces biotic and abiotic stress responses via a novel cis-element. PLoS Genet 3 1800–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Zhang XC, Neece D, Ramonell KM, Clough S, Kim SY, Stacey MG, Stacey G (2008) A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen MC, Innes RW, Bent AF, Staskawicz BJ (1991) Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell 3 49–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalamanchili RD, Stratmann JW (2002) Ultraviolet-B activates components of the systemin signaling pathway in Lycopersicon peruvianum suspension-cultured cells. J Biol Chem 277 28424–28430 [DOI] [PubMed] [Google Scholar]
- Yoon HS, Hackett JD, Ciniglia C, Pinto G, Bhattacharya D (2004) A molecular timeline for the origin of photosynthetic eukaryotes. Mol Biol Evol 21 809–818 [DOI] [PubMed] [Google Scholar]
- Yuan Q, Ouyang S, Wang A, Zhu W, Maiti R, Lin H, Hamilton J, Haas B, Sultana R, Cheung F, et al (2005) The Institute for Genomic Research Osa1 rice genome annotation database. Plant Physiol 138 18–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Gaut BS (2003) Does recombination shape the distribution and evolution of tandemly arrayed genes (TAGs) in the Arabidopsis thaliana genome? Genome Res 13 2533–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou A, Wang H, Walker JC, Li J (2004) BRL1, a leucine-rich repeat receptor-like protein kinase, is functionally redundant with BRI1 in regulating Arabidopsis brassinosteroid signaling. Plant J 40 399–409 [DOI] [PubMed] [Google Scholar]
- Zhou J, Loh YT, Bressan RA, Martin GB (1995) The tomato gene Pti1 encodes a serine/threonine kinase that is phosphorylated by Pto and is involved in the hypersensitive response. Cell 83 925–935 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125 749–760 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.