Abstract
Arabidopsis (Arabidopsis thaliana) siliques synthesize high levels of benzoic acid (BA), which is incorporated into several glucosinolate compounds. The origin of BA in the siliques has not yet been determined. Here, we show that siliques have higher levels of benzaldehyde (BD)-oxidizing activity relative to leaves. The BD-oxidizing activity was purified from siliques in several chromatographic steps, and a 145-kD protein was identified as the enzyme most likely to possess this activity. The protein was trypsinized, and the sequence of the resulting peptides was determined by mass spectrometry, identifying it as the product of gene At1g04580, also designated as AAO4 (for ARABIDOPSIS ALDEHYDE OXIDASE4). AAO4 had previously been shown to be highly and specifically expressed in developing seeds, and its protein was shown to belong to a family of aldehyde oxidases. Here, we show that the AAO4 protein is an aldehyde oxidase that can use several substrates but that, among the substrates tested, has the lowest Km value (23 μm) with BD. AAO4 is able to oxidize BD without NAD+, but its activity increases by 50% when this cofactor is added. The pH optimum of AAO4 is 7.0. Plants homozygous for a null allele in AAO4 showed a reduction of 30% to 45% in the total levels of BA in seeds as well as 7% to 9% and 32% to 38% decreases in the levels of 3-benzoyloxypropylglucosinolate and 4-benzoyloxybutylglucosinolate, respectively. Expressing AAO4 in Escherichia coli resulted in a 3-fold increase of BD-oxidizing activity in crude bacterial extracts over endogenous levels. These findings indicate that in Arabidopsis seeds, oxidation of BD contributes in part to the synthesis of BA.
The aromatic metabolites benzoic acid (BA) or benzyl alcohol constitute the backbone of numerous compounds in plants, including taxol and cocaine (Bjorklund and Leete, 1992; Walker and Croteau, 2000), methylbenzoate, and benzylbenzoate (Raguso and Pichersky, 1995; Negre et al., 2003). Despite numerous investigations into the synthesis of BA and benzyl alcohol, as well as benzaldehyde (BD), no consensus has yet emerged and no specific enzymes and pathways have been conclusively demonstrated to yield these compounds (for review, see Wildermuth, 2006).
Arabidopsis (Arabidopsis thaliana) plants synthesize a set of defense compounds of the glucosinolate class throughout the plant (Graser et al., 2001). Two such glucosinolates, 3-benzoyloxypropylglucosinolate (3BZO) and 4-benzoyloxybutylglucosinolate (4BZO), contain a benzoyl moiety. 3BZO and 4BZO are very abundant in developing seeds and are also found in germinating seedlings (Kliebenstein et al., 2007). The enzyme that adds the benzoyl moiety to these glucosinolates in Arabidopsis is most likely an acyltransferase that uses BA-CoA (Graser et al., 2001), although this putative enzyme has not yet been identified. BA-CoA is made from BA, CoA, and ATP in a reaction catalyzed by benzoyl-CoA ligase (BZL) in Arabidopsis (Kliebenstein et al., 2007) as well as in other plants (Beuerle and Pichersky, 2002). However, the source of BA for the modification of glucosinolates in Arabidopsis seeds is not clear. Some BA may be derived by degradation of cinnamoyl-CoA through the β-oxidation pathway (Orlova et al., 2006). An enzyme that can nonoxidatively convert hydroxycinnamoyl-CoA directly to hydroxybenzaldehyde has been identified in bacteria (Gasson et al., 1998), and an analogous reaction that can convert cinnamoyl-CoA to BD may occur in plants (Abd El-Mawla and Beerhues, 2002), but a definite demonstration of the existence of an enzyme in plants capable of catalyzing this reaction is still lacking (Wildermuth, 2006).
If BD is produced in plants directly and nonoxidatively from cinnamoyl-CoA (or analogously from cinnamic acid, another hypothesized nonoxidative reaction; Beuerle and Pichersky, 2002), it might be converted to BA through oxidation. It has previously been shown that certain oxidases and dehydrogenases can convert BD to BA in plants (Koiwai et al., 2000; Liu and Schnable, 2002; Skibbe et al., 2002; Kirch et al., 2004), in animals (Yoshida et al., 1998; Huang et al., 1999; Maia and Mira, 2002), and in bacteria (Yasuhara et al., 2002; Hirano et al., 2007).
Here, we report that Arabidopsis siliques contain an enzymatic activity capable of converting BD to BA. We further show that at least some of this activity resides in a 145-kD protein, designated ARABIDOPSIS ALDEHYDE OXIDASE4 (AAO4), that belongs to a previously characterized family of aldehyde oxidases, some of which may be involved in the biosyntheses of indole-3-acetic acid (Koiwai et al., 2000) and abscisic acid (Seo et al., 1998). The gene encoding AAO4, At1g04580, is preferentially expressed in developing seeds. T-DNA insertions that eliminate functional transcripts of At1g04580 cause decreases in the levels of total BA and in the levels of benzoylated glucosinolates in seeds.
RESULTS
Purification of BD-Oxidizing Activity from Siliques
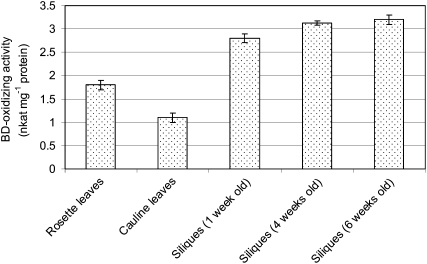
Protein extract from Arabidopsis siliques tested for BD-oxidizing activity showed no such activity (data not shown). However, when the crude extracts were first loaded onto an ion-exchange column (HiTrap-Q HP) and batch eluted, BD-oxidizing activity could be detected. The BD-oxidizing activity was enhanced 1.5-fold when 1 mm NAD+ was included in the assay. Subsequently, all enzyme assays were carried out in the presence of NAD+. Therefore, we prepared partially purified protein extracts from leaves and siliques and tested their activity. Extracts from 1-, 4-, and 6-week-old siliques converted BD to BA, in the presence of 1 mm NAD+, at a rate of 2.8 to 3.2 nkat mg−1 protein, while similar preparations from mature rosette and cauline leaves did so at rates that were 1.5- to 3-fold lower (Fig. 1).
Figure 1.
Detection of BD-oxidizing activity (with NAD+ as a cosubstrate) from protein extracts of leaves and developing siliques of Arabidopsis.
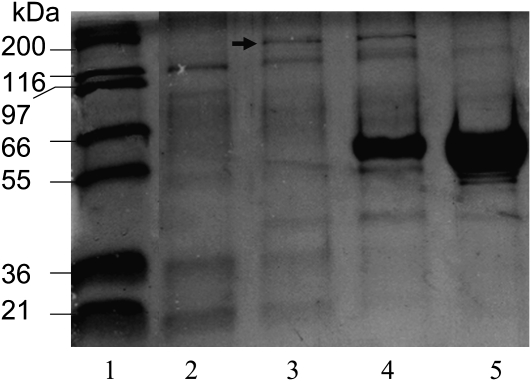
These preliminary results prompted us to purify this activity from the 4- to 6-week-old siliques. After four chromatographic steps (Table I), the fraction with peak BD oxidase activity exhibited a specific activity of 290 nkat mg−1 protein, with a total increase in specific activity of more than 2,000-fold. It was noted that the total activity also increased by approximately 15-fold, rather than decreased, and this unusual increase (see “Discussion”) means that the overall increase in specific activity during the purification procedure is actually substantially lower than 2,000-fold. The proteins in the fractions with peak BD-oxidizing activity and neighboring fractions were analyzed by SDS-PAGE. The fractions with the peak activity (Fig. 2, lanes 3 and 4) contained several proteins, but the only protein whose concentration (judged by the intensity of staining with Coomassie Brilliant Blue) correlated with BD-oxidizing activity was a 145-kD protein.
Table I.
Purification of BD oxidase activity from Arabidopsis siliques
| Chromatographic Step | Protein | Total Activity | Specific Activity | Purification |
|---|---|---|---|---|
| mg | nkat | nkat mg−1 | fold | |
| DEAE-cellulose (DE53) | 300 | 40 | 0.13 | 1 |
| HiTrap-Q HP | 69 | 110 | 1.59 | 12.2 |
| Superdex 200 10/300 GL | 23 | 150 | 6.52 | 50.1 |
| Mono-Q HR 10/10 | 2 | 580 | 290 | 2,230 |
Figure 2.
Identification of the protein responsible for BD-oxidizing activity in Arabidopsis siliques. Coomassie Brilliant Blue-stained SDS-PAGE gel of four consecutive protein fractions after Mono-Q anion-exchange chromatography. Lane 1, Protein marker; lane 2, a fraction with no detectable BD-oxidizing activity; lane 3, the next fraction, with 220 nkat mg−1 protein BD-oxidizing activity; lane 4, the next fraction, with 290 nkat mg−1 protein BD-oxidizing activity; lane 5, the next fraction, with no detectable BD-oxidizing activity. BD and NAD+ were used as substrates. The candidate protein is marked with an arrow.
This protein band, therefore, was excised from the gel and trypsinized, and the resulting peptides were separated by HPLC and analyzed by mass spectrometry. Analysis of peptide sequences determined in this way identified this protein as encoded by the gene At1g04580.
Seven unique peptides obtained from this 145-kD protein band (TEIIR, IGVHMEK, GFHPIHK, LPPYNPEK, RINLHTYESLR, QFNVQILNSGHHK, and CDLGFELPVPATMPVVK) matched the protein sequences encoded by AAO4. An eighth peptide, SMPVAAACALAASK, was only 85% identical to AAO4 but 100% identical to the corresponding region in AAO1.
Substrate Specificity of AAO4
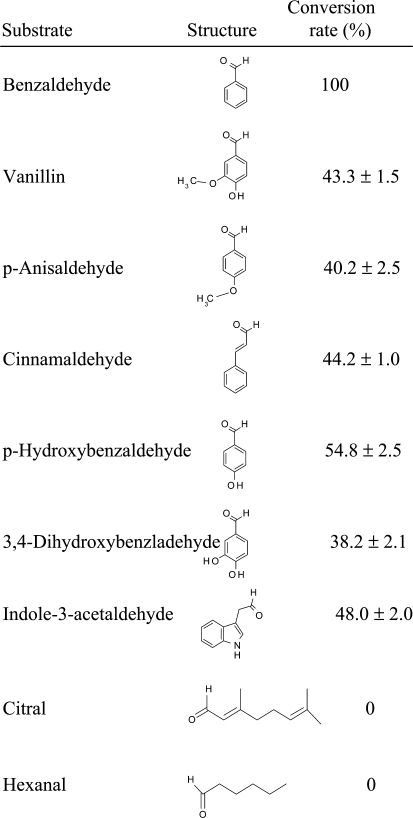
At1g04580 encodes a protein of 1,337 amino acids, with a calculated molecular mass of 145 kD. This gene was previously annotated as AAO4 because the protein it encodes shares significant sequence similarity with three aldehyde oxidases encoded by genes AAO1, AAO2, and AAO3 (Koiwai et al., 2000; Mendel and Bittner, 2006). AAO1, AAO2, and AAO3 were previously shown to be present mostly in vegetative parts of the plant and to possess oxidase activity with several different aldehydes, most notably indole-3-acetaldehyde, but AAO4 has not previously been biochemically characterized (Seo et al., 2000). We tested the partially purified BD-oxidizing activity eluting from the Mono-Q fraction with a wide array of potential substrates to determine substrate preference (Fig. 3). The enzyme had the highest activity with BD. However, the enzyme also oxidizes several BD derivates as well as indole-3-acetaldehyde. The enzyme had no oxidase activity against aliphatic aldehyde substrates (citral and hexanal; Fig. 3).
Figure 3.
Substrate specificity of partially purified AAO4 with various substrates. The activity with BD was arbitrarily set at 100%, and this level of activity represented 5 nkat mg−1 protein. The enzyme assays were carried out in buffer containing 50 mm Tris-HCl, pH 7.0, 1 mm NAD+, and 0.5 mm substrate.
The kinetic parameters of partially purified AAO4 with a selected set of substrates in the presence of NAD+ were also determined (Table II). AAO4 had the lowest Km value (23.8 μm) with BD. AAO4 showed a temperature maximum at 30°C and a broad pH optimum between pH 6 and pH 9. The divalent cations Mg2+, Fe2+, and Mn2+ as well as EDTA (at 1.0 mm) did not significantly affect enzyme activity, while Cu2+ at this concentration significantly inhibited more than 95% of AAO4 enzyme activity.
Table II.
Kinetic parameters of Arabidopsis AAO4, obtained with partially purified protein
| Substrate | Km | Vmax | kcat | kcat/Km |
|---|---|---|---|---|
| μm | nmol s−1 mg−1 | s−1 | μm−1 s−1 | |
| BD | 23.8 | 1.2 | 95.2 | 4.0 |
| NAD+ (with BD) | 58.1 | 3.2 | 253.9 | 4.37 |
| Indole-3-acetaldehyde | 2,070 | 1,501 | 1,904 | 0.92 |
| Cinnamylaldehyde | 103.9 | 5.5 | 436.5 | 4.2 |
Seeds of Arabidopsis Plants Homozygous for Null aao4 Alleles Show Reduced Levels of Total BA and of Benzoylated Glucosinolates
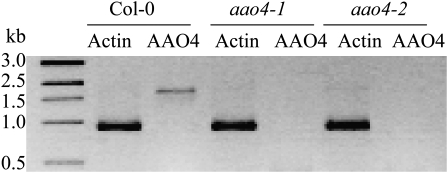
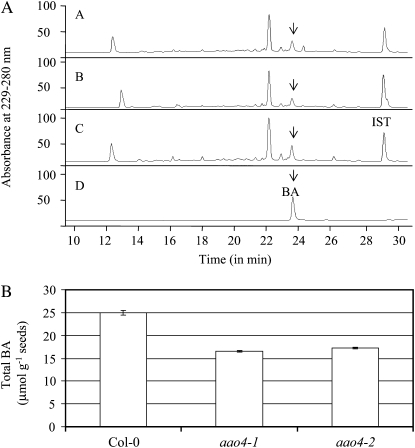
We have previously shown that BA can be detected in base-hydrolyzed methanolic extracts of seeds but not in methanolic extracts that have not been hydrolyzed, suggesting that BA is present in the seeds in a bound form (Ibdah and Pichersky, 2009). To examine the effect of silencing the AAO4 gene on levels of BA in seeds, we obtained two Arabidopsis ecotype Columbia-0 (Col-0) lines with T-DNA insertions in the AAO4 gene (SALK_047520, designated aao4-1, and SALK_057531, designated aao4-2) and determined that both insertions occur in the second exon of this gene. AAO4 transcripts from siliques of plants homozygous for the aao4-1 and aao4-2 alleles could not be detected (Fig. 4). Next, the seeds of aao4-1 and aao4-2 lines were analyzed for total BA levels (following extraction and hydrolysis). The seeds of the aao4-1 line contained 66% ± 1.7% (16.6 ± 0.17 μmol mg−1 seeds), and the seeds of the aao4-2 lines contained 69% ± 2.0% (17.3 ± 0.2 μmol mg−1 seeds) of the total BA levels present in wild-type plants (25 ± 0.5 μmol mg−1 seeds; Fig. 5).
Figure 4.
RT-PCR analysis of mRNA extracted from siliques of wild-type and aao4-1 and aao4-2 homozygous mutant lines. Actin was used as a control.
Figure 5.
Seeds of plants homozygous for null mutations in the AAO4 gene show altered BA accumulation. A, An example of HPLC analysis of total BA found in Arabidopsis seeds of Col-0, aao4-1, and aao4-2 lines after extraction and hydrolysis. Scan A, aao4-2 mutant line; scan B, aao4-1 mutant line; scan C, Col-0; scan D, BA standard. Relative abundance is shown in arbitrary units, based on absorbance at 229 to 280 nm, normalized to the added internal standard (IST) cinnamic acid. BA was identified by absorption profile and retention time in comparison with an authentic standard. B, Comparisons of the total BA levels found in wild-type and AAO4 mutant lines. Each bar represents three independent measurements.
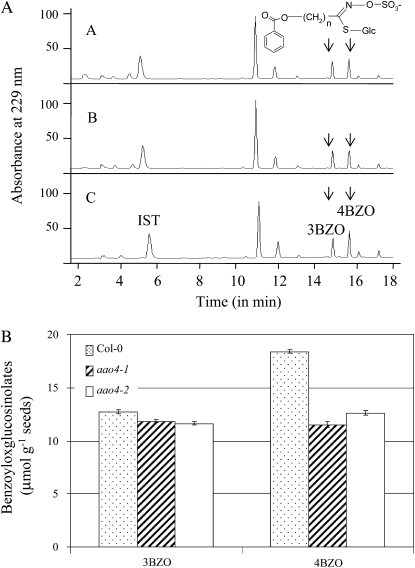
Seeds of plants from homozygous aao4-1 and aao4-2 lines were next analyzed for the levels of benzoylated glucosinolates. The seeds of aao4-1 and aao4-2 were found to accumulate just 93% ± 2.0% and 91% ± 2.6% (11.8 ± 1.5 and 11.6 ± 1.52 μmol g−1 seeds) of 3BZO compared with the levels present in wild-type plants (12.7 ± 2.1 μmol g−1 seeds; Fig. 6). The levels of 4BZO in the seeds of aao4-1 and aao4-2 were reduced to 64% ± 1.9% (11.8 ± 2.5 μmol g−1 seeds) and 68% ± 1.4% (12.6 ± 2.0 μmol g−1 seeds), respectively, compared with the levels found in the seeds of Arabidopsis Col-0 wild-type plants (18.4 ± 1.5 μmol g−1 seeds; Fig. 6).
Figure 6.
Mutants in the AAO4 gene show changes in benzoyloxyalkylglucosinolate accumulation in the seeds. A, HPLC analysis of benzoyloxyalkylglucosinolates extracted from Arabidopsis seeds of Col-0, aao4-1, and aao4-2 lines. Scan A, aao4-2 mutant line; scan B, aao4-1 mutant line; scan C, Col-0. Relative abundance is shown in arbitrary units, based on absorbance at 229 to 280 nm, and normalized to the added internal standard (IST) sinigrin. 3BZO (n = 3) and 4BZO (n = 4) were identified by absorption profiles and retention times in comparison with standards. B, Comparisons of the levels of 3BZO and 4BZO found in wild-type and AAO4 mutant lines. Each bar represents three independent measurements.
Reduced BD-Oxidizing Activity in the Siliques of aao4 Mutants
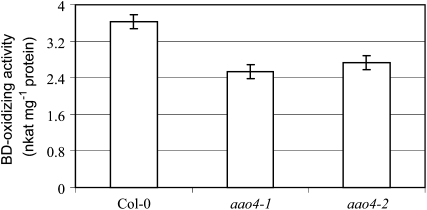
To determine if the protein encoded by AAO4 contributes to the BD-oxidizing activity observed in protein extracts of siliques, such extracts from 6-week-old siliques of Col-0, aao4-1, and aao4-2 were loaded onto an ion-exchange column (HiTrap-Q HP) and batch eluted and the BD-oxidizing activity was measured. Extracts from siliques of aao4-1 and aao4-2 were found to convert BD to BA at a rate of 2.5 ± 0.15 and 2.7 ± 0.15 nkat mg−1 protein, respectively, while the extracts from Arabidopsis wild-type plants did so at a rate that was 1.4-fold higher (3.6 ± 0.15 nkat mg−1 protein; Fig. 7).
Figure 7.
Detection of BD-oxidizing activity from protein extracts of 6-week-old siliques of Arabidopsis Col-0 and aao4 mutant lines. Each bar represents three independent measurements.
Expression of AAO4 in Escherichia coli
A full-length cDNA of AAO4 was inserted into the bacterial expression vector pEXP5-CT/TOPO, and the recombinant plasmid was transferred to E. coli cells. Bacterial cells expressing AAO4 were harvested and lysed, and the crude protein extract was assayed for BD-oxidizing activity. Assays of lysed control E. coli cells not expressing AAO4 indicated that BD-oxidizing activity occurs normally in such cells (20 ± 0.82 nkat mg−1 protein), as has been reported previously (Yasuhara et al., 2002). Cells expressing AAO4 had BD-oxidizing activity of 60 ± 0.62 nkat mg−1 protein, a 3-fold increase over the endogenous levels. Attempts to purify the 145-kD AAO4 protein (with or without a His tag) from E. coli proved unsuccessful, most likely due to proteolysis, since fractions with recovered activity showed increased concentration of several smaller peptides. Other aldehyde oxidases, for example xanthine oxidase, are known to be proteolytically cleaved while maintaining activity (Maia and Mira, 2002). The BD-oxidizing activity from cells expressing AAO4 (as well as from control E. coli cells) was found to be enhanced 1.5-fold when 1 mm NAD+ was included in the assay.
DISCUSSION
Enzymes that can catalyze the conversion of BD to BA have been reported from bacteria and animals (Yoshida et al., 1998; Hirano et al., 2007). Unlike in animals and bacteria, however, plant enzymes whose main function is to convert BD to BA have not yet been reported, although several cytoplasmic aldehyde oxidases (AOs; EC 1.2.3.1) from several plant species have been shown to be able to oxidize BD as well as a variety of other aromatic and nonaromatic aldehydes (Seo et al., 2004; Mendel and Bittner, 2006; Schwarz and Mendel, 2006). For example, a purified AO from maize (Zea mays) coleoptiles was shown to oxidize indole-3-acetaldehyde and other aliphatic and aromatic aldehydes, including BD (Koshiba et al., 1996). A barley (Hordeum vulgare) AO present in leaves, roots, and seeds was capable of oxidizing a number of aliphatic and aromatic aldehydes (Omarov et al., 1999). Additionally, pea (Pisum sativum) leaves possessed AO activity with indole-3-acetaldehyde and abscisic aldehyde (Zdunek-Zastocka et al., 2004).
Recently, four related AO genes were identified in the Arabidopsis genome and designated as AAO1 to AAO4 (Koiwai et al., 2000; Seo et al., 2004). Both AAO1 and AAO2 are expressed specifically in seedlings, and the enzymes they encode target a similarly wide range of substrates, including BD (Koiwai et al., 2000; Seo et al., 2000). The expression of AAO1, AAO2, and AAO3 (as well as possibly other BD-oxidizing enzymes) in vegetative organs explains our observation that BD-oxidizing activity is found in these organs as well as in siliques (Fig. 1). However, specific biochemical roles for AAO1 and AAO2 have not yet been assigned. In contrast, based on the substrate specificity of the AAO3 protein and its tissue distribution, as well as the phenotypic effects of mutations in the AAO3 gene, it was concluded that AAO3 catalyzes the conversion of abscisic aldehyde to abscisic acid (Seo et al., 2000; Koiwai et al., 2004).
Seo et al. (2004) showed that unlike AAO1, AAO2, and AAO3, AAO4 is highly expressed in developing siliques, and they concluded, based on mutational analysis, that it is unlikely to be involved in ABA biosynthesis, although the biochemical properties of AAO4 were not determined in that study. Our purification protocol for BD-oxidizing activity identified AAO4 as the protein responsible for most of the activity, consistent with the known high level of expression of its gene in the silique, although we cannot exclude that small amounts of AAO1 (or even AAO2 and AAO3) contribute to this activity. We further show that AAO4 can use a variety of aldehyde substrates but that, of the substrates tested, it has the highest affinity to BD (Table II; Fig. 3). The Km value of AAO4 for BD is comparable to the values reported for Arabidopsis AAO3 and for the maize coleoptile AO (Koshiba et al., 1996; Koiwai et al., 2000).
Although AAO4 does not possess an NAD-binding site (Schwarz and Mendel, 2006), the addition of NAD+ increased AAO4 activity by about 50%. Mammalian xanthine dehydrogenase is known to switch from the dehydrogenase (using NAD+ as the electron acceptor) to the oxidase mode of activity after undergoing either oxidation of Cys residues or partial proteolysis (Nishino et al., 2008). While we did not investigate such a transition with AAO4, it is possible that a similar mechanism operates on AAO4 in planta. AAO4, when expressed in E. coli, did appear to under partial proteolytic cleavage with the retention of at least some activity. The unusual observation that total AAO4 activity increased from one purification step to the next (typically, total activity might increase only after the first purification step, due to the removal of inhibitors) may also be related to a switch between dehydrogenase and oxidase activities, perhaps due to oxidation of Cys residues of the enzyme during the purification procedure, but this too remains to be examined in detail.
Protein extracts from Arabidopsis siliques of aao4-1 and aao4-2 showed a decrease BD-oxidizing activity by 30% to 45% as compared with protein extract of wild-type plants (Fig. 7). In addition, siliques of these mutant lines showed a decrease in the amount of total BA (Fig. 5), and the level of the benzoylated glucosinolate 3BZO was reduced by 7% to 9% while the level of the benzoylated glucosinolate 4BZO was reduced by 32% to 38% as compared with seeds of wild-type plants (Fig. 6). Since no free BA was detected in the siliques, and the reduction in the level of hydrolyzable BA was similar to the reduction in the levels of 3BZO and 4BZO, it is likely that all of the BA we detected after hydrolysis came from the benzoylated glucosinolates. Taken together, these results suggest that some, but not all, of the BA synthesized in siliques is derived from BD by the action of AAO4. However, it is clear that the siliques have other enzymes capable of catalyzing this reaction, and it is also equally likely that a substantial amount of BA is derived from a source other than BD. The reason for the difference in the magnitude of the reduction in the levels of 3BZO and 4BZO in the mutants is not clear, although it is possible that the enzymes responsible for their formation are affected differently by the reduced levels of the BA-containing precursors.
BA biosynthesis in developing Arabidopsis seeds constitutes a system that is amenable to genetic analysis. A mutant that completely lacks benzoylated glucosinolates has recently been found, and the analysis identified the mutated gene as BZL1 (At1g65880; Kliebenstein et al., 2007). Interestingly, BZL1 has a peroxisomal targeting sequence at its C terminus, although the synthesis of benzoylated glucosinolates is believed to be in the cytosol, not the peroxisomes (Chen and Andreasson, 2001). Our data indicate that some of the benzoyl moiety of benzoylated glucosinolates may be synthesized in the cytosol, since the AAO4 protein lacks any identifiable sorting signal and therefore is likely to reside in the cytosol. Whether Arabidopsis also has other BD-oxidizing enzymes that reside in the cytosol or elsewhere in the cell, as well as the total contribution of such enzymes to the synthesis of BA, remain to be determined.
MATERIALS AND METHODS
Plant Materials
Arabidopsis (Arabidopsis thaliana Col-0) plants were grown on soil at 23°C under 16 h of light/8 h of dark for up to 4 to 6 weeks. The cDNAs and the SALK T-DNA knockout mutant lines of AAO4 (SALK_057531 and SALK_047520) in the Col-0 background were obtained from the Arabidopsis Biological Resource Center.
Screening of T-DNA Insertion Mutants
The T-DNA insertion sites in the AAO4 gene were verified first by PCR. For AAO4, we used the genomic primers AAO4 forward (5′-GGCAACAACCAACCCTGGCAAAGATAG-3′) and AAO4 reverse (5′-TGCATTTCAGCTCCAAACTTCACCAACAGG-3′) to characterize the insertion line SALK_057531 and AAO4 forward (5′-GGAGAAGGTGGCTAACCATTTTATCAGAAACTC-3′) and AAO4 reverse (5′-CACAAGAGCAATTATTTGTCCGGCAG-3′) to characterize the insertion line SALK_047520. In these experiments, we also used the T-DNA-specific primer LBb1 (5′-CGTGGACCGCTTGCTGCAACT-3). All PCR products were further verified by sequencing.
Reverse Transcription-PCR Analyses
The abolition of steady-state accumulation of AAO4 transcripts was verified by reverse transcription (RT)-PCR, using the gene-specific forward primer 5′-CTGACCCTTCTTTGCAGCTTAAATGGGTG-3′ and the gene-specific reverse primer 5′-GGCTTTGTGCTATAAATGAATGCTCCATG-3′. For RT-PCR analysis of the aao4-1 and aao4-2 mutants, 4-week-old siliques were used for extraction of total RNA (Maes and Messens, 1992). Total RNA (5 μg) was reversed transcribed using oligo(dT) primer and Moloney murine leukemia virus reverse transcriptase (Stratagene). The following actin primers were used as positive controls: actin forward primer (5′-GCTGTGTTTCCTAGTATTGTGGGTCGTCCT-3′) and the reverse actin primer (5′-GTATTTCCTCTCTGGAGGGGCAACGA-3′).
Enzyme Purification and Amino Acid Sequence Analysis
The BD-oxidizing activity was extracted from 50 g of Arabidopsis siliques (4–6 weeks old) that were grown under normal conditions, ground in liquid nitrogen with extraction buffer, and stored at −80°C. All extraction and purification steps were performed at 4°C. Extraction buffer contained 50 mm Tris-HCl (pH 7.0), 5% (w/v) polyvinylpolypyrrolidone, 1 mm 2-mercaptoethanol, 5 mm Na2S2O5, 0.2 mm phenylmethylsulfonyl fluoride, and 10% (v/v) glycerol. The crude protein extract was passed through two layer of Micracloth (Calbiochem) and centrifuged for 20 min at 12,000g at 4°C. The pellet was discarded, the supernatant (300 mg of protein) was passed over DE53 beads (preequilibrated with 50 mm Tris-HCl, pH 7.0) to remove residual impurities, and proteins were eluted with a stepwise KCl gradient (200, 300, and 500 mm KCl in 50 mm Tris-HCl, pH 7.0) with monitoring of enzyme activity. Active protein fractions were pooled together and dialyzed overnight at 4°C in 50 mm Tris-HCl (pH 7.0) buffer containing 1 mm 2-mercaptoethanol, 5 mm Na2S2O5, 0.2 mm phenylmethylsulfonyl fluoride, 10% (v/v) glycerol, and 5 mm KCl. After dialysis, the sample was applied to a HiTrap-Q HP column (Amersham) preequilibrated with 50 mm Tris-HCl (pH 7.0), and proteins were eluted with a linear KCl gradient from 50 to 500 mm KCl with monitoring of the enzyme activity. Fractions with peak activity were pooled and applied to a Superdex 200 10/300 GL column (Amersham) equilibrated with 50 mm Tris-HCl (pH 7.0) and were eluted with 50 mm KCl with monitoring of the enzyme activity. Active fractions were combined (2 mg), diluted to 50 mm Tris-HCl (pH 7.0), directly applied to a Mono-Q column (HR 10/10; Amersham) equilibrated with 50 mm Tris-HCl (pH 7.0), and then eluted with a linear KCl gradient from 50 to 500 mm KCl with monitoring of the enzyme activity.
Protein Measurements
Protein concentrations were determined by the method of Bradford (1976) using reagents obtained from Bio-Rad. Bovine serum albumin was used as a standard for a calibration curve.
Protein Sequencing
Samples of the enzyme preparations were concentrated by StrataClean resin (Stratagene) and than subjected to SDS-PAGE. The 145-kD protein band (Fig. 2) was eluted from the gel, trypsinized, and subjected to liquid chromatography-tandem mass spectrometry analysis as described previously (Chen et al., 2005), followed by a search of the National Center for Biotechnology Information database using the program MASCOT (Perkins et al., 1999).
Enzyme Activity Measurements
Enzyme assays were carried out in a buffer containing 50 mm Tris-HCl, pH 7.0, 1 mm NAD+, 500 μm substrate, and 1 to 500 ng of total protein in a total volume of 100 μL. The assays were incubated at 30°C for 30 min, after which the reaction mixture was terminated by the addition of 20 μL of 3.5% (w/v) TCA in 50% (v/v) acetonitrile. The reaction products were analyzed by reverse-phase liquid chromatography on a Inertsil ODS-2 5-μm C18 column (150 × 4.6 mm i.d., 5 μm particle size) operated at 0.8 mL min−1 and 30°C and eluted with a gradient (solvent A, 1.5% [v/v] phosphoric acid in water; solvent B, acetonitrile) of 0% to 10% B (2 min), 10% to 45% B (12 min), 45% to 50% B (16 min), 50% to 95% B (17 min), 95% B (1 min), and 95 to 10% B (25 min). Compound elution was monitored at 200 to 400 nm with a Waters 996 UV/visible photodiode array detector.
For Km value determination, substrate concentrations were varied between 50 and 30,000 μm. The Km and Vmax values were calculated from Lineweaver-Burk plots. All enzyme assays were recorded in triplicate. For determination of kinetic data, we adjusted the total protein quantity to 5 μg per assay.
Extraction and Analysis of Benzoylglucosinolates
Extracts were made using methods adapted from previously published protocols (Reichelt et al., 2002). Seeds (20 mg) were extracted in a 1-mL solution of 80% methanol to which 30 μL of 10 mm sinigrin (Sigma-Aldrich) was added as an internal standard (final concentration, 300 μm). The extracts were centrifuged in a microcentrifuge at 4,300 rpm for 10 min, and the supernatant was loaded onto a small (50-mg) column of DEAE Sephadex A25. DEAE Sephadex A25 was swelled in water. The column was rinsed successively with 1 mL of 80% (aqueous) methanol, 1 mL of deionized water, and 1 mL of 20 mm NaOAc, pH 5.5. After washing, 25 μL of sulfatase (Sigma) solution prepared according to Graser et al. (2001) was loaded on the column, and the column was left standing overnight. The resulting desulfoglucosinolates were eluted from the column with 0.5 mL of 20% (aqueous) methanol.
Extraction and HPLC Analysis of BA
The protocol for extraction of BA was modified from Chong et al. (2001). Plant material (10–20 mg of seeds) was extracted with 2 mL of 90% methanol containing 10 μL of 10 mm cinnamic acid as an internal standard. After centrifugation in a microcentrifuge for 5 min at 14,000 rpm, the residue was extracted again with 2 mL of 100% methanol. The combined extracts were reduced to dryness under nitrogen. Samples were either analyzed directly by HPLC for BA after resuspension in methanol or the dried extracts were resuspended in 1 mL of water at 50°C, saponified with 100 μL of 1 n NaOH for 30 min at room temperature, neutralized with 30 μL of 1 n HCl and centrifuged in a microcentrifuge for 5 min at 14,000 rpm, and then injected into the HPLC apparatus.
Isolation and Heterologous Expression of an AAO4 cDNA
The AAO4 open reading frame was amplified by PCR using the forward primer 5′-ATGGCGGGTGACGATTTGGTG-3′ and the reverse primer 5′-AGGATATGTTTTCCATTCTAAGTACTTCTCTATGCTTTCAAGGCC-3′. The resulting PCR fragment was cloned into the pEXP5-CT/TOPO TA expression vector (Invitrogen). All PCR procedures were performed using KOD DNA polymerase (Novagen) to enhance fidelity. All constructs were verified by DNA sequencing. RNA from the siliques of Arabidopsis Col-0 plants was isolated using methods adapted from previously published protocols (Maes and Messens, 1992). Total RNA (5 μg) was reverse transcribed using oligo(dT) primer and Moloney murine leukemia virus reverse transcriptase (Stratagene). The corresponding cDNA was amplified to yield a 4,014-bp fragment, which was ligated into the vector Topo pEX5-CT/TOPO TA, verified by sequencing, and the recombinant plasmid was transferred into Escherichia coli BL21(DE3)pLysS (Stratagene). AAO4 gene expression in E. coli was induced with 1 mm isopropyl-1-thio-β-d-galactopyranoside, and cells were grown at 18°C for 24 h. Bacteria were lysed, and soluble protein was assayed with BD as a substrate and NAD+ as a cosubstrate and in parallel by SDS-PAGE.
This work was supported by the National Science Foundation (grant no. MCB 0331353 to E.P.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Eran Pichersky (lelx@umich.edu).
Open Access articles can be viewed online without a subscription.
References
- Abd El-Mawla AM, Beerhues L (2002) Benzoic acid biosynthesis in cell cultures of Hypericum androsaemum. Planta 214 727–733 [DOI] [PubMed] [Google Scholar]
- Beuerle T, Pichersky E (2002) Purification and characterization of benzoate:coenzyme A ligase from Clarkia breweri. Arch Biochem Biophys 400 258–264 [DOI] [PubMed] [Google Scholar]
- Bjorklund J, Leete E (1992) Biosynthesis of the benzoyl moiety of cocaine from cinnamic acid via (R)-(+)-3-hydroxy-3-phenylpropanoic acid. Phytochemistry 3 357–360 [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Chen H, Wilkerson CG, Kuchar JA, Phinney BS, Howe GA (2005) Jasmonate-inducible plant enzymes degrade essential amino acids in the herbivore midgut. Proc Natl Acad Sci USA 102 19237–19242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Andreasson E (2001) Update on glucosinolate metabolism and transport. Plant Physiol Biochem 39 743–758 [Google Scholar]
- Chong J, Pierrel MA, Atanassova R, Werck-Reichhart D, Fritig B, Saindrenan P (2001) Free and conjugated benzoic acid in tobacco plants and cell cultures: induced accumulation upon elicitation of defense responses and role as salicylic acid precursors. Plant Physiol 125 318–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasson MJ, Kitamura Y, McLauchlan WR, Narbad A, Parr AJ, Parsons ELH, Payne J, Rhodes MJC, Walton NJ (1998) Metabolism of ferulic acid to vanillin. J Biol Chem 273 4163–4170 [DOI] [PubMed] [Google Scholar]
- Graser G, Oldham NJ, Brown PD, Temp U, Gershenzon J (2001) The biosynthesis of benzoic acid glucosinolate esters in Arabidopsis thaliana. Phytochemistry 57 23–32 [DOI] [PubMed] [Google Scholar]
- Hirano JI, Miyamoto K, Ohta H (2007) Purification and characterization of aldehyde dehydrogenase with a broad substrate specificity originated from 2 phenylethanol-assimilating Brevibacterium sp. KU1309. Appl Microbiol Biotechnol 76 357–363 [DOI] [PubMed] [Google Scholar]
- Huang DY, Furukawa A, Ichikawa Y (1999) Molecular cloning of retinal/aldehyde oxidase cDNAs from rabbit and mouse livers and functional expression of recombinant mouse retinal oxidase cDNA in E. coli. Arch Biochem Biophys 364 264–272 [DOI] [PubMed] [Google Scholar]
- Ibdah M, Pichersky E (2009) Arabidopsis Chy1 null mutants are deficient in benzoic acid-containing glucosinolates in the seeds. Plant Biol (submitted) [DOI] [PubMed]
- Kirch HH, Bartels D, Wei Y, Schnable PS, Wood AJ (2004) The ALDH gene superfamily of Arabidopsis. Trends Plant Sci 9 371–377 [DOI] [PubMed] [Google Scholar]
- Kliebenstein D, D'Auria J, Behere A, Kim J, Gunderson K, Breen J, Lee G, Gershenzon J, Last R, Jander G (2007) Characterization of seed-specific benzoyloxyglucosinolate mutations in Arabidopsis thaliana. Plant J 51 1062–1076 [DOI] [PubMed] [Google Scholar]
- Koiwai H, Akaba S, Seo M, Komano T, Koshiba T (2000) Functional expression of two Arabidopsis aldehyde oxidases in the yeast Pichia pastoris. J Biochem 127 659–664 [DOI] [PubMed] [Google Scholar]
- Koiwai H, Nakaminami K, Seo M, Mitsuhashi W, Toyomasu T, Koshiba T (2004) Tissue-specific localization of an abscisic acid biosynthetic enzyme, AAO3, in Arabidopsis. Plant Physiol 134 1697–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiba T, Saito E, Ono N, Yamamoto N, Sato M (1996) Purification and properties of flavin- and molybdenum containing aldehyde oxidase from coleoptiles of maize. Plant Physiol 110 781–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Schnable PS (2002) Functional specialization of maize mitochondrial aldehyde dehydrogenases. Plant Physiol 130 1657–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes M, Messens M (1992) Phenol as grinding material in RNA preparations. Nucleic Acids Res 20 4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maia L, Mira L (2002) Xanthine oxidase and aldehyde oxidase: a simple procedure for the simultaneous purification from rat liver. Arch Biochem Biophys 44 48–53 [DOI] [PubMed] [Google Scholar]
- Mendel RR, Bittner F (2006) Cell biology of molybdenum. Biochim Biophys Acta 1763 621–635 [DOI] [PubMed] [Google Scholar]
- Negre F, Kish CM, Boatright J, Underwood B, Shibuya K, Wagner C, Clark DG, Dudareva N (2003) Regulation of methylbenzoate emission after pollination in snapdragon and petunia flowers. Plant Cell 15 2992–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T, Okamoto K, Eger BT, Pai EF, Nishino T (2008) Mammalian xanthine oxidoreductase: mechanism of transition from xanthine dehydrogenase to xanthine oxidase. FEBS J 275 3278–3289 [DOI] [PubMed] [Google Scholar]
- Omarov R, Akaba S, Koshiba T, Lips H (1999) Aldehyde oxidase in roots, leaves and seeds of barley (Hordeum vulgare L.). J Exp Bot 50 63–69 [Google Scholar]
- Orlova I, Marshall-Colón A, Schnepp J, Wood B, Varbanova M, Fridman E, Blakeslee JJ, Peer WA, Murphy AS, Rhodes D, et al (2006) Reduction of benzenoid synthesis in petunia flowers reveals multiple pathways to benzoic acid and enhancement in auxin transport. Plant Cell 18 3458–3475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20 3551–3567 [DOI] [PubMed] [Google Scholar]
- Raguso RA, Pichersky E (1995) Floral volatiles from Clarkia breweri and C. concinna (Onagraceae): recent evolution of floral scent and moth pollination. Plant Syst Evol 194 55–67 [Google Scholar]
- Reichelt M, Brown MD, Schneider B, Oldham NJ, Stauber E, Tokuhisa J, Kliebenstein DJ, Mitchell-Olds T, Gershenzon J (2002) Benzoic acid glucosinolate esters and other glucosinolates from Arabidopsis thaliana. Phytochemistry 59 663–671 [DOI] [PubMed] [Google Scholar]
- Schwarz G, Mendel RR (2006) Molybdenum cofactor biosynthesis and molybdenum enzymes. Annu Rev Plant Biol 57 623–647 [DOI] [PubMed] [Google Scholar]
- Seo M, Akaba S, Oritani T, Delarue M, Bellini C, Caboche M, Koshiba T (1998) Higher activity of an aldehyde oxidase in the auxin-overproducing superroot mutant of Arabidopsis thaliana. Plant Physiol 116 687–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M, Aoki H, Koiwai H, Kamiya Y, Nambara E, Koshiba T (2004) Comparative studies on the Arabidopsis aldehyde oxidase (AAO) gene family revealed a major role of AAO3 in ABA biosynthesis in seeds. Plant Cell Physiol 45 1694–1703 [DOI] [PubMed] [Google Scholar]
- Seo M, Koiwai H, Ahaba S, Komano T, Oritani T, Kamiya T, Koshiba T (2000) Abscisic aldehyde oxidase in leaves of Arabidopsis thaliana. Plant J 23 481–488 [DOI] [PubMed] [Google Scholar]
- Skibbe DS, Liu F, Wen TJ, Yandeau MD, Cui X, Cao J, Simmons CR, Schnable PS (2002) Characterization of the aldehyde dehydrogenase gene families of Zea mays and Arabidopsis. Plant Mol Biol 48 751–764 [DOI] [PubMed] [Google Scholar]
- Walker K, Croteau R (2000) Taxol biosynthesis: molecular cloning of a benzoyl-CoA:taxane 2a-O-benzoyltransferase cDNA from Taxus and functional expression in Escherichia coli. Proc Natl Acad Sci USA 97 13591–13596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth MC (2006) Variations on a theme: synthesis and modification of plant benzoic acids. Curr Opin Plant Biol 9 288–296 [DOI] [PubMed] [Google Scholar]
- Yasuhara A, Akiba-Goto M, Fujishiro K, Uchida H, Uwajima T, Aisaka K (2002) Production of aldehyde oxidases by microorganisms and their enzymatic properties. J Biosci Bioeng 94 124–129 [DOI] [PubMed] [Google Scholar]
- Yoshida A, Rzhetsky A, Hsu LC, Chang C (1998) Human aldehyde dehydrogenase gene family. Eur J Biochem 251 549–557 [DOI] [PubMed] [Google Scholar]
- Zdunek-Zastocka E, Omarov RT, Koshiba T, Lips HS (2004) Activity and protein level of AO isoforms in pea plants (Pisum sativum L.) during vegetative development and in response to stress conditions. J Exp Bot 55 1361–1369 [DOI] [PubMed] [Google Scholar]