Abstract
Protein phosphatase 1 (PP1) is a eukaryotic serine/threonine protein phosphatase, and mediates diverse cellular processes in animal systems via the association of a catalytic subunit (PP1c) with multiple regulatory subunits that determine the catalytic activity, the subcellular localization, and the substrate specificity. However, no regulatory subunit of PP1 has been identified in plants so far. In this study, we identified inhibitor-3 (Inh3) as a regulatory subunit of PP1 and characterized a functional role of Inh3 in Vicia faba and Arabidopsis (Arabidopsis thaliana). We found Inh3 as one of the proteins interacting with PP1c using a yeast two-hybrid system. Biochemical analyses demonstrated that Arabidopsis Inh3 (AtInh3) bound to PP1c via the RVxF motif of AtInh3, a consensus PP1c-binding sequence both in vitro and in vivo. AtInh3 inhibited the PP1c phosphatase activity in the nanomolar range in vitro. AtInh3 was localized in both the nucleus and cytoplasm, and it colocalized with Arabidopsis PP1c in these compartments. Disruption mutants of AtINH3 delayed the progression of early embryogenesis, arrested embryo development at the globular stage, and eventually caused embryo lethality. Furthermore, reduction of AtINH3 expression by RNA interference led to a decrease in fertility. Transformation of the lethal mutant of inh3 with wild-type AtINH3 restored the phenotype, whereas that with the AtINH3 gene having a mutation in the RVxF motif did not. These results define Inh3 as a regulatory subunit of PP1 in plants and suggest that Inh3 plays a crucial role in early embryogenesis in Arabidopsis.
Reversible protein phosphorylation, which is regulated by the activities of protein kinase and phosphatase, is an important mechanism of the posttranslational regulation of cellular and developmental processes. Protein phosphatase 1 (PP1) is a major member of the PPP family of Ser/Thr protein phosphatases (PP1, PP2A, PP2B, PP4, PP5, PP6, and PP7) and is ubiquitously distributed in higher eukaryotes (Barton et al., 1994; Cohen, 1997, 2002). In animals, PP1 has been shown to regulate diverse cellular processes, such as cell cycle progression, transcription, protein synthesis, carbohydrate metabolism, muscle contraction, and neuronal signaling (Bollen, 2001; Cohen, 2002; Ceulemans and Bollen, 2004).
Recently, we have demonstrated that PP1 functions as a positive regulator in the signaling of blue light-dependent stomatal opening in plants, a process essential for photosynthetic CO2 fixation and transpiration (Takemiya et al., 2006; Shimazaki et al., 2007). Transient expressions of either dominant negative forms of the PP1 catalytic subunit (PP1c) or the mammalian PP1c-specific inhibitor protein in guard cells impaired blue light-specific stomatal opening. However, aside from their role in stomatal opening, little is known about the physiological function of PP1 in plant cells, although a number of pharmacological studies have suggested that PP1 or PP2A is involved in various processes, such as gene expression, phytohormone signaling, regulation of ion channels, and pathogen resistance (Smith and Walker, 1996; Luan, 2003).
PP1 consists of a PP1c and multiple distinct regulatory subunits that specify the catalytic activity, substrate specificity, and subcellular localization of the enzyme (Bollen, 2001; Cohen, 2002). PP1c is an approximately 37-kD protein and is highly conserved throughout eukaryotes, including plants. In most eukaryotes, several isoforms of PP1c with high similarities have been identified, e.g. three in Homo sapiens, at least four in Vicia faba, five in Oryza sativa, and nine in Arabidopsis (Arabidopsis Genome Initiative, 2000; Kerk et al., 2002; International Rice Genome Sequencing Project, 2005; Takemiya et al., 2006). By contrast, the regulatory subunits are multiple and structurally quite different, but most of these regulatory subunits in animals possess a common consensus-binding motif for PP1c, which is referred to as the RVxF motif, with short degenerate sequences (K/R/H/N/S V/I/L X F/W/Y; Egloff et al., 1997; Zhao and Lee, 1997; Barford et al., 1998). This RVxF motif acts as a primary binding site to PP1c and also serves as an anchor for building secondary low-affinity binding sites, which determines the specific function of individual regulatory subunits (Wakula et al., 2003).
In animals, at least 45 genes that encode genuine or putative PP1 regulatory subunits have been identified, and this large set of regulatory subunits forms a variety of distinct holoenzymes with PP1c and enables PP1 to be involved in diverse cellular processes (Bollen, 2001; Cohen, 2002). Surprisingly, however, no regulatory subunits of PP1 have been identified in plants yet (Smith and Walker, 1996; Luan, 2003; DeLong, 2006). Therefore, it is very important to identify and characterize the regulatory subunits of PP1 to elucidate the regulatory mechanisms and physiological functions of plant PP1.
In this study, we identified inhibitor-3 (Inh3) in V. faba and Arabidopsis as a regulatory subunit for PP1. Inh3 interacted specifically with PP1c, inhibited the PP1c catalytic activity, and was localized in both the nucleus and the cytosol. We also demonstrated that Inh3 plays an essential role in embryo development.
RESULTS
Isolation of a Regulatory Subunit of PP1 by Yeast Two-Hybrid Screening
We carried out a yeast two-hybrid screen using VfPP1c-1, a PP1c expressed in V. faba, as bait (Takemiya et al., 2006), and a cDNA expression library constructed from Vicia guard cell protoplasts as prey (Emi et al., 2005). We obtained 20 positive clones from 20 × 106 transformants, and nine of them exhibited overlapping cDNA sequences. A full-length cDNA was obtained by inverse PCR and 3′ RACE. The cDNA contained an open reading frame of 375 bp and encoded a small polypeptide of 124 amino acids with a predicted molecular mass of 13.8 kD (Fig. 1A). We named this novel protein V. faba PP1 Regulatory Subunit1 (VfPRS1).
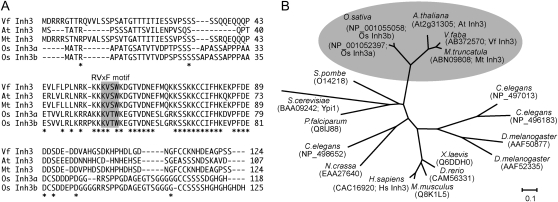
Figure 1.
Sequence alignment of plant Inh3. A, Alignment of the deduced amino acid sequences of plant Inh3 from V. faba (VfInh3), Arabidopsis (AtInh3), M. truncatula (MtInh3), and O. sativa (OsInh3a and OsInh3b). Asterisks show identical amino acids. Dashes indicate gaps to allow for optimal alignment of sequences. The sequences were aligned using the ClustalW program (Thompson et al., 1994). Regions corresponding to the RVxF motif are highlighted with a gray background. B, Phylogenetic analysis of Inh3 proteins in plants (V. faba, Arabidopsis, M. truncatula, and O. sativa), protozoa (Plasmodium falciparum), fungi (Saccharomyces cerevisiae, Schizosaccharomyces pombe, and Neurospora crassa), insects (Drosophila melanogaster), nematode (Caenorhabditis elegans), vertebrates (Danio rerio and Xenopus laevis), and mammals (Mus musculus and H. sapiens). The names and accession numbers for individual proteins are shown. Multiple alignments and bootstrap values with 1,000 replicates for this tree are shown in Supplemental Figures S1 and S2, respectively. The bar represents substitutions per site.
Database searches revealed that this protein is a homolog of human Inh3 and yeast phosphatase inhibitor 1 (Ypi1), which are PP1 regulatory subunits (Zhang et al., 1998; García-Gimeno et al., 2003). The homologous proteins were found in Arabidopsis, Medicago truncatula, and O. sativa (Fig. 1A). They contained conserved regions in the middle of the sequences, where we identified two putative RVxF motifs, KVSW in Vicia, Arabidopsis, and Medicago and KVTW in rice. These proteins are conserved in eukaryotes, including fungi, protozoa, insects, nematode, vertebrates, and mammals (Fig. 1B). Therefore, the PP1c-interactive protein of VfPRS1 was renamed VfInh3.
Inh3 Specifically Interacted with PP1c Both in Vitro and in Vivo
The interaction of VfInh3 with VfPP1c-1 was revealed by the β-galactosidase activity in the yeast two-hybrid system. Strong β-galactosidase activity was observed when VfInh3 was coexpressed with VfPP1c-1, but not with VfPP2Ac-1, a PP2A catalytic subunit from Vicia (Fig. 2A). Human inhibitor-2, a known PP1 regulatory subunit in mammals, interacted with VfPP1c-1 to the same degree (Fig. 2A). To determine whether the interaction between VfInh3 and PP1c is mediated by KVSW (as RVxF motif), we substituted Ala for the Trp-59 in VfInh3. The resulting mutant protein of VfInh3-W59A lost the ability to bind to VfPP1c-1 (Fig. 2A).
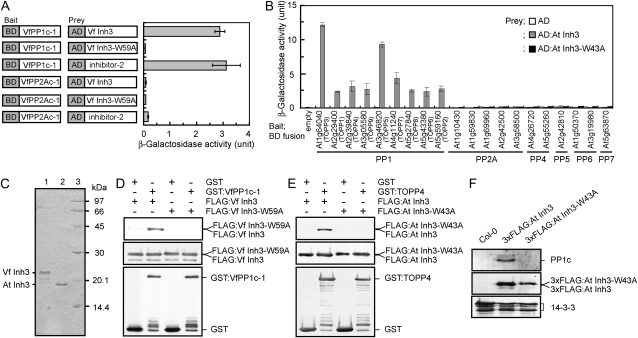
Figure 2.
Binding of Vicia and Arabidopsis Inh3 to PP1c in vitro and in vivo. A, Interaction of VfInh3 with VfPP1c-1 as determined by a yeast two-hybrid system. β-Galactosidase activity was measured in a liquid culture using ONPG as a substrate. Gene constructs for each measurement are indicated on the left. One unit of β-galactosidase activity was defined as the amount that hydrolyzed 1 μmol of ONPG to o-nitrophenol and d-Gal per min per cell at 28°C. Each value represents the means of three independent experiments ± se. BD, GAL4 DNA-binding domain; AD, GAL4 activation domain. B, Specific interaction of AtInh3 with Arabidopsis PP1cs. β-Galactosidase activity was measured as described in A. Members of the Arabidopsis PPP family of protein phosphatase catalytic subunits that includes PP1 (At1g64040, TOPP3; At2g29400, TOPP1; At2g39840, TOPP4; At3g05580, TOPP9; At3g46820, TOPP5; At4g11240, TOPP7; At5g27840, TOPP8; At5g43380, TOPP6; and At5g59160, TOPP2), PP2A (At1g10430, At1g59830, At1g69960, At2g42500, and At3g58500), PP4 (At4g26720 and At5g55260), PP5 (At2g42810), PP6 (At1g50370 and At3g19980), and PP7 (At5g63870) were used as bait. Yeast expressing a bait protein was transformed with AD alone (white bars), AD:AtInh3 (gray bars), or AD:AtInh3-W43A (black bars). Each value represents the means of three independent experiments ± se. C, Coomassie Brilliant Blue staining of recombinant VfInh3 and AtInh3 proteins. Inh3 was expressed in E. coli and purified as described in “Materials and Methods.” The purified proteins (2 μg) were subjected to SDS-PAGE on a 15% acrylamide gel and stained with Coomassie Brilliant Blue. Lane 1, VfInh3; lane 2, AtInh3; lane 3, protein standards. The numbers on the right indicate the molecular masses (kD). D and E, In vitro pull-down assays of Inh3 and PP1c in Vicia (D) and Arabidopsis (E). Recombinant GST or GST:PP1c (GST-VfPP1c-1 and GST-TOPP4; 1 μg protein) was preincubated with glutathione Sepharose beads, and then bacterial crude extracts (20 μg protein) expressing FLAG:Inh3 (FLAG:VfInh3 and FLAG:AtInh3) or FLAG:Inh3 with a single amino acid substitution (FLAG:VfInh3-W59A and FLAG:AtInh3-W43A) were added to the remaining beads. Coprecipitated proteins were immunodetected with antibodies to FLAG (top panel) and to GST (bottom panel). Proteins from bacterial crude extracts were also detected as internal controls (middle panel). F, Coimmunoprecipitation assays of AtInh3 and PP1c in vivo. Total proteins (1 mg) from wild-type (Col-0), 3xFLAG:AtINH3, and 3xFLAG:AtINH3-W43A transgenic plants were used for in vivo coimmunoprecipitation with antibodies against FLAG. Immunoprecipitated proteins were probed with antibodies for PP1c (top panel) and for FLAG (middle panel). The 14-3-3 proteins (40 μg protein) were used as loading controls (bottom panel).
We cloned various catalytic subunits of the PPP family of Ser/Thr protein phosphatases from Arabidopsis and investigated the interaction of these subunits with Arabidopsis Inh3 (AtInh3; At2g31305) using the yeast two-hybrid system (Fig. 2B). AtInh3 bound to all nine PP1c isoforms (Type One Protein Phosphatase [TOPP]) but did not bind to any of the other catalytic subunits (i.e. PP2A, PP4, PP5, PP6, or PP7; Fig. 2B). Substitution of Ala for Trp-43 in the RVxF motif of AtInh3 abolished the binding ability of AtInh3 to PP1cs (Fig. 2B).
The interaction between Inh3 and PP1c was further tested by in vitro pull-down assays using recombinant Inh3 and PP1c (Fig. 2C). That the molecular masses of recombinant VfInh3 (22.3 kD) and AtInh3 (19.0 kD) were higher than the predicted values of 13.8 and 10.5 kD, respectively, was considered to be due to the high hydrophilicity of the two proteins (Zhang et al., 1998). FLAG-tagged Inh3 (FLAG:VfInh3 and FLAG:AtInh3) was coprecipitated with glutathione S-transferase (GST)-tagged-PP1c (GST-VfPP1c-1 and GST-TOPP4) but not with GST alone (Fig. 2, D and E). The disruption of putative RVxF motifs in Inh3 abolished the binding between Inh3 and PP1c.
We also observed a binding between Inh3 and PP1c in vivo. Wild-type Columbia-0 (Col-0) plants were transformed with a full-length AtINH3 or AtINH3-W43A fused with a 3xFLAG under a native promoter. The PP1c was coimmunoprecipitated with 3xFLAG:AtInh3 but not with 3xFLAG:AtInh3-W43A by anti-FLAG antibodies (Fig. 2F). Based on these results in vitro and in vivo, we concluded that Inh3 functions as a PP1 regulatory subunit in plant cells.
Inh3 Inhibited PP1c Catalytic Activities
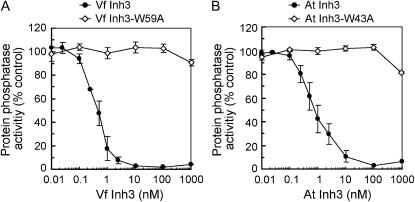
We investigated the effect of Inh3 on PP1c catalytic activities using a [32P]labeled myelin basic protein as a substrate. Both VfInh3 and AtInh3 strongly inhibited the dephosphorylation activities of PP1c in a concentration-dependent manner, with half-inhibitory concentrations (IC50) of 0.50 and 0.75 nm for VfInh3 and AtInh3, respectively (Fig. 3). When the same assay was performed using VfInh3-W59A and AtInh3-W43A, which lost the binding ability to PP1c (Fig. 2), the dephosphorylation activities were not greatly affected by these mutant proteins (Fig. 3). These results suggest that plant Inh3 functions as an activity modulator of PP1c.
Figure 3.
Inhibition of PP1c activity by plant Inh3. A and B, Phosphatase activities were measured using [32P]labeled myelin basic protein as a substrate in the presence or absence of the indicated concentrations of Inh3 protein. Phosphatase activities were expressed as percentages of the values of VfPP1c-1 (A) and TOPP4 (B) measured without inh3 protein. Error bars represent the means ± se (n = 3). The full activities correspond to 63,895 ± 1,889 and 70,078 ± 789 units/mg protein (± se, n = 3) for VfPP1c-1 and TOPP4, respectively. One unit represents the release of 1 nmol phosphate per minute at 30°C.
Inh3 Was Localized in the Nucleus and the Cytoplasm
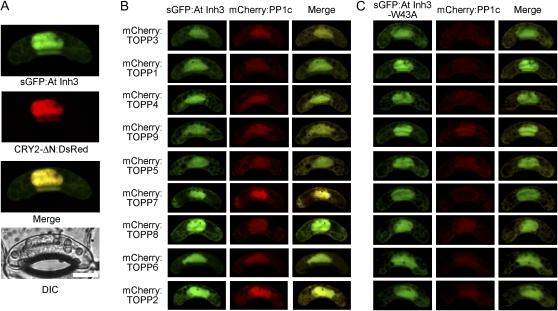
We next studied the subcellular localization of Inh3 by transient expression of AtInh3 fused to a green fluorescent protein (sGFP:AtINH3) in Vicia guard cells after particle bombardment as well as the coexpression of the nuclear localization site of cryptochrome 2 (CRY2) fused to a red fluorescent protein (CRY2-ΔN:DsRed; Pollmann et al., 2006). sGFP:AtInh3 was localized in both the nucleus and the cytoplasm (Fig. 4A).
Figure 4.
Subcellular localization of plant Inh3. A, Gene constructs encoding sGFP:AtINH3 and CRY2-ΔN:DsRed were cotransformed into Vicia guard cells by particle bombardment, and fluorescent images were obtained by confocal laser-scanning microscopy. The individual fluorescent image, merged image, and corresponding differential interference contrast (DIC) image are presented. Bar = 10 μm. B and C, sGFP:AtINH3 (B) or sGFP:AtINH3-W43A (C) were cotransformed with the mCherry fusion Arabidopsis PP1c (mCherry:TOPP) in Vicia guard cells.
If AtInh3 binds to PP1c in vivo, these two proteins should be colocalized. To examine this, nine PP1c isoforms of Arabidopsis were fused to mCherry (mCherry:TOPP) and transiently coexpressed with AtInh3 or AtInh3-W43A. All nine PP1c isoforms were colocalized with AtInh3 in both the nucleus and cytoplasm (Fig. 4B). In contrast, the efficient nuclear localization of PP1c was diminished when PP1c was coexpressed with AtInh3-W43A, although AtInh3-W43A was localized in both the nucleus and the cytoplasm (Fig. 4C). These results support an in vivo interaction of AtInh3 with PP1c via the RVxF motif.
Tissue-Specific Expression of Plant INH3
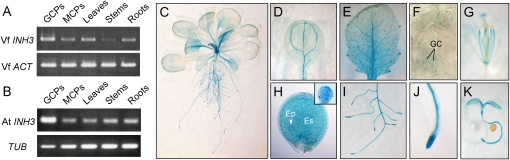
Expression of Vicia and Arabidopsis INH3 genes was determined by reverse transcription (RT)-PCR in guard cell protoplasts, mesophyll cell protoplasts, leaves, stems, and roots. Transcripts of VfINH3 and AtINH3 were found in all cell/tissue types and particularly in guard cell protoplasts (Fig. 5, A and B).
Figure 5.
Expression of plant INH3 in various tissues. A and B, Expression of VfINH3 (A) or AtINH3 (B) mRNA determined by RT-PCR in guard cell protoplasts (GCPs), mesophyll cell protoplasts (MCPs), leaves, stems, and roots from 5-week-old V. faba and Arabidopsis. Vicia ACT and Arabidopsis TUB were used as standards for A and B, respectively. C to K, Expression of AtINH3Pro:GUS in Arabidopsis transgenic plants. Results are shown for a 3-week-old whole seedling (C), cotyledon leaf (D), young leaf (E), abaxial epidermis (F), 6-week-old flower (G), developing seed (H), root tissue (I), root tip (J), and 3-d-old seedling (K). Inset in H represents the globular stage embryo removed from the developing seed. GC, guard cells; Ep, embryo proper; Es, endosperm.
We generated transgenic plants expressing the GUS reporter gene driven by the AtINH3 promoter (the 2.2-kb region upstream from the initiator ATG). Blue staining was detected in several organs, including roots and aerial parts (Fig. 5C). In aerial parts, the staining was found in photosynthetic and reproductive organs and was particularly strong in trichomes and the veins of cotyledons and leaves (Fig. 5, D and E). In agreement with the results of the RT-PCR analysis, the staining was found in guard cells (Fig. 5F). The staining was also found in the filaments and stigma of flowers (Fig. 5G), in both the embryo proper (globular stage) and endosperm of developing seeds (Fig. 5H) and in the root tips and steles of root (Fig. 5, I and J). Finally, the staining was visible in young seedlings (Fig. 5K).
Isolation and Characterization of T-DNA Insertion Mutants of AtINH3
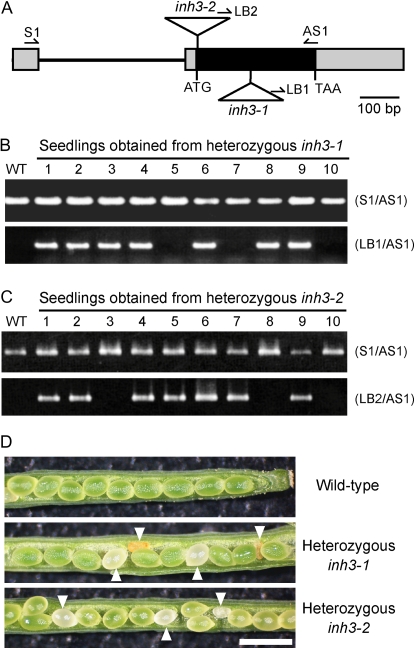
Since the expression of AtINH3 was observed preferentially in guard cells, we initially attempted to characterize the physiological function of Inh3 in guard cells using Arabidopsis T-DNA insertion lines. We obtained two T-DNA insertion lines from the SALK and SAIL collections and designated the corresponding alleles inh3-1 (SALK_044593) and inh3-2 (SAIL_806_C02), respectively. The insertions were located in the second exon of AtINH3, 146 and 1 bp downstream of the ATG initiation codon in inh3-1 and inh3-2, respectively (Fig. 6A). A PCR-based screen using gene-specific primers and the T-DNA left border primer revealed that all the plants from the T3 or T4 generation were the heterozygous or wild-type strains, and there were no homozygous plants (Fig. 6, B and C). All the heterozygous plants were morphologically similar to the wild-type plants and did not show any impairment in stomatal responses (data not shown). Segregation analyses of >100 self-pollinated heterozygous (parent) plants revealed that there was a 2:1 ratio of T-DNA heterozygous plants to wild-type plants (Table I), suggesting that a homozygous insertion confers the lethality and/or reduced gametophytic transmission. To test whether the insertion was transmitted normally through both male and female gametophytes, we reciprocally crossed inh3-1 with the wild type and screened F1 plants for the T-DNA insertion by PCR. Transmission of the T-DNA insertion occurred through both gametophytes (Table I).
Figure 6.
Functional characterization of the AtINH3 disruption mutants. A, Positions of T-DNA insertions in Arabidopsis inh3 mutants. Boxes indicate exons, with black representing the coding sequence and gray the untranslated region. Lines represent introns. Triangles indicate the T-DNA insertion sites of inh3-1 and inh3-2. Arrows represent the gene-specific primers (S1 and AS1) and the T-DNA border primers (LB1 and LB2) used for PCR-based screens. B and C, PCR-based screens of the genotypes in seedlings from heterozygous inh3-1 (B) and inh3-2 (C). PCR was conducted with genomic DNA using a pair of gene-specific primers (top panel) or by the combination of the T-DNA left border primer and the gene-specific primer (bottom panel). The first lane represents the results for the wild-type (WT) control. D, Morphology of seeds from self-pollinated wild-type, heterozygous inh3-1, and heterozygous inh3-2 plants. The aborted seeds are marked with arrowheads. Bar = 1 mm.
Table I.
Segregation ratio of the T-DNA insertion in inh3 mutants
| Cross | Insertion+ | Insertion− | Total |
|---|---|---|---|
| inh3-1 selfed | 66.0% | 34.0% | n = 100 |
| inh3-2 selfed | 66.7% | 33.3% | n = 108 |
| inh3-1/− × wild type | 41.2% | 58.8% | n = 97 |
| Wild type × inh3-1/− | 50.0% | 50.0% | n = 98 |
In Arabidopsis, seeds within a single silique develop at a similar rate. If the homozygous insertion in AtINH3 causes embryo lethality, the self-pollinated seeds from heterozygous inh3 mutants would be expected to include aborted seeds. In wild-type siliques, all seeds developed normally and were green in color (Fig. 6D). However, the heterozygous lines of inh3-1 and inh3-2 produced 24.6% and 22.2% white or brown shriveled seeds, respectively (Table II), and the segregation ratio of normal to abnormal seeds was approximately 3:1, suggesting that these aborted seeds might have been the missing homozygous mutants. Furthermore, the mature dried seeds from wild-type plants exhibited a light brown color and were uniform in shape, whereas the seeds from heterozygous inh3 mutants contained a number of shriveled and dark brown seeds (Supplemental Fig. S3). Taken together, these results suggest that the disruption of AtINH3 causes embryo lethality in Arabidopsis. We thus focused our attention on the role of Inh3 in early embryogenesis.
Table II.
Percentages of aborted seeds in wild-type and inh3 mutants
| Allele | Normal Seeds | Aborted Seeds | Total |
|---|---|---|---|
| Wild type | 96.5% | 3.5% | n = 85 |
| inh3-1 selfed | 75.4% | 24.6% | n = 135 |
| inh3-2 selfed | 77.8% | 22.2% | n = 99 |
AtINH3, But Not AtINH3-W43A, Complemented the Embryo-Lethal Phenotype of the inh3 Mutant
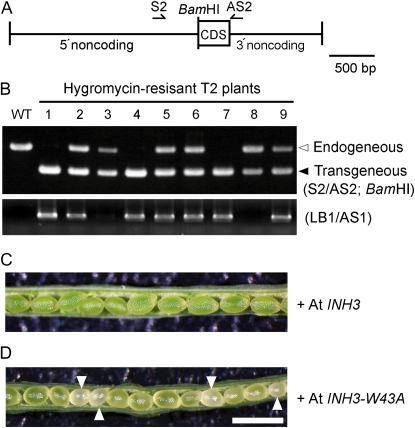
We complemented the inh3-1 mutant to demonstrate homozygous lethality using a construct containing both the full-length AtINH3 and the hygromycin resistance gene as a marker (Fig. 7A). We selected T1 plants that contained the heterozygous inh3-1 T-DNA by PCR-based screen (data not shown). The complementation was tested on the hygromycin-resistant T2 plants by PCR using sense (S2) and antisense (AS2) primers, which amplified both endogenous and transgenous AtINH3. The individual PCR products were distinguished by digesting them at the BamHI site that was introduced in the transgene. Among the nine randomly selected hygromycin-resistant T2 plants, three did not possess the endogenous AtINH3 (Fig. 7B, lanes 1, 4, and 7); thus, these three lines were homozygous for the inh3-1 allele. The rest were either heterozygous inh3-1 plants having both endogenous and transgenous AtINH3 (Fig. 7B, lanes 2, 5, 6, and 9) or wild-type plants having the transgene (Fig. 7B, lanes 3 and 8). The siliques of heterozygous inh3-1 plants possessing the transgenous AtINH3 produced almost no white or shriveled brown seeds, and nearly all the seeds exhibited fully developed and uniformly sized embryos (Fig. 7C). Therefore, the embryo-lethal phenotype of the homozygous inh3-1 mutant was fully complemented by the AtINH3 transgene.
Figure 7.
Complementation of the inh3-1 mutation with AtINH3 genomic DNA. A, Diagram of the AtINH3 transgene for complementation analysis. The full-length AtINH3 coding sequence (CDS) was fused with the native promoter and terminator of AtINH3 and introduced into heterozygous inh3-1 plants by gene transfer. The BamHI digestion site was inserted after the initiation codon to discriminate transgenous AtINH3 from the endogenous one. Arrows show the gene-specific primers (S2 and AS2) used to amplify the genomic region of AtINH3. B, PCR-based screens of the genotypes in hygromycin-resistant T2 seedlings obtained from T1 plants having a heterozygous inh3-1 mutation. A partial AtINH3 sequence was amplified using a pair of gene-specific primers and digested with BamHI (top panel). The T-DNA insertion for the inh3-1 mutation was verified by PCR using the T-DNA left border primer and the gene-specific primer (bottom panel). The first lane represents the results for the wild-type (WT) control. C and D, Morphology of seeds from self-pollinated hygromycin-resistant T2 plants having a heterozygous inh3-1 mutation. These transformants express the wild-type AtINH3 (C) and mutant AtINH3-W43A (D), respectively. Bar = 1 mm.
The complementation of the inh3-1 embryo lethality by the AtINH3 gene was expected to be due to restoration of the regulation of PP1c via Inh3. It is also possible that Inh3 itself has an essential role in embryogenesis that is independent of PP1c regulation. To test this alternative, we introduced AtINH3-W43A, which lost the ability to bind to PP1c (Fig. 2), into the inh3-1 mutant and inspected the embryo phenotype. We randomly selected nine individual T2 lines and found that there was no homozygous mutant for inh3-1 (data not shown) and that all these heterozygous inh3-1 plants possessing the transgenous AtINH3-W43A produced the aborted seeds (Fig. 7D). The results may support the possibility that the PP1c-Inh3 complex has an important role in early embryogenesis.
Embryo Development Was Arrested at the Globular Stage in the inh3 Mutant
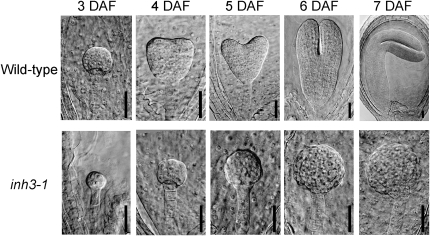
We next compared the development of the wild-type plants with that of the mutants during seed maturation. Siliques on 4 to 7 d after flowering (DAF) from wild-type and heterozygous inh3-1 plants were dissected, and the cleared seeds were inspected by light microscopy. The wild-type embryos developed from the globular to the bent cotyledon stage (Table III). The majority of the embryos were in the early and late heart developmental stages at 4 DAF, the late heart and torpedo stages at 5 DAF, the torpedo stage at 6 DAF, and the bent cotyledon stage at 7 DAF. In heterozygous inh3-1 embryos, approximately 25% of the plants were arrested at the octant-dermatogen and globular stages, whereas the rest of the embryos developed similarly to the wild type (Table III). The seeds of arrested embryos displayed a white color in the early developmental stages due to the depletion of green embryos and later degenerated to show a dark brown color. Figure 8 presents images of defective embryos that appeared in the silique of heterozygous inh3-1 and normal embryos in the wild-type plants. In heterozygous inh3-1, a quarter of embryos grew much slower than the rest and remained in the eight-cell stage at 3 DAF and the globular stage at 4 DAF, whereas the wild-type embryos were in the globular stage at 3 DAF and the early heart stage at 4 DAF. Although embryo development was delayed in the mutant in these stages, embryo patterning appeared not to be largely changed. From 5 to 6 DAF, while the defective embryos continued to grow, they never developed beyond the globular stage and began to show irregular surfaces. At 7 DAF, the defective embryos stopped developing altogether.
Table III.
Developmental progression of embryogenesis in wild-type and heterozygous inh3-1 plants
| DAF | Octant-Dematogen Stage | Globular Stage | Early Heart Stage | Late Heart Stage | Torpedo Stage | Bent Cotyledon Stage | Total |
|---|---|---|---|---|---|---|---|
| Wild type | |||||||
| 4 | – | 7.4% | 59.6% | 28.7% | 4.3% | – | n = 94 |
| 5 | – | 5.2% | 15.8% | 31.6% | 47.4% | – | n = 114 |
| 6 | – | 6.9% | 3.4% | 19.3% | 65.5% | 4.8% | n = 145 |
| 7 | – | 5.6% | 1.2% | 3.1% | 14.9% | 75.2% | n = 161 |
| inh3-1 | |||||||
| 4 | 4.4% | 25.7% | 64.0% | 5.9% | – | – | n = 136 |
| 5 | 1.9% | 21.7% | 15.9% | 25.5% | 35.0% | – | n = 157 |
| 6 | 1.0% | 20.5% | 3.6% | 11.3% | 56.4% | 7.2% | n = 195 |
| 7 | 0.7% | 22.2% | 0.7% | 5.9% | 19.6% | 51.0% | n = 153 |
Figure 8.
Embryos of wild-type and inh3-1 defective mutants in various developmental stages of seeds. Each seed was obtained from immature siliques at 3 to 7 DAF. Cleared seeds were inspected by a microscope equipped with differential interference contrast optics. Bars = 30 μm.
Suppression of AtINH3 by RNA Interference Resulted in Reduced Fertility
To further elucidate the function of Inh3, we generated transgenic plants expressing AtINH3-RNA interference (RNAi) constructs. A full-length coding region of the AtINH3 was cloned in both a sense and an antisense orientation and was used for transforming Arabidopsis Col-0 plants. We obtained only three transformants, probably due to the embryo lethality in the plants. All these transformants displayed normal vegetative growth and development, but they also exhibited reduced fertility. The siliques of RNAi plants were much shorter than those of wild-type plants and produced a reduced number of seeds (Supplemental Fig. S4A). In accord with the impaired phenotypes, the AtINH3 transcript was detected in immature siliques of wild-type plants, whereas the expression levels were greatly reduced in the AtINH3-RNAi lines (Supplemental Fig. S4B).
DISCUSSION
Identification of Inh3 as a Regulatory Subunit for PP1
In animals, the function of PP1 is determined by PP1c activity through the control of regulatory subunits, and a large number of regulatory subunits for PP1c have been identified (Bollen, 2001; Cohen, 2002; Ceulemans and Bollen, 2004). Similar regulatory subunits have been suggested to be present in plant cells. For example, Stubbs et al. (2001) identified the proteins that bound to PP1c by protein blot analysis in Arabidopsis suspension cells. MacKintosh et al. (1991) reported the complex formation between PP1c and unknown proteins in carrot (Daucus carota) cells. Recent database searches have identified putative Arabidopsis homologs of animal PP1 regulatory subunits (Farkas et al., 2007). However, no conclusive evidence for the existence of plant PP1 regulatory subunits has been obtained so far. In this study, we identified genes that encode PP1c-binding proteins from V. faba, and one of these genes was Inh3. Disruptions of the putative RVxF motif of KVSW in both VfInh3 and AtInh3 by substitution of Ala for Trp abolished the interactions between PP1c and Inh3 in vitro and in vivo (Fig. 2), indicating that the identical mechanisms of regulatory subunits for PP1c operate in plant cells. The results suggest that Inh3 acts as a PP1 regulatory subunit, and the mechanisms by which PP1c functions are controlled are highly conserved in eukaryotes.
Biochemical and Cell Biological Properties of Inh3
We investigated the biochemical properties of plant Inh3 and elucidated that Inh3 regulates the catalytic activity of PP1c. Human Inh3 was initially isolated by its physical interaction with PP1c, and biochemical analysis has shown that the recombinant protein is a heat-stable inhibitor protein of PP1c (Zhang et al., 1998). Subsequently, the yeast Inh3 homolog Ypi1 was identified and shown to possess an inhibitory effect on the PP1c activity (García-Gimeno et al., 2003). In accord with these findings, our results demonstrated that plant Inh3 inhibited the catalytic activity of PP1c at subnanomolar ranges (Fig. 3), and this inhibitory potency was comparable to that of human Inh3 (Zhang et al., 1998). Furthermore, the substitution of Ala for Trp in the RVxF motif of Inh3 eliminated the inhibitory action on PP1c (Fig. 3), supporting the notion that one of the roles of AtInh3 is to regulate the PP1c activity. Since the RVxF motif functions as an initial binding site for PP1c and does not affect the PP1c catalytic activity (Wakula et al., 2003), the secondary low affinity binding of AtInh3 to PP1c, which is not mediated by the RVxF motif, might bring the conformational changes of PP1c and inhibit its catalytic activity.
In animals, a number of PP1 regulatory subunits that regulate PP1c activity (i.e. other than Inh3) have been identified. Heat-stable inhibitory activities toward PP1c have also been detected in the extracts of Arabidopsis and Zea mays root (Lin et al., 1998; Stubbs et al., 2001). The existence of a number of activity modulator proteins that interact with PP1c suggests that PP1c activity is strictly controlled in vivo. Some activity modulator proteins are regulated by a reversible phosphorylation. DARPP-32 and inhibitor-1, for example, inhibit the PP1c activity only when they are phosphorylated by PKA (Shenolikar and Nairn, 1991; Wang et al., 1995). Inhibitor-2 inhibits in its dephosphorylated form and activates the PP1c activity via the phosphorylation by Cdk5 and glycogen synthase kinase-3 (Woodgett and Cohen, 1984; Agarwal-Mawal and Paudel, 2001). Interestingly, the plant Inh3 identified in this study possesses several putative phosphorylation sites (data not shown). Thus, Inh3 may be regulated by a reversible phosphorylation. Further studies will be needed to elucidate the regulatory mechanism for the PP1c-Inh3 complex in plants.
Studies investigating human Inh3 by immunostaining and the GFP fusion system have shown that Inh3 is localized in the nucleoli and the centrosomes (Huang et al., 2005) and further identified two basic clusters in the N terminus (RKRK) and the C terminus (HRKGRRR) of human Inh3 that function as a nuclear localization signal and a nucleolar targeting signal, respectively. In contrast, AtInh3 does not appear to have a basic cluster acting as a nucleolar targeting signal and has only the putative nuclear localization signal from amino acid residues 37 to 60 as predicted by the PredictNLS program (Cokol et al., 2000). In accord with this, this study suggested that AtInh3 was localized in the nucleus and cytoplasm (Fig. 4A), and this localization pattern of AtInh3 was consistent with that of yeast Ypi1 (Pedelini et al., 2007). Furthermore, the PP1c isoforms were colocalized with AtInh3 in all these compartments, but the nuclear localization of PP1cs was eliminated when the PP1cs were coexpressed with mutant AtInh3-W43A (Fig. 4, B and C). This result can be simply interpreted as indicating that AtInh3 plays an essential role in targeting the PP1c to the nucleus, as suggested for Ypi1 (Bharucha et al., 2008). However, the possibility cannot be ruled out that AtInh3 stabilizes the PP1c by forming the AtInh3-PP1c complex and that the complex efficiently accumulates in the nucleus.
Functional Role of Inh3 in Plants
Inh3 was initially discovered as an inhibitor of PP1c catalytic activity in humans (Zhang et al., 1998) and was later shown to inhibit the same activity in yeast (García-Gimeno et al., 2003); however, the precise functions of Inh3 in these organisms have remained unclear. Therefore, we attempted to elucidate the function of Inh3 in plant cells. We found that the self-pollinated seeds from heterozygous inh3 mutants with T-DNA insertion were aborted at a rate of 25% in their immature siliques, and no homozygous plants were obtained (Fig. 6D; Table II). The aborted seeds produced embryos that remained at an earlier developmental stage. The majority of these seeds were arrested at the globular stage, whereas the wild-type seeds developed from the early heart to the bent cotyledon stage in a silique (Table III; Fig. 8). Furthermore, RNAi-mediated suppression of AtINH3 gene expression inhibited the seed maturation (Supplemental Fig. S4). Thus, the disruption of the AtINH3 gene in plant cells appeared to cause embryo lethality. In support of these results, transformation of the heterozygous inh3 mutant by the wild-type AtINH3 gene rescued the embryo lethality phenotype (Fig. 7C). Taken together, these results indicate that the plant Inh3 plays a crucial role in early embryogenesis. We note here that Inh3 may exert its function as a regulatory subunit of PP1, but direct evidence will be required to confirm this.
Molecular Mechanisms of the Function of Inh3 in Embryogenesis
An important finding in our study was that the lethal phenotype of the inh3 mutant was rescued by wild-type AtINH3, but not by the mutant AtINH3-W43A, which lost its ability to bind and inhibit the PP1c. This result suggests that Inh3 exerts its function by binding to PP1c in embryogenesis, and it is also possible that the PP1c catalytic activity may be regulated by Inh3 via the binding. We tried to measure the PP1c activity in the wild-type and inh3 mutant embryos, but we could not determine the phosphatase activities because of the scarcity of mutant embryo preparations. We also identified the isoforms of Arabidopsis PP1c expressed during early embryogenesis and found that eight of nine PP1c isoforms were expressed in young siliques (Supplemental Fig. S5). This made it difficult to identify the functional role of specific isoforms of PP1c in embryogenesis. Further studies will be needed to determine the regulatory role of Inh3 in embryogenesis.
What, then, is the role of the PP1c-Inh3 complex in early embryogenesis? Both the inh3 mutant and the wild-type showed similar early embryo pattern formation, but then embryo development in the mutant was retarded relative to that in the wild type, and the former ultimately ceased at the globular stage (Table III; Fig. 8). This inh3 embryo showed a phenotype similar to that of the mutant, i.e. a defect in the control of cell division and the cell cycle, similar to that observed in rpn1a (Brukhin et al., 2005). In fact, PP1 and/or PP2A have been proposed to play a role in mitosis in several plant species based on studies using pharmacological methods. In alfalfa (Medicago sativa) suspension cells, endothall, an inhibitor of PP1 and PP2A, increased the appearance of hypercondensed early and late prophase chromosomes that could not enter metaphase (Ayaydin et al., 2000). Treatment of tobacco (Nicotiana tabacum) suspension cells with a similar inhibitor, okadaic acid, resulted in a similar chromosome hypercondensation and blockade of the cell cycle at G2 phase (Zhang et al., 1992). Therefore, the PP1c-Inh3 complex might regulate the chromatin state and mitotic progression.
Furthermore, recent characterizations of yeast Ypi1 led us to speculate that AtInh3 plays a role in the regulation of mitosis. Deletion of YPI1 is lethal in yeast, and conditional suppression of YPI1 results in the inhibition of cell growth of mid-mitosis (García-Gimeno et al., 2003; Pedelini et al., 2007). Moreover, Ypi1 has been shown to interact with Sds22, a PP1 regulatory subunit required for mitosis, and to form a ternary complex with PP1c (Pedelini et al., 2007; Bharucha et al., 2008). Since the suppression of Ypi1 mimics the phenotype caused by the inactivation of Sds22, Ypi1 and Sds22 are expected to function cooperatively in regulating the PP1c activity (Pedelini et al., 2007). Considering the similar biochemical properties of Inh3 among eukaryotes, it is conceivable that plant Inh3 may regulate the function of PP1c required for mitosis. Identification of the substrate and the binding proteins of the PP1c-Inh3 complex will be helpful for our understanding of the function of PP1 in plant embryogenesis.
MATERIALS AND METHODS
Plant Materials
Plants of Vicia faba ‘Ryosai Issun’ were cultured hydroponically in a greenhouse as described previously (Shimazaki et al., 1992). Arabidopsis (Arabidopsis thaliana) plants (ecotype Col) were grown from seeds at 22°C ± 3°C with a photoperiod of 14 h. T-DNA insertion lines SALK_044593 (inh3-1) and SAIL_806_C02 (inh3-2) were obtained from the SALK collection via the Arabidopsis Biological Resource Center (ABRC) at Ohio State University and from the SAIL collection (Sessions et al., 2002; Alonso et al., 2003). Information on the insertional mutant was obtained from the SIGnAL website at http://signal.salk.edu.
Yeast Two-Hybrid Screen
The yeast two-hybrid screen was performed using the MATCHMAKER Two-Hybrid System 2 (CLONTECH). The full-length VfPP1c-1 was fused in-frame to the GAL4 DNA-binding domain in the bait plasmid pAS2-1 (CLONTECH). A cDNA library was constructed from Vicia guard cell protoplasts into the phagemid vector pAD-GAL4-2.1 (Stratagene) as described previously (Emi et al., 2005). Yeast strain Y190 was transformed with the bait construct and subsequently with the cDNA library phagemid constructs. Positive transformants (His+ and LacZ+) were selected in the presence of 25 mm 3-aminotriazole to eliminate false-positive results. Phagemid vectors containing the cDNA clones were recovered by back transformation into Escherichia coli strain JM109.
Phylogenetic Analysis
Full-length amino acid sequences were used for phylogenetic analyses. Sequence alignment was performed using the ClustalW program with default parameters (gap opening penalty, 10.00; gap extension penalty, 0.20; delay divergent cutoff, 30%; Supplemental Fig. S1). Phylogenetic analysis was done using MEGA software, version 4, with the neighbor-joining method (Tamura et al., 2007). All positions containing gaps and missing data were eliminated from the data set. The number of bootstrap replicates was 1,000.
Determination of Full-Length VfINH3
3′ RACE was done using a GeneRacer kit (Invitrogen) with two primers (5′-TCTCGTGGAAAGATGGCACTGTGGA-3′ and 5′-TGAAGCTGGCCCGAGCAGTTAGGT-3′). Full-length VfINH3 was obtained by inverse PCR with circular cDNA prepared from the first-strand cDNA using T4 polynucleotide kinase and T4 RNA ligase (TaKaRa). The first PCR was carried out with the primers 5′-GCAAGAATCACGATGAAGCTG-3′ and 5′-CTGCATGAACTCATTGTCCAC-3′, and the nested PCR was performed with the primers 5′-CCGAGCAGTTAGGTTCTGATTG-3′ and 5′-AGTGCCATCTTTCCACGAGAC-3′. The resulting PCR products were subcloned into a pCR4-TOPO vector (Invitrogen). Sequences were determined from both strands of the cDNA (ABI PRISM 3100; Applied Biosystems).
Quantitative β-Galactosidase Assay
The bait and prey constructs were prepared using pAS2-1 and pACT2 (CLONTECH) and transformed into the yeast strain Y190. The β-galactosidase liquid culture assay was carried out using o-nitrophenyl β-d-galactopyranoside (ONPG) as a substrate. Briefly, overnight cultures grown at 30°C in synthetic complete media lacking Trp and Leu were centrifuged at 12,000g for 10 s, and the resulting pellet was washed with Z buffer (pH 7.0) containing 60 mm Na2HPO4, 40 mm NaH2PO4, 10 mm KCl, and 1 mm MgSO4. The samples were resuspended in Z buffer and incubated for 5 min at 28°C. Thirty-two microliters of chloroform and 56 μL of 0.1% (w/v) SDS were added to 800 μL of the samples and further incubated for 5 min at 28°C. The assays were started by adding ONPG 160 μL of 0.4% (w/v) ONPG, and the reaction was performed for 120 min at 28°C. The reactions were stopped by adding 400 μL of 1 m Na2CO3. The samples were centrifuged at 10,000g for 10 min, and the OD420 of the supernatant was measured.
Point Mutation of the VfInh3 and AtInh3 Amino Acid Sequences
Site-directed mutagenesis was performed using a QuikChange site-directed mutagenesis kit (Stratagene). The primer sets used were 5′-AGAAGAAGAAGGTCTCGGCGAAAGATGGCACTGTGG-3′ and 5′-CCACAGTGCCATCTTTCGCCGAGACCTTCTTCTTCT-3′ for VfINH3-W59A, and 5′-GGAAGAAGAAGAAAGTTTCAGCGAAAGATGGGACTGTAGACA-3′ and 5′-TGTCTACAGTCCCATCTTTCGCTGAAACTTTCTTCTTCTTCC-3′ for AtINH3-W43A.
Expression and Purification of Recombinant Inh3 in E. coli
Full-length VfINH3 and AtINH3 were fused to a polyhistidine tag and thrombin cleavage sequence and subcloned into pFLAG-MAC expression vector (Sigma-Aldrich). E. coli strain BL21 was used as the host for protein expression. Cell growth and induction conditions were as described previously (Zhang et al., 1998). Cells were harvested and resuspended in 50 mm sodium phosphate (pH 7.4), 0.3 m NaCl, 10 mm imidazole, 2 mm phenylmethane sulfonyl fluoride, and 200 μm leupeptin and then disrupted by sonication. The lysate obtained by sonication was heated at 95°C for 20 min and centrifuged at 10,000g for 30 min. The supernatant was mixed with HIS-Select Nickel Affinity Gel (Sigma-Aldrich) and incubated for 1 h at 4°C. The nickel affinity gel was washed three times with extraction buffer and once with 1× PBS buffer. To remove the polyhistidine tag from Inh3, thrombin (GE Healthcare) was added and incubated for 16 h at room temperature. The suspension was centrifuged at 10,000g for 1 min, and the supernatant was saved. The recombinant protein concentrations were determined by Bradford assays using BSA as a standard (Bradford, 1976).
Preparation of Recombinant PP1c and Determination of PP1c Activity
Full-length VfPP1c-1 and TOPP4 were subcloned into pGEX-2T (GE Healthcare), and the recombinant proteins were prepared as described previously (Takemiya et al., 2006). Ser/Thr phosphatase activity was assayed using [32P]labeled myelin basic protein (MyBP) as a substrate according to the manufacturer's protocol (New England Biolabs). The assay buffer contained 50 mm Tris-HCl (pH 7.0), 0.1 mm Na2EDTA, 5 mm dithiothreitol, 0.01% (w/v) Brij 35, 10 mm [32P]MyBP, purified PP1c (0.25 ng), and various concentrations of Inh3 with a molar ratio of 1:0.074 to 7,400 (PP1c:Inh3). PP1c was preincubated with Inh3 for 30 min on ice, followed by the incubation for 5 min at 30°C, and the reaction was started by adding [32P]labeled MyBP. The final reaction volume was 50 μL. The reaction was terminated by adding cold trichloroacetic acid after 10 min, and the amount of [32P]phosphate released was measured in the supernatant obtained by centrifugation. All assays were performed within the linear ranges of activities (<20% substrate dephosphorylation).
In Vitro Pull-Down Assay
Pull-down assays were carried out using recombinant PP1c and Inh3. One microgram of GST fusion proteins (GST:VfPP1c-1 and GST:TOPP4) was allowed to bind to glutathione Sepharose 4B (GE Healthcare) for 1 h at 4°C with gentle shaking. The beads were incubated with E. coli lysate (20 μg protein) containing FLAG:VfInh3, FLAG:VfInh3-W59A, FLAG:AtInh3, or FLAG:AtInh3-W43A for 1 h at 4°C. The proteins on beads were solubilized and subjected to SDS-PAGE on 12.5% acrylamide gel. The proteins were immunodetected by anti-GST polyclonal (GE Healthcare) or anti-FLAG (Sigma-Aldrich) monoclonal antibodies.
Coimmunoprecipitation Assay
The AtINH3 promoter region (2,151 bp) including the start codon was amplified by PCR from Arabidopsis genomic DNA using the primer pair of 5′-CCCAAGCTTAAGAACCAGAAAAATTAAACATTTACC-3′ and 5′-CGGGATCCCATAGCTGAAGATTAGCTTTCAAAATCTG-3′. The fragment was subcloned into the HindIII and BamHI sites of pCAMBIA1300 binary vector (CAMBIA) with the nopaline synthase terminator. Full-length AtINH3 or AtINH3-W43A cDNA was also amplified using the primers 5′-CCCAAGCTTATGAGCACAGCAACAAGG-3′ and 5′-CCCAAGCTTGGATCCTTAGTCAACGGCTTTAGAATC-3′ and cloned into a pCRII vector (Invitrogen) containing the 3xFLAG sequence. The resulting 3xFLAG:AtINH3 fragment was introduced into the BamHI site of the pCAMBIA1300-AtINH3pro vector. The construct was transformed into Arabidopsis Col-0 plants using an Agrobacterium tumefaciens-mediated method.
Total proteins of 3-week-old Arabidopsis Col-0 and transgenic plants were extracted by homogenizing the seedlings (0.4 g) in extraction buffer containing 50 mm MOPS-KOH (pH 7.5), 2.5 mm EDTA, 100 mm NaCl, 2 mm phenylmethane sulfonyl fluoride, and 20 μm leupeptin. Triton X-100 was added to the extracts (1 mg protein) at 1% (w/v) and incubated at 4°C for 1 h. After centrifugation at 10,000g for 10 min, the supernatants were mixed with anti-FLAG M2 Agarose Affinity Gel (Sigma-Aldrich) and incubated at 4°C for 2 h. After washing four times, the proteins were eluted with 3xFLAG peptide (Sigma-Aldrich), solubilized, and subjected to SDS-PAGE on a 12.5% acrylamide gel. Proteins immunoprecipitated by anti-FLAG antibodies were visualized using the antibodies against PP1c, which were raised against the full-length TOPP4 prepared in E. coli. The 14-3-3 protein was immunodetected in the same supernatants as described previously (Kinoshita and Shimazaki, 1999).
Transient Expression of Fusion Proteins with sGFP, DsRed, or mCherry in Vicia Guard Cells
Full-length AtINH3 and AtINH3-W43A were subcloned into the CaMV35SΩ-sGFP(S65T)-nos3′ (Niwa et al., 1999). The C-terminal part of Arabidopsis CRY2 was cloned into the 35S promoter of Cauliflower mosaic virus (CaMV35S)-DsRed vector (Takemiya et al., 2006). Full-length mCherry, a fluorescent protein derived from DsRed, was amplified from the pmCherry vector (CLONTECH) and subcloned into the pCR4-TOPO vector having both a CaMV35S promoter and a nopaline synthase terminator, and the resulting vector was designated CaMV35S-mCherry. Arabidopsis PP1c isoforms were cloned into CaMV35S-mCherry vectors. The constructs were introduced into Vicia guard cells by particle bombardment (PDS-1000/He particle-delivery system; Bio-Rad), and fluorescent images were obtained with a confocal laser-scanning fluorescent microscope (Digital Eclipse C1; Nikon) as described by Takemiya et al. (2006). The wavelengths of excitation and emission for sGFP were 488 nm and 515 to 530 nm, respectively, and those for DsRed and mCherry were 543 nm and 578 to 632 nm, respectively.
RNA Isolation and RT-PCR Analysis
Total RNA was extracted from guard cell protoplasts, mesophyll cell protoplasts, leaves, stems, roots, green siliques, and flowers of V. faba and Arabidopsis using ISOGEN (Nippon Gene). The first-strand cDNA was synthesized from 1 μg of total RNA using a SuperScript III first-strand synthesis system (Invitrogen). The primer sets used for PCR were 5′-GATCCTCTTCATAAGAAATCGTGT-3′ and 5′-AAACCGTGTTTTATTCCGGAATGT-3′ for VfINH3; 5′-GTAGGTGATGAAGCCCAATC-3′ and 5′-GGAATCCAACACAATACCAG-3′ for V. faba ACTIN (VfACT); 5′-AGTTTCTCGATTCAACAAGCGTC-3′ and 5′-TTAGTCAACGGCTTTAGAATCATTAG-3′ for AtINH3; 5′-ACTTTACGCCAGTGGTCGTACAAC-3′ and 5′-AAGGACTTCTGGGCACCTGAATCT-3′ for ACT8; and 5′-CTCAAGAGGTTCTCAGCAGTA-3′ and 5′-TCACCTTCTTCATCCGCAGTT-3′ for β-TUBULIN (TUB), respectively. The PCR reaction mixture was denatured at 95°C for 2 min followed by 27 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min for VfACT, ACT8, and TUB or 30 cycles of the same protocol for VfINH3 and AtINH3.
Construction of the Promoter-GUS Fusion Vector and GUS Activity Assay
The AtINH3 promoter region (2,148 bp) including the start codon was amplified by PCR from Arabidopsis genomic DNA using the primers 5′-CCCAAGCTTAAGAACCAGAAAAATTAAACATTTACC-3′ and 5′-CATGCCATGGCTGAAGATTAGCTTTCAAAATCTG-3′. The fragment was subcloned into the HindIII and NcoI sites of the pCAMBIA1303 vector (CAMBIA), leading to a fusion between the AtINH3 promoter and GUS. The construct was transformed into Arabidopsis Col-0 plants using A. tumefaciens-mediated methods, and the resulting transformants were selected on agar plates with hygromycin. The GUS assay was done for homozygous T3 plants. The plant materials were incubated with GUS staining solution containing 100 mm sodium phosphate (pH 7.0), 10 mm EDTA, 0.5 mm potassium ferricyanide, 0.5 mm potassium ferrocyanide, 0.1% (w/v) Triton X-100, and 0.5 mg mL−1 X-Gluc (5-bromo-4-chloro-3-indolyl-β-glucuronide) for 6 h at 37°C. Stained samples were fixed in the solution containing 85% (v/v) ethanol and 15% (v/v) acetic acid for 2 h at room temperature.
Microscopy Observation of Arabidopsis Embryos
Wild-type and heterozygous mutant siliques were fixed in 90% (v/v) ethanol and 10% (v/v) acetic acid overnight, washed with a graded ethanol series (90%, 70%, 50%, and 30% ethanol) for 20 min each, and cleared in a derivative of Hoyer's solution (chloral hydrate/glycerol/water, 8 g:1 mL:2 mL) for 2 to 24 h at room temperature. Cleared seeds were observed using Digital Eclipse C1 (Nikon) equipped with differential interference contrast optics.
DNA Isolation and Genotype Analysis
Leaves were homogenized in extraction buffer containing 0.1 m Tris, 2 m NaCl, 25 mm EDTA, and 2% (w/v) cetyltrimethylammonium bromide and incubated at 60°C for 30 min. After centrifugation at 10,000g for 5 min, the supernatant was mixed with chloroform and centrifuged again. The DNA in the aqueous phase was precipitated with 2-propanol, washed with 70% (v/v) ethanol, and used for PCR. Genotype analyses of inh3-1 and inh3-2 were conducted by PCR using the gene-specific primers S1 (5′-AGTTTCTCGATTCAACAAGCGTC-3′) and AS1 (5′-TTAGTCAACGGCTTTAGAATCATTAG-3′) and the T-DNA border primers LB1 (5′-TGGTTCACGTAGTGGGCCATCG-3′) and LB2 (5′-GCCTTTTCAGAAATGGATAAATAGCCTTGCTTCC-3′). For complementation lines, a pair of gene-specific primers, S2 (5′-GTTTCTCGATTCAACAAGCGTCCC-3′) and AS2 (5′-GAAGGAGTTAACATTGCAATGAATCTGC-3′), were used for PCR, and the resulting products were digested with BamHI.
Construction of Complementation and Silencing Plasmids
For complementation of inh3-1, a 3,463-bp fragment of AtINH3 was amplified from Arabidopsis genomic DNA. First, a 2,151-bp fragment corresponding to the promoter region of AtINH3 was amplified using the primers 5′-CCCAAGCTTAAGAACCAGAAAAATTAAACATTTACC-3′ and 5′-CGGGATCCCATAGCTGAAGATTAGCTTTCAAAATCTG-3′, digested with HindIII and BamHI, and subcloned into pCAMBIA1300 binary vector (CAMBIA). The remaining 1,315-bp fragment including the coding sequence and the 3′-untranslated region of AtINH3 was amplified using the primers 5′-CGGGATCCATGAGCACAGCAACAAGGCCTTC-3′ and 5′-CGGGATCCAGAGAGACCCATGATTATCCAGCC-3′, digested with BamHI, and introduced into the pCAMBIA1300 with a 2,151-bp partial fragment. The resulting construct was used to transform heterozygous inh3-1 plants, and the transformants were selected on the agar plates supplemented with hygromycin.
For silencing of AtINH3 expression, full-length AtINH3 cDNA (321 bp) was amplified by PCR using the primer pairs (5′-GGGGTACCATGAGCACAGCAACAAGG-3′ and 5′-CCGCTCGAGGATCCGTCAACGGCTTTAGAATCATTAG-3′) and (5′-CCCAAGCTTATGAGCACAGCAACAAGG-3′ and 5′-GCTCTAGAGGATCCGTCAACGGCTTTAGAATCATTAG-3′) and subcloned in both the sense and antisense directions into a pKANNIBAL vector (Wesley et al., 2001). The RNAi cassette was digested with BamHI and introduced into a pPZP211 binary vector with a CaMV35S promoter and the nopaline synthase terminator. The construct was transformed into Arabidopsis Col-0 plants, and the transgenic plants were identified by kanamycin selection.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers At2g31305 (AtINH3), AB372570 (VfINH3), NP_001052397 (OsINH3a), NP_001055058 (OsINH3b), and ABN09808 (MtINH3).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Multiple alignments of the Inh3 protein sequences by ClustalW.
Supplemental Figure S2. Neighbor-joining phylogenetic tree of Inh3 protein sequences with bootstrap values (1,000 replicates).
Supplemental Figure S3. Dry mature seeds from siliques of wild-type and heterozygous inh3 mutants.
Supplemental Figure S4. Seed development defect by RNAi-mediated suppression of AtINH3 expression.
Supplemental Figure S5. RT-PCR analyses of the expression of the Arabidopsis PP1c isoforms in young green siliques.
Supplementary Material
Acknowledgments
We thank M. Tasaka and M. Furutani (Nara Institute of Science and Technology) for their valuable suggestions and N. Nishihara and M. Inoue in our laboratory for their helpful technical assistance. We are also grateful to the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutants and the ABRC at Ohio State University for providing the mutant seeds.
This work was supported by the Japanese Ministry of Education, Science, Sports and Culture, a Grant-in-Aid for Scientific Research on Priority Areas (grant no. 17084005), and a Grant-in-Aid for Scientific Research (B) (grant no. 19370020) to K.S.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Ken-ichiro Shimazaki (kenrcb@mbox.nc.kyushu-u.ac.jp).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Agarwal-Mawal A, Paudel HK (2001) Neuronal Cdc2-like proteinkinase (Cdk5/p25) is associated with protein phosphatase 1 and phosphorylates inhibitor-2. J Biol Chem 276 23712–23718 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815 [DOI] [PubMed] [Google Scholar]
- Ayaydin F, Vissi E, Mészáros T, Miskolczi P, Kovács I, Fehér A, Dombrádi V, Erdödi F, Gergely P, Dudits D (2000) Inhibition of serine/threonine-specific protein phosphatases causes premature activation of cdc2MsF kinase at G2/M transition and early mitotic microtubule organisation in alfalfa. Plant J 23 85–96 [DOI] [PubMed] [Google Scholar]
- Barford D, Das AK, Egloff MP (1998) The structure and mechanism of protein phosphatases: insights into catalysis and regulation. Annu Rev Biophys Biomol Struct 27 133–164 [DOI] [PubMed] [Google Scholar]
- Barton GJ, Cohen PTW, Barford D (1994) Conservation analysis and structure prediction of the protein serine/threonine phosphatases: Sequence similarity with diadenosine tetraphosphatase from E. coli suggests homology to the protein phosphatases. Eur J Biochem 220 225–237 [DOI] [PubMed] [Google Scholar]
- Bharucha JP, Larson JR, Gao L, Daves LK, Tatchell K (2008) Ypi1, a positive regulator of nuclear protein phosphatase type 1 activity in Saccharomyces cerevisiae. Mol Biol Cell 19 1032–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollen M (2001) Combinational control of protein phosphatase-1. Trends Biochem Sci 26 426–431 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Brukhin V, Gheyselinck J, Gagliardini V, Genschik P, Grossniklaus U (2005) The RPN1 subunit of the 26S proteasome in Arabidopsis is essential for embryogenesis. Plant Cell 17 2723–2737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceulemans H, Bollen M (2004) Functional diversity of protein phosphatase-1, a cellular economizer and reset button. Physiol Rev 84 1–39 [DOI] [PubMed] [Google Scholar]
- Cohen PTW (1997) Novel protein serine/threonine phosphatases: Variety is the spice of life. Trends Biochem Sci 22 245–251 [DOI] [PubMed] [Google Scholar]
- Cohen PTW (2002) Protein phosphatase 1: targeted in many directions. J Cell Sci 115 241–256 [DOI] [PubMed] [Google Scholar]
- Cokol M, Nair R, Rost B (2000) Finding nuclear localization signals. EMBO Rep 1 411–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong A (2006) Switching the flip: protein phosphatase roles in signaling pathways. Curr Opin Plant Biol 9 470–477 [DOI] [PubMed] [Google Scholar]
- Egloff MP, Johnson F, Moorhead G, Cohen PTW, Cohen P, Barford D (1997) Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. EMBO J 16 1876–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emi T, Kinoshita T, Sakamoto K, Mineyuki Y, Shimazaki K (2005) Isolation of a protein interacting with Vfphot1a in guard cells of Vicia faba. Plant Physiol 138 1615–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas I, Dombrádi V, Miskei M, Szabados L, Koncz C (2007) Arabidopsis PPP family of serine/threonine phosphatases. Trends Plant Sci 12 169–176 [DOI] [PubMed] [Google Scholar]
- García-Gimeno MA, Muñoz I, Ariño J, Sanz P (2003) Molecular characterization of Ypi1, a novel Saccharomyces cerevisiae type 1 protein phosphatase inhibitor. J Biol Chem 278 47744–47752 [DOI] [PubMed] [Google Scholar]
- Huang HS, Pozarowski P, Gao Y, Darzynkiewicz Z, Lee EYC (2005) Protein phosphatase-1 inhibitor-3 is co-localized to the nucleoli and centrosomes with PP1γ1 and PP1α, respectively. Arch Biochem Biophys 443 33–44 [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436 793–800 [DOI] [PubMed] [Google Scholar]
- Kerk D, Bulgrien J, Smith DW, Barsam B, Veretnik S, Gribskov M (2002) The complement of protein phosphatase catalytic subunits encoded in the genome of Arabidopsis. Plant Physiol 129 908–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Shimazaki K (1999) Blue light activates the plasma membrane H+-ATPase by phosphorylation of the C-terminus in stomatal guard cells. EMBO J 18 5548–5558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Li J, Smith RD, Walker JC (1998) Molecular cloning and chromosomal mapping of type one serine/threonine protein phosphatases in Arabidopsis thaliana. Plant Mol Biol 37 471–481 [DOI] [PubMed] [Google Scholar]
- Luan S (2003) Protein phosphatases in plants. Annu Rev Plant Biol 54 63–92 [DOI] [PubMed] [Google Scholar]
- MacKintosh C, Coggins J, Cohen P (1991) Plant protein phosphatases: subcellular distribution, detection of protein phosphatase 2C and identification of protein phosphatase 2A as the major quinate dehydrogenase phosphatase. Biochem J 273 733–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa Y, Hirano T, Yoshimoto K, Shimizu M, Kobayashi H (1999) Non-invasive quantitative detection and applications of non-toxic, S65T-type green fluorescent protein in living plants. Plant J 18 455–463 [DOI] [PubMed] [Google Scholar]
- Pedelini P, Marquina M, Ariño J, Casamayor A, Sanz L, Bollen M, Sanz P, Garcia-Gimeno MA (2007) YPI1 and SDS22 proteins regulate the nuclear localization and function of yeast type 1 phosphatase Glc7. J Biol Chem 282 3282–3292 [DOI] [PubMed] [Google Scholar]
- Pollmann S, Neu D, Lehmann T, Berkowitz O, Schafer T, Weiler EW (2006) Subcellular localization and tissue specific expression of amidase 1 from Arabidopsis thaliana. Planta 224 1241–1253 [DOI] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenolikar S, Nairn AC (1991) Protein phosphatases: recent progress. Adv Second Messenger Phosphoprotein Res 23 1–121 [PubMed] [Google Scholar]
- Shimazaki K, Doi M, Assmann SM, Kinoshita T (2007) Light regulation of stomatal movement. Annu Rev Plant Biol 58 219–247 [DOI] [PubMed] [Google Scholar]
- Shimazaki K, Kinoshita T, Nishimura M (1992) Involvement of calmodulin and calmodulin-dependent myosin light chain kinase in blue light-dependent H+ pumping by guard cell protoplasts from Vicia faba L. Plant Physiol 99 1416–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RD, Walker JC (1996) Plant protein phosphatases. Annu Rev Plant Physiol Plant Mol Biol 47 101–125 [DOI] [PubMed] [Google Scholar]
- Stubbs MD, Tran HT, Atwell AJ, Smith CS, Olson D, Moorhead GBG (2001) Purification and properties of Arabidopsis thaliana type 1 protein phosphatase (PP1). Biochim Biophys Acta 1550 52–63 [DOI] [PubMed] [Google Scholar]
- Takemiya A, Kinoshita T, Asanuma M, Shimazaki K (2006) Protein phosphatase 1 positively regulates stomatal opening in response to blue light in Vicia faba. Proc Natl Acad Sci USA 103 13549–13554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24 1596–1599 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakula P, Beullens M, Ceulemans H, Stalmans W, Bollen M (2003) Degeneracy and function of the ubiquitous RVXF-motif that mediates binding to protein phosphatase-1. J Biol Chem 278 18817–18823 [DOI] [PubMed] [Google Scholar]
- Wang QM, Guan KL, Roach PJ, DePaoli-Roach AA (1995) Phosphorylation and activation of the ATP-Mg-dependent protein phosphatase by the mitogen-activated protein kinase. J Biol Chem 270 18352–18358 [DOI] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang M, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27 581–590 [DOI] [PubMed] [Google Scholar]
- Woodgett JR, Cohen P (1984) Multisite phosphorylation of glycogen synthase. Molecular basis for the substrate specificity of glycogen synthase kinase-3 and casein kinase-II (glycogen synthase kinase-5). Biochim Biophys Acta 788 339–347 [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang L, Zhao S, Lee EYC (1998) Identification and characterization of human HCG V gene product as a novel inhibitor of protein phosphatase-1. Biochemistry 37 16728–16734 [DOI] [PubMed] [Google Scholar]
- Zhang K, Tsukitani Y, John PCL (1992) Mitotic arrest in tobacco caused by the phosphoprotein phosphatases inhibitor okadaic acid. Plant Cell Physiol 33 677–688 [Google Scholar]
- Zhao S, Lee EYC (1997) A protein phosphatase-1-binding motif identified by the panning of a random peptide display library. J Biol Chem 272 28368–28372 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.