Abstract
Nicotianamine chelates and transports micronutrient metal ions in plants. It has been speculated that nicotianamine is involved in seed loading with micronutrients. A tomato (Solanum lycopersicum) mutant (chloronerva) and a tobacco (Nicotiana tabacum) transgenic line have been utilized to analyze the effects of nicotianamine loss. These mutants showed early leaf chlorosis and had sterile flowers. Arabidopsis (Arabidopsis thaliana) has four NICOTIANAMINE SYNTHASE (NAS) genes. We constructed two quadruple nas mutants: one had full loss of NAS function, was sterile, and showed a chloronerva-like phenotype (nas4x-2); another mutant, with intermediate phenotype (nas4x-1), developed chlorotic leaves, which became severe upon transition from the vegetative to the reproductive phase and upon iron (Fe) deficiency. Residual nicotianamine levels were sufficient to sustain the life cycle. Therefore, the nas4x-1 mutant enabled us to study late nicotianamine functions. This mutant had no detectable nicotianamine in rosette leaves of the reproductive stage but low nicotianamine levels in vegetative rosette leaves and seeds. Fe accumulated in the rosette leaves, while less Fe was present in flowers and seeds. Leaves, roots, and flowers showed symptoms of Fe deficiency, whereas leaves also showed signs of sufficient Fe supply, as revealed by molecular-physiological analysis. The mutant was not able to fully mobilize Fe to sustain Fe supply of flowers and seeds in the normal way. Thus, nicotianamine is needed for correct supply of seeds with Fe. These results are fundamental for plant manipulation approaches to modify Fe homeostasis regulation through alterations of NAS genes.
One of the breeding goals for high-quality nutrition food crops is the production of micronutrient (iron,zinc [Fe,Zn])-enriched plants. The relevance of Fe for human nutrition is evident from the most severe human micronutrient deficiency disease, which is Fe deficiency anemia (see the 2004 report of the World Health Organization; de Benoist et al., 2008). In plants, Fe deficiency is also the most widespread micronutrient deficiency; it is seen frequently on calcareous and alkaline soils, where Fe is almost unsoluble. On the other hand, beneficial biochemical properties can render this same metal potentially toxic. Free Fe is a catalyst in the formation of hydroxyl radicals, which can unspecifically damage biological molecules. To balance these effects, Fe uptake and homeostasis are tightly controlled.
Genetic and transgenic approaches targeting specific Fe homeostasis genes are under way to yield micronutrient-enriched crops (Goto et al., 1999; Takahashi et al., 2001; Sautter et al., 2006; Uauy et al., 2006). It is obvious that a precise breeding approach would be more efficient if more genetic components were known controlling Fe acquisition, transport, and storage.
Among the key elements for Fe homeostasis are small chelators of metals, both to render the metals soluble and to detoxify them (Colangelo and Guerinot, 2006; Briat et al., 2007). For example, organic acids such as citric acid can bind free metal ions. This mechanism is primarily used for the transport of Fe in the xylem, where it is considered that the majority of Fe is bound to citrate. On the other hand, phloem Fe is bound by nicotianamine or other amino acids such as part of proteins (von Wiren et al., 1999; Kruger et al., 2002).
The plant metal chelator nicotianamine is a free nonproteinogenic amino acid (Scholz et al., 1992; Stephan, 2002; Hell and Stephan, 2003). Nicotianamine is mobile in the plant and has been detected in root and leaf cells as well as in phloem sap. It can bind metal ions like Fe, Zn, copper (Cu), and nickel (Ni; Scholz et al., 1992; Schmiedeberg et al., 2003). Nicotianamine is synthesized by a one-step condensation reaction of three molecules of S-adenosyl-Met by the enzyme NICOTIANAMINE SYNTHASE (NAS; Shojima et al., 1989; Herbik et al., 1999; Higuchi et al., 1999). In graminaceous plants like barley (Hordeum vulgare), rice (Oryza sativa), and maize (Zea mays), nicotianamine has a prominent role in Fe acquisition from the soil. It is the direct precursor of phytosiderophores of the mugineic acid family (Mori, 1999). Phytosiderophores are strong Fe3+ chelators that are extruded to the rhizosphere, where they serve to chelate and solubilize Fe3+ (Römheld and Marschner, 1986a, 1986b). NAS genes form gene families in grasses that cover a range of expression patterns (Higuchi et al., 2001; Inoue et al., 2003; Mizuno et al., 2003). NAS genes involved in phytosiderophore production are induced by Fe deficiency in the root. Phytosiderophore-Fe3+ complexes are imported into the rhizodermis by the transporter of the type YELLOW STRIPE1 (YS1) from maize that is up-regulated by Fe deficiency in roots (Curie et al., 2001). YELLOW STRIPE-LIKE (YSL) genes are encoded by multigene families in grasses as well as in dicot plants (Curie et al., 2001; DiDonato et al., 2004). This indicates that NAS and YSL genes have diversified roles throughout plant development in addition to Fe uptake into the root.
In solanaceous plants, nicotianamine was shown to act in Fe homeostasis, which has been mostly found through analysis of the nas tomato (Solanum lycopersicum) mutant chloronerva and of a transgenic tobacco (Nicotiana tabacum) line with ectopic NICOTIANAMINE AMINOTRANSFERASE (NAAT) activity that consumed the available nicotianamine (Scholz et al., 1992; Ling et al., 1999; Takahashi et al., 2003). Tomato and tobacco showed leaf chlorosis as a result of reduced nicotianamine and sterility. Solanaceous plants and other dicots rely on a Fe reduction-based mechanism to solubilize and acquire Fe2+ (Römheld and Marschner, 1986a). Fe is reduced by a membrane-bound ferric chelate reductase (FRO2 in Arabidopsis [Arabidopsis thaliana]; Robinson et al., 1999). Fe2+ is taken up into the root epidermis by a membrane-bound divalent metal transporter (IRON-REGULATED TRANSPORTER1 [IRT1] in Arabidopsis; Eide et al., 1996; Vert et al., 2002). FRO2 and IRT1-type genes are induced by an Fe deficiency-dependent basic helix-loop-helix transcription factor protein (FER in tomato, FIT in Arabidopsis; Ling et al., 2002; Colangelo and Guerinot, 2004; Jakoby et al., 2004; Yuan et al., 2005). In tomato, it was found that chloronerva plants up-regulate FER, IRT1, and FRO1 even upon sufficient Fe supply (Bereczky et al., 2003; Li et al., 2004; Brumbarova and Bauer, 2005). Since chloronerva plants can accumulate Fe in leaves but not Cu (Pich and Scholz, 1996), nicotianamine is not directly needed for Fe uptake and translocation into leaves in solanaceous plants (Scholz et al., 1992). Most likely, nicotianamine plays a role in intracellular and intercellular distribution of Fe, whereas it may be involved in Cu translocation to leaves. However, contrasting effects were found in tobacco NAAT plants, which had reduced Fe levels in leaves compared with the wild type, as well as reduced levels of Cu and Zn (Takahashi et al., 2003). Hence, there were clear distinctions between tomato and tobacco mutant nicotianamine lines. Another disadvantage of the two solanaceous plant studies was that only mutants with severe defects were available. Thus, it remains unknown which phenotypes were pleiotropic in nature and which phenotypes were directly caused by nicotianamine loss. For example, chloronerva and NAAT tobacco both had abnormal flowers.
A function of nicotianamine in seed metal loading has been inferred from studies of the presumptive nicotianamine-Fe transporter family YSL (Curie et al., 2001; Schaaf et al., 2004). Loss of YSL1 resulted in decreased nicotianamine and Fe levels in ysl1 mutant seeds (Le Jean et al., 2005). YSL genes have clearly redundant functions. ysl1ysl3 double mutants are chlorotic and have lower Fe levels in leaves than the wild type (Waters et al., 2006). Their seeds also contain less Fe, and fertility is reduced (Waters et al., 2006). ysl3 and ysl2 single mutants have no apparent phenotype (DiDonato et al., 2004; Waters et al., 2006). Expression patterns of the three YSL genes suggest that the encoded transporters are involved in exiting nicotianamine-Fe from the vascular system to leaves upon sufficient Fe supply, whereas reduced exit upon Fe deficiency may serve to translocate Fe toward the growing apex (DiDonato et al., 2004; Le Jean et al., 2005; Schaaf et al., 2005; Waters et al., 2006).
Indeed, various families of metal transporters and regulators were found and are to date best described in Arabidopsis. The Arabidopsis genome has four NAS genes (Suzuki et al., 1999; Bauer et al., 2004). Thanks to the reverse genetic resources, it was possible to identify mutant T-DNA alleles of all four genes. To be able to study nicotianamine function in a broad genetic network of known players in metal homeostasis, we decided to develop Arabidopsis as a model system for investigating nicotianamine function. Here, we present the construction and analysis of quadruple nas mutants, one of which had an intermediate phenotype, named nas4x-1. In contrast to chloronerva, the tobacco NAAT line, and nas4x-2, which is the second nas mutant we identified that had full loss of NAS function, nas4x-1 had strongly reduced but not fully eliminated nicotianamine content. Due to its survival and maintained seed production, we could analyze nicotianamine involvement in Fe allocation to reproductive organs and seeds.
RESULTS
Generation and Initial Characterization of Multiple nas Mutants
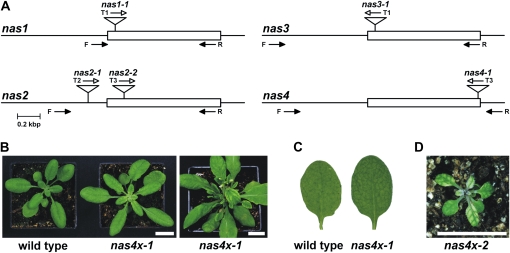
The four members of the NAS gene family are distributed on both arms of chromosomes I and V. All NAS genes have a single exon of 963 bp (NAS1, NAS2, and NAS3) and 975 bp (NAS4). With the aim of ultimately studying nas mutants, we identified and confirmed nas T-DNA insertion mutants for the four NAS genes (Fig. 1A). For the genes NAS1, NAS3, and NAS4, we could confirm that the positions of T-DNA insertions were in the exon-coding sequences (alleles nas1-1, nas3-1, and nas4-1, respectively). For the NAS2 gene, we could confirm the mutant nas2-1 allele harboring a T-DNA insertion 70 bp upstream of the start codon in the 5′ untranslated region of NAS2 and the mutant nas2-2 allele with an insertion 144 bp after the start codon in the exon.
Figure 1.
Genetic composition and phenotype of nas4x-1 mutant plants. A, Location of T-DNA insertions in nas alleles: nas1-1 (GABI_223A09), nas2-1 (SAIL_156C08), nas2-2 (SALK_066962), nas3-1 (GABI_010A10), and nas4-1 (SALK_135507). Arrows indicate the positions of primers used for genotyping by PCR. B, Morphological appearance of plants on soil; from left to right, Col-0 (wild type), nas4x-1 (vegetative stage), nas4x-1 (reproductive stage). C, Transmission light microscopic images of wild-type and nas4x-1 rosette leaves. Note that nas4x-1 rosette leaf veins are darker green than the intercostal leaf areas. D, Morphological appearance of nas4x-2 (vegetative stage).
Since single T-DNA insertion lines did not show any visible phenotype on soil (Bauer et al., 2004), we crossed together different nas mutants. First, we generated homozygous double mutants by crossing and selfing T-DNA insertion mutants of genes located on the same chromosome, namely nas1-1nas2-1 and nas3-1nas4-1. Homozygous double mutants were confirmed by genotyping. We found that the crossing-over events took place with a recombination rate of approximately 5% on the two chromosomes. Homozygous nas1-1nas2-1 double mutants were then crossed with nas3-1nas4-1 to generate heterozygous quadruple mutants. Quadruple heterozygous plants were selfed. We used genomic PCR to identify and select the quadruple homozygous mutant as well as double and triple mutants. A quadruple homozygous mutant that was fertile was identified as nas1-1nas2-1nas3-1nas4-1 and termed the nas4x-1 mutant. We observed that the nas4x-1 mutant had minor leaf chlorosis during vegetative growth that intensified during the transition to reproductive growth on soil (Fig. 1B). Nonchlorotic leaves from the vegetative stage of the nas4x-1 mutant had darker green veins than wild-type leaves when observed at transmitted light conditions (Fig. 1C). The nas4x-1 mutant was crossed with a homozygous nas2-2 mutant. The resulting multiple heterozygous plant was selfed and its progeny genotyped. We identified a homozygous quadruple nas1-1nas2-2nas3-1nas4-1 mutant that we termed nas4x-2. nas4x-2 plants had strong leaf chlorosis already upon vegetative growth and were sterile (Fig. 1D). Therefore, we suspected that nas4x-2 was a strong loss-of-function mutant similar to chloronerva, while nas4x-1 had an intermediate nas phenotypic expression.
As a first experiment, we studied whether any full-length NAS gene products were made in the quadruple homozygous mutants (data not shown). We found that in nas4x-1 and nas4x-2 mutant plants, full-length coding transcripts were not produced for nas1-1, nas2-2, nas3-1, and nas4-1. However, in the case of the nas2-1 allele present in the nas4x-1 mutant, it was possible to amplify a DNA fragment that contained the full-length coding NAS2 region after 35 PCR cycles, indicating that despite the insertion in the 5′ untranslated region a PCR product was made. Very interestingly, nas2-1-derived transcripts were found expressed not only in roots, as was the case in the wild type, but also in leaves (data not shown; see Fig. 3A below). This result indicated that the NAS2 gene was aberrantly expressed from the nas2-1 allele.
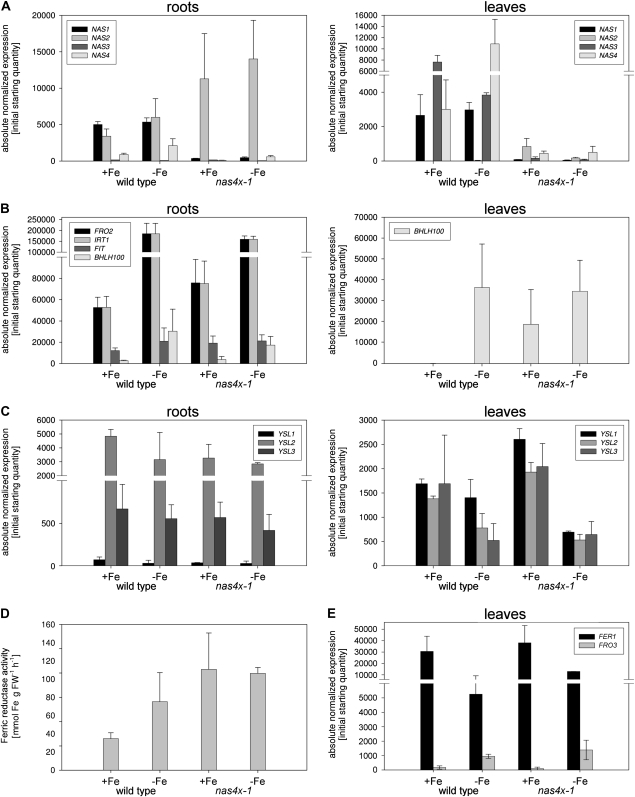
Figure 3.
Molecular-physiological analysis of nas4x-1 mutant and wild-type plants grown in a hydroponic system and exposed for 5 d to 0 μm Fe (−Fe) and control 50 μm Fe (+Fe). A to C and E, Quantitative reverse transcription-PCR analysis of root and leaf samples (n = 2). A, NAS gene expression. B, Fe deficiency response gene expression. C, YSL transporter gene expression in leaves and roots. E, FER1 and FRO3 gene expression. D, Ferric chelate reductase activity in roots (n = 3). FW, Fresh weight. Student's t test showed that significant differences (P < 0.05) were found for the comparisons wild-type +Fe versus −Fe, wild-type +Fe versus nas4x-1 + Fe, and wild-type +Fe versus nas4x-1 −Fe.
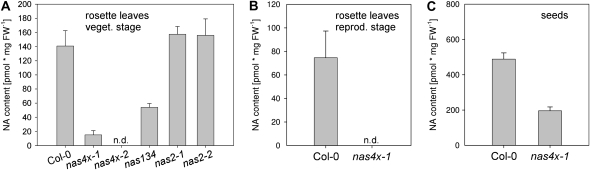
Next, we analyzed nicotianamine contents. We found that nas4x-1 mutant rosette leaves harvested in the vegetative phase had 10% nicotianamine levels compared with the wild type (11-h light period; Fig. 2A). We were not able to detect any nicotianamine in nas4x-1 rosette leaves of the reproductive stage when plants were grown with a 16-h light period (Fig. 2B). In nas4x-1 seeds, nicotianamine levels were about 40% of those in the wild type (Fig. 2C).
Figure 2.
Nicotianamine (NA) contents. A, Rosette leaves harvested during the vegetative phase. Plants were grown in a hydroponic system with quarter-strength Hoagland medium and 50 μm Fe under an 11-h light period. Six-week-old plants were used for analysis (Col-0/nas4x-1, n = 8; other mutants, n = 4). FW, Fresh weight; n.d., nondetectable. B, Rosette leaves harvested during the reproductive phase. Plants were grown as in A but under a 16-h light period. Six-week-old plants were used for analysis (n = 8). C, Seeds harvested from plants grown in a hydroponic system as described in B (n = 3).
Since vegetative growth resulted in the highest ratio of nicotianamine levels in nas4x-1 mutant versus wild-type plants, we analyzed nas4x-2 nicotianamine contents at the vegetative stage. We found that in the strong nas4x-2 mutant, nicotianamine was not detectable in rosette leaves, demonstrating full loss of NAS function (Fig. 2A). This finding explained the strong nas4x-2 phenotype.
Arabidopsis has four functional NAS genes that are differentially expressed (Bauer et al., 2004). Therefore, one might assume that NAS genes may have acquired specific functions during their evolution. The genetic data, however, indicate that NAS genes are functionally largely redundant. Therefore, we assessed the contribution of individual nas alleles for nicotianamine production by analyzing nicotianamine contents in the four single mutants with a single nas gene knockout and in triple mutants containing only a single viable NAS gene. We found that all nas single mutants had wild-type nicotianamine levels (Fig. 2A; Supplemental Fig. S1A). All triple mutants analyzed had about 30% to 50% of the nicotianamine content compared with wild-type plants (Fig. 2A; Supplemental Fig. S1B). No single NAS genes, therefore, may play any predominant role for overall nicotianamine synthesis. We also utilized these studies to refine the function of the nas2-1 allele in the nas4x-1 mutant. We found that nas1-1nas3-1nas4-1 had 35% nicotianamine compared with the wild type. On the other hand, the nas2-1 and nas2-2 mutants had the same amount of nicotianamine as the wild type (Fig. 2A). Thus, nas2-1 does not lead to increased nicotianamine production. Also, NAS2 loss of function could be compensated by other functional NAS genes so that overall nicotianamine content remained constant.
To test whether the redundancy could be due to compensatory effects of NAS gene transcription levels, we tested NAS gene expression levels in the single or triple mutant background (Supplemental Fig. S2). We observed that the presence of other mutant nas alleles did not affect gene expression of NAS2 and NAS3. NAS1 expression levels did not seem to be influenced by other nas mutations; however, due to high variability of NAS1 expression in these same plant samples, no clear conclusions were possible (see comment below on variable NAS1 expression in response to Fe deficiency). The only case where NAS gene expression may have been influenced was that of NAS4 wild-type allelic expression in the background of a nas2-1 allele. These control experiments indicate that redundancy of the four NAS genes was not generally due to compensation of defective NAS genes by increased expression of intact NAS genes.
The reduced nicotianamine content of nas4x-1 during the reproductive phase could be due to removal of nicotianamine through remobilization and transport and to reduced production. To test this, we grew plants in a hydroponic system for 6 weeks under either short-day conditions (8-h light period, harvest at the vegetative stage) or long-day conditions (16-h light period, harvest at the reproductive stage) and analyzed NAS gene expression (Supplemental Fig. S3). We found that NAS1 and NAS2 were expressed at 8- to 10-fold higher levels in roots at the vegetative stage than in the reproductive stage. NAS3, on the other hand, was expressed at about 4-fold higher levels in leaves at the reproductive stage than at the vegetative stage. NAS4 was expressed at a similar level in the two stages. These results are in agreement with the microarray data available from public databases (data not shown). In the nas4x-1 plants, NAS2 was expressed 2-fold higher upon the vegetative stage than the reproductive stage, whereas all other NAS genes were down-regulated due to the mutations (Supplemental Fig. S3). Thus, NAS genes were differentially expressed with respect to the growth stage. At every stage, at least two NAS genes were expressed in the wild type. nas4x-1 had NAS expression in roots at a higher level than in leaves, whereas in roots, it was lower upon the reproductive phase than the vegetative stage. Therefore, reduced nicotianamine production along with increased transport away from leaves may explain the leaf chlorosis phenotype upon reproduction.
This prompted us to speculate that nicotianamine plays an important role in seed metal homeostasis at this growth phase. With the intermediate fertile nas mutant in hand, we could investigate this point further. To better understand the nas4x-1 plants, we analyzed it in more detail at the molecular-physiological level.
Effect of Fe, Zn, and Cu Deficiency on nas4x-1 Mutants
Nicotianamine chelates Fe, Cu, and Zn. Therefore, we tested first whether deficiency of these metals may play a role in the expression of the mutant phenotype.
We grew wild-type and nas4x-1 plants in a hydroponic system and then exposed 5-week-old plants for 5 d to control medium (10 μm Fe) or Fe deficiency (0 Fe). We observed that nas4x-1 mutants had developed leaf chlorosis after the first 5 weeks. The leaf chlorosis became severe when plants were transferred to 0 Fe (Supplemental Fig. S4).
When we tested 0 Cu and 0 Zn supply, we did not observe any strong leaf chlorosis in the mutant after 7 d of exposure to deficiency. Cu and Zn contents in leaves of 0 Cu-grown or 0 Zn-grown mutant and wild-type plants, however, were reduced, demonstrating that the plants had indeed been Cu and Zn deficient (Supplemental Fig. S5).
The results show that nas4x-1 mutants were sensitive to Fe deficiency but not to deficiency of Zn and Cu. Hence, in nas4x-1 plants, nicotianamine is present in insufficient amounts to properly distribute the low Fe, whereas no problem exists for distribution of low Cu and Zn.
Gene Expression of nas4x-1 Plants in Response to Fe Deficiency
To analyze the susceptibility of nas4x-1 plants to Fe deficiency further, we harvested roots and leaves from hydroponically grown nas4x-1 and wild-type plants and studied gene expression in response to Fe deficiency by quantitative reverse transcription-PCR (Fig. 3).
As a control, NAS gene expression is first shown. NAS1, NAS2, and NAS4 were expressed in roots, whereas NAS3 was not expressed in roots (Fig. 3A). NAS2 and NAS4 were induced by Fe deficiency in roots to about three times. NAS1 was not found to be generally induced by Fe deficiency in the hydroponic growth system, whereas on agar it was found induced (compare with Fig. 4A; note that in the various nas mutant backgrounds, NAS1 also showed the most variable gene expression levels). In leaves, we found expression of NAS1, NAS3, and NAS4. NAS4 was induced by Fe deficiency in leaves, whereas NAS3 was repressed under Fe deficiency. NAS1 was not Fe regulated in leaves. In the nas4x-1 plants, NAS1, NAS3, and NAS4 were down-regulated due to the respective nas mutations, whereas NAS2 was found to be expressed in roots and leaves due to the nas2-1 allele (Fig. 3A). Hence, in roots, NAS genes are regulated by Fe deficiency, suggesting a functional involvement of these genes in Fe deficiency responses. It was confirmed that NAS genes are deregulated in nas4x-1 due to T-DNA insertions.
Figure 4.
Analysis of NAS and FIT gene interactions. A, Quantitative reverse transcription-PCR analysis of NAS1, FRO2, and FIT gene expression in roots and leaves of fit-3 mutant and p2x35S∷FIT overexpression plants. B, Ferric chelate reductase activity in roots of nas4x-1fit-3 plants. FW, Fresh weight. C, Quantitative reverse transcription-PCR analysis of FRO2 gene expression in roots of nas4x-1fit-3 mutant plants.
Next, we tested whether the susceptibility of the mutant to Fe deficiency was due to altered expression of Fe uptake genes (Fig. 3B). We investigated expression of the transcription factor genes FIT, encoding an essential regulator for Fe acquisition (Colangelo and Guerinot, 2004; Jakoby et al., 2004; Yuan et al., 2005), and BHLH100, which is induced by Fe deficiency in roots and leaves (Wang et al., 2007). Furthermore, we studied gene expression of the Fe acquisition genes FRO2 (Fe reductase) and IRT1 (Fe transporter), which are up-regulated by Fe deficiency dependent on FIT (Eide et al., 1996; Robinson et al., 1999; Vert et al., 2002). We found that FIT, IRT1, FRO2, and BHLH100 were induced by Fe deficiency in roots of wild-type plants (Fig. 3B). In wild-type leaves, BHLH100 was also induced by Fe deficiency. Therefore, the marker genes were up-regulated by Fe deficiency in the hydroponic system, as shown previously for the agar plate system (Jakoby et al., 2004; Wang et al., 2007). The levels of induction of IRT1 and FRO2 were lower in hydroponically grown plants than in agar plate-grown plants, due to the different growth conditions and ages of the plants. Interestingly, we found that in the nas4x-1 mutant plants, Fe deficiency response genes were expressed at higher levels than in the wild-type plants upon sufficient Fe supply in roots and in leaves (Fig. 3B). Upon 0 Fe, the Fe deficiency response genes were expressed at lower or equal levels in nas4x-1 plants than in wild-type plants. Hence, nas4x-1 plants experienced Fe deficiency upon sufficient Fe supply, but the mutant phenotype was not explained by the inability to induce Fe deficiency response genes.
We then studied whether altered distribution of Fe might cause the phenotype. Therefore, we studied gene expression of YSL genes YSL1, YSL2, and YSL3 encoding potential transporters for nicotianamine-metal complexes (DiDonato et al., 2004; Le Jean et al., 2005; Schaaf et al., 2005; Waters et al., 2006). We confirmed that in the wild type, all three YSL genes were down-regulated by Fe deficiency in roots and leaves compared with +Fe plants (Fig. 3C). In nas4x-1 mutant roots grown at +Fe, the three genes were expressed at comparable levels as in wild-type −Fe. The same was found upon −Fe. In leaves of nas4x-1 mutants, the three YSL genes were up-regulated or equally expressed upon sufficient Fe supply, whereas they were rather down-regulated upon Fe deficiency. Hence, YSL gene expression suggested that upon Fe sufficiency, nas4x-1 mutant roots behaved like Fe-deficient roots, whereas leaves of nas4x-1 plants had characteristics of Fe-sufficient leaves despite the weak leaf chlorosis.
The previous results were contradictory, as nas4x-1 leaf cells were on the one hand Fe sufficient (as suggested by YSL up-regulation) and on the other hand Fe deficient (as revealed by BHLH100 up-regulation). To further investigate this, we analyzed expression of the two leaf marker genes for either Fe sufficiency or Fe deficiency. One marker gene was the ferritin gene FER1, which encodes a Fe storage protein in leaf chloroplasts expressed upon sufficient Fe (Petit et al., 2001). The other was the Fe deficiency-inducible ferric reductase gene FRO3 (Mukherjee et al., 2006). As expected, FER1 was up-regulated in Fe-sufficient wild-type plants (Fig. 3E). FRO3, on the other hand, was up-regulated in wild-type leaves upon −Fe (Fig. 3E). However, in nas4x-1 mutant plants, FER1 and FRO3 were surprisingly regulated as in the wild type upon both +Fe and −Fe, demonstrating that the leaf Fe regulatory system perceived Fe sufficiency in nas4x-1 leaves upon +Fe (Fig. 3E). Up-regulation of BHLH100 and up-regulation of Fe deficiency response genes in the root, therefore, must be linked with another signaling system that is defective in nas4x-1.
In order to assess a physiological marker for Fe deficiency, we determined Fe reductase activity of nas4x-1 plants. We found that, as expected, root Fe reductase activity was induced in the wild type upon transfer to Fe deficiency in the hydroponic system. In nas4x-1 roots, Fe reductase activity was increased compared with the wild type upon sufficient Fe supply (Fig. 3D).
Taken together, we conclude from these molecular and physiological results that nas4x-1 mutants suffer from Fe deficiency when sufficient Fe is supplied. Roots showed symptoms of Fe deficiency (up-regulation of FIT, IRT1, FRO2, and BHLH100) upon sufficient Fe. Leaves show at the same time symptoms for Fe deficiency (BHLH100 induction) and Fe sufficiency (YSL and FER1 up-regulation and FRO3 down-regulation), which can be explained by cellular or subcellular mislocalization of Fe. Obviously, low nicotianamine resulted in an altered regulation of Fe deficiency responses.
Genetic Interaction of NAS and FIT Genes
The gene expression analysis supports a role of nicotianamine in the regulation of Fe deficiency responses. We analyzed this further by studying the phenotypes of nas4x-1 mutations in the background of a fit-3 mutant. nas4x-1fit-3 multiple mutants were not able to reduce Fe in the ferric reductase assay, as was also the case for fit-3 mutants (Fig. 4B), nor could they induce FRO2 gene expression (Fig. 4C). nas4x-1fit-3 multiple mutants showed a similar yellow leaf chlorosis phenotype as fit-3 mutants (data not shown).
Since NAS genes were regulated by Fe supply, we investigated whether NAS gene expression in roots might require the Fe deficiency response regulator FIT. fit loss-of-function mutants are chlorotic and show down-regulation of IRT1 and FRO2. We grew fit-3 mutants and p2x35S∷FIT overexpression plants for 2 weeks on sufficient Fe supply, transferred them for 3 d to Fe deficiency as described previously (Jakoby et al., 2004), and analyzed NAS gene expression. We found that among the four NAS genes tested, only NAS1 was down-regulated in roots of fit-3 mutant plants compared with wild-type plants. However, it was up-regulated in leaves of FIT overexpression plants exposed to Fe deficiency (Fig. 4A). From previous experiments, we know that in FIT overexpression plants, FRO2 and IRT1 are ectopically expressed (Jakoby et al., 2004), Hence, NAS1 is partially under the control of the FIT transcription factor as well (Colangelo and Guerinot, 2004). The results also show that NAS1 can be induced by −Fe in roots upon agar growth (Fig. 4A), whereas in the hydroponic system, this was not the case (Fig. 3A).
Taken together, these results show that fit-3 is epistatic to nas4x-1 and that FIT controls the expression of NAS1. Hence, NAS and FIT genes act in the same pathway of regulating Fe mobilization.
Gene Expression during Reproduction
Since nas4x-1 leaf chlorosis phenotypes became severe upon transition to flowering, we reasoned that most likely increased nicotianamine levels were needed at this stage in the growing inflorescence.
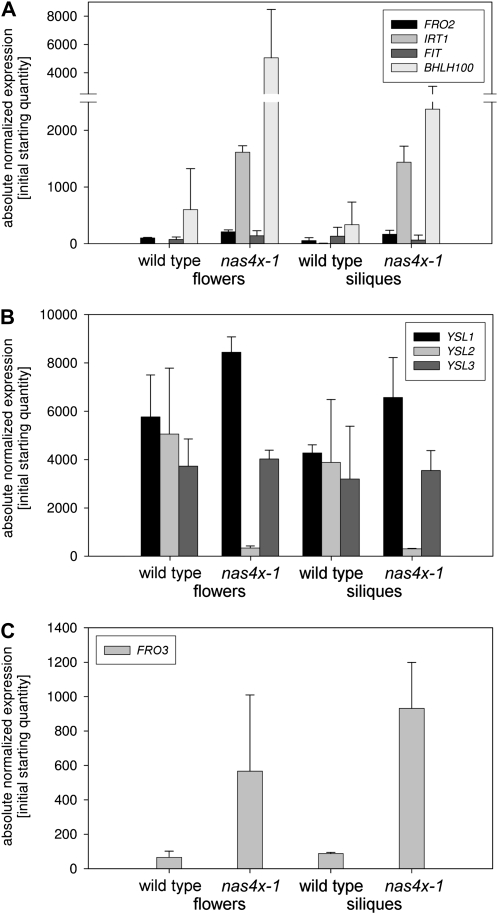
We investigated whether in flowers the nas4x-1 mutation had an influence on the expression of Fe-regulated genes (Fig. 5A). IRT1 was found previously to be expressed in flowers, whereas loss of irt1 function had no effect on reproduction (Vert et al., 2002). Interestingly, we found that IRT1 and BHLH100 were strongly up-regulated in nas4x-1 flowers and siliques in contrast to the wild type. As expected, FIT and FRO2 were not expressed significantly in these organs in either genetic background. Hence, this could be a hint that IRT1 plays a role in Fe transport in flowers.
Figure 5.
Quantitative reverse transcription-PCR analysis of nas4x-1 mutant flowers and siliques. A, Fe deficiency response gene expression. B, YSL gene expression. C, FRO3 gene expression.
We studied the expression of YSL transporter genes (Fig. 5B). Whereas YSL3 was expressed at similar levels between the mutant and the wild type, YSL1 was up-regulated to about 30% in nas4x-1 flowers and siliques. YSL2 expression was drastically suppressed in nas4x-1 mutant flowers and siliques to about 10- to 15-fold. Hence, YSL1 and YSL3 expression patterns behaved in flowers and siliques similar to the leaves in the wild type compared with nas4x-1. If YSL1 and YSL3 expression was indeed correlated with Fe sufficiency to promote lateral removal of Fe, this observation may indicate that despite Fe deficiency, flowers and siliques were also Fe sufficient. YSL2 down-regulation in nas4x-1 flowers and siliques was very surprising, and in contrast to the expression in leaves. Perhaps down-regulation of YSL2 in nas4x-1 reproductive organs was advantageous for Fe supply, for example of seeds while nicotianamine was reduced.
Finally, we tested whether FRO3 and FER1 might also be regulated in reproductive organs. FRO3 was found up-regulated in nas4x-1 flowers and siliques compared with the wild type (Fig. 5C). This was an indication that in contrast to leaves, the reproductive organs were rather Fe deficient. FER1 expression was highly variable in both the wild type and nas4x-1, so no conclusions were drawn (data not shown).
In summary, gene expression in nas4x-1 mutant flowers and siliques indicated that these organs suffered from Fe deficiency and suggested that an alternative rescue pathway for Fe uptake of cells might have been switched on.
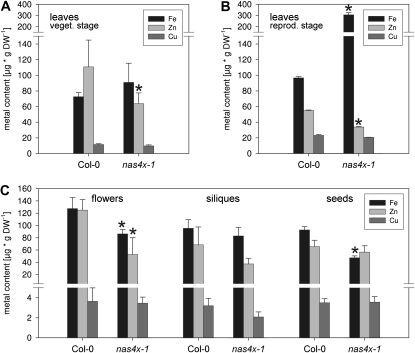
Metal Contents of nas4x-1 Plants
It was then interesting to analyze whether low nicotianamine levels influenced the transport of metals (Fe, Cu, and Zn) into various organs. In roots, where metals are found in the vascular system, inside root cells, and in the apoplast, we did not detect changes between nas4x-1 and the wild type (data not shown). Differences in metal contents were observed in aboveground organs. Vegetative leaves of nas4x-1 plants contained about the same Fe as the wild type, whereas Zn was significantly reduced by about 38% (Fig. 6A). Cu contents were unchanged. In rosette leaves in the reproductive stage, Fe contents were significantly increased by 216% in nas4x-1, whereas Zn remained significantly decreased by half (Fig. 6B). Cu was if at all slightly reduced (approximately 11%) in these nas4x-1 leaves. On the other hand, in flowers, siliques, and seeds, Fe contents were decreased by 32% (significantly), 13%, and 46% (significantly), respectively (Fig. 6C). Zn contents were also decreased in nas4x-1 flowers (significantly) and siliques, whereas it was similar in seeds when compared with the wild type. Cu contents were slightly reduced in nas4x-1 siliques but not in the other plant parts tested. Thus, significance tests of metal determination showed that nicotianamine was not needed to take up and transport Fe to leaves but it was required to efficiently mobilize Fe from leaves during the reproductive phase to flowers and seeds. Nicotianamine was also beneficial for Zn uptake and transport to leaves and flowers.
Figure 6.
Metal contents of nas4x-1 mutant and wild-type plants. A, Leaves harvested during the vegetative phase from plants grown under an 11-h light period. B, Leaves harvested during the reproductive phase from plants grown under a 16-h light period. C, Flowers, siliques, and seeds. Plants were grown in a regular hydroponic system. The indicated plant organs were dissected and freeze dried. Metal contents (Fe, Zn, and Cu) were determined by graphite furnace atomic absorption spectroscopy with direct solid sampling. For leaves, n = 8; for flowers/siliques, n = 5; for seeds, n = 6. DW, Dry weight. * P < 0.01 using the t test.
Phenotypes of nas4x-1 Plants in Response to Toxic Metal Supply
Our results showed that reduced nicotianamine caused reduced Fe transport to reproductive plant parts and reduced Zn in the plant. Hence, we wondered whether the loss of the ability to translocate metals inside the plant could lead to an altered response to metal toxicity. To test this, we studied the responses of nas4x-1 plants to supply of excess concentrations of Fe (up to 200 μm), Zn (up to 200 μm), and Cu (up to 28 μm). Fe, Zn, and Cu caused shorter roots and smaller shoots than control conditions, and Zn and Cu also caused yellow leaves (data not shown). In none of the three cases did we observed a significant phenotypic difference between nas4x-1 and wild-type plants (data not shown). We conclude, therefore, that wild-type nicotianamine levels did not alleviate Fe, Zn, and Cu toxicity.
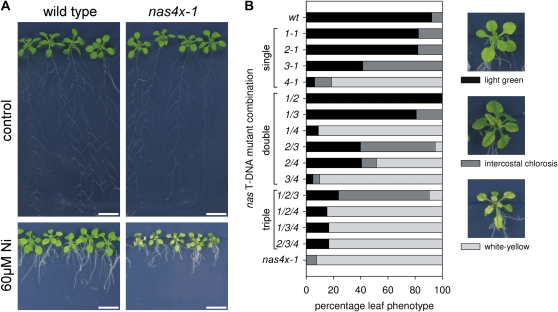
As a control, we tested the responses to Ni, which is known to be chelated by nicotianamine. Elevated nicotianamine levels have recently been correlated with tolerance to Ni (Becher et al., 2004; Weber et al., 2004; Douchkov et al., 2005; Kim et al., 2005; Pianelli et al., 2005). Interestingly, nas4x-1 seedlings were highly sensitive to 60 μm Ni, in contrast to wild-type plants. nas4x-1 mutants developed strongly chlorotic leaves and short roots at 2 weeks of age and older, whereas wild-type plants still remained green under these conditions (Fig. 7A). Multiple mutant analysis showed epistatic interactions of nas mutant alleles. The single and multiple mutant combinations with nas4-1 were most severely affected, the single and multiple mutants with nas3-1 (but not nas4-1) were second most affected by Ni, whereas nas1-1, nas2-1, and nas1-1nas2-1 double mutants were not affected more than the wild type (Fig. 7B).
Figure 7.
Phenotypes of nas mutants to Ni treatment. A, Growth assay showing 17-d-old seedlings grown on 60 μm Ni and control Hoagland agar medium. Note the short root phenotypes of Ni-treated plants and the severe white-yellow leaf chlorosis phenotype of nas4x-1 mutants compared with the wild type on Ni. B, Distribution of leaf chlorosis phenotypes among multiple nas mutant lines grown for 17 d on 60 μm Ni and control Hoagland agar medium (n = 10–35). Three leaf phenotypic classes were defined: light green leaf appearance of mildly affected plants, intermediate intercostal leaf chlorosis of Ni-sensitive plants, and the most severe white-yellow leaf chlorosis of severely Ni-affected plants. The number of plants belonging to each phenotypic class was calculated in percentage. [See online article for color version of this figure.]
We also tested whether NAS genes were affected by Ni treatment. NAS1, NAS2, and NAS4 genes but not NAS3 were induced by Ni treatment in roots, although only to a 2- to 3-fold level, whereas general transcript levels were low for NAS4 compared with NAS1 and NAS2 (Supplemental Fig. S6). Interestingly, FRO2 as a marker for Fe deficiency was also up-regulated in Ni-treated roots, suggesting that Ni-treated plants might experience Fe deficiency. In leaves, NAS3 and NAS4 were up-regulated 2- to 3-fold by Ni treatment (Supplemental Fig. S6). Hence, NAS gene expression was affected by Ni but induction was quite low. NAS3 and NAS4 responses to Ni were in agreement with Kim et al. (2005).
Hence, wild-type levels of nicotianamine were used for detoxification of Ni but not of Zn, Cu, and Fe. Moreover, NAS4 and NAS3 conferred higher Ni resistance than NAS1 and NAS2, suggesting that at the level of Ni toxicity there was not full redundancy of NAS genes.
DISCUSSION
Our analysis of the Arabidopsis nas mutant may form the ground for targeted improvement of the Fe nutritional value of crops. Our results support that by way of altering expression of NAS genes using transgenic and classical breeding approaches, the seed and leaf Fe contents can be manipulated.
We have shown here that complete loss of nicotianamine resulted in a chloronerva phenotype in Arabidopsis (nas4x-2 mutant), namely in severe leaf chlorosis and sterility. Since Arabidopsis and solanaceous plants belong to two different subclasses of the eudicots, namely Rosidae and Asteridae, we conclude that nicotianamine function is conserved in most if not all dicots.
Our study concentrated on the analysis of a knockdown mutant of nicotianamine synthase function, nas4x-1, that had residual nicotianamine. This mutant had chloronerva-like features, such as leaf chlorosis, susceptibility to Fe deficiency, increased Fe uptake and Fe levels in leaves, and fit/nas epistatic interactions. The mutant also was susceptible to Ni supply. Remaining nicotianamine contents in nas4x-1 still allowed the plants to complete their life cycle and produce seeds. Mutants with intermediate phenotypes yielded interesting results in plant biology and were often discovered in forward genetic screens. Interpretation of null mutants is frequently limited by the onset of pleiotropic responses and/or a precocious block in development. Examples for intermediate phenotypes are found in many of the developmental mutants, and they have also been valuable in the Fe homeostasis field, where for most gene families only single mutant analysis was performed.
The four NAS genes act in a functionally redundant manner, since the enzyme product nicotianamine is a mobile compound in the plant. Despite the redundancy, individual NAS enzymes have gained specific properties, for example due to differential gene expression patterns such as in different tissues and by regulation through metal supply. Functional specification of NAS1 can also explain the variable NAS1 gene expression we have observed while analyzing multiple nas mutants and Fe deficiency responses. NAS1 may have responded specifically to certain developmental or environmental cues that occurred in a variable manner in these latter growth conditions.
We have used this mutant to ask several questions.
Why have nas4x-1 leaves been more chlorotic during the reproductive stage than the vegetative stage? The late leaf chlorosis was the first remarkable phenotype of nas4x-1 plants. We found that rosette leaves of the reproductive stage of nas4x-1 plants did not have any nicotianamine, whereas during the vegetative stage such leaves contained nicotianamine, although only at 10% of wild-type levels. We deduce from these observations that nicotianamine had disappeared from the rosette leaves during the reproductive phase and was no longer replenished through synthesis or transport. The removal of nicotianamine caused a Fe distribution problem in leaf cells, which was the cause for the leaf chlorosis. Clearly, some cells or compartments of nas4x-1 plants were Fe starved, so that Fe deficiency signaling took place in the leaves. In support of this is the fact that leaves turned chlorotic and that expression of BHLH100 was induced. The transcription factor gene BHLH100 responds to local Fe deficiency, as shown by previous grafting studies (Wang et al., 2007). It is very interesting that in nas4x-1 plants, BHLH100 regulation was decoupled from the presence of Fe in the leaves. It could indeed have been that Fe deficiency-mediated stress responses that had occurred in nas4x-1 leaves up-regulated BHLH100. Also in support of Fe deficiency of nas4x-1 plants was the fact that root Fe acquisition responses had been switched on, such as IRT1 and FRO2 induction. On the other hand, nas4x-1 plants contained Fe during the reproductive phase at even more than twice the amount compared with the wild type. Obviously, this Fe did not enter all target places. However, the Fe in the leaves was sensed, so that FRO3, a ferric reductase gene normally induced by −Fe in leaves (Mukherjee et al., 2006), was not induced. Therefore, FRO3 was probably not required for uptake and transport of Fe into leaves of nas4x-1 plants. Similarly, nas4x-1 leaves had similar FER1 expression levels as Fe-sufficient wild-type leaves. Therefore, the signaling machinery that regulated FRO3 and FER1 expression in response to Fe operated and sensed Fe in nas4x-1 leaves.
The genetics of the nas4x-1 mutant also support the idea that in leaves nicotianamine had disappeared during the reproductive phase. Gene expression studies and mutant T-DNA analysis had shown that only the nas2-1 allele was responsible for residual nicotianamine production in the mutant. This allele was essentially expressed in roots, whereas it was expressed to a low level in leaves. Moreover, its gene expression levels were higher in the vegetative phase than in the reproductive phase. For example, in the reproductive phase, the total number of NAS transcripts of nas4x-1 plants was 25% higher in roots compared with wild-type plants; on the other hand, it was 88% lower in leaves. This suggests that roots were able to continuously supply nicotianamine in nas4x-1. We can exclude that the higher and partially ectopic NAS2 gene expression caused higher nicotianamine production from nas2-1 than from the wild-type NAS2. In fact, we found that a nas1-1nas3-1nas4-1 mutant with wild-type NAS2 had three to four times higher nicotianamine levels than the mutant with the nas2-1 allele (which was nas4x-1). Hence, nas2-1 resulted in a reduced function of NAS2. The NAS leaf isoforms are normally encoded by NAS1, NAS3, and NAS4. Since NAS3 and NAS4 were mutated in nas4x-1, they could not replenish nicotianamine in leaves, especially during reproduction, when NAS3 was induced in leaves.
The question remained, what happened to the nicotianamine that had been depleted from leaves? The answer led to a second striking phenotype of nas4x-1 plants: reduced Fe contents in seeds. Our results show that wild-type nicotianamine levels were essential for normal Fe supply of seeds. Nicotianamine was present in wild-type seeds. nas4x-1 seeds contained nicotianamine to a lower level than wild-type seeds, and they also contained Fe to a lower level. However, neither in flowers nor in developing siliques of the wild type or mutant could we detect NAS gene transcripts above background levels in our experiments (data not shown). In both the wild type and mutant, therefore, it seemed impossible that reproductive parts had their own nicotianamine synthase activity. Hence, any nicotianamine that was detected in seeds must have been produced elsewhere in the plant and transported to the flowers before its deposition into developing seeds. The disappearance of nicotianamine from leaves, accompanied by the onset of leaf chlorosis during reproduction, suggests an important function of nicotianamine at this stage. Therefore, this explains our results such that nicotianamine was transported away from leaves toward developing seeds. Presumably, nicotianamine was transported as Fe chelate. Since the nas4x-1 mutant had less nicotianamine, a consequence was that less Fe reached the developing seeds and more Fe remained in leaves. In addition, due to diminished NAS activity in nas4x-1 roots, we assume that less nicotianamine-Fe reached the reproductive organs directly from the roots but may have ended up in leaves in a nicotianamine-independent manner.
ysl1 and ysl1ysl3 mutants shared phenotypes of nas4x-1, such as low Fe content in seeds, Fe deficiency responses, and leaf chlorosis, which had suggested that YSL genes were involved in seed Fe partitioning (Le Jean et al., 2005; Waters et al., 2006; Waters and Grusak, 2008). We found that YSL gene expression was affected in nas4x-1 plants. These results strongly suggest that YSL and NAS genes indeed acted in similar biological processes. The best explanation is that YSL proteins are indeed the transporters for nicotianamine-Fe.
Although seeds were clearly reduced in Fe, they were only slightly reduced in Zn content and not reduced in Cu. This shows that high amounts of nicotianamine were needed for Fe deposition but low amounts were sufficient for Zn and Cu transport into seeds. An interesting phenotype was observed in naat mutant rice plants. These mutants had increased nicotianamine levels due to dysfunctional phytosiderophore synthesis, which led to reduced Fe mobilization via strategy II. But in return, these plants could take up Fe through an IRT-like transporter, which may have been aided by the increased nicotianamine (Cheng et al., 2007). Since these plants had higher seed Fe contents, it may have been possible that in these rice plants increased nicotianamine was implicated in the loading of seeds with Fe, the opposite case of nas4x-1.
Why did the nas4x-1 plants increase Fe acquisition during the reproductive phase while Fe accumulated in leaves? Previous systemic signaling studies of a number of mutants had suggested that systemic Fe deficiency signals must exist that stem from leaves and that regulate Fe acquisition in the root (Grusak and Pezeshgi, 1996; Vert et al., 2002; Enomoto et al., 2007; Wang et al., 2007). As discussed above, nas4x-1 leaves showed signs of Fe sufficiency but also of Fe deficiency. It is possible that the FER1/FRO3 regulatory system was simply not connected with the systemic signaling mechanism for root Fe acquisition responses, whereas BHLH100 might have been. Another explanation for the induction of Fe acquisition in roots could be the high demand for Fe in the plants. Very interestingly, we have observed that IRT1, BHLH100, and FRO3 were all induced in flowers and siliques, showing that these organs not only had low Fe levels but also responded to that. It is possible that due to reduced nicotianamine-Fe transport in flowers and siliques, alternative routes for Fe uptake were switched on based on FRO3/IRT1. Perhaps these reproductive organs with their Fe deficiency were capable of producing a systemic signal that overruled the potential Fe sufficiency of leaves so that they acted as sink organs for Fe.
Could the reduced Fe content of nas4x-1 seeds result in a germination defect? The answer is that a 60% reduction of Fe in nas4x-1 seeds did not result in any germination defect on regular medium. However, upon germination on Fe-deficient medium, nas4x-1 seedlings had stronger chlorotic leaves than wild-type seedlings (M. Klatte and M. Schuler, unpublished data). Hence, nicotianamine helped in Fe mobilization during germination when external Fe supply was low. Interestingly, this susceptibility to Fe deficiency was also apparent later in development, when plants were transferred from regular to Fe-deficient medium for several days. These plants had been able to take up Fe during their postgermination growth, yet Fe deficiency was more severe than in the wild type. Since NAS genes and FIT acted in the same pathway of Fe acquisition (epistatic interactions of FIT over NAS in the multiple mutants), nicotianamine was needed during the Fe acquisition process. One possible explanation could be that nicotianamine is used to translocate Fe, such as from the epidermis to the vascular system in roots upon Fe deficiency.
The final question for discussion is, why have nas4x-1 plants been sensitive to Ni but not to elevated Fe, Zn, or Cu? Obviously, the normal nicotianamine levels were not used in prevention of the Fenton reaction and other toxic effects of Fe, Cu, or Zn. If they were used for preventing toxic effects of these metals, we would have expected the mutant to react in an oversensitive manner to these metals. On the one hand, this finding is surprising. In fact, hyperaccumulators have mostly strongly up-regulated nicotianamine synthesis (Becher et al., 2004; Weber et al., 2004; Mari et al., 2006) and NAS overexpression can lead to increased tolerance to metals, including Zn (Kim et al., 2005). The function of high nicotianamine for Zn detoxification, therefore, remains dubious. Interestingly, overexpression of Thlaspi TcNAS1 in Arabidopsis led to increased sensitivity to Fe deficiency (Cassin et al., 2009). This was also surprising, as Fe deficiency is normally conferred by a loss of nas function. As discussed by Cassin et al. (2009), the apparent contradictory results between nas loss of function and NAS overexpression could be explained by the altered location of nicotianamine. Our metal determination analyses have shown that nas4x-1 leaves have lower Zn content than wild-type leaves. Thus, nas4x-1 mutants translocate less Zn to leaves than the wild type, which may partially protect the mutants from Zn toxicity. On the other hand, less nicotianamine was available in nas4x-1 leaves and roots to chelate Zn and prevent toxic effects in the cells, and in addition, stronger Fe deficiency was present. In turn, this may explain why nas4x-1 mutants and the wild type reacted in the end similarly to high Zn, while only NAS overexpression was protective, as shown by Kim et al. (2005).
Indeed, we have confirmed that nicotianamine can be used for detoxification of Ni through induced Ni sensitivity in nas4x-1, whereas other groups showed Ni tolerance upon increased nicotianamine in Arabidopsis or tobacco (Douchkov et al., 2005; Kim et al., 2005; Pianelli et al., 2005). With respect to this heavy metal, we also found nonredundant functions of NAS genes. NAS4 and to a lower degree NAS3 contributed essentially to Ni tolerance, whereas NAS1 and NAS2 could be neglected. Ni may directly affect NAS function, so perhaps NAS3 and NAS4 could be the Ni-tolerant NAS isoforms. Upon Ni treatment, increased nicotianamine might have been needed for Fe mobilization. Indeed, our results based on FRO2 expression indicate that Ni treatment caused Fe deficiency, perhaps through competition of binding sites to nicotianamine and other compounds in the cells. Interestingly, previous studies have shown that increased nicotianamine in Arabidopsis not only resulted in tolerance to Ni but also in tolerance to Fe deficiency (Douchkov et al., 2005; Kim et al., 2005). Hence, the two processes could be physiologically linked.
MATERIALS AND METHODS
Plant Material and Genotyping
The Arabidopsis (Arabidopsis thaliana) ecotype Columbia (Col-0) was used as the wild-type control. nas T-DNA insertion mutants were obtained from various stock centers (nas1-1 = GABI-kat 223A09, nas2-1 = SAIL_156_C08, nas2-2 = SALK_066962, nas3-1 = GABI-kat 010A10, and nas4-1 = SALK_135507) and tested for the presence of the respective T-DNA insertions using genomic PCR (primer sequences are given in Supplemental Table S1). The inheritance of T-DNA insertions was analyzed by segregation analysis. Multiple mutants were obtained through crossing as indicated in the text. Genotypes of multiple mutants were confirmed by genomic PCR and subsequent genotyping in the progeny of the plants. fit mutants and the p2x35S∷FIT overexpression line were described by Jakoby et al. (2004).
Plant Growth
Seeds were surface sterilized and stratified for 2 to 3 d at 4°C. The agar plate system using 1× Hoagland agar square plates was as described by Jakoby et al. (2004). Fe-deficient agar medium was devoid of Fe or supplemented with 50 μm ferrozine. For Ni stress conditions, Hoagland medium was modified to contain 60 μm Ni (NiCl2). Zn stress medium contained 200 μm ZnSO4 (instead of the regular 2 μm Zn), and Cu stress medium contained 28 μm CuSO4 (instead of the regular 1.5 μm Cu). For the hydroponic system, seedlings were germinated on quarter-strength Hoagland agar medium in 500-μL support tubes containing a hole at the bottom for root growth. After 2 weeks, plants were placed into aerated quarter-strength Hoagland liquid medium for another 2 weeks. Medium was exchanged weekly. Quarter-strength Hoagland medium contained one-quarter of the Hoagland salts and 10 μm FeEDTA (control). Solid germination medium of the hydroponic system contained no Suc and 0.6% plant agar. Metal deficiency in the hydroponic solution was achieved using quarter-strength Hoagland medium lacking Fe, Cu, or Zn salts. Cultivation took place at 21°C/19°C and 16-h-light/8-h-dark cycles or as indicated in the text.
Physiological Plant Analysis
Leaf chlorosis phenotypes were determined by manual inspection as indicated in the text. Ferric reductase activity was measured as described by Jakoby et al. (2004). Metal contents were determined on dried plant samples using graphite furnace atomic absorption spectroscopy with direct solid sampling (AAS vario 6, graphite furnace and flame technique; Analytik Jena).
Gene Expression Analysis
Gene expression analysis was performed by reverse transcription-quantitative real-time PCR as described previously (Wang et al., 2007). Briefly, DNase-treated RNA was used for cDNA synthesis. SYBR Green I-based real-time PCR analysis was performed using ExTaq R-PCR (TaKaRa) in a Mx3000P real-time PCR cycler (Stratagene). For each gene, the absolute quantity of initial transcript was determined by standard curve analysis. Absolute expression data were normalized against the averaged expression values of the internal control genes EF1BALPHA2 and UBP6. Primer sequences are provided in Supplemental Table S1. Specific PCR oligonucleotides were designed using Primer3 software. NAS primer specificity was thoroughly verified in test amplifications.
Determination of Nicotianamine Contents
Nicotianamine was extracted using a modified protocol (Neumann et al., 1999). The identity of nicotianamine was evidenced by spiking of chemically synthesized nicotianamine to plant samples. The nicotianamine-free tomato (Solanum lycopersicum) mutant chloronerva was used as a negative control. In leaves of Arabidopsis, a recovery rate of 98% ± 7% was achieved after addition of external standard to the plant sample prior to nicotianamine extraction (Supplemental Fig. S7). The detailed protocol of the modified method is available as Supplemental Materials S1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Nicotianamine contents of single and triple nas mutants.
Supplemental Figure S2. NAS gene expression in single and triple nas mutants.
Supplemental Figure S3. NAS gene expression during vegetative and reproductive growth.
Supplemental Figure S4. Effect of Fe deficiency treatment in nas4x-1 mutants.
Supplemental Figure S5. Zn and Cu contents of nas4x-1 and wild-type plants.
Supplemental Figure S6. NAS gene expression in response to Ni.
Supplemental Figure S7. Recovery rate of nicotianamine.
Supplemental Table S1. Gene-specific primers used for real-time quantitative PCR analysis.
Supplemental Materials S1. Determination of nicotianamine contents.
Supplementary Material
Acknowledgments
We thank the Institute of Plant Genetics and Crop Plant Research Gatersleben for support in the initial stages of this project.
This work was supported by the Deutsch Forschungsgemeinschaft in the Arabidopsis Functional Genomics Program, Arabidopsis Functional Genomics Network (grant nos. Ba1610/4–1 and Ba1610/4–3).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Petra Bauer (p.bauer@mx.uni-saarland.de).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Bauer P, Thiel T, Klatte M, Bereczky Z, Brumbarova T, Hell R, Grosse I (2004) Analysis of sequence, map position, and gene expression reveals conserved essential genes for iron uptake in Arabidopsis and tomato. Plant Physiol 136 4169–4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher M, Talke IN, Krall L, Krämer U (2004) Cross-species microarray transcript profiling reveals high constitutive expression of metal homeostasis genes in shoots of the zinc hyperaccumulator Arabidopsis halleri. Plant J 37 251–268 [DOI] [PubMed] [Google Scholar]
- Bereczky Z, Wang HY, Schubert V, Ganal M, Bauer P (2003) Differential regulation of nramp and irt metal transporter genes in wild type and iron uptake mutants of tomato. J Biol Chem 278 24697–24704 [DOI] [PubMed] [Google Scholar]
- Briat JF, Curie C, Gaymard F (2007) Iron utilization and metabolism in plants. Curr Opin Plant Biol 10 276–282 [DOI] [PubMed] [Google Scholar]
- Brumbarova T, Bauer P (2005) Iron-mediated control of the basic helix-loop-helix protein FER, a regulator of iron uptake in tomato. Plant Physiol 137 1018–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassin G, Mari S, Curie C, Briat JF, Czernic F (2009) Increased sensitivity to iron deficiency in Arabidopsis thaliana overaccumulating nicotianamine. J Exp Bot 60 1249–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng LJ, Wang F, Shou HX, Huang FL, Zheng LQ, He F, Li JH, Zhao FJ, Ueno D, Ma JF, et al (2007) Mutation in nicotianamine aminotransferase stimulated the Fe(II) acquisition system and led to iron accumulation in rice. Plant Physiol 145 1647–1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colangelo EP, Guerinot ML (2004) The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16 3400–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colangelo EP, Guerinot ML (2006) Put the metal to the petal: metal uptake and transport throughout plants. Curr Opin Plant Biol 9 322–330 [DOI] [PubMed] [Google Scholar]
- Curie C, Panaviene Z, Loulergue C, Dellaporta SL, Briat JF, Walker EL (2001) Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature 409 346–349 [DOI] [PubMed] [Google Scholar]
- de Benoist B, McLean E, Egli I, Cogswell M, editors (2008) Worldwide Prevalence of Anaemia 1993-2005. World Health Organization Press, Geneva [DOI] [PubMed]
- DiDonato RJJ, Roberts LA, Sanderson T, Eisley RB, Walker EL (2004) Arabidopsis Yellow Stripe-Like2 (YSL2): a metal-regulated gene encoding a plasma membrane transporter of nicotianamine-metal complexes. Plant J 39 403–414 [DOI] [PubMed] [Google Scholar]
- Douchkov D, Gryczka C, Stephan UW, Hell R, Baumlein H (2005) Ectopic expression of nicotianamine synthase genes results in improved iron accumulation and increased nickel tolerance in transgenic tobacco. Plant Cell Environ 28 365–374 [Google Scholar]
- Eide D, Broderius M, Fett J, Guerinot ML (1996) A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci USA 93 5624–5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto Y, Hodoshima H, Shimada H, Shoji K, Yoshihara T, Goto F (2007) Long-distance signals positively regulate the expression of iron uptake genes in tobacco roots. Planta 227 81–89 [DOI] [PubMed] [Google Scholar]
- Goto F, Yoshihara T, Shigemoto N, Toki S, Takaiwa F (1999) Iron fortification of rice seed by the soybean ferritin gene. Nat Biotechnol 17 282–286 [DOI] [PubMed] [Google Scholar]
- Grusak MA, Pezeshgi S (1996) Shoot-to-root signal transmission regulates root Fe(III) reductase activity in the dgl mutant of pea. Plant Physiol 110 329–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell R, Stephan UW (2003) Iron uptake, trafficking and homeostasis in plants. Planta 216 541–551 [DOI] [PubMed] [Google Scholar]
- Herbik A, Koch G, Mock HP, Dushkov D, Czihal A, Thielmann J, Stephan UW, Baumlein H (1999) Isolation, characterization and cDNA cloning of nicotianamine synthase from barley: a key enzyme for iron homeostasis in plants. Eur J Biochem 265 231–239 [DOI] [PubMed] [Google Scholar]
- Higuchi K, Suzuki K, Nakanishi H, Yamaguchi H, Nishizawa NK, Mori S (1999) Cloning of nicotianamine synthase genes, novel genes involved in the biosynthesis of phytosiderophores. Plant Physiol 119 471–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi K, Watanabe S, Takahashi M, Kawasaki S, Nakanishi H, Nishizawa NK, Mori S (2001) Nicotianamine synthase gene expression differs in barley and rice under Fe-deficient conditions. Plant J 25 159–167 [DOI] [PubMed] [Google Scholar]
- Inoue H, Higuchi K, Takahashi M, Nakanishi H, Mori S, Nishizawa NK (2003) Three rice nicotianamine synthase genes, OsNAS1, OsNAS2, and OsNAS3 are expressed in cells involved in long-distance transport of iron and differentially regulated by iron. Plant J 36 366–381 [DOI] [PubMed] [Google Scholar]
- Jakoby M, Wang HY, Reidt W, Weisshaar B, Bauer P (2004) FRU (BHLH029) is required for induction of iron mobilization genes in Arabidopsis thaliana. FEBS Lett 577 528–534 [DOI] [PubMed] [Google Scholar]
- Kim S, Takahashi M, Higuchi K, Tsunoda K, Nakanishi H, Yoshimura E, Mori S, Nishizawa NK (2005) Increased nicotianamine biosynthesis confers enhanced tolerance of high levels of metals, in particular nickel, to plants. Plant Cell Physiol 46 1809–1818 [DOI] [PubMed] [Google Scholar]
- Kruger C, Berkowitz O, Stephan UW, Hell R (2002) A metal-binding member of the late embryogenesis abundant protein family transports iron in the phloem of Ricinus communis L. J Biol Chem 277 25062–25069 [DOI] [PubMed] [Google Scholar]
- Le Jean M, Schikora A, Mari S, Briat JF, Curie C (2005) A loss-of-function mutation in AtYSL1 reveals its role in iron and nicotianamine seed loading. Plant J 44 769–782 [DOI] [PubMed] [Google Scholar]
- Li L, Cheng X, Ling HQ (2004) Isolation and characterization of Fe(III)-chelate reductase gene LeFRO1 in tomato. Plant Mol Biol 54 125–136 [DOI] [PubMed] [Google Scholar]
- Ling HQ, Bauer P, Bereczky Z, Keller B, Ganal M (2002) The tomato fer gene encoding a bHLH protein controls iron-uptake responses in roots. Proc Natl Acad Sci USA 99 13938–13943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling HQ, Koch G, Baumlein H, Ganal MW (1999) Map-based cloning of chloronerva, a gene involved in iron uptake of higher plants encoding nicotianamine synthase. Proc Natl Acad Sci USA 96 7098–7103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mari S, Gendre D, Pianelli K, Ouerdane L, Lobinski R, Briat JF, Lebrun M, Czernic P (2006) Root-to-shoot long distance circulation of nicotianamine and nicotianamine-nickel chelates in the metal hyperaccumulator Thlaspi caerulescens. J Exp Bot 57 4111–4122 [DOI] [PubMed] [Google Scholar]
- Mizuno D, Higuchi K, Sakamoto T, Nakanishi H, Mori S, Nishizawa NK (2003) Three nicotianamine synthase genes isolated from maize are differentially regulated by iron nutritional status. Plant Physiol 132 1989–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S (1999) Iron acquisition by plants. Curr Opin Plant Biol 2 250–253 [DOI] [PubMed] [Google Scholar]
- Mukherjee I, Campbell NH, Ash JS, Connolly EL (2006) Expression profiling of the Arabidopsis ferric chelate reductase (FRO) gene family reveals differential regulation by iron and copper. Planta 223 1178–1190 [DOI] [PubMed] [Google Scholar]
- Neumann G, Haake C, Römheld V (1999) Improved HPLC method for determination of phytosiderophores in root washings and tissue extracts. J Plant Nutr 22 1389–1402 [Google Scholar]
- Petit JM, Briat JF, Lobreaux S (2001) Structure and differential expression of the four members of the Arabidopsis thaliana ferritin gene family. Biochem J 359 575–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pianelli K, Mari S, Marquès L, Lebrun M, Czernic P (2005) Nicotianamine over-accumulation confers resistance to nickel in Arabidopsis thaliana. Transgenic Res 14 739–748 [DOI] [PubMed] [Google Scholar]
- Pich A, Scholz G (1996) Translocation of copper and other micronutrients in tomato plants (Lycopersicon esculentum Mill.): nicotianamine-stimulated copper transport in the xylem. J Exp Bot 47 41–47 [Google Scholar]
- Robinson NJ, Procter CM, Connolly EL, Guerinot ML (1999) A ferric-chelate reductase for iron uptake from soils. Nature 397 694–697 [DOI] [PubMed] [Google Scholar]
- Römheld V, Marschner H (1986. a) Different strategies in higher plants in mobilization and uptake of iron. J Plant Nutr 9 695–713 [Google Scholar]
- Römheld V, Marschner H (1986. b) Evidence for a specific uptake system for iron phytosiderophores in roots of grasses. Plant Physiol 80 175–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sautter C, Poletti S, Zhang P, Gruissem W (2006) Biofortification of essential nutritional compounds and trace elements in rice and cassava. Proc Nutr Soc 65 153–159 [DOI] [PubMed] [Google Scholar]
- Schaaf G, Ludewig U, Erenoglu BE, Mori S, Kitahara T, von Wiren N (2004) ZmYS1 functions as a proton-coupled symporter for phytosiderophore- and nicotianamine-chelated metals. J Biol Chem 279 9091–9096 [DOI] [PubMed] [Google Scholar]
- Schaaf G, Schikora A, Haberle J, Vert G, Ludewig U, Briat JF, Curie C, von Wiren N (2005) A putative function for the Arabidopsis Fe-phytosiderophore transporter homolog AtYSL2 in Fe and Zn homeostasis. Plant Cell Physiol 46 762–774 [DOI] [PubMed] [Google Scholar]
- Schmiedeberg L, Krüger C, Stephan UW, Bäumlein H, Hell R (2003) Synthesis and proof-of-function of a [14C]-labelled form of the plant iron chelator nicotianamine using recombinant nicotianamine synthase from barley. Physiol Plant 118 430–438 [Google Scholar]
- Scholz G, Becker R, Pich A, Stephan UW (1992) Nicotianamine: a common constituent of strategy-I and strategy-II of iron acquisition by plants. A review. J Plant Nutr 15 1647–1665 [Google Scholar]
- Shojima S, Nishizawa NK, Mori S (1989) Establishment of a cell free system for the biosynthesis of nicotianamine. Plant Cell Physiol 30 673–677 [Google Scholar]
- Stephan UW (2002) Intra- and intercellular iron trafficking and subcellular compartmentation within roots. Plant Soil 241 19–25 [Google Scholar]
- Suzuki K, Higuchi K, Nakanishi H, Nishizawa NK, Mori S (1999) Cloning of nicotianamine synthase genes from Arabidopsis thaliana. Soil Sci Plant Nutr 45 993–1002 [Google Scholar]
- Takahashi M, Nakanishi H, Kawasaki S, Nishizawa NK, Mori S (2001) Enhanced tolerance of rice to low iron availability in alkaline soils using barley nicotianamine aminotransferase genes. Nat Biotechnol 19 466–469 [DOI] [PubMed] [Google Scholar]
- Takahashi M, Terada Y, Nakai I, Nakanishi H, Yoshimura E, Mori S, Nishizawa NK (2003) Role of nicotianamine in the intracellular delivery of metals and plant reproductive development. Plant Cell 15 1263–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uauy C, Distelfeld A, Fahima T, Blechl A, Dubcovsky J (2006) A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314 1298–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dedaldechamp F, Gaymard F, Guerinot ML, Briat JF, Curie C (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wiren N, Klair S, Bansal S, Briat JF, Khodr H, Shioiri T, Leigh RA, Hider RC (1999) Nicotianamine chelates both Fe-III and Fe-II: implications for metal transport in plants. Plant Physiol 119 1107–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HY, Klatte M, Jakoby M, Baumlein H, Weisshaar B, Bauer P (2007) Iron deficiency-mediated stress regulation of four subgroup Ib BHLH genes in Arabidopsis thaliana. Planta 226 897–908 [DOI] [PubMed] [Google Scholar]
- Waters BM, Chu HH, DiDonato RJ, Roberts LA, Eisley RB, Lahner B, Salt DE, Walker EL (2006) Mutations in Arabidopsis Yellow Stripe-Like1 and Yellow Stripe-Like3 reveal their roles in metal ion homeostasis and loading of metal ions in seeds. Plant Physiol 141 1446–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters BM, Grusak MA (2008) Whole-plant mineral partitioning throughout the life cycle in Arabidopsis thaliana ecotypes Columbia, Landsberg erecta, Cape Verde Islands, and the mutant line ysl1ysl3. New Phytol 177 389–405 [DOI] [PubMed] [Google Scholar]
- Weber M, Harada E, Vess C, Roepenack-Lahaye E, Clemens S (2004) Comparative microarray analysis of Arabidopsis thaliana and Arabidopsis halleri roots identifies nicotianamine synthase, a ZIP transporter and other genes as potential metal hyperaccumulating factors. Plant J 37 269–281 [DOI] [PubMed] [Google Scholar]
- Yuan YX, Zhang J, Wang DW, Ling HQ (2005) AtbHLH29 of Arabidopsis thaliana is a functional ortholog of tomato FER involved in controlling iron acquisition in strategy I plants. Cell Res 15 613–621 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.