Abstract
Although the final size of plant organs is influenced by environmental cues, it is generally accepted that the primary size determinants are intrinsic factors that regulate and coordinate cell proliferation and cell expansion. Here, we show that optimal proteasome function is required to maintain final shoot organ size in Arabidopsis (Arabidopsis thaliana). Loss of function of the subunit regulatory particle AAA ATPase (RPT2a) causes a weak defect in 26S proteasome activity and leads to an enlargement of leaves, stems, flowers, fruits, seeds, and embryos. These size increases are a result of increased cell expansion that compensates for a reduction in cell number. Increased ploidy levels were found in some but not all enlarged organs, indicating that the cell size increases are not caused by a higher nuclear DNA content. Partial loss of function of the regulatory particle non-ATPase (RPN) subunits RPN10 and RPN12a causes a stronger defect in proteasome function and also results in cell enlargement and decreased cell proliferation. However, the increased cell volumes in rpn10-1 and rpn12a-1 mutants translated into the enlargement of only some, but not all, shoot organs. Collectively, these data show that during Arabidopsis shoot development, the maintenance of optimal proteasome activity levels is important for balancing cell expansion with cell proliferation rates.
The 26S proteasome (26SP) is a multisubunit, multicatalytic, 2.4-MD protease responsible for the degradation of proteins involved in various biological processes (Varshavsky, 2005; DeMartino and Gillette, 2007; Hanna and Finley, 2007; Kurepa and Smalle, 2008). Prior to their degradation, most 26SP target proteins are covalently modified with a polyubiquitin chain in a three-step enzymatic reaction (Smalle and Vierstra, 2004). In addition to its central function in recognizing and degrading polyubiquitinated proteins, the 26SP can also degrade proteins that were not modified by polyubiquitination (Benaroudj et al., 2001; Asher et al., 2006; Asher and Shaul, 2006; Lee et al., 2006; Pande et al., 2007).
The 26SP consists of a cylindrical 20S core complex and two 19S regulatory particles that cap the 20S core on both ends. The 20S proteasome (20SP) is composed of seven related α-subunits and seven related β-subunits arranged in a stack of four heptameric rings. The outer rings are composed of α-subunits and the inner rings are composed of β-subunits, of which three have proteolytic activities described as caspase-, trypsin-, and chymotrypsin-like (Kurepa and Smalle, 2008). The regulatory particles (RPs) serve as highly restrictive gatekeepers for the core protease. Each RP is composed of a lid subcomplex, which contains at least nine non-ATPase subunits designated RPN3, RPN5 to RPN9, RPN11, RPN12, and RPN15 and a base subcomplex that contains RPN1, RPN2, RPN13, and RPT1 to RPT6 subunits. The RPN10 and RPN13 subunits have been shown to recognize polyubiquitinated proteins; thus, they define the main interaction points of the 26SP with its target proteins (Young et al., 1998; Smalle and Vierstra, 2004; Husnjak et al., 2008; Kurepa and Smalle, 2008; Schreiner et al., 2008). Ubiquitinated proteins can also interact with the 26SP by first binding to the carrier proteins RAD23 (for radiation sensitive 23), DSK2 (for dominant suppressor of kar1 2), and DDI1 (for DNA damage inducible 1), which then bind the RP base subunit RPN1 (Elsasser et al., 2002, 2004; Elsasser and Finley, 2005). The RPT base subunits belong to the class of AAA ATPases and are thought to be important for the unfolding and translocation of substrate proteins through the central pore in the 20SP base called the 20SP gate (Vale, 2000; Bajorek and Glickman, 2004). Similar to all other AAA ATPases, the non-AAA modules of RPTs confer functional specificity to these proteins, and in yeast, the functions of the six RP ATPases have been shown to differ (Smith et al., 2007). For example, the main function of yeast RPT2 and RPT5 subunits is to regulate the opening of the 20SP gate (Smith et al., 2007).
Despite the many recent advances in proteasome research (Husnjak et al., 2008; Kurepa and Smalle, 2008; Schreiner et al., 2008), the specific functions of most of the RP subunits remain unknown. Because of the complex quaternary structure of the 26SP and the feedback mechanisms that coregulate the expression of proteasome subunit genes in response to temporal or spatial demands, the overexpression of individual subunits in most cases does not lead to a change in total proteasome activity (Kurepa and Smalle, 2008). Thus, the main strategy used to study the specific functions of a particular RP subunit involves the analyses of mutants in which the expression of the corresponding subunit gene is down-regulated or abolished (Kurepa and Smalle, 2008). However, assigning a specific function to an RP subunit based on the analyses of loss-of-function mutants can be difficult. The major challenge in this approach has been to discern between the effects of a mutation on subunit function in particular and on total proteasome function in general. For example, the halted root (hlr) mutant was isolated as a short-root mutant that has an expanded root tip, and this phenotype was shown to be a result of a 13-bp-long deletion in the RPT2a gene (Ueda et al., 2004). However, the majority of RP mutants described to date have shorter roots, which suggests that it is the down-regulation of 26SP function and not specifically the RPT2a subunit that leads to the change in root growth (Kurepa et al., 2008). In another example, the ae3-1 mutant, isolated as an enhancer of asymmetric leaves1 (as1) and as2, was shown to carry a mutation in the RPN8a gene (Huang et al., 2006). The ae3-1 plants have a reduced rosette size, altered leaf phyllotaxy, and lanceolated, abaxialized leaves (Huang et al., 2006). Double mutant analyses of as2 with other proteasome subunit mutants showed that all double mutant lines have comparable leaf phenotypes and, thus, that the originally observed effect of ae3-1 on as1/as2 phenotypes is the result of a general alteration in 26SP function (Huang et al., 2006). In conclusion, most of the phenotypes of the RP mutants described to date seem to reflect a general alteration in proteasome activity and not a specific defect that relates to a specific function of a particular subunit (Huang et al., 2006; Kurepa et al., 2008).
Comparative analyses of the RP mutants rpn10-1, rpn12a-1, and rpt2a-2 revealed that the strongest proteasome defect was caused by rpn10-1 and the weakest defect was caused by the rpt2a-2 mutation (Kurepa et al., 2008). The rpt2a-2 mutation induces a phenotype that sets this line apart from the other RP mutants: it leads to an increase in size of the rosette and other shoot organs. In this study, we tested the hypothesis that the enlargement of aerial organs in rpt2a-2 reflects a specific function of the RPT2 subunit. To this end, we determined the cellular phenotypes of two rpt2a mutant alleles and compared them with the phenotypes of rpn10-1 and rpn12a-1 mutants. We demonstrate that any decrease in 26SP function leads to a reduction in cell number that is accompanied by increased cell sizes. This suggests that optimal proteasome function is required for the proper execution of preprogrammed cell proliferation and expansion rates.
RESULTS
Isolation of Arabidopsis rpt2 Mutants
In contrast to Saccharomyces cerevisiae, Caenorhabditis elegans, and Drosophila melanogaster, which have one RPT2 gene, the RPT2 subunit in Arabidopsis (Arabidopsis thaliana) is encoded by a two-member gene family (Rubin et al., 1998; Takahashi et al., 2002; Wójcik and DeMartino, 2002; Shibahara et al., 2004; Ueda et al., 2004; Sönnichsen et al., 2005; Kurepa et al., 2008). AtRPT2a (At4g29040) and AtRPT2b (At2g20140; Fig. 1A) encode 444-amino acid proteins sharing 98.8% identity at the protein level. Analyses of public microarray expression data showed that RPT2a is consistently expressed at a higher level than RPT2b and that the expression of both genes is developmentally regulated (Fig. 1B).
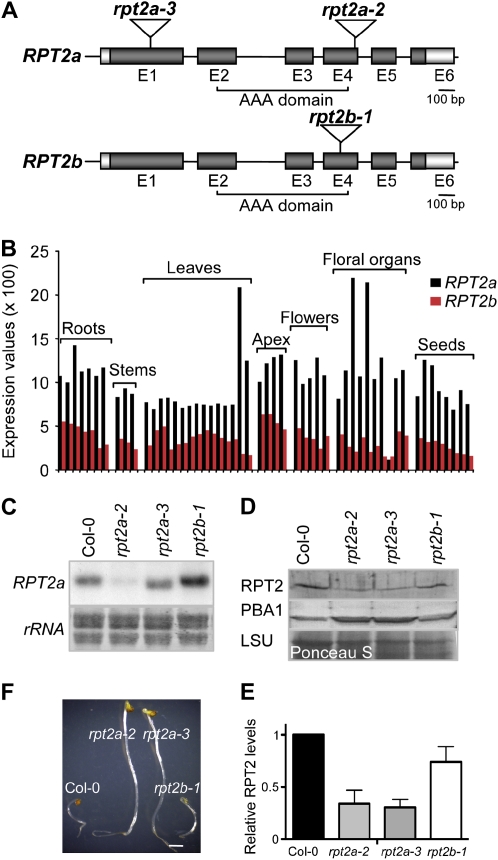
Figure 1.
Molecular analyses of rpt2 mutants. A, RPT2a and RPT2b gene structures and the positions of the T-DNA insertions. Exons (E) and introns are represented by boxes (dark gray, coding region; light gray, untranslated region) and lines, respectively. Insertion positions of the T-DNA in the rpt2a-2 (SALK_005596), rpt2a-3 (SALK_130019), and rpt2b-1 (SALK_043450C) alleles are shown. B, RPT2 expression levels during Arabidopsis development. The developmental data set of the AtGenExpress project was used (Schmid et al., 2005). Different bars in the same category denote the different developmental stages as specified by Schmid et al. (2005). C, Expression analyses. RNA gel blots were probed with an RPT2a antisense probe. The region of a methylene blue-stained membrane encompassing ribosomal RNAs (rRNA) is shown as a loading control. D, RPT2 protein level in rpt2a mutants. Protein extracts from 10-d-old Col-0 and rpt2 mutant plants were separated by SDS-PAGE, blotted, and probed with anti-RPT2 and anti-PBA1 antisera. A region of the Ponceau S-stained membrane encompassing the large subunit of Rubisco (LSU) is shown as a loading control. E, Relative levels of RPT2 protein were assessed by densitometry from four immunoblots. The signal intensity of Col-0 was normalized to 1, and mean values ± sd are shown. F, MG132 tolerance of rpt2 mutants. Seeds were sown and grown for 5 d in darkness on water/agar containing 50 μm MG132. Bar = 1 mm. [See online article for color version of this figure.]
To analyze the effects of the loss of RPT2 function on shoot growth, we isolated a second ecotype Columbia (Col-0) T-DNA insertion mutation in RPT2a, rpt2a-3, and an insertion mutation in RPT2b. Gel-blot analysis of total RNA revealed that the RPT2a transcript level was altered in all tested rpt2 mutants (Fig. 1C). The weak signal at the migration position of the RPT2a transcript in rpt2a-2 likely represents a cross-reaction of the probe with the RPT2b mRNA. In the rpt2a-3 allele, the transcript was shorter than in the wild type, and sequence analyses of RT-PCR products revealed that it contains the RPT2a coding region downstream of the insertion mutation, suggesting that the expression of this transcript is driven by a promoter within the T-DNA (Fig. 1C; data not shown). In the rpt2b-1 mutant, the RPT2a transcript was more abundant than in the wild type, which is in agreement with previously described findings that a potential decrease in proteasome activity leads to a compensatory up-regulation of the expression of 26SP subunit genes (Yang et al., 2004; Huang et al., 2006; Kurepa et al., 2008). We then tested the levels of immunoreactive RPT2 protein in 10-d-old seedlings in all rpt2 alleles (Fig. 1, D and E). Because RPT2a and RPT2b are nearly identical at the amino acid level, the polyclonal antibodies generated against RPT2a are expected to recognize both isoforms. In homozygous rpt2a mutants, the RPT2 levels were reduced to 30% ± 10% of the wild-type levels, and in rpt2b-1, they were reduced to 70% ± 10%. This suggests that all lines are indeed null mutants and that the RPT2b protein represents 30% or less of the total RPT2 pool in wild-type seedlings. Both rpt2a mutants displayed an up-regulation in the abundance of the 20SP subunit PBA1, which is indicative of a loss in 26SP function (Fig. 1D; Yang et al., 2004).
In 26SP RP mutants, decreased 26SP assembly rates lead to an increase in the abundance of the free 20SP, which is further enhanced by the compensatory up-regulation of the proteasome subunit gene set (Kurepa et al., 2008). The increased 20SP activity in rpt2a-2 and other tested RP mutants leads to an elevated tolerance to the proteasome inhibitor MG132 in a growth response assay (Kurepa et al., 2008). The hypocotyls of 5-d-old etiolated rpt2a seedlings were approximately 20% longer than those of wild-type and rpt2b-1 seedlings (data not shown). Similar to rpt2a-2 (Kurepa et al., 2008), the rpt2a-3 mutant was more tolerant to MG132 (Fig. 1F). The hypocotyl elongation of Col-0 seedlings grown on 50 μm MG132 was inhibited by approximately 80% compared with the untreated control, while the same treatment reduced rpt2a hypocotyl growth only approximately 50% compared with the untreated mutants. Two-way nonparametric ANOVA with the Bonferroni posttest confirmed that the effects of the treatment on wild-type and mutant lines were significantly different (P < 0.0001). The rpt2b-1 mutant was not more tolerant to the MG132-induced growth inhibition, however (Fig. 1F). Thus, we concluded that the rpt2a-2 and rpt2a-3 mutants share proteasome-related phenotypes both qualitatively and quantitatively.
Morphological Characterization of rpt2a Mutants
Due to the low expression level of RPT2b compared with RPT2a and the relatively weak proteasome defect caused by rpt2a mutations, we predicted that the rpt2b-1 mutant would not show a strong developmental phenotype. Indeed, rpt2b-1 mutant seedlings were indistinguishable from the wild type (Fig. 2A; data not shown for later developmental stages). In contrast, we noticed that 3-d-old light-grown rpt2a mutants had enlarged cotyledons (Fig. 2A). The length and thickness of rpt2a roots at this developmental stage were also increased compared with the wild type, but at later stages of development, root growth ceased, similar to the Wassilewskija mutant line hlr-1/rpt2a-1 (Ueda et al., 2004). Analyses of rpt2a embryos revealed that the increased organ size was established already during embryogenesis. In mature embryos, the cotyledon area was 30% ± 10% larger and the radicle was 30% ± 10% longer than in the wild type (Fig. 2B; Supplemental Table S1). Since light is an important factor in the control of cell expansion and overall plant morphology, we also measured the dimensions of cotyledons, roots, and hypocotyls of 3-d-old dark-grown rpt2a seedlings (Supplemental Table S1). All parts of the etiolated rpt2a seedlings were larger than those in the wild type (approximately 30% increase for root length, approximately 20% for hypocotyl length, approximately 30% for hypocotyl width, and approximately 70% for cotyledon area).
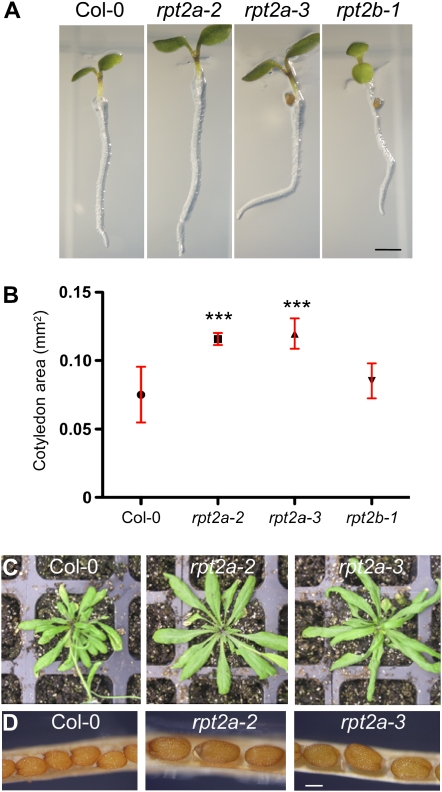
Figure 2.
Organ sizes in rpt2 mutants. A, Phenotypes of 3-d-old rpt2 mutants grown on vertically positioned MS/2 agar plates. Bar = 1 mm. B, Cotyledon area of mature embryos. Dry seeds were imbibed overnight, cleared with lactophenol, and photographed. A minimum of 20 cotyledons per line was measured. Symbols on the graph represent average values, and the error bars correspond to the sd. *** Significantly different from the wild type (P ≤ 0.0001). C, Rosette phenotype of rpt2a mutants. Seedlings were germinated and grown for 10 d on MS/2 and then transferred to soil. Plants were grown under a long-day photoperiod and photographed 30 d after germination. D, Seed size in rpt2a mutants. Mature siliques were dissected, and the central parts of the fruits are shown. Bar = 200 μm.
In the rpt2a mutants, all shoot organs remained larger than in the wild type throughout the life cycle (Fig. 2, C and D; Supplemental Table S1). For example, the rosette diameters of 30-d-old rpt2a-2 and rpt2a-3 plants were 30% ± 10% larger than in Col-0 (7.8 ± 0.3, 10.3 ± 0.4, and 10.4 ± 0.2 cm for Col-0, rpt2a-2, and rpt2a-3, respectively). Mature rpt2a siliques were approximately 30% longer and contained seeds with a 1.4-fold larger area compared with the wild type (Fig. 2D; Supplemental Table S1). The seed number per silique, however, was reduced in both mutants by approximately 20% (51 ± 4, 40 ± 3, and 38 ± 3 for Col-0, rpt2a-2, and rpt2a-3, respectively; n ≥ 8, P ≤ 0.005) due to a reduction in the number of ovules to approximately 80% of the wild-type level (52 ± 2, 40 ± 7, and 41 ± 5 for Col-0, rpt2a-2, and rpt2a-3, respectively; n ≥ 3, P ≤ 0.005). Analyses of the morphometric data showed not only that the size of aerial organs in the rpt2a mutants was increased but also that organ shapes differed from those of the wild type. In cotyledons, leaves, and petals, for example, the length-to-width ratio in the mutants was larger than in the wild type (Supplemental Table S1). The length-to-width ratios for juvenile leaves (leaves 1 and 2) of 30-d-old plants were 1.1 ± 0.1, 1.7 ± 0.3, and 1.4 ± 0.2 for Col-0, rpt2a-2, and rpt2a-3, respectively. Taken together, these results indicate that loss of function of the RPT2a gene affected the mechanisms that determine the final organ size. In addition, the data suggest that the rpt2a mutations affect differently those mechanisms that govern proximodistal (i.e. length-related) and mediolateral (i.e. width-related) control of leaf expansion. Since the establishment of organ polarity was affected in a similar manner in cotyledons, leaves, and flower organs, we also concluded that the affected mechanisms are general and not organ specific.
It should be noted that the increase in rpt2a rosette size is not observed when plants are grown on half-strength Murashige and Skoog medium (MS/2 medium; Kurepa et al., 2008). Although we currently do not know what causes this growth difference, one explanation relates to the difference in salt concentrations between the MS/2 medium and soil and the hypersensitivity of 26SP mutants to stresses that cause protein misfolding, such as salt stress.
Kinematic Analyses of Cotyledon Growth in the rpt2a Mutants
The final size of an organ depends on the number and size of its cells (Mizukami, 2001). Cell number and cell volume depend on the rate and the duration of cell proliferation and elongation phases but also on compensation, a still unknown mechanism that coordinates cell proliferation and elongation during organ morphogenesis (Tsukaya, 2008). To determine which of the organ size determinants are affected in the rpt2a mutants, we first measured the length of the growth period of cotyledons (Fig. 3). The postgermination growth of cotyledons is mostly based on cell expansion. Total cell number per cotyledon has been shown to increase by approximately 30% after 3 d of growth, after which no further cell division has been detected (Tsukaya et al., 1994; Stoynova-Bakalova et al., 2004). Thus, analyses of cotyledon growth rates should allow us to distinguish between proliferation and elongation and to determine whether the elongation rate or the duration of the elongation phase contributes to the increase in final cotyledon size in the mutants. The time course analyses suggested that the duration of longitudinal growth of wild-type and mutant cotyledons was nearly identical; in all lines, the growth curves reached a plateau after 8 d (Fig. 3A). Thus, the increased final cotyledon length in the mutants was not due to extended postgermination expansion growth and reflected the size differences already established during embryogenesis. The lateral growth period for the wild type and the rpt2a-2 mutant was also nearly identical, ending around day 8, whereas the rpt2a-3 mutant cotyledons displayed a modest expansion beyond this time point (Fig. 3B). The longitudinal and especially lateral growth rates differed between the mutants and the wild type. The HillSlope coefficients that describe the steepness of the growth curves were 0.24 ± 0.05, 0.36 ± 0.07, and 0.36 ± 0.05 for longitudinal growth and 0.31 ± 0.05, 0.18 ± 0.07, and 0.15 ± 0.06 for lateral growth curves of Col-0, rpt2a-2, and rpt2a-3, respectively. The faster proximodistal expansion and reduced mediolateral expansion in rpt2a mutants explain the increased length-to-width ratio of their cotyledons (Supplemental Table S1).
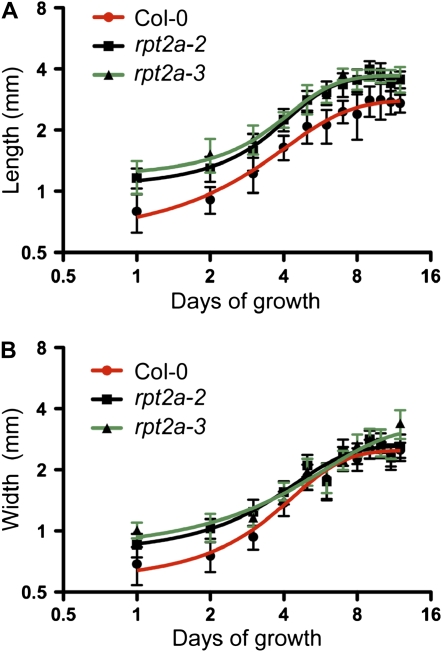
Figure 3.
Cotyledon growth in the wild type and rpt2a mutants. Plants were grown on MS/2 plates for 2 weeks under a long-day photoperiod. Length (A) and width (B) of 20 or more cotyledons were measured daily. Data are plotted on a log2-log2 scale. All data are presented as means ± sd, and the curves represent nonlinear curve fitting of data using a sigmoidal model with variable slope.
To test if the increased cotyledon size in the mutants is a result of increased cell size, cell number, or both, we analyzed the cell sizes and numbers in cotyledons of mature embryos and 3- and 10-d-old light-grown seedlings (Fig. 4). In mature rpt2a embryos, the cells of the adaxial palisade layer from the central part of the cotyledons were larger than in the wild type (Fig. 4, A and B). The overall cell numbers per cotyledon, however, were not significantly different (Fig. 4C), which would account for the increased cotyledon area. Measurement of cell sizes and cell numbers in cotyledons of 3- and 10-d-old plants showed that while the cells remained larger, their number was reduced in both mutants when compared with the wild type (Fig. 4, D–G). Thus, the rpt2a mutations affect the postgerminative cell proliferation in cotyledons on the one hand and cell sizes during all phases of development on the other. The increase in cell size in an organ accompanied by a reduced cell number is a characteristic of compensation mutants, and because the final organ size in rpt2a alleles is increased compared with the wild type, the rpt2a mutants can be classified as large-leaf compensation mutants, as defined by Horiguchi et al. (2006).
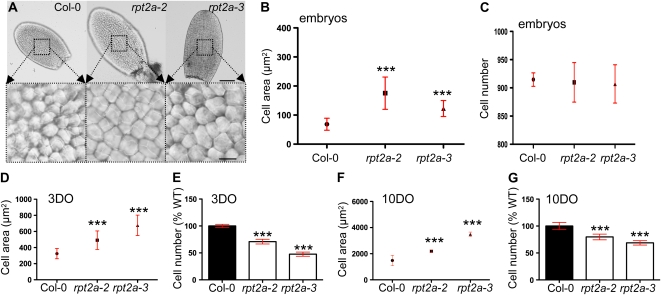
Figure 4.
Cell sizes and cell numbers in cotyledons of the wild type and rpt2a mutants. A, Cleared cotyledons of mature embryos and close-ups of the central regions showing the sizes of the adaxial subepidermal cells. Bars = 100 μm and 10 μm for top and bottom panels, respectively. B, Average area ± sd of adaxial subepidermal cells of mature embryos. C, Cell number per cotyledon. The number of palisade cells was counted from micrographs of cleared cotyledons. A minimum of five cotyledons per line was analyzed, and the mean ± sd is shown. D and E, Cell area (D) and cell number (E) in unexpanded 3-d-old (3DO) cotyledons. The number of adaxial subepidermal cells per cotyledon is presented as mean ± sd, and the cell number is presented relative to the wild-type (WT) values ± sd. F and G, Cell area (F) and cell number (G) in fully expanded, 10-d-old (10DO) cotyledons. *** P < 0.0001. [See online article for color version of this figure.]
Cell Sizes and Cell Numbers in Petals of rpt2a Mutants
It has been shown that the compensation phenomenon occurs in determinate organs such as cotyledons, leaves, and petals but not in indeterminate organs such as roots (Ferjani et al., 2007). To test if other enlarged determinate organs in rpt2a mutants also display compensation of the decreased cell number by increased cell volumes, we analyzed the cells of mature petals. The ridged cells of the adaxial surface of mature petal blades of the mutants had an approximately 3-fold larger area (Fig. 5, A and B), and their total number per petal was reduced to 70% ± 10% of the wild type (Fig. 5C). Interestingly, the decrease in total cell number was due solely to a decrease in the cell numbers of the mediolateral plane by approximately 30% (data not shown). In addition to the petal cells, and excepting pollen grains, we observed cell size increases in all other organs of the Arabidopsis shoot that were analyzed (i.e. rosette leaves, siliques, and stems), suggesting that this is a general cellular phenotype associated with rpt2a mutants (data not shown).
Figure 5.
Cell sizes in petals and trichome branch numbers. A, Mature petals of the wild type and rpt2a mutants were analyzed using the method of Horiguchi et al. (2006). Ridged epidermal cells from the central region of petals are shown. Bar = 20 μm. B, The average cell area was measured in petals dissected from 10 flowers at stage 15. *** P < 0.0001. C, The cell number per petal was counted from at least three petals. The data are presented as means ± sd. *** P < 0.0001. D, Increased trichome branching in rpt2a mutants. A minimum of 120 trichomes per line were analyzed in rosette leaves 3 and 4, and the frequency of trichomes with three, four, five, six, and seven branches is presented as the percentage of the total number of trichomes counted. [See online article for color version of this figure.]
Polyploidization of Cells in Determinate Organs of rpt2a Mutants
The final size of plant cells is often positively correlated with their ploidy level (Mizukami, 2001). Another parameter that has been shown to positively correlate with ploidy levels is the number of trichome branches (Hulskamp, 2004). In Col-0 plants, most trichomes on the adaxial side of the first leaf pair have three branches and a small number (approximately 5%) have four (Fig. 5D). In contrast, more than 40% of trichomes in rpt2a mutants had four or more branches (Fig. 5D).
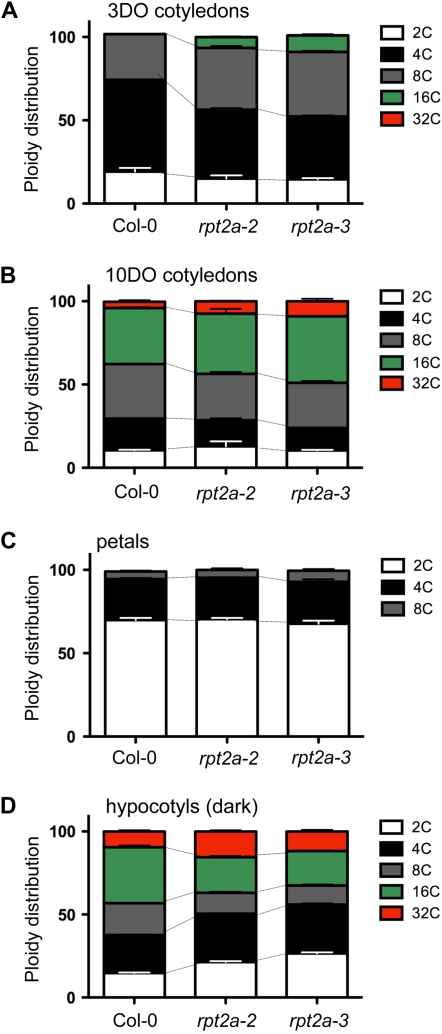
Thus, both the cell size and the trichome morphology of rpt2a mutants suggested an overall increase in ploidy levels. To test the ploidy levels in cells of an organ at different stages in development, we isolated nuclei from unexpanded and mature cotyledons (Fig. 6, A and B). In immature cotyledons dissected from 3-d-old seedlings, we detected a significant increase in 8C and 16C nuclei in both rpt2a mutants (Fig. 6A). It has been shown that the overall proportion of cells with higher ploidy levels (4C, 8C, 16C, and 32C) increases with the age of Arabidopsis tissues (Galbraith et al., 1991). Indeed, in fully expanded cotyledons dissected from 10-d-old seedlings, we observed nuclei in five ploidy classes, from 2C to 32C, in both wild-type and mutant lines (Fig. 6B). In mutants, however, the number of cells that contained highly polyploid nuclei was significantly higher than in the wild type (Fig. 6B). The endoreduplication factor (EF), which measures the average number of endocycles per 100 cells (Cookson et al., 2006), was 109.1 ± 1.1, 135.3 ± 2.4, and 145.2 ± 0.9 for 3-d-old Col-0, rpt2a-2, and rpt2a-3 cotyledons, respectively, and 200.5 ± 4.5, 227.5 ± 2.5, and 223.6 ± 4 for cotyledons of 10-d-old Col-0, rpt2a-2, and rpt2a-3 plants. The differences between the Col-0 EF and the EFs of the rpt2a mutants were statistically significant (P < 0.005), suggesting that there is a positive correlation between ploidy level, cell size, and organ size in cotyledons.
Figure 6.
Quantification of the distribution of ploidy levels in rpt2a mutants. A and B, Relative ratio of ploidy classes in unexpanded 3-d-old (3DO) and mature 10-d-old (10DO) cotyledons. Twenty cotyledons were pooled for each measurement, and measurements were repeated at least two times. Error bars indicate sd. C, Ploidy distribution in petals was determined by isolating nuclei from petals dissected from at least 10 mature flowers. Mean values ± sd are shown. D, Ploidy distribution in hypocotyls of 3-d-old etiolated seedlings.
To further test if the cell size increases in the rpt2a mutants are correlated with increased ploidy levels, we measured the DNA content of cells of two other organs that are enlarged in both mutants. First, we measured the DNA content of petal cells (Fig. 6C). Previous studies have shown that cells at the tip of the petal blade are predominantly diploid (Hase et al., 2005), while DNA content analyses of the entire organ (i.e. the petal blade and claw) showed some polyploidization (Dewitte et al., 2007). We measured the ploidy distribution in whole petals of flowers at stage 15 (Smyth et al., 1990). Surprisingly, the distribution of ploidy classes in the mutants was almost identical to that of the wild type (Fig. 6C). The EFs were 33.3 ± 2.1, 35.4 ± 1.5, and 38.9 ± 1.9 for Col-0, rpt2a-2, and rpt2a-3, respectively. In petals, therefore, there was no significant correlation between cell size and ploidy level. Finally, we also analyzed the relationship between the overall size and ploidy level of cells in hypocotyls of the wild type and mutants. Hypocotyls of 3-d-old rpt2a mutant seedlings grown in the dark were longer and thicker compared with the wild type (Supplemental Table S1) and also contained larger cells (data not shown). However, although the distributions of ploidy classes were altered in both mutants, the overall ploidy levels were not increased (Fig. 6D). In fact, the EFs of the mutants were lower than that in the wild type (201.7 ± 3.5, 179.1 ± 3.8, and 163.4 ± 6.2 for Col-0, rpt2a-2, and rpt2a-3, respectively). We concluded that in rpt2a mutants, similar to fugu compensation mutants (Ferjani et al., 2007), cell and organ enlargement was not correlated with increased endoreduplication.
Cell Sizes, Cell Numbers, and Ploidy Levels in Other RP Mutants
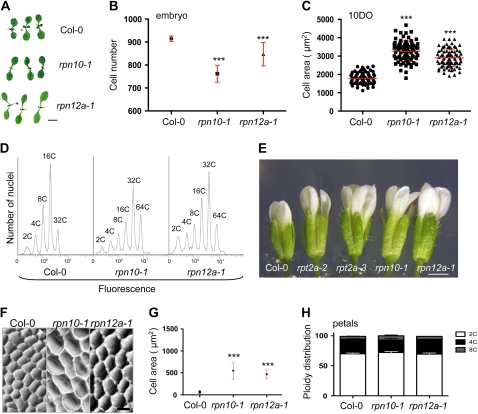
While the general size increases of rpt2a shoot organs sets these mutants apart from other 26SP RP mutants, it remained possible that this phenotype merely reflects a different degree in proteasome malfunction than a specific function of RPT2 within the particle. To test this possibility, we analyzed two other RP mutants that carry more severe defects in proteasome function. The proteasome activity levels and development of the RP mutants rpn10-1 and rpn12-1 have been described previously (Smalle et al., 2002, 2003; Kurepa et al., 2008). The 26SP activity is decreased to approximately 30% of the wild-type level in rpn10-1 and to approximately 40% in rpn12a-1 (Kurepa et al., 2008). Thus, if the cellular phenotypes that we observed for rpt2a mutants are a result of a general decrease in 26SP function, they should be even more pronounced in the rpn10-1 and rpn12a-1 lines, given that the rpt2a-2 mutation only caused a reduction to approximately 60% of the wild-type 26SP activity (Kurepa et al., 2008). The most obvious developmental phenotypes of rpn10-1 and rpn12a-1 are shorter roots and smaller rosettes (Smalle et al., 2002, 2003; Kurepa et al., 2008). However, some shoot organs in rpn10-1 and rpn12a-1 are larger than in the wild type. In rpn12a-1, flowers and cotyledons are larger, while in rpn10-1, only flowers are enlarged (Fig. 7, A and E; Smalle et al., 2003). Thus, any level of decrease in 26SP function leads to an inhibition of root development, while the effects on shoot organ size appeared to be dose dependent and organ specific.
Figure 7.
Organ and cell sizes and ploidy levels in rpn10-1 and rpn12a-1 mutants. A, Ten-day-old wild-type, rpn10-1, and rpn12a-1 seedlings. Bar = 1 mm. B, The cell number of cotyledons was counted from photographs of lactophenol-cleared mature embryos. Five cotyledons per line were analyzed. C, The size of adaxial parenchymatic cells was determined in 10-d-old (10DO) cotyledons. A minimum of 40 cells were counted from each cotyledon, and 10 cotyledons per line were analyzed. The horizontal line represents the mean, and error bars indicate sd. D, Representative histogram of propidium iodide fluorescence intensity in nuclei isolated from cotyledons of wild-type, rpn10-1, and rpn12a-1 plants grown on MS/2 for 16 d. E, Flowers of RP mutants. Bar = 1 mm. F, Adaxial epidermal cells of wild-type, rpn10-1, and rpn12a-1 petals. Bar = 20 μm. G, Cell area of adaxial epidermal cells of petals from flowers at floral stage 15. Data are represented as means ± sd (n ≥ 50). Asterisks indicate significant differences from the wild type (*** P < 0.0001). H, Ploidy distribution in petals was determined as described in the legend to Figure 6C. Mean values ± sd are shown. [See online article for color version of this figure.]
Next, we tested whether organ size in both RP mutants is correlated with cell size and ploidy level. We analyzed cotyledons, which are similar to wild-type size in rpn10-1 and larger in rpn12a-1 seedlings, and flowers, which are larger than the wild type in both RP mutants (Fig. 7, A and E). In contrast to rpt2a (Fig. 4B), the total number of palisade cells was also reduced in cotyledons of rpn10-1 and rpn12a-1 embryos (Fig. 7B) and remained lower than in the wild type throughout cotyledon development (data not shown). Similar to rpt2a mutants, cells of rpn10-1 and rpn12a-1 were enlarged at all stages of cotyledon development (Fig. 7C; data not shown). Because the rpn10-1 mutation leads to a stronger defect in proteasome function, a likely explanation for the absence of any size increase in mature cotyledons is that the cell proliferation rate both during embryogenesis and during postgermination development is more affected than in rpn12a-1. In both mutants, we also detected an increase in polyploid nuclei (Fig. 7D). In 16-d-old Col-0 cotyledons, 12% ± 2% of cells underwent four or more endoreduplication cycles, while in the mutants, 46% ± 5% (rpn10-1) and 44% ± 3% (rpn12a-1) of cells had a 32C or 64C content. In flowers of both mutants, epidermal cells in petals were larger than those in the wild type (Fig. 7, F and G). In addition, the cells of rpn10-1 were larger than those of rpn12a-1. Similar to the rpt2a mutants, the increased cell sizes in rpn10-1 and rpn12a-1 petals were accompanied by decreases in cell numbers (51% ± 6% and 63% ± 9% of the wild type for rpn10-1 and rpn12a-1, respectively). Flow cytometric analyses of the DNA content, however, showed that the ploidy distribution in wild-type and mutant petals also did not differ significantly (Fig. 7H). We conclude that the rpn10-1 and rpn12a-1 mutants, similar to rpt2a mutants, display a combined decrease in cell proliferation and increase in cell size that is not correlated with nuclear DNA content.
DISCUSSION
Compensation Mechanism of Organ Development and the 26SP
The goals of this study were to determine the cellular basis of shoot organ enlargement in rpt2a mutants and to analyze whether the underlying mechanisms reflect a specific function of RPT2a or a general defect in proteasome activity. The mechanisms that control shoot organ size are still largely unknown, in spite of their fundamental importance and economic potential. The current view is that final organ size is controlled by internal developmental signals and modulated by environmental cues (Mizukami, 2001; Tsukaya, 2008). To control final organ size, the intrinsic signaling networks need to control the initiation, duration, and termination of cell proliferation and cell expansion. This implies that these signaling networks are complex and include both promoters and inhibitors of cell division and cell expansion, many of which have been identified (Mizukami and Fischer, 2000; Mizukami, 2001; Hu et al., 2003; Jofuku et al., 2005; Ohto et al., 2005; Disch et al., 2006; Inzé and De Veylder, 2006; Szécsi et al., 2006; Song et al., 2007; Busov et al., 2008). In addition to the networks that regulate cell proliferation and cell expansion, final organ size is controlled by compensation, an integrative system that leads to cell enlargement when the cell number in a determinate organ is reduced (Tsukaya, 2008). Mature cotyledons and petals of the rpt2a-2, rpt2a-3, rpn10-1, and rpn12a-1 mutants contain fewer cells than those in the wild type, while the average cell volumes are increased. Thus, all tested regulatory particle mutants exhibit compensation. Similar to other mutant or transgenic lines for which compensation phenomena have been described (Inzé and De Veylder, 2006; Ferjani et al., 2007; Tsukaya, 2008), the effects of compensation on final organ size varied considerably within the RP mutant series. For example, petals in all lines were larger than those of the wild type, but the rosette leaves were larger only in the rpt2a mutants and smaller in rpn10-1 and rpn12a-1.
The role of ubiquitin/26SP-dependent degradation in the control of cell division has been amply documented (Callis and Vierstra, 2000; Hansen et al., 2002; Smalle and Vierstra, 2004; Gutierrez and Ronai, 2006; Inzé and De Veylder, 2006; Pines, 2006; Dreher and Callis, 2007; Stone and Callis, 2007). It has also been shown that the strength of the proteasome defect varies considerably within the RP mutant series used in this study (Kurepa et al., 2008). This suggests that in the weakest RP mutants, the rpt2a lines, the cell division controls are affected less than in stronger RP mutants, such as rpn10-1 and rpn12a-1. Indeed, while the cell number in expanding cotyledons of young rpt2a seedlings was reduced, no reductions were found in cotyledons of mature rpt2a embryos, in contrast to the rpn10-1 and rpn12a-1 mutants that already showed a lower cell number at this early stage in development (Figs. 4C and 7B). Furthermore, the reduction in cell number in mature petals of all RP mutants was strongest in rpn12a-1 and rpn10-1 (Fig. 5C). The compensation phenomenon tends to be observed only in lines in which cell division is decreased below a critical threshold (Tsukaya, 2008). Our results suggest that this compensation threshold has been reached in all RP mutants and that it is predominantly the strength of the cell division defect that determines the final outcome of the compensation effect on organ size. In the weak rpt2a mutant lines, combined effects of the decreased cell numbers and compensative cell enlargement led to a general increase in final organ size. In the strong rpn10-1 mutant, the compensative cell size increases did not lead to enlargement of most shoot organs, probably because of the more severe reductions in cell division activity. This suggests that the growth differences observed between the RP mutants derive from the different strengths of the proteasome defect and are not related to any specific functions that the respective subunits might have within the 26SP particle.
This also suggests that during Arabidopsis shoot development, total 26SP activity needs to be maintained above a critical threshold to ensure the coordinated execution of cell proliferation and expansion programs. This critical threshold must be higher than approximately 60% of the wild-type level, because this activity level in the rpt2a mutants was sufficient to trigger a compensation mechanism. This further suggests that fluctuations in total proteasome activity may contribute to the regulation of plant growth. Although our current knowledge of the regulation of proteasome activity in plants is insufficient to support this hypothesis, there is evidence to suggest that proteasome abundance is not maintained at a constant level during the plant life cycle (Kurepa and Smalle, 2008). Recent reports show that proteasome abundance and activity vary during plant development and in response to changes in environmental conditions (Smalle et al., 2002; Itoh et al., 2003; Kim et al., 2003; Shibahara et al., 2004; Yang et al., 2004; Lorenzo and Solano, 2005; Kurepa and Smalle, 2008). These developmental changes in 26SP activity could be an integral component of the regulatory network that controls plant stature, form, and size and may involve, in addition to effects on cell division and expansion, also developmentally activated plant cell death programs (Kim et al., 2003).
Polyploidy and the 26SP
Similar to its role in cell cycle progression, the role of the ubiquitin/26SP system in the switch from a mitotic cycle to an endoreduplication cycle has been amply documented (Kominami and Toda, 1997; Sigrist and Lehner, 1997; Genschik et al., 1998; Kominami et al., 1998; Cebolla et al., 1999; Szlanka et al., 2003; Neuburger et al., 2006). Here, we show that all RP mutants have altered ploidy profiles in some but not all shoot organs. Because the effect of decreased proteasome function on endoreduplication was organ specific (i.e. it led to an increase in ploidy in cotyledons but not in petals and to a change in ploidy class distribution in etiolated hypocotyls), it confirms previous observations that cells of different tissues and organs express distinct factors that control the mitotic/endoreduplication cycle switch (Churchman et al., 2006). In addition, because the observed changes in cellular DNA content were not correlated with the increased cell and organ sizes, we concluded that the general increase in cell expansion in RP mutants was not driven by increased polyploidization.
26SP- Versus 20SP-Dependent Proteolysis
Numerous studies in plants and animals have described the essential role for 26SP-dependent turnover of cell cycle regulators in controlling the various stages of cell division (Callis and Vierstra, 2000; Hansen et al., 2002; Smalle and Vierstra, 2004; Gutierrez and Ronai, 2006; Inzé and De Veylder, 2006; Pines, 2006; Dreher and Callis, 2007; Stone and Callis, 2007). Accordingly, the cell cycle-related changes in RP mutants can be readily linked to their decreased 26SP activity. However, loss of 26SP function in RP mutants also resulted in increased activity of the free 20SP (Kurepa et al., 2008), and the contribution of this proteolytic pathway to the cellular phenotypes of RP mutants is more difficult to assess. Although some target proteins for the free 20SP have been identified in mammals and yeast, it is currently unclear to what extent this proteolytic pathway contributes to the protein turnover in eukaryotic cells (Asher et al., 2006; Asher and Shaul, 2006). Future studies will have to address to what extent the plant 20SP contributes to the stability control of regulatory proteins, including factors involved in the regulation of cell proliferation, cell expansion, and the mitotic/endoreduplication cycle switch.
MATERIALS AND METHODS
Plant Materials, Growth Conditions, and Treatments
The rpt2a-3 (SALK_130019) and rpt2b-1 (SALK_043450C) T-DNA insertion mutants of Arabidopsis (Arabidopsis thaliana) were identified in the SIGNAL collection (Alonso et al., 2003). The rpt2a-2/hlr-2 (SALK_005596) as well as rpn10-1 and rpn12a-1 mutants in the Col-0 background were described previously (Ueda et al., 2004; Kurepa et al., 2008). All plants were grown on MS/2 medium (Sigma), pH 5.7, with 1% Suc and 0.8% phytoagar (RPI Corporation). For soil growth, 7-day-old seedlings were transplanted from MS/2 plates to pots containing 50% Promix and 50% vermiculite. All plants were grown in Conviron growth chambers at 22°C under long-day photoperiods and light of 120 μmol m−2 s−1, unless indicated otherwise. MG132 treatments were done as described (Kurepa et al., 2008).
RNA and Protein Analyses
RNA was isolated from 7-d-old seedlings grown in liquid cultures using TRIzol reagent (Invitrogen). Total RNA (10 μg) was separated on 1% agarose-formaldehyde gels, transferred to nitrocellulose membranes (Hybond N+; GE Healthcare), and probed with [32P]UTP-labeled riboprobes synthesized from linearized plasmids using the Riboprobe Combination System (Promega). The antisense RPT2a probe was synthesized from PstI-linearized plasmid obtained from the Arabidopsis Biological Resource Center (stock no. 145D18). For immunoblot analyses, plants were weighed, frozen in liquid nitrogen, and ground in two volumes of 2× Laemmli sample buffer. Proteins were separated by SDS-PAGE, transferred to nitrocellulose membranes (Hybond C-Extra; GE Healthcare), and probed as described previously (Smalle et al., 2002) using anti-AtRPT2a antibodies purchased from Biomol.
Organ and Cell Analyses
For all morphometric and kinematic analyses, plants or plant organs were photographed and the relevant parameter was measured on digital images using ImageJ (Abramoff et al., 2004). For cotyledon growth curves, a minimum of 30 in vitro-grown seedlings were analyzed. To determine organ size, a minimum of 10 organs were analyzed. For microscopic analyses of embryonic cells and cotyledon palisade cells, tissues were cleared with lactophenol (1:1:1:1 phenol:water:100% glycerol:lactic acid) for 12 h before analyses. For the analyses of cell sizes and numbers in petals, the printing technique of Horiguchi et al. (2006) was used. A minimum of five cotyledons per line were used to determine cell numbers and cell sizes. Trichome branch numbers were determined using an SZX12 microscope (Olympus).
Flow Cytometry
Nuclei were prepared and stained using the CyStain PI Absolute P kit following the manufacturer's protocol (Partec) and analyzed on a PAS flow cytometer (Partec). A minimum of 20 cotyledons, petals, or hypocotyls per line were pooled for each measurement. The data were analyzed using FlowJo 8.7.5 (Tree Star). Data were derived from at least 10,000 events, and the same gating hierarchy was used for all samples. The propidium iodide-stained nuclei were initially selected on the fluorescence versus the side scatter plot. The nuclear aggregates were then excluded on the forward scatter width versus the side scatter plot. The resulting population was used to analyze DNA content (fluorescence versus number of events).
Statistical Analyses
Descriptive statistics, hypothesis testing, and curve fitting were done using Prism 5.0a software (GraphPad Software). All data are presented as means ± sd of at least two independent experiments. When means of more than two samples were compared, we used one-way nonparametric ANOVA with the null hypothesis that the value measured in Col-0 equals the value measured in the mutant. When the ANOVA P value was less than 0.05, we used the Tukey-Kramer posttest to find a significant difference between pairs of means. The significance levels, indicated by one (P < 0.05), two (P < 0.001), or three (P < 0.0001) asterisks in the figures, illustrate the results of the Tukey-Kramer posttest.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Morphometric characterization of rpt2a mutants.
Supplementary Material
Acknowledgments
We thank Dr. Tim Phillips (Department of Plant and Soil Science, University of Kentucky) for the use of his Partec PA flow cytometer, Angela Schoergendorfer (Department of Statistics, University of Kentucky) for help with statistical analyses, and the Salk Institute and the Arabidopsis Biological Resource Center for providing the seeds of the rpt2 mutant lines used in this study.
This work was supported by the Kentucky Tobacco Research and Development Center, the U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service (grant no. 2005–35304–16043), and the Kentucky Science and Engineering Foundation (grant no. 148–502–06–189).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Jan A. Smalle (jsmalle@uky.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abramoff MD, Magelhaes PJ, Ram SJ (2004) Image processing with ImageJ. Biophotonics Int 11 36–42 [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Asher G, Reuven N, Shaul Y (2006) 20S proteasomes and protein degradation “by default.” Bioessays 28 844–849 [DOI] [PubMed] [Google Scholar]
- Asher G, Shaul Y (2006) Ubiquitin-independent degradation: lessons from the p53 model. Isr Med Assoc J 8 229–232 [PubMed] [Google Scholar]
- Bajorek M, Glickman MH (2004) Keepers at the final gates: regulatory complexes and gating of the proteasome channel. Cell Mol Life Sci 61 1579–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benaroudj N, Tarcsa E, Cascio P, Goldberg AL (2001) The unfolding of substrates and ubiquitin-independent protein degradation by proteasomes. Biochimie 83 311–318 [DOI] [PubMed] [Google Scholar]
- Busov VB, Brunner AM, Strauss SH (2008) Genes for control of plant stature and form. New Phytol 177 589–607 [DOI] [PubMed] [Google Scholar]
- Callis J, Vierstra RD (2000) Protein degradation in signaling. Curr Opin Plant Biol 3 381–386 [DOI] [PubMed] [Google Scholar]
- Cebolla A, Vinardell JM, Kiss E, Olah B, Roudier F, Kondorosi A, Kondorosi E (1999) The mitotic inhibitor ccs52 is required for endoreduplication and ploidy-dependent cell enlargement in plants. EMBO J 18 4476–4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman ML, Brown ML, Kato N, Kirik V, Hülskamp M, Inzé D, De Veylder L, Walker JD, Zheng Z, Oppenheimer DG, et al (2006) SIAMESE, a plant-specific cell cycle regulator, controls endoreplication onset in Arabidopsis thaliana. Plant Cell 18 3145–3157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson SJ, Radziejwoski A, Granier C (2006) Cell and leaf size plasticity in Arabidopsis: what is the role of endoreduplication? Plant Cell Environ 29 1273–1283 [DOI] [PubMed] [Google Scholar]
- DeMartino GN, Gillette TG (2007) Proteasomes: machines for all reasons. Cell 129 659–662 [DOI] [PubMed] [Google Scholar]
- Dewitte W, Scofield S, Alcasabas AA, Maughan SC, Menges M, Braun N, Collins C, Nieuwland J, Prinsen E, Sundaresan V, et al (2007) Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc Natl Acad Sci USA 104 14537–14542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disch S, Anastasiou E, Sharma VK, Laux T, Fletcher JC, Lenhard M (2006) The E3 ubiquitin ligase BIG BROTHER controls Arabidopsis organ size in a dosage-dependent manner. Curr Biol 16 272–279 [DOI] [PubMed] [Google Scholar]
- Dreher K, Callis J (2007) Ubiquitin, hormones and biotic stress in plants. Ann Bot (Lond) 99 787–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsasser S, Chandler-Militello D, Müller B, Hanna J, Finley D (2004) Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J Biol Chem 279 26817–26822 [DOI] [PubMed] [Google Scholar]
- Elsasser S, Finley D (2005) Delivery of ubiquitinated substrates to protein-unfolding machines. Nat Cell Biol 7 742–749 [DOI] [PubMed] [Google Scholar]
- Elsasser S, Gali RR, Schwickart M, Larsen CN, Leggett DS, Müller B, Feng MT, Tübing F, Dittmar GA, Finley D (2002) Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat Cell Biol 4 725–730 [DOI] [PubMed] [Google Scholar]
- Ferjani A, Horiguchi G, Yano S, Tsukaya H (2007) Analysis of leaf development in fugu mutants of Arabidopsis reveals three compensation modes that modulate cell expansion in determinate organs. Plant Physiol 144 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith DW, Harkins KR, Knapp S (1991) Systemic endopolyploidy in Arabidopsis thaliana. Plant Physiol 96 985–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschik P, Criqui MC, Parmentier Y, Derevier A, Fleck J (1998) Cell cycle-dependent proteolysis in plants: identification of the destruction box pathway and metaphase arrest produced by the proteasome inhibitor MG132. Plant Cell 10 2063–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez GJ, Ronai Z (2006) Ubiquitin and SUMO systems in the regulation of mitotic checkpoints. Trends Biochem Sci 31 324–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Finley D (2007) A proteasome for all occasions. FEBS Lett 581 2854–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen DV, Hsu JY, Kaiser BK, Jackson PK, Eldridge AG (2002) Control of the centriole and centrosome cycles by ubiquitination enzymes. Oncogene 21 6209–6221 [DOI] [PubMed] [Google Scholar]
- Hase Y, Fujioka S, Yoshida S, Sun G, Umeda M, Tanaka A (2005) Ectopic endoreduplication caused by sterol alteration results in serrated petals in Arabidopsis. J Exp Bot 56 1263–1268 [DOI] [PubMed] [Google Scholar]
- Horiguchi G, Fujikura U, Ferjani A, Ishikawa N, Tsukaya H (2006) Large-scale histological analysis of leaf mutants using two simple leaf observation methods: identification of novel genetic pathways governing the size and shape of leaves. Plant J 48 638–644 [DOI] [PubMed] [Google Scholar]
- Hu Y, Xie Q, Chua NH (2003) The Arabidopsis auxin-inducible gene ARGOS controls lateral organ size. Plant Cell 15 1951–1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Pi L, Liang W, Xu B, Wang H, Cai R, Huang H (2006) The proteolytic function of the Arabidopsis 26S proteasome is required for specifying leaf adaxial identity. Plant Cell 18 2479–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulskamp M (2004) Plant trichomes: a model for cell differentiation. Nat Rev Mol Cell Biol 5 471–480 [DOI] [PubMed] [Google Scholar]
- Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I (2008) Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 453 481–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inzé D, De Veylder L (2006) Cell cycle regulation in plant development. Annu Rev Genet 40 77–105 [DOI] [PubMed] [Google Scholar]
- Itoh H, Matsuoka M, Steber CM (2003) A role for the ubiquitin-26S-proteasome pathway in gibberellin signaling. Trends Plant Sci 8 492–497 [DOI] [PubMed] [Google Scholar]
- Jofuku KD, Omidyar PK, Gee Z, Okamuro JK (2005) Control of seed mass and seed yield by the floral homeotic gene APETALA2. Proc Natl Acad Sci USA 102 3117–3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Ahn JW, Jin UH, Choi D, Paek KH, Pai HS (2003) Activation of the programmed cell death pathway by inhibition of proteasome function in plants. J Biol Chem 278 19406–19415 [DOI] [PubMed] [Google Scholar]
- Kominami K, Ochotorena I, Toda T (1998) Two F-box/WD-repeat proteins Pop1 and Pop2 form hetero- and homo-complexes together with cullin-1 in the fission yeast SCF (Skp1-Cullin-1-F-box) ubiquitin ligase. Genes Cells 3 721–735 [DOI] [PubMed] [Google Scholar]
- Kominami K, Toda T (1997) Fission yeast WD-repeat protein Pop1 regulates genome ploidy through ubiquitin-proteasome-mediated degradation of the CDK inhibitor Rum1 and the S-phase initiator Cdc18. Genes Dev 11 1548–1560 [DOI] [PubMed] [Google Scholar]
- Kurepa J, Smalle J (2008) Structure, function and regulation of plant proteasomes. Biochimie 90 324–335 [DOI] [PubMed] [Google Scholar]
- Kurepa J, Toh-e A, Smalle J (2008) 26S proteasome regulatory particle mutants have increased oxidative stress tolerance. Plant J 53 102–114 [DOI] [PubMed] [Google Scholar]
- Lee H, Zeng SX, Lu H (2006) UV induces p21 rapid turnover independently of ubiquitin and Skp2. J Biol Chem 281 26876–26883 [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Solano R (2005) Molecular players regulating the jasmonate signalling network. Curr Opin Plant Biol 8 532–540 [DOI] [PubMed] [Google Scholar]
- Mizukami Y (2001) A matter of size: developmental control of organ size in plants. Curr Opin Plant Biol 4 533–539 [DOI] [PubMed] [Google Scholar]
- Mizukami Y, Fischer RL (2000) Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc Natl Acad Sci USA 97 942–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuburger PJ, Saville KJ, Zeng J, Smyth KA, Belote JM (2006) A genetic suppressor of two dominant temperature-sensitive lethal proteasome mutants of Drosophila melanogaster is itself a mutated proteasome subunit gene. Genetics 173 1377–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto MA, Fischer RL, Goldberg RB, Nakamura K, Harada JJ (2005) Control of seed mass by APETALA2. Proc Natl Acad Sci USA 102 3123–3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pande AH, Scaglione P, Taylor M, Nemec KN, Tuthill S, Moe D, Holmes RK, Tatulian SA, Teter K (2007) Conformational instability of the cholera toxin A1 polypeptide. J Mol Biol 374 1114–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines J (2006) Mitosis: a matter of getting rid of the right protein at the right time. Trends Cell Biol 16 55–63 [DOI] [PubMed] [Google Scholar]
- Rubin DM, Glickman MH, Larsen CN, Dhruvakumar S, Finley D (1998) Active site mutants in the six regulatory particle ATPases reveal multiple roles for ATP in the proteasome. EMBO J 17 4909–4919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37 501–506 [DOI] [PubMed] [Google Scholar]
- Schreiner P, Chen X, Husnjak K, Randles L, Zhang N, Elsasser S, Finley D, Dikic I, Walters KJ, Groll M (2008) Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature 453 548–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibahara T, Kawasaki H, Hirano H (2004) Mass spectrometric analysis of expression of ATPase subunits encoded by duplicated genes in the 19S regulatory particle of rice 26S proteasome. Arch Biochem Biophys 421 34–41 [DOI] [PubMed] [Google Scholar]
- Sigrist SJ, Lehner CF (1997) Drosophila fizzy-related down-regulates mitotic cyclins and is required for cell proliferation arrest and entry into endocycles. Cell 90 671–681 [DOI] [PubMed] [Google Scholar]
- Smalle J, Kurepa J, Yang P, Babiychuk E, Kushnir S, Durski A, Vierstra RD (2002) Cytokinin growth responses in Arabidopsis involve the 26S proteasome subunit RPN12. Plant Cell 14 17–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Kurepa J, Yang P, Emborg TJ, Babiychuk E, Kushnir S, Vierstra RD (2003) The pleiotropic role of the 26S proteasome subunit RPN10 in Arabidopsis growth and development supports a substrate-specific function in abscisic acid signaling. Plant Cell 15 965–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55 555–590 [DOI] [PubMed] [Google Scholar]
- Smith DM, Chang SC, Park S, Finley D, Cheng Y, Goldberg AL (2007) Docking of the proteasomal ATPases' carboxyl termini in the 20S proteasome's α ring opens the gate for substrate entry. Mol Cell 27 731–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song XJ, Huang W, Shi M, Zhu MZ, Lin HX (2007) A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet 39 623–630 [DOI] [PubMed] [Google Scholar]
- Sönnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, Brehm M, Alleaume AM, Artelt J, Bettencourt P, Cassin E, et al (2005) Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434 462–469 [DOI] [PubMed] [Google Scholar]
- Stone SL, Callis J (2007) Ubiquitin ligases mediate growth and development by promoting protein death. Curr Opin Plant Biol 10 624–632 [DOI] [PubMed] [Google Scholar]
- Stoynova-Bakalova E, Karanov E, Petrov P, Hall MA (2004) Cell division and cell expansion in cotyledons of Arabidopsis seedlings. New Phytol 162 471–479 [Google Scholar]
- Szécsi J, Joly C, Bordji K, Varaud E, Cock JM, Dumas C, Bendahmane M (2006) BIGPETALp, a bHLH transcription factor is involved in the control of Arabidopsis petal size. EMBO J 25 3912–3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szlanka T, Haracska L, Kiss I, Deák P, Kurucz E, Andó I, Virágh E, Udvardy A (2003) Deletion of proteasomal subunit S5a/Rpn10/p54 causes lethality, multiple mitotic defects and overexpression of proteasomal genes in Drosophila melanogaster. J Cell Sci 116 1023–1033 [DOI] [PubMed] [Google Scholar]
- Takahashi M, Iwasaki H, Inoue H, Takahashi K (2002) Reverse genetic analysis of the Caenorhabditis elegans 26S proteasome subunits by RNA interference. Biol Chem 383 1263–1266 [DOI] [PubMed] [Google Scholar]
- Tsukaya H (2008) Controlling size in multicellular organs: focus on the leaf. PLoS Biol 6 e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukaya H, Tsuge T, Uchimiya H (1994) The cotyledon: a superior system for studies of leaf development. Planta 195 309–312 [Google Scholar]
- Ueda M, Matsui K, Ishiguro S, Sano R, Wada T, Paponov I, Palme K, Okada K (2004) The HALTED ROOT gene encoding the 26S proteasome subunit RPT2a is essential for the maintenance of Arabidopsis meristems. Development 131 2101–2111 [DOI] [PubMed] [Google Scholar]
- Vale RD (2000) AAA proteins: lords of the ring. J Cell Biol 150 F13–F19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A (2005) Regulated protein degradation. Trends Biochem Sci 30 283–286 [DOI] [PubMed] [Google Scholar]
- Wójcik C, DeMartino GN (2002) Analysis of Drosophila 26S proteasome using RNA interference. J Biol Chem 277 6188–6197 [DOI] [PubMed] [Google Scholar]
- Yang P, Fu H, Walker J, Papa CM, Smalle J, Ju YM, Vierstra RD (2004) Purification of the Arabidopsis 26 S proteasome: biochemical and molecular analyses revealed the presence of multiple isoforms. J Biol Chem 279 6401–6413 [DOI] [PubMed] [Google Scholar]
- Young P, Deveraux Q, Beal RE, Pickart CM, Rechsteiner M (1998) Characterization of two polyubiquitin binding sites in the 26 S protease subunit 5a. J Biol Chem 273 5461–5467 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.