Abstract
MYB transcription factors play central roles in plant responses to abiotic stresses. How stress affects development is poorly understood. Here, we show that OsMYB3R-2 functions in both stress and developmental processes in rice (Oryza sativa). Transgenic plants overexpressing OsMYB3R-2 exhibited enhanced cold tolerance. Cold treatment greatly induced the expression of OsMYB3R-2, which encodes an active transcription factor. We show that OsMYB3R-2 specifically bound to a mitosis-specific activator cis-element, (T/C)C(T/C)AACGG(T/C)(T/C)A, a conserved sequence that was found in promoters of cyclin genes such as OsCycB1;1 and OsKNOLLE2. In addition, overexpression of OsMYB3R-2 in rice led to higher transcript levels of several G2/M phase-specific genes, including OsCycB1;1, OsCycB2;1, OsCycB2;2, and OsCDC20.1, than those in OsMYB3R-2 antisense lines or wild-type plants in response to cold treatment. Flow cytometry analysis revealed an increased cell mitotic index in overexpressed transgenic lines of OsMYB3R-2 after cold treatment. Furthermore, resistance to cold stress in the transgenic plants overexpressing OsCycB1;1 was also enhanced. The level of cellular free proline was increased in the overexpressed rice lines of OsMYB3R-2 and OsCycB1;1 transgenic plants compared with wild-type plants under the cold treatment. These results suggest that OsMYB3R-2 targets OsCycB1;1 and regulates the progress of the cell cycle during chilling stress. OsCPT1, which may be involved in the dehydration-responsive element-binding factor 1A pathway, showed the same transcription pattern in response to cold as did OsCycB1;1 in transgenic rice. Therefore, a cold resistance mechanism in rice could be mediated by regulating the cell cycle, which is controlled by key genes including OsMYB3R-2.
Rice (Oryza sativa), a plant normally grown in tropical and temperate climate zones, is often threatened by cold stress and is especially sensitive to chilling stress at the seedling and reproductive stages (Mukhopadhyay et al., 2004). Staged cold can result in poor germination, stunted seedlings, yellowing or withering, and reduced tillering. Unpredictable cold snaps at the reproductive stage delay heading and result in pollen sterility, which was thought to be one of the key factors responsible for the reduction in grain yield of rice (Kaneda, 1974; Mackill, 1997; Andaya and Tai, 2006; Suzuki et al., 2008). Transgenic approaches can be used to improve rice cold tolerance, and screening for genes involved in cold tolerance is an important initial step for crop improvement strategy using engineering (Hsieh et al., 2002; Dubouzet et al., 2003; Choi et al., 2004, 2005; Ohnishi et al., 2005; Ito et al., 2006).
Transcription factors, including the Myb family, which has been widely studied in both animals and plants, play essential roles in regulating gene expression in response to environmental and developmental changes. According to the number of tandem repeats of the SANT (for SWI3, ADA2, N-CoR, and TFIIIB) DNA-binding domains (Rosinski and Atchley, 1998; Jin and Martin, 1999; Stracke et al., 2001), MYB proteins can be divided into three subfamilies: MYB-like proteins (MYB1R factors), R2R3-type MYB factors, and R1R2R3 MYB (MYB3R) factors, with one, two, and three repeats, respectively (Rosinski and Atchley, 1998).
The Arabidopsis (Arabidopsis thaliana) MYB family consists of 198 genes and is one of the largest families among all transcription factor families (Kranz et al., 2000; Yanhui et al., 2006; Pasquali et al., 2008). In Arabidopsis, large-scale insertional mutagenesis (Meissner et al., 1999; Stracke et al., 2001) and expression profiling analysis (Kranz et al., 2000; Yanhui et al., 2006) have been used to analyze comprehensive functions and to explore the roles of R2R3 MYB proteins in cell cycle control, secondary metabolism, cellular morphogenesis, meristem formation, hormonal signaling, and stress responses (Salomoni et al., 1997; Meissner et al., 1999). Several R2R3 MYB genes involved in the responses to environmental stimuli such as drought, salt, and cold have been studied (Yanhui et al., 2006). AtMYB2 and AtMYC2 are transcriptional activators of RESPONSIVE TO DEHYDRATION22 (RD22). Expression of RD22 is induced by drought and abscisic acid (ABA; Urao et al., 1993; Ito et al., 1997). AtMYB102 is a component of the regulatory complex that directs signaling pathways for responses to wounding, osmotic stress, and ABA (Denekamp and Smeekens, 2003). AtMYB60, a guard cell-specific transcription factor, regulates stomatal movement in response to drought stress (Cominelli et al., 2005). HOS10, encoding an R2R3-type MYB transcription factor, is essential for acclimation to cold stress and may affect tolerance against dehydration by controlling ABA biosynthesis (Zhu et al., 2005). MYB15 controls the expression of C-repeat-binding factors (CBFs) and other genes responding to cold stress (Agarwal et al., 2006). Overexpression of AtMYB44 can enhance tolerance to drought and salt stresses by reducing the expression of genes encoding PP2Cs, including ABI1, ABI2, AtPP2CA, HAB1, and HAB2, which negatively regulate ABA signaling (Jung et al., 2008).
In rice, numerous transcription factors have been found to play important roles in response to cold stress. Overexpression of zinc finger genes such as OsISAP8, OsCOIN, and OsISAP1 confers cold stress tolerance at the seedling stage (Mukhopadhyay et al., 2004; Liu et al., 2007; Kanneganti and Gupta, 2008). Overexpression of OsbHLH1, OsDREB1/CBF, ROs-bZIP, SNAC2, and OsNAC6 also enhanced transgenic seedling resistance to chilling stress (Wang et al., 2003; Ohnishi et al., 2005; Ito et al., 2006; Cheng et al., 2007; Nakashima et al., 2007; Hu et al., 2008). Overexpression of OsMYB4 significantly increased tolerance to freezing stress in transgenic Arabidopsis (Vannini et al., 2004; Pasquali et al., 2008).
Signaling components and metabolic regulators have also been shown to be involved in stress responses. OsTPP1, a gene encoding a trehalose-6-P phosphatase, confers cold stress tolerance in rice and activated stress-responsive genes (Pramanik and Imai, 2005; Shima et al., 2007; Ge et al., 2008). OsLti6 genes (OsLti6a and OsLti6b) encoding hydrophobic proteins homologous to Arabidopsis RCI2 enhanced tolerance to chilling stress in rice seedlings (Morsy et al., 2005). Stress responses in eukaryotic organisms can be mediated by the mitogen-activated protein kinase (MAPK) cascades. Overexpression of OsMAPK5 conferred tolerance to drought, salt, and cold stresses in rice seedlings (Xiong and Yang, 2003). Stress-responsive CIPK genes encoding calcineurin B-like protein-interacting protein kinases such as OsCIPK03 and OsCIPK12 also play important roles in improving the tolerance to chilling stress in rice (Xiang et al., 2007). OsCDPK7 and OsCDPK13 encoding Ca2+-dependent protein kinases are positive regulators that enhance cold and salt stress tolerance (Saijo et al., 2000, 2001; Wan et al., 2007; Wang et al., 2008).
The cell cycle is an intrinsic part of plant growth and development. However, less is known about how the cell cycle may be affected upon cold stress and how this may affect the plant's survival. Evidence suggests that cell cycle activities are involved in the stress response mediated by transcription factors (Morano et al., 1999; Luft et al., 2001; Nakai and Ishikawa, 2001; Santilli et al., 2005). MYB proteins, such as NtmybA1, NtmybA2, and NtmybB, may bind specifically to the core motif AACGG of DNA sequences, the M phase-specific activator (MSA) cis-element, in tobacco (Nicotiana tabacum) in vitro (Ito et al., 2001; Araki et al., 2004; Suzuki et al., 2006; Haga et al., 2007). NtmybA1 and NtmybA2 mediate the transcription of a G2/M phase-specific gene in tobacco cells, whereas NtmybB functions as a transcription repressor. MYB3R1 and MYB3R4, homologs of NtmybA1 and NtmybA2, respectively, positively regulate cytokinesis mainly through transcriptional activation of the KNOLLE gene in Arabidopsis (Haga et al., 2007).
Our previous studies have indicated that OsMYB3R-2 transgenic Arabidopsis plants are more resistant to freezing, drought, and salt stresses (Dai et al., 2007), leading us to speculate whether the regulation of the cell cycle is involved in the OsMYB3R-2-modulated stress response. Here, we show that OsMYB3R-2 is involved in regulating the responses to cold stress in rice. We demonstrate that OsMYB3R-2 functions as a MYB3R transcription factor targeting to OsCycB1;1, which is involved in the G2/M phase transition at low temperature. The transcript level of OsCPT1, a putative member of the dehydration-responsive element-binding factor 1 (DREB1)/CBF pathway, is also enhanced by OsMYB3R-2, accompanied by an increased Pro level. Our data indicate that in rice, OsMYB3R-2 may play an important role in the cold stress signaling pathway modulated by the cell cycle and a putative DREB/CBF pathway.
RESULTS
Molecular Characteristics and Phenotypes of OsMYB3R-2 Transgenic Rice
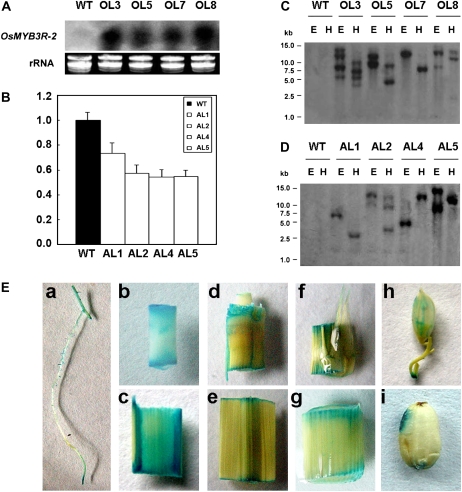
Transgenic lines of OsMYB3R-2 in rice were confirmed by hygromycin selection and GUS staining. The results of northern blot and real-time PCR showed that expression of OsMYB3R-2 was increased in the four independent overexpressing lines but decreased in the four antisense lines (Fig. 1, A and B). Southern-blot analysis with a specific GUS probe showed diverse expression patterns in the four overexpressed lines as well as in the four antisense lines (Fig. 1, C and D).
Figure 1.
Identification of OsMYB3R-2 transgenic rice and its expression pattern. A, Northern-blot assay of rice transgenic plants. Total RNA isolated from wild-type (WT) or transformed plants underwent hybridization with a [α-32P]dCTP-labeled probe of OsMYB3R-2 cDNA as described in “Materials and Methods.” B, Real-time RT-PCR of the expression of OsMYB3R-2 in antisense lines. C and D, Southern-blot assay of transformed rice plants. Genomic DNA isolated from wild-type or transformed plants was digested with EcoRI (E) or HindIII (H). The blot was hybridized with the open reading frame of the GUS gene labeled with [α-32P]dCTP. OL3, OL5, OL7, and OL8 and AL1, AL2, AL4, and AL5 represent overexpression (O) and antisense (A) lines of OsMYB3R-2 transgenic rice. E, Expression pattern of OsMYB3R-2 in vivo. GUS staining shows expression pattern of OsMYB3R-2 in vivo in various tissues from the T1 generation of OsMYB3R-2 promoter∷GUS transgenic rice. a, Root; b, young internode; c, mature internode; d, node; e, mature leaf; f, lamina joint; g, leaf sheath; h, flower; i, immature seed.
To examine the expression patterns of OsMYB3R-2 in vivo, transgenic rice lines were generated with a GUS expression construct driven by a putative OsMYB3R-2 promoter of 1,285 bp in length. GUS staining assay of T1 rice plants showed strong signals in nearly all tissues examined, including roots, internodes, nodes, leaf blades, lamina joints, sheaths, glumes of flower organs, and young embryos of immature seeds (Fig. 1E), suggesting that OsMYB3R-2 is a constitutively expressed gene.
Transgenic rice seeds of either the OsMYB3R-2 overexpressors or the antisense lines did not differ from wild-type seeds in germination (Fig. 2). However the overexpressing plants showed growth retardation in comparison with wild-type plants under normal conditions (Fig. 2, B and C). The length of the root cell resulted in shorter roots in the overexpression transgenic lines (Supplemental Fig. S3). When wild-type plants reached the tetraphyllous stage, more than 80% of the OsMYB3R-2 overexpression plants were still at the trefoil stage (Fig. 2B). Growth retardation was observed in transgenic plants up to the heading stage.
Figure 2.
Phenotype analysis of the T2 generation of OsMYB3R-2 transgenic rice. A, The germination and growth of OsMYB3R-2 transgenic rice at 30°C. Shelled seeds of rice at 0 d; germinating seeds at 1, 2, and 3 d; young seedlings transferred to light (12 h of light/12 h of dark, 30°C/26°C) at 4 and 6 d. B, Two-week-old seedlings (T2 generation) of OsMYB3R-2 transgenic rice. C, Transgenic overexpressing seedlings of OsMYB3R-2 at the tillering stage 35 d after germination. AL1, OsMYB3R-2-antisense rice; OL5, OsMYB3R-2-overexpressing rice; WT, wild type. Bars = 10 cm.
Overexpression of OsMYB3R-2 Enhanced Tolerance to Chilling Stress in Rice
To test the possible effect of OsMYB3R-2 expression on tolerance to chilling, the T2 transgenic and wild-type seedlings at the trefoil stage were exposed to reduced temperature (2°C) for various durations, followed by incubation at a normal growth condition in a greenhouse for 2 weeks. Fewer than 20% of the wild-type seedlings survived a cold treatment of 72 h, but none of them was able to resume growth when transferred to normal growth conditions. However, more than 50% of the OsMYB3R-2-overexpressing seedlings could survive and grow normally (Fig. 3, B and C). The survival ratio of antisense seedlings was less than that of the wild type. A time course of treatment showed drastic differences as the process was extended (Fig. 3D). The wild-type and transgenic seedlings showed no growth differences after chilling treatment for up to 48 h. In contrast, when the time of treatment was extended up to 60 h, more than 80% of the OsMYB3R-2-overexpressing plants grew normally, as compared with 55% of the wild-type seedlings and 45% of the antisense seedlings. Finally, after 84 h, neither wild-type nor antisense plants survived; in contrast, 20% of the overexpression lines were still healthy. Therefore, OsMYB3R-2 plays an important role in regulating tolerance against chilling stress in rice.
Figure 3.
Tolerance response of the OsMYB3R-2 transgenic lines to cold stress. A, Wild-type and OsMYB3R-2 transgenic 2-week-old rice seedlings at the same stage before the 2°C treatment. B, Seedlings were grown in the greenhouse for 2 weeks after 2°C treatment for 72 h. C, Survival rate of seedlings grown for 2 weeks in the greenhouse after 2°C treatment for 72 h. D, Time course for cold treatment in survival rate of seedlings grown for 2 weeks in the greenhouse. In C and D, the error bars show se and are from three independent replications in the same experiment. The phenotype was confirmed by further experiments that were repeated more than four times. AL1 and AL4, OsMYB3R-2-antisense lines; OL3, OL5, and OL7, OsMYB3R-2-overexpressing lines; WT, wild type.
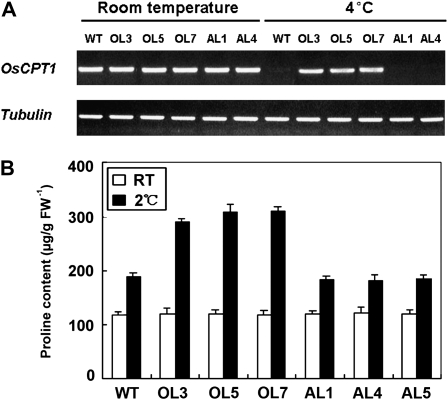
To investigate the functions of OsMYB3R-2 in DREB/CBF stress pathways, we tested the expression patterns of more than 10 genes by reverse transcription (RT)-PCR in wild-type and transgenic rice plants. One of the genes, OsCPT1, was activated by OsMYB3R-2 under cold stress, a deduced target gene of the DREB pathway with the DRE/CRT cis-elements (Fig. 4A). A DRE/CRT cis-element, CCGACCT, appeared in the upstream sequence (602–596 bp) of the OsCPT1 promoter. The transcription levels of rice DREB genes, including OsDREB1A, OsDREB2A, and OsCBF, did not show any changes in OsMYB3R-2-overexpressing transgenic lines under the cold conditions compared with the wild type. For the expression patterns of other cold-regulated (COR) genes such as OsCORTM1 and OsMAT1, which are the rice homologs of target genes of Arabidopsis DREBs (Dubouzet et al., 2003; Chen et al., 2008; Supplemental Fig. S1; Supplemental Table S1), there were no differences between the transgenic lines and the wild type. These data suggest that OsMYB3R-2 may regulate the plant response to cold stress through the deduced OsCPT1-involved DREB/CBF pathway in rice.
Figure 4.
Gene expression patterns and free Pro levels of transgenic rice plants in response to cold. A, Expression pattern of OsCPT1 in wild-type and OsMYB3R-2 transgenic rice at room temperature (25°C) or low temperature (4°C). Tubulin was used as an internal control. The expression pattern of OsCPT1 to cold stress was confirmed with two independent experiments. B, Cellular free Pro level. Data represent means and se of experiments performed in triplicate. Error bars show se and are from three independent replications of the same experiment. The Pro content determination was confirmed by experiments that were repeated twice. AL1, AL4, and AL5, OsMYB3R-2-antisense lines; FW, fresh weight of materials; OL3, OL5, and OL7, OsMYB3R-2-overexpressing lines; RT, room temperature (25°C); WT, wild type.
Under normal growth conditions (25°C), the levels of cellular free Pro did not differ between wild-type and transgenic rice in the range of 112 to 118 μg fresh weight of material (Fig. 4B). In contrast, after cold treatment (2°C), the levels of free Pro in OsMYB3R-2-overexpressing transgenic rice increased substantially, with more than 300 μg g−1 fresh weight compared with 188 μg g−1 fresh weight in the wild-type plants. These results were similar to the alterations observed in other transgenic plants overexpressing the resistant genes, such as OsDREB1, OsCOIN, OsCIPK03, and OsCIPK12, which showed resistance to cold stress in rice (Ito et al., 2006; Liu et al., 2007; Xiang et al., 2007). Thus, cellular free Pro level is involved in enhanced resistance to cold regulated by OsMYB3R-2 via a putative DREB/CBF-CPT pathway in rice.
OsMYB3R-2 Protein Showed Transcription Activation
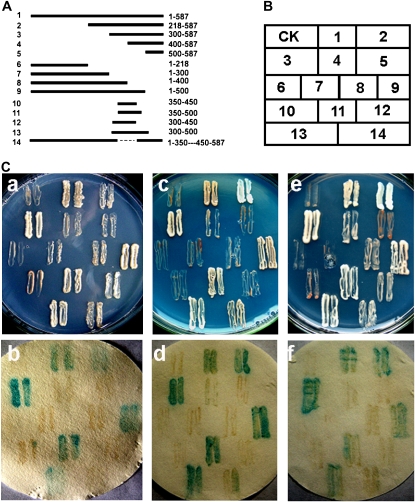
A yeast GAL4 system was used to investigate the transcription activation of OsMYB3R-2 (Fig. 5A). OsMYB3R-2 mutants deleted in various domains were tested. The N and C termini of OsMYB3R-2 were truncated and the products were termed numbers 2 to 14, with the full-length protein termed number 1. Figure 5 shows a yeast growth analysis on screened medium with SD/-Trp/-Ade, SD/-Trp/-His, or SD/-Trp/-His/-Ade (see “Materials and Methods”) as well as in the galactosidase assay. Stronger blue signals corresponding to good growth of yeast on both media appeared in numbers 1, 2, 3, 9, 11, and 13 compared with the control empty vector and the remaining constructs. A common region among constructs was the region of 350 to 500 amino acids at the C terminus. These results suggest that the OsMYB3R-2 protein has transcriptional activation activity, and the core region with the activity was from 350 to 450 at the N terminus.
Figure 5.
Transcription activation analysis of OsMYB3R-2 protein. A, Different pGBKT7-OsMYB3R-2 vector constructs. The truncated cDNA fragments of OsMYB3R-2 were sequenced and inserted into the NdeI-PstI sites, with ATG added at the end of the NdeI site in every forward primer. For numbers at left, 1 represents full-length OsMYB3R-2 protein and 2 to 14 represent different truncated OsMYB3R-2 protein fragments; numbers at right represent the positions of different truncated OsMYB3R-2 protein fragments. The broken line represents the deleted fragment (amino acids 351 to 449) of OsMYB3R-2. The transcription activation of OsMYB3R-2 was confirmed twice. B, The corresponding positions of transformed yeast thalli daubed on the plates. CK, pGBKT7 vector used as a control. C, a, The transformed yeast thalli grew on the SD/-His/-Trp plates with solid SD medium. C, b, X-Gal activation detection of transformed yeast thalli on the SD/-His/-Trp plates with solid SD medium shown in C, a. C, c, The transformed yeast thalli grew on the SD/-Ade/-Trp plates with solid SD medium. C, d, X-Gal activation detection of transformed yeast thalli on the SD/-Ade/-Trp plates with solid SD medium shown in C, c. C, e, The transformed yeast thalli grew on the SD/-His/-Ade/-Trp plates with solid SD medium. C, f, X-Gal activation detection of transformed yeast thalli on the SD/-His/-Ade/-Trp plates with solid SD medium shown in C, e.
OsMYB3R-2 Targeted the Type B Cyclin Gene OsCycB1;1
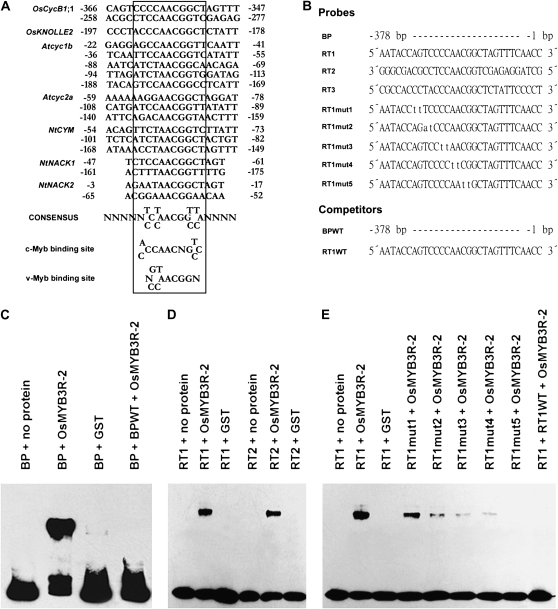
Bioinformatic analysis showed MSA-like sequences in the promoters of cyclin genes in rice (La et al., 2006). Two MSA-like sequences of OsCycB1;1 appeared between −200 to −400 bp upstream of the transcription start site. A fully conserved central core pentamer, AACGG, was found in the MSA-like elements. There is a 3-bp less conserved sequence at each side of the core motif. The MSA consensus sequence (T/C)C(T/C)AACGG(T/C)(T/C)A is shown in Figure 6A (Ferreira et al., 1994; Day et al., 1996; Ito et al., 1997, 1998). It matches the consensus sequences of c-Myb and v-Myb binding sites (Howe and Watson, 1991; Grotewold et al., 1994; Ito et al., 1998), which suggests that the Myb transcription factors may bind the MSA motif.
Figure 6.
DNA binding affinity of OsMYB3R-2 protein. A, The alignment of MSA-like sequences shown in the promoters of type B cyclin genes: rice OsCycB1;1 and OsKNOLLE2, Arabidopsis cyc1bAt (Day et al., 1996) and cyc2aAt (Ferreira et al., 1994), and tobacco NtCYM (Ito et al., 1997), NACK1, and NACK2 (Ito et al., 1998) encoding kinesin-like proteins. The boxed 11-bp sequences share high homology with each other. The nucleotide positions are numbered from the transcription start sites. The motifs of binding sites of c-Myb (Howe and Watson, 1991) and v-Myb (Grotewold et al., 1994) are also shown. B, Probes and competitors used in EMSA. BP, A 378-bp fragment of OsCycB1;1 promoter upstream of the transcription start site ATG; RT1, an MSA cis-acting element of BP; RT2, an MSA trans-acting element of BP; RT3, an MSA cis-acting element of OsKNOLLE2 promoter upstream of the transcription start site ATG; RT1mut, mutant of the RT1 motif; BPWT and RT1WT, competitors of biotin-labeled probes of BP and RT1, respectively. C to E, EMSA assays of OsMYB3R-2 protein. Abbreviations are the same as in B. Binding reaction mixtures were incubated with the probes and mock-translated product (mock = probe + no protein) or in vitro-synthesized OsMYB3R-2, with GST as a control, in the presence or absence of a 200-fold molar excess of unlabeled oligonucleotide competitors. DNA binding affinity of OsMYB3R-2 was confirmed experimentally twice. AL1 and AL4, OsMYB3R-2-antisense lines; OL5 and OL7, OsMYB3R-2-overexpressing lines.
To test whether OsMYB3R-2 interacts with the MSA motif in the promoter of OsCycB1;1, electrophoretic mobility shift assay (EMSA) was carried out. EMSA showed that OsMYB3R-2 can specifically bind the OsCycB1;1 promoter of a 378-bp fragment in rice (Fig. 6, B and C). EMSA analysis of two MSA elements (RT1 and RT2) from the OsCycB1;1 promoter (Fig. 6, B and D) showed that the mobility of OsMYB3R-2 specifically shifted with the MSA elements from type B cyclin genes on the membrane map (Fig. 6E).
Assay of the mutated RT1 sequence of the OsCycB1;1 promoter showed that any mutation could weaken the DNA-binding ability of OsMYB3R-2 protein (Fig. 6E), whereas the DNA-binding ability was abolished by base substitution of RT1mut5. Therefore, we concluded that the CCCAACGG sequence in the OsCycB1;1 promoter was recognized by OsMYB3R-2 protein.
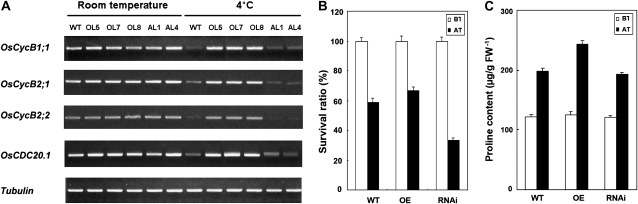
We further examined the expression patterns of genes related to the cell cycle (Supplemental Table S2). Compared with the expression levels of types A and D cyclins, the expression levels of the type B cyclin genes OsCycB1;1, OsCycB2;1, OsCycB2;2, and OsCDC20.1 were suppressed by cold treatment in both the wild-type and antisense plants. In the OsMYB3R-2-overexpressed lines under cold treatment, expression patterns of those type B cyclin genes were the same as those at room temperature (25°C; Fig. 7A).
Figure 7.
Expression patterns of cyclin genes in the OsMYB3R-2 transgenic rice, cold response and survival ratio, and cellular free Pro in the OsCycB1;1 transgenic rice lines. A, Expression patterns of cyclin-responsive genes in wild-type (WT) and OsMYB3R-2 transgenic rice under room temperature (25°C) or cold treatment (4°C) for 24 h. The method of obtaining seedlings at the same stage is described in “Materials and Methods.” Tubulin was used as an internal control. AL1 and AL4, OsMYB3R-2-antisense lines; OL5, OL7, and OL8, OsMYB3R-2-overexpressing lines. The experiments on the response of cyclin genes to cold were repeated at least twice. B, The response of the OsCycB1;1 transgenic rice lines to cold stress. AT, Grown for 1 week after the cold treatment (3°C) for 72 h; BT, before the cold treatment (3°C), which is a control without cold treatment; OE, OsCycB1;1-overexpressing lines; RNAi, OsCycB1;1-RNAi lines. C, The determination of cellular free Pro in OsCycB1;1 transgenic rice lines. AT, After the cold treatment (2°C) for 24 h; BT, before the cold treatment (2°C); FW, fresh weight of materials. Error bars in B and C show se and are from three independent replications. Data represent means and se of experiments performed in triplicate.
To test whether the cold tolerance phenotype could be reproduced by overexpressing OsCycBs, transgenic rice overexpressing OsCycB1;1 as well as RNA interference (RNAi) lines (Supplemental Fig. S2) were tested for cold stress. The results showed that less than 58% of wild-type seedlings could survive after the treatment for 72 h, whereas more than 67% of overexpressed OsCycB1;1 seedlings could survive and grow normally (Fig. 7B). In contrast, less than 33% of OsCycB1;1-RNAi seedlings could resume growth under normal growth conditions. Our data showed that the overexpressing lines of OsCycB1;1 enhanced the tolerance to chilling stress compared with the wild type. This suggests that OsCycB1 is likely to be one of the downstream genes regulated by OsMYB3R-2 under chilling stress.
To investigate whether there is any relationship between Pro level and resistance to cold stress in OsCycB1;1-overexpressing transgenic rice plants, the level of cellular free Pro was monitored. The results showed that under normal growth conditions (25°C), the levels of cellular free Pro were similar in both wild-type and OsCycB1;1 transgenic rice at a range of 120 to 124 μg g−1 fresh weight of material (Fig. 7C). In contrast, after cold treatment (2°C), the level of free Pro in OsCycB1;1-overexpressing transgenic rice increased substantially, with 243 μg g−1 fresh weight compared with 197 μg g−1 fresh weight in the wild type and 185 μg g−1 fresh weight in OsCycB1;1-RNAi plants. These results of the changed pattern for cellular free Pro were similar to those in OsMYB3R-2 transgenic plants. Taken together, the data suggested that OsCycB1;1 was directly regulated by OsMYB3R-2, which was involved in the tolerance to cold in rice.
Cell Cycle Progression in Transgenic Rice Lines
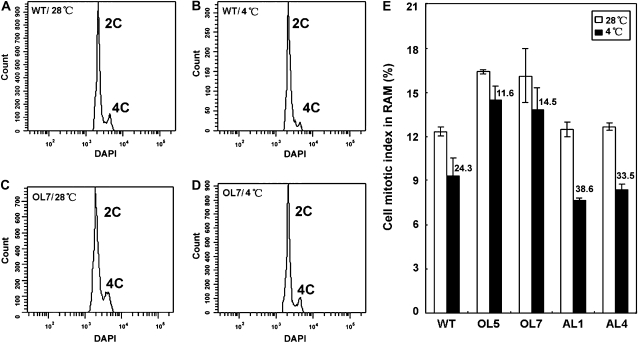
Based on the results of the expression levels of cyclins, we monitored the mitotic index of OsMYB3R-2-overexpressed lines under cold conditions. Mitotic index is defined as the ratio between the number of cells in mitosis and the total number of cells, which is used as a measure for the proliferation status of a cell population. Flow cytometry revealed that the DNA content of the OsMYB3R-2-overexpressed lines increased at 4°C compared with the wild type under normal (28°C) and cold (4°C) conditions (Fig. 8, A–D). Thus, the overexpressing lines possessed more cells in the G2/M phase, especially under the cold conditions. Under the normal conditions, the overexpressing lines showed a higher mitotic index than wild-type and antisense lines. Under cold conditions, in contrast, the mitotic index in the overexpressing lines was markedly higher than that of the wild type, and the index of the antisense lines was notably lower than that of the wild type under cold stress (Fig. 8E). The decreased percentage of the mitotic index under cold stress compared with normal conditions (28°C) was 24.3%, 11.6% to 14.5%, and 33.5% to 38.6% in the wild type, the overexpressing lines, and the antisense lines, respectively. The changes in the mitotic index correlated to the expression pattern of OsMYB3R-2. Therefore, we conclude that OsMYB3R-2-overexpressing lines possess more cells at the G2/M cell cycle phase, which promoted an increased mitosis.
Figure 8.
Cell cycle progression response to cold in flow cytometry assay in the OsMYB3R-2 transgenic lines. A, The wild type at 28°C. B, The wild type at 4°C. C, Overexpressing line 7 (OL7) of OsMYB3R-2 transgenic rice at 28°C. D, Overexpressing line 7 at 4°C. Seedlings 5 d after germination were treated with low temperature (4°C) or room temperature (control, 28°C) for 24 h. Cell nuclei (10,000) taken from the root apical meristem were stained with DAPI (1 μg mL−1) and analyzed by flow cytometry. 2C and 4C represent the DAPI signals that correspond to nuclei with different DNA contents. E, Cell mitotic index in root apical meristem (RAM) in rice. Numbers above the black histograms represent the percentage of decrease in the mitotic index at 4°C. The error bars show se and are from three independent replications of the same experiment. Flow cytometry determination was repeated twice. AL1 and AL4, OsMYB3R-2-antisense lines; OL5 and OL7, OsMYB3R-2-overexpressing lines; WT, wild type.
DISCUSSION
OsMYB3R-2 Is a Positive Regulator for a Subset of G2/M Phase-Specific Genes in Rice
MYB3R genes constitute a small gene family of transcription factors in plants that play regulatory roles in the cell cycle and in response to environmental stresses (Ito et al., 2001; Araki et al., 2004; Dai et al., 2007; Haga et al., 2007). MYB3R genes exert their functions through binding to MSA elements, which mediate the G2/M phase of the cell cycle (Ito et al., 2001; Araki et al., 2004). In Arabidopsis, five R1R2R3-type Myb genes have been described (Braun and Grotewold, 1999; Kranz et al., 2000; Haga et al., 2007). MYB3R1 and MYB3R4 are two genes homologous to NtmybA and NtmybA2 (Haga et al., 2007), which positively regulate cytokinesis by activating the transcription of several G2/M phase-specific genes in Arabidopsis. The defects of multinucleate cells and cell wall stubs in the myb3r1 myb3r4 double mutant are caused by the selective reduction of transcript levels of several type B2 cyclin genes, including CYCB2, CDC20.1, and KNOLLE (Haga et al., 2007).
OsMYB3R-2 protein functions as an R1R2R3-type MYB transcription factor. It has three tandem SANT DNA-binding domains, is localized to the nucleus, and binds to specific cis-elements (Dai et al., 2007). OsMYB3R-2 can recognize the consensus sequence (T/C)C(T/C)AACGG(T/C)(T/C)A with the core motif CCAGG (Fig. 6). Although the promoters of OsCycB1;1 and OsKNOLLE2 contain the core motif, only OsCycB1;1 (a type B cyclin gene) can be activated to a high transcriptional level under cold conditions (Figs. 6 and 7). Therefore, our evidence supports the notion that OsMYB3R-2 functions as a positive regulator to modulate the G2/M phase of the cell cycle via OsCycB1;1 in rice.
OsMYB3R-2 Coordinates the Cell Cycle and a Deduced DREB/CBF Pathway to Increase Cold Tolerance in Rice
OsMYB3R-2 functions as a transcription factor with a specific DNA-binding characteristic (Fig. 6). The increased mitosis index in transgenic rice of overexpressing OsMYB3R-2 indicates that OsMYB3R-2 probably regulates the process of the cell cycle (Fig. 8), showing a similar function to that of its homologs such as NtmybA1 and NtmybA2 and MYB3R1 and MYB3R4 (Ito et al., 2001; Araki et al., 2004; Suzuki et al., 2006; Haga et al., 2007). Under cold conditions, we found that some genes of cyclin B type, including OsCycB1;1, one target gene of OsMYB3R-2, were activated to high levels of transcription in the OsMYB3R-2-overexpressing transgenic lines (Figs. 6 and 7). These results suggested that OsMYB3R-2 plays an essential role in maintaining a high progression of the cell cycle under cold stress.
In Arabidopsis, the transcript levels of DREB2A, COR15a, and RCI2A are induced in 35S∷OsMYB3R-2 plants (Dai et al., 2007). There is a correlation between the enhanced tolerance to environmental stress in 35S∷OsMYB3R-2 plants and the up-regulation of stress-responsive genes. Our data suggest that OsMYB3R-2 may play an important role in the complex gene network controlling the stress signaling pathways (Dai et al., 2007). OsDREB genes, encoding transcription activators, function in response to drought, high salt, and cold stress in rice (Dubouzet et al., 2003; Chen et al., 2008). The expression of DREB2A is enhanced in OsMYB3R-2-overexpressed plants (Dai et al., 2007). In contrast, in OsMYB3R-2 transgenic rice, the transcripts of rice DREB genes and their putative target genes did not show any changes compared with that in the wild type except for OsCPT1 (Supplemental Fig. S1; Supplemental Table S1). The expression of OsCPT1, the homolog of At2g02100, was activated by OsMYB3R-2 in overexpressed transgenic lines under cold stress (Fig. 4). In Arabidopsis, At2g02100, which encodes a putative protease inhibitor II, is a target gene of DREB1A/CBF3 with unknown function (Dubouzet et al., 2003; Chen et al., 2008). Furthermore, a DRE/CRT cis-element (CCGACCT) was involved upstream of the OsCPT1 promoter 602 to 596 bp from the transcription start site in rice, indicating that OsCPT1 might be involved in the DREB/CBF pathway. Therefore, overexpression of OsMYB3R-2 activates the expression of OsCPT1 in the cold response of rice plants, which is putatively targeted by DREB genes.
Two pieces of the core motif (CCAGG) of MSA and a consensus MYB-binding site (TAACTG; Urao et al., 1993) were found in the promoters of DREB2A and RCI2A in Arabidopsis. And a core motif (CCAGG) of MSA and a consensus MYB-binding site (TAACTG) also appeared in the promoter of COR15A. There is a report that MYB proteins can specially bind to specific DNA sequences, including their consensus MYB binding sites (Xue, 2005). Therefore, it is suggested that OsMYB3R-2 can bind to multiple elements and activate a diverse set of genes.
Tolerance to cold stress is controlled by complex mechanisms involving many changes, including membrane lipid composition, accumulation of compatible solutes, and expression of COR genes. A downstream change is the up-regulation of cellular Pro levels, with overexpression of genes showing resistance to cold stress (Thomashow, 1999; Korenjak et al., 2004). We found a remarkable increase in cellular free Pro levels with OsMYB3R-2 overexpression or OsCycB1;1 overexpression after cold treatment (Figs. 4B and 7C). This pattern is similar to that of other genes that enhance resistance to cold stress, such as OsDREB1, OsCOIN, OsCIPK03, and OsCIPK12 (Ito et al., 2006; Liu et al., 2007; Xiang et al., 2007). Therefore, the up-regulated level of cellular free Pro may be one of the common characteristics of genes conferring resistance to cold stress.
Our experimental observations of rice seedlings with either cold-sensitive traits or resistant traits showed no morphological differences in growth at the early stages under cold conditions. When plants were returned to normal conditions, 25°C to 28°C, the leaves rapidly withered in both kinds of plants. However, newly differentiated leaves were formed in OsMYB3R-2-overexpressing cold-resistant plants. Cold-resistant plants may have higher competence in maintaining cell division under cold stress than cold-sensitive plants, which is supported by our observations on the cell cycle (Fig. 8).
Taken together, we conclude that OsMYB3R-2 is a MYB3R transcription factor and OsCycB1;1 is one of its target genes regulated under cold conditions. The enhanced cold stress tolerance of 35S∷OsMYB3R-2 and 35S∷OsCycB1;1 rice plants reveals that OsMYB3R-2 can mediate cold stress signal transduction and regulate some stress-responsive genes involved in the cell cycle or a deduced DREB/CBF pathway. Although it is still not clear what the sensing mechanism of OsMYB3R-2 to chilling stress in rice is, the functions of OsMYB3R-2 in the cell cycle and cold tolerance will provide new insights into cold stress pathways. The information gained will be beneficial for directing molecular breeding strategies to generate rice varieties with enhanced tolerance to cold stress.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The plant material used was rice (Oryza sativa subsp. japonica ‘Zhonghua 10’). Rice seeds were germinated in small plastic boxes filled with a mixture of flower nutrimental soil and vermiculite (8:1) for at least 14 d. The rice seedlings at the trifoliate stage were transferred to grow in big plastic buckets at 30°C during the day and 20°C during the night in a greenhouse.
Plasmid Construction and Plant Transformation
Total RNA of rice seedlings was isolated by use of TRIzol Reagent (Invitrogen). The cDNA of rice was synthesized using AMV Reverse Transcriptase (Promega). Full-length OsMYB3R-2 was amplified by RT-PCR with Pyrobest DNA Polymerase (TaKaRa), ligated into pGEM-T Easy vector (Promega), and sequenced (Dai et al., 2007). The digestion product of OsMYB3R-2 from pT Easy-OsMYB3R-2 was directionally cloned into the KpnI-BamHI sites of a pUN1301 vector to create the pUN1301-OsMYB3R-2 construct, which carried a GUS marker, with the forward primer 5′-GGATCCATGGGGGCCATGGCGATGGTG-3′ and the reverse primer 5′-GGTACCGGTTACATCAAATTGGTTGT-3′. A pUN1301-antisense-OsMYB3R-2 construct was created with the forward primer 5′-GGTACCATGGGGGCCATGGCGATGGTG-3′ and the reverse primer 5′-GGATCCGGTTACATCCAAATTGGTTGT-3′. OsMYB3R-2 was driven by a ubiquitin promoter in the construct. The pUN1301-OsMYB3R-2 construct was electroporated into Agrobacterium tumefaciens EHA105. Rice embryonic calli were induced on scutella from germinated seeds and transfected with A. tumefaciens EHA105 containing the desired binary vector as described previously (Ge et al., 2004). OsMYB3R-2 transgenic plants were screened in half-strength Murashige and Skoog medium containing 75 mg L−1 hygromycin (Sigma). Transgenic plants of the T0 generation from calli with hygromycin-resistant plants were transplanted into soil and grown in a greenhouse.
Construction of 35S∷OsCycB1;1 (accession no. AY647458) vector and RNAi plasmid were done according to the methods of Jiang et al. (2007) and Wang et al. (2004). Gene transformation used the method described above.
Construction of OsMYB3R-2 Promoter∷GUS Vector and Transformation
The OsMYB3R-2 promoter of 1,285 bp was amplified by PCR from the rice genome and inserted upstream of GUS at the KpnI-BamHI sites of the pGUS1301 vector (Ge et al., 2004). The OsMYB3R-2 promoter-pGUS1301 was constructed with the primer pair 5′-GGGTACCCCAACTCGTATTGCTCCTCTT-3′ and 5′-CGGATCCAGGCACAAGCACATCCTCA-3′. The construct of the OsMYB3R-2 promoter-pGUS1301 was electroporated into A. tumefaciens EHA105 and transfected into rice embryonic calli as described previously (Ge et al., 2004). GUS staining was used to investigate the OsMYB3R-2 expression in the T1 generation of OsMYB3R-2 promoter∷GUS transgenic rice.
RT-PCR and Real-Time PCR
Total RNA was isolated from rice seedlings by use of TRIzol Reagent (Invitrogen). The cDNA of rice was synthesized using AMV Reverse Transcriptase (Promega) in a 25-μL reaction containing 2 μg of total RNA. RT reactions were carried out at 42°C for 60 min followed by chilling on ice for 5 min. An amount of 2 μL of 5-fold-diluted cDNA was used as a PCR template in a 20-μL RT reaction mixture. All PCR products were loaded onto 0.8% agarose gel to visualize the amplified cDNAs. RT-PCR was repeated three times. RT-PCR of OsCPT1 involved the forward primer 5′-CGGTGGCAGTAGGAAAGTAG-3′ and the reverse primer 5′-CATGAACAACAGACAAAGGAGA-3′ with 28 cycles. Tubulin with the forward primer 5′-TCAGATGCCCAGTGACAGGA-3′ and the reverse primer 5′-TTGGTGATCTCGGCAACAGA-3′ was used as a control for 25 cycles. Real-time PCR was used to investigate the level of OsMYB3R-2 in antisense lines with the primers 5′-AGGTCCACCAATCATTCTCC-3′ and 5′-GTAAATTCACAAAGTGCAGCG-3′, which were designed in the 3′ untranslated region to identify the knockdown efficiency of the endogenous OsMYB3R-2 gene. The methods for real-time PCR were described previously in detail (Dai et al., 2007).
Southern- and Northern-Blot Assays
Genomic DNA was isolated from 2-week-old rice seedlings and digested with EcoRI or HindIII. DNA of 20 μg was used for Southern-blot analysis as described previously (Ge et al., 2004). The fractioned DNA underwent electrophoresis on a 0.7% (w/v) agarose gel and was then blotted onto a nylon membrane (Amersham Pharmacia Biotech). The membrane was prehybridized at 65°C for 4 h and hybridized in the same solution containing [α-32P]dCTP-labeled GUS for 20 h at 65°C. The GUS of 680 bp was amplified by PCR with the forward primer 5′-CAACTGGACAAGGCACTAGC-3′ and the reverse primer 5′-AGCGTCGCAGAACATTACAT-3′. The membrane was washed with washing buffer (2× SSC plus 0.1% SDS) at 65°C for 20 min after hybridization and then washed twice with 1× SSC plus 0.1% SDS at 65°C for 15 min. The membrane was stored at −70°C for 3 to 7 d and then exposed to x-ray film (Eastman-Kodak).
Northern blotting was performed as described previously (Ge et al., 2004). Total RNA was extracted from 2-week-old rice seedlings with use of TRIzol Reagent (Invitrogen). Each lane was loaded with 20 μg of total RNA isolated from 2-week-old seedlings. Ethidium bromide-stained rRNA was used as an RNA-loading control. Total RNA was loaded in each lane on a 1% agarose gel containing 0.4 m formaldehyde and transferred to Hybond-N+ membrane (Amersham Pharmacia Biotech). A probe of OsMYB3R-2 cDNA labeled with [α-32P]dCTP was prepared by PCR for hybridization. After hybridization for 20 h at 65°C, the membrane was washed and exposed to x-ray film. The loading control was ribosomal RNA stained with ethidium bromide.
Treatment of Rice Seedlings with Chilling Stress (2°C)
To analyze the response to cold stress, T2 generation transgenic plants with positive GUS staining were used. To ensure that the seedlings were at the same morphological stage as wild-type and OsMYB3R-2 transgenic lines, the transgenic seeds from OsMYB3R-2-overexpressing plants were sown 36 h earlier than those of wild-type and antisense lines under the same conditions of 12 h of light/12 h of dark (30°C/26°C). The seeds of wild-type and antisense lines were sown at the same time because they showed no difference in germination and plant growth. All seeds were germinated in a mixture of nutrimental soil and vermiculite (8:1). Two-week-old seedlings at the early period of the tetraphyllous leaf stage were subjected to treatment at 2°C in the Low-temperature Biochemical Incubator (BTI100; LEAD TECH). At 0, 24, 48, 60, 72, and 84 h, cold-treated seedlings were moved to a greenhouse for 2 weeks. Surviving seedlings were photographed and analyzed to investigate the response of OsMYB3R-2 transgenic plants to chilling stress.
To analyze the response of OsCycB1;1 to cold stress, T1 generation transgenic plants with positive GUS staining were used. Two-week-old seedlings at the early period of the tetraphyllous leaf stage were treated at 3°C in the low-temperature cultivation room (7 m long and 3.5 m wide), and cold-treated seedlings were moved to a greenhouse for 1 week. Surviving seedlings were analyzed to investigate the response of OsCycB1;1 transgenic plants to chilling stress. The method of cold treatment was similar to that described in detail as shown above.
Transactivation Assay Based on the Yeast GAL4 System
The cDNA fragments of OsMYB3R-2 were generated by PCR amplification, cloned into NdeI and PstI sites, and fused in-frame to the GAL4 DNA-binding domain in the pGBKT7 vector. ATG bases were added after the NdeI site to all forward primers, except for the forward primer for PCR amplification of full-length OsMYB3R-2. A transactivation assay was performed as described (Choi et al., 2004). The constructs of OsMYB3R-2-pGBKT7 were transformed into AH109 cells by the lithium acetate-mediated method (Gietz et al., 1992), and transformants were selected on synthetic medium plates (SD medium) lacking Trp (SD/-Trp) at 28°C for 2 d. Yeast transformants from SD medium lacking Trp were then transferred and streaked onto solid SD agar medium lacking Trp/His (SD/-Trp/-His), Trp/adenine (SD/-Trp/-Ade), or Trp/His/adenine (SD/-Trp/-His/-Ade) to score the growth response after 3 d. For the β-galactosidase assay, the transformants were blotted on Whatman filter paper, and the cells imprinted on the filter were lysed by freezing in liquid nitrogen, then thawed at room temperature. The filter was then incubated in 2.5 mL of Z buffer containing 0.8 mg of 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside (X-Gal), which consisted of 16.1 g L−1 Na2HPO4·7H2O, 5.5 g L−1 NaH2PO4·H2O, 0.7 g L−1 KCl, and 0.246 g L−1 MgSO4·7H2O (pH 7.0) at 30°C, and monitored for color reaction.
Determination of Cellular Pro Levels
Fresh material (0.5 g) of 2-week-old seedlings was taken from wild-type and OsMYB3R-2 transgenic rice (T2 generation) or OsCycB1;1 transgenic rice (T1 generation) at the same stage with or without cold treatment (4°C) for 24 h. The method of obtaining rice seedlings at the same morphological stage was described in detail above. The samples were homogenized in 2 mL of 3% aqueous sulfosalicylic acid and centrifuged. The content of cellular free Pro was measured using of a spectrophotometer at λ = 520 nm as described previously (Wang et al., 2006).
EMSA
The coding sequence of OsMYB3R-2 with R1R2R3 tandem copies of DNA-binding domains was amplified by PCR and cloned into pGEX-4T-1 vector with a glutathione S-transferase (GST) gene and sequenced. A forward primer, 5′-CGCGGATCCATGGGTTGGGGCGCGGTGG-3′, and a reverse primer, 5′-CCGGAATTCTCAATCAATTGGGTGCTTGTCTG-3′, were used in PCR. The OsMYB3R-2-GST fusion protein was induced in strain BL21 (DE3) of Escherichia coli and purified as described previously (Han et al., 2005).
A 378-bp fragment upstream of the OsCycB1;1 promoter was amplified by PCR from the rice genome and sequenced. The primer pair used was 5′-AGCATTTCTGAGGAAGAAGT-3′ and 5′-ATACAACTTTATTCTTCCCT-3′. The PCR product of the 378-bp fragment was purified with use of the TIAN Quick Oligo Purification Kit (Tiangen) and labeled with use of the Biotin 3′ End DNA Labeling Kit (Pierce). Eight pairs of synthetic oligonucleotides containing optimal and mutant derivatives of the binding site for OsMYB3R-2 were labeled with use of the Biotin 3′ End DNA Labeling Kit (Pierce). The 50-μL reaction mixture was mixed gently with the following components: ultrapure water (25 μL), 5× TdT reaction buffer (10 μL), unlabeled oligonucleotide (1 μm, 5 μL), biotin-11-dUTP (5 μm, 5 μL), and diluted TdT (2 units μL−1, 5 μL). Anneal oligonucleotides were mixed with equal amounts of labeled complementary oligonucleotides and denatured at 90°C for 1 min, then slowly cooled and incubated at melting temperature for 30 min, and stored at −20°C. Frozen annealed oligonucleotides were thawed on ice for immediate use. The oligonucleotides were as follows: RT1WT (5′-AATACCAGTCCCCAACGGCTAGTTTCAACC-3′ and 5′-GGTTGAAACTAGCCGTTGGGGACTGGTATT-3′); RT2WT (5′-GCTAGGAGAGCTGGCAACCTCCGCAGCGGG-3′ and 5′-CGATCCTCTCGACCGTTGGAGGCGTCGCCC-3′); RT3WT (5′-CGCCACCCTACCCAACGGCTCTATTCCCCT-3′ and 5′-AGGGGAATAGAGCCGTTGGGTAGGGTGGCG-3′); RT1WTmut1 (5′-AATACCTTTCCCCAACGGCTAGTTTCAACC-3′ and 5′-GGTTGAAACTAGCCGTTGGGGAAAGGTATT-3′); RT1WTmut2 (5′-AATACCAGATCCCAACGGCTAGTTTCAACC-3′ and 5′-GGTTGAAACTAGCCGTTGGGATCTGGTATT-3′); RT1WTmut3 (5′-AATACCAGTCCTTAACGGCTAGTTTCAACC-3′ and 5′-GGTTGAAACTAGCCGTTAAGGACTGGTATT-3′); RT1WTmut4 (5′-AATACCAGTCCCCTTCGGCTAGTTTCAACC-3′ and 5′-GGTTGAAACTAGCCGAAGGGGACTGGTATT-3′); and RT1WTmut5 (5′-AATACCAGTCCCCAATTGCTAGTTTCAACC-3′ and 5′-GGTTGAAACTAGCAATTGGGGACTGGTATT-3′).
Standard reaction mixtures (20 μL) for EMSA contained 2 μL of purified proteins, 2 μL of biotin-labeled annealed oligonucleotides, 2 μL of 10× binding buffer (100 mm Tris, 500 mm KCl, and 10 mm dithiothreitol, pH 7.5), 1 μL of 50% glycerol, 1 μL of 1% Nonidet P-40, 1 μL of 1 m KCl, 1 μL of 100 mm MgCl2, 1 μL of 200 mm EDTA, 1 μL of 1 μg μL−1 poly(dI-dC), and 8 μL of ultrapure water. The reactions were incubated at room temperature (25°C) for 20 min. The 10% native polyacrylamide gel was prepared and prerun in 0.5× TBE buffer (45 mm Tris, 45 mm boric acid, and 1 mm EDTA, pH 8.3) for 30 to 60 min at 100 V before loading the samples, then the gel was run at room temperature in 0.5× TBE buffer at 100 V for 60 min until the bromphenol blue dye reached three-fourths of the gel. The gels were sandwiched and transferred to a N+ nylon membrane (Millipore) in 0.5× TBE buffer at 380 mA in a 4°C refrigerator for 60 min. When the transfer was complete, the membrane was placed with the bromphenol blue side up on a dry paper towel until the buffer on the membrane surface absorbed into the membrane and then was cross-linked for 10 to 15 min with the membrane face down on a transilluminator equipped with 312-nm bulbs. The detection of biotin-labeled DNA by chemiluminescence followed the manual of the LightShift Chemiluminecent EMSA Kit (20148; Pierce).
Flow Cytometry of Cell Cycle Progression
T2 generation seeds of OsMYB3R-2 transgenic rice were sterilized with 0.15% mercuric chloride and germinated on the filter with sterilized water at 28°C in the dark for 7 d. To obtain seedlings at the same stage as that of wild-type and the OsMYB3R-2 transgenic lines, the transgenic seeds with overexpressed OsMYB3R-2 were germinated 16 h earlier than those of wild-type and antisense lines under the same conditions in petri dishes (diameter, 20 cm). All of the GUS-positive seedlings of wild-type or OsMYB3R-2 transgenic rice were assigned in equal quantity and subjected to 28°C or 4°C for 24 h, respectively. Samples of cell nuclei were prepared as described by Galbraith et al. (1983). Root apical tips (1 mm) were excised, immediately chilled on ice, and chopped with a single-edged razor blade in a glass petri dish (diameter, 5 cm). Chopping buffer (45 mm MgCl2, 30 mm sodium citrate, 20 mm 4-morpholinepropane sulfonate, and 1 mg mL−1 Triton X-100, pH 7.0) was used to release the cells from the chopped tissues. The DNA content of individual transgenic cells was determined by flow cytometry. Cell nuclei were prepared for FACSAria by staining with 2 μg mL−1 2-(4-amidinophenyl)-6-indolecarbamidine dihydrochloride (DAPI). Each sample was prepared three times and subjected to FACS Caliber cytometry three times (BD Corporation). A total of 10,000 nuclei were designed to be measured per analysis.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number BAD81765.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression patterns of the DREB genes and their target genes in OsMYB3R-2 transgenic rice plants.
Supplemental Figure S2. Molecular identification of OsCycB1;1 transgenic rice.
Supplemental Figure S3. The length of the root cell in OsMYB3R-2 transgenic rice.
Supplemental Table S1. The information of rice DREB genes and the rice homologs of target genes of Arabidopsis DREBs.
Supplemental Table S2. Primer sets used for RT-PCR of cell cycle genes.
Supplementary Material
Acknowledgments
We are grateful to Ms. Rongxi Jiang and Yuan Zhao (Institute of Botany, Chinese Academy of Sciences) for their help in rice transformation and Prof. Yikun He (Capital Normal University) for his help with the equipment. We thank Dr. Jia Li (University of Oklahoma) for critically modifying the manuscript, and Dr. Ruth Gordon-Weeks and Dr. Hai-Chun Jing (Rothamsted Research) and Ms. Laura Heraty (BioMedEditing) for their help with language checking.
This work was supported by the Major State Basic Research Program of the People's Republic of China (grant no. 2005CB120806), the National Natural Science Foundation of China (grant nos. 30525026 and 30470866), and the State High-Tech Project (grant no. 2006 AA10Z169).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Kang Chong (chongk@ibcas.ac.cn).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Agarwal M, Hao Y, Kapoor A, Dong CH, Fujii H, Zheng X, Zhu JK (2006) A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J Biol Chem 281 37636–37645 [DOI] [PubMed] [Google Scholar]
- Andaya VC, Tai TH (2006) Fine mapping of the qCTS12 locus, a major QTL for seedling cold tolerance in rice. Theor Appl Genet 113 467–475 [DOI] [PubMed] [Google Scholar]
- Araki S, Ito M, Soyano T, Nishihama R, Machida Y (2004) Mitotic cyclins stimulate the activity of c-Myb-like factors for transactivation of G2/M phase-specific genes in tobacco. J Biol Chem 279 32979–32988 [DOI] [PubMed] [Google Scholar]
- Braun EL, Grotewold E (1999) Newly discovered plant c-myb-like genes rewrite the evolution of the plant myb gene family. Plant Physiol 121 21–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JQ, Meng XP, Zhang Y, Xia M, Wang XP (2008) Over-expression of OsDREB genes lead to enhanced drought tolerance in rice. Biotechnol Lett 30 2191–2198 [DOI] [PubMed] [Google Scholar]
- Cheng C, Yun KY, Ressom HW, Mohanty B, Bajic VB, Jia Y, Yun SJ, de los Reyes BG (2007) An early response regulatory cluster induced by low temperature and hydrogen peroxide in seedlings of chilling-tolerant japonica rice. BMC Genomics 8 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi D, Kim JH, Kende H (2004) Whole genome analysis of the OsGRF gene family encoding plant-specific putative transcription activators in rice (Oryza sativa L.). Plant Cell Physiol 45 897–904 [DOI] [PubMed] [Google Scholar]
- Choi MS, Kim MC, Yoo JH, Moon BC, Koo SC, Park BO, Lee JH, Koo YD, Han HJ, Lee SY, et al (2005) Isolation of a calmodulin-binding transcription factor from rice (Oryza sativa L.). J Biol Chem 280 40820–40831 [DOI] [PubMed] [Google Scholar]
- Cominelli E, Galbiati M, Vavasseur A, Conti L, Sala T, Vuylsteke M, Leonhardt N, Dellaporta SL, Tonelli C (2005) A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr Biol 15 1196–1200 [DOI] [PubMed] [Google Scholar]
- Dai X, Xu Y, Ma Q, Xu W, Wang T, Xue Y, Chong K (2007) Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol 143 1739–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day IS, Reddy AS, Golovkin M (1996) Isolation of a new mitotic-like cyclin from Arabidopsis: complementation of a yeast cyclin mutant with a plant cyclin. Plant Mol Biol 30 565–575 [DOI] [PubMed] [Google Scholar]
- Denekamp M, Smeekens SC (2003) Integration of wounding and osmotic stress signals determines the expression of the AtMYB102 transcription factor gene. Plant Physiol 132 1415–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J 33 751–763 [DOI] [PubMed] [Google Scholar]
- Ferreira P, Hemerly A, de Almeida Engler J, Bergounioux C, Burssens S, Van Montagu M, Engler G, Inze D (1994) Three discrete classes of Arabidopsis cyclins are expressed during different intervals of the cell cycle. Proc Natl Acad Sci USA 91 11313–11317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith DW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, Firoozabady E (1983) Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220 1049–1051 [DOI] [PubMed] [Google Scholar]
- Ge L, Chen H, Jiang JF, Zhao Y, Xu ML, Xu YY, Tan KH, Xu ZH, Chong K (2004) Overexpression of OsRAA1 causes pleiotropic phenotypes in transgenic rice plants, including altered leaf, flower, and root development and root response to gravity. Plant Physiol 135 1502–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge LF, Chao DY, Shi M, Zhu MZ, Gao JP, Lin HX (2008) Overexpression of the trehalose-6-phosphate phosphatase gene OsTPP1 confers stress tolerance in rice and results in the activation of stress responsive genes. Planta 228 191–201 [DOI] [PubMed] [Google Scholar]
- Gietz D, St Jean A, Woods RA, Schiestl RH (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res 20 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E, Drummond BJ, Bowen B, Peterson T (1994) The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 76 543–553 [DOI] [PubMed] [Google Scholar]
- Haga N, Kato K, Murase M, Araki S, Kubo M, Demura T, Suzuki K, Muller I, Voss U, Jurgens G, et al (2007) R1R2R3-Myb proteins positively regulate cytokinesis through activation of KNOLLE transcription in Arabidopsis thaliana. Development 134 1101–1110 [DOI] [PubMed] [Google Scholar]
- Han Y, Wang X, Jiang J, Xu Y, Xu Z, Chong K (2005) Biochemical character of the purified OsRAA1, a novel rice protein with GTP-binding activity, and its expression pattern in Oryza sativa. J Plant Physiol 162 1057–1063 [DOI] [PubMed] [Google Scholar]
- Howe KM, Watson RJ (1991) Nucleotide preferences in sequence-specific recognition of DNA by c-myb protein. Nucleic Acids Res 19 3913–3919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh TH, Lee JT, Charng YY, Chan MT (2002) Tomato plants ectopically expressing Arabidopsis CBF1 show enhanced resistance to water deficit stress. Plant Physiol 130 618–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, You J, Fang Y, Zhu X, Qi Z, Xiong L (2008) Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol Biol 67 169–181 [DOI] [PubMed] [Google Scholar]
- Ito M, Araki S, Matsunaga S, Itoh T, Nishihama R, Machida Y, Doonan JH, Watanabe A (2001) G2/M-phase-specific transcription during the plant cell cycle is mediated by c-Myb-like transcription factors. Plant Cell 13 1891–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Iwase M, Kodama H, Lavisse P, Komamine A, Nishihama R, Machida Y, Watanabe A (1998) A novel cis-acting element in promoters of plant B-type cyclin genes activates M phase-specific transcription. Plant Cell 10 331–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Marie-Claire C, Sakabe M, Ohno T, Hata S, Kouchi H, Hashimoto J, Fukuda H, Komamine A, Watanabe A (1997) Cell-cycle-regulated transcription of A- and B-type plant cyclin genes in synchronous cultures. Plant J 11 983–992 [DOI] [PubMed] [Google Scholar]
- Ito Y, Katsura K, Maruyama K, Taji T, Kobayashi M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2006) Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol 47 141–153 [DOI] [PubMed] [Google Scholar]
- Jiang J, Xu Y, Chong K (2007) Overexpression of OsJAC1, a lectin gene, suppresses the coleoptile and stem elongation in rice. J Integr Plant Biol 49 230–237 [Google Scholar]
- Jin H, Martin C (1999) Multifunctionality and diversity within the plant MYB-gene family. Plant Mol Biol 41 577–585 [DOI] [PubMed] [Google Scholar]
- Jung C, Seo JS, Han SW, Koo YJ, Kim CH, Song SI, Nahm BH, Choi YD, Cheong JJ (2008) Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol 146 623–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda CBH (1974) Response of indica-japonica rice hybrids to low temperatures. SABRAO J 6 17–32 [Google Scholar]
- Kanneganti V, Gupta AK (2008) Overexpression of OsiSAP8, a member of stress associated protein (SAP) gene family of rice confers tolerance to salt, drought and cold stress in transgenic tobacco and rice. Plant Mol Biol 66 445–462 [DOI] [PubMed] [Google Scholar]
- Korenjak M, Taylor-Harding B, Binne UK, Satterlee JS, Stevaux O, Aasland R, White-Cooper H, Dyson N, Brehm A (2004) Native E2F/RBF complexes contain Myb-interacting proteins and repress transcription of developmentally controlled E2F target genes. Cell 119 181–193 [DOI] [PubMed] [Google Scholar]
- Kranz H, Scholz K, Weisshaar B (2000) c-MYB oncogene-like genes encoding three MYB repeats occur in all major plant lineages. Plant J 21 231–235 [DOI] [PubMed] [Google Scholar]
- La H, Li J, Ji Z, Cheng Y, Li X, Jiang S, Venkatesh PN, Ramachandran S (2006) Genome-wide analysis of cyclin family in rice (Oryza sativa L.). Mol Genet Genomics 275 374–386 [DOI] [PubMed] [Google Scholar]
- Liu K, Wang L, Xu Y, Chen N, Ma Q, Li F, Chong K (2007) Overexpression of OsCOIN, a putative cold inducible zinc finger protein, increased tolerance to chilling, salt and drought, and enhanced proline level in rice. Planta 226 1007–1016 [DOI] [PubMed] [Google Scholar]
- Luft JC, Benjamin IJ, Mestril R, Dix DJ (2001) Heat shock factor 1-mediated thermotolerance prevents cell death and results in G2/M cell cycle arrest. Cell Stress Chaperones 6 326–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackill DJLX (1997) Genetic variation for traits related to temperate adaptation of rice cultivars. Crop Sci 37 1340–1346 [Google Scholar]
- Meissner RC, Jin H, Cominelli E, Denekamp M, Fuertes A, Greco R, Kranz HD, Penfield S, Petroni K, Urzainqui A, et al (1999) Function search in a large transcription factor gene family in Arabidopsis: assessing the potential of reverse genetics to identify insertional mutations in R2R3 MYB genes. Plant Cell 11 1827–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morano KA, Santoro N, Koch KA, Thiele DJ (1999) A trans-activation domain in yeast heat shock transcription factor is essential for cell cycle progression during stress. Mol Cell Biol 19 402–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morsy MR, Almutairi AM, Gibbons J, Yun SJ, de Los Reyes BG (2005) The OsLti6 genes encoding low-molecular-weight membrane proteins are differentially expressed in rice cultivars with contrasting sensitivity to low temperature. Gene 344 171–180 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay A, Vij S, Tyagi AK (2004) Overexpression of a zinc-finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgenic tobacco. Proc Natl Acad Sci USA 101 6309–6314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai A, Ishikawa T (2001) Cell cycle transition under stress conditions controlled by vertebrate heat shock factors. EMBO J 20 2885–2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Tran LS, Van Nguyen D, Fujita M, Maruyama K, Todaka D, Ito Y, Hayashi N, Shinozaki K, Yamaguchi-Shinozaki K (2007) Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J 51 617–630 [DOI] [PubMed] [Google Scholar]
- Ohnishi T, Sugahara S, Yamada T, Kikuchi K, Yoshiba Y, Hirano HY, Tsutsumi N (2005) OsNAC6, a member of the NAC gene family, is induced by various stresses in rice. Genes Genet Syst 80 135–139 [DOI] [PubMed] [Google Scholar]
- Pasquali G, Biricolti S, Locatelli F, Baldoni E, Mattana M (2008) Osmyb4 expression improves adaptive responses to drought and cold stress in transgenic apples. Plant Cell Rep 27 1677–1686 [DOI] [PubMed] [Google Scholar]
- Pramanik MH, Imai R (2005) Functional identification of a trehalose 6-phosphate phosphatase gene that is involved in transient induction of trehalose biosynthesis during chilling stress in rice. Plant Mol Biol 58 751–762 [DOI] [PubMed] [Google Scholar]
- Rosinski JA, Atchley WR (1998) Molecular evolution of the Myb family of transcription factors: evidence for polyphyletic origin. J Mol Evol 46 74–83 [DOI] [PubMed] [Google Scholar]
- Saijo Y, Hata S, Kyozuka J, Shimamoto K, Izui K (2000) Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J 23 319–327 [DOI] [PubMed] [Google Scholar]
- Saijo Y, Kinoshita N, Ishiyama K, Hata S, Kyozuka J, Hayakawa T, Nakamura T, Shimamoto K, Yamaya T, Izui K (2001) A Ca(2+)-dependent protein kinase that endows rice plants with cold- and salt-stress tolerance functions in vascular bundles. Plant Cell Physiol 42 1228–1233 [DOI] [PubMed] [Google Scholar]
- Salomoni P, Perrotti D, Martinez R, Franceschi C, Calabretta B (1997) Resistance to apoptosis in CTLL-2 cells constitutively expressing c-Myb is associated with induction of BCL-2 expression and Myb-dependent regulation of bcl-2 promoter activity. Proc Natl Acad Sci USA 94 3296–3301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santilli G, Schwab R, Watson R, Ebert C, Aronow BJ, Sala A (2005) Temperature-dependent modification and activation of B-MYB: implications for cell survival. J Biol Chem 280 15628–15634 [DOI] [PubMed] [Google Scholar]
- Shima S, Matsui H, Tahara S, Imai R (2007) Biochemical characterization of rice trehalose-6-phosphate phosphatases supports distinctive functions of these plant enzymes. FEBS J 274 1192–1201 [DOI] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4 447–456 [DOI] [PubMed] [Google Scholar]
- Suzuki K, Nagasuga K, Okada M (2008) The chilling injury induced by high root temperature in the leaves of rice seedlings. Plant Cell Physiol 49 433–442 [DOI] [PubMed] [Google Scholar]
- Suzuki K, Nishiuchi T, Nakayama Y, Ito M, Shinshi H (2006) Elicitor-induced down-regulation of cell cycle-related genes in tobacco cells. Plant Cell Environ 29 183–191 [DOI] [PubMed] [Google Scholar]
- Thomashow MF (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol 50 571–599 [DOI] [PubMed] [Google Scholar]
- Urao T, Yamaguchi-Shinozaki K, Urao S, Shinozaki K (1993) An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5 1529–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini C, Locatelli F, Bracale M, Magnani E, Marsoni M, Osnato M, Mattana M, Baldoni E, Coraggio I (2004) Overexpression of the rice Osmyb4 gene increases chilling and freezing tolerance of Arabidopsis thaliana plants. Plant J 37 115–127 [DOI] [PubMed] [Google Scholar]
- Wan B, Lin Y, Mou T (2007) Expression of rice Ca(2+)-dependent protein kinases (CDPKs) genes under different environmental stresses. FEBS Lett 581 1179–1189 [DOI] [PubMed] [Google Scholar]
- Wang JW, Yang FP, Chen XQ, Liang RQ, Zhang LQ, Geng DM, Zhang XD, Song YZ, Zhang GS (2006) Induced expression of DREB transcriptional factor and study on its physiological effects of drought tolerance in transgenic wheat. Yi Chuan Xue Bao 33 468–476 [DOI] [PubMed] [Google Scholar]
- Wang L, Cai H, Bai X, Li LW, Li Y, Zhu YM (2008) Cultivation of transgenic rice plants with OsCDPK7 gene and its salt tolerance. Yi Chuan 30 1051–1055 [DOI] [PubMed] [Google Scholar]
- Wang YJ, Zhang ZG, He XJ, Zhou HL, Wen YX, Dai JX, Zhang JS, Chen SY (2003) A rice transcription factor OsbHLH1 is involved in cold stress response. Theor Appl Genet 107 1402–1409 [DOI] [PubMed] [Google Scholar]
- Wang ZCC, Xu Y, Jiang R, Xu Z, Chong K (2004) A practical vector for efficient knockdown of gene expression in rice (Oryza sativa L.). Plant Mol Biol Rep 22 409–417 [Google Scholar]
- Xiang Y, Huang Y, Xiong L (2007) Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant Physiol 144 1416–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Yang Y (2003) Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid-inducible mitogen-activated protein kinase. Plant Cell 15 745–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue GP (2005) A CELD-fusion method for rapid determination of the DNA-binding sequence specificity of novel plant DNA-binding proteins. Plant J 41 638–649 [DOI] [PubMed] [Google Scholar]
- Yanhui C, Xiaoyuan Y, Kun H, Meihua L, Jigang L, Zhaofeng G, Zhiqiang L, Yunfei Z, Xiaoxiao W, Xiaoming Q, et al (2006) The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol 60 107–124 [DOI] [PubMed] [Google Scholar]
- Zhu J, Verslues PE, Zheng X, Lee BH, Zhan X, Manabe Y, Sokolchik I, Zhu Y, Dong CH, Zhu JK, et al (2005) HOS10 encodes an R2R3-type MYB transcription factor essential for cold acclimation in plants. Proc Natl Acad Sci USA 102 9966–9971 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.