Abstract
Transgenerational epigenetic inheritance has been defined by the study of relatively few loci. We examined a population of recombinant inbred lines with epigenetically mosaic chromosomes consisting of wild-type and CG methylation-depleted segments (epiRILs). Surprisingly, transposons that were immobile in the parental lines displayed stochastic movement in 28% of the epiRILs. Although analysis after eight generations of inbreeding, supported by genome-wide DNA methylation profiling, identified recombined parental chromosomal segments, these were interspersed with unexpectedly high frequencies of nonparental methylation polymorphism. Hence, epigenetic inheritance in hybrids derived from parents with divergent epigenomes permits long-lasting epi-allelic interactions that violate Mendelian expectations. Such persistently “unstable” epigenetic states complicate linkage-based epigenomic mapping. Thus, future epigenomic analyses should consider possible genetic instabilities and alternative mapping strategies.
Keywords: DNA methylation, epigenetics, epigenome maps, methylation profiling
The term “epigenome” refers to the genome-wide distribution of epigenetic marks such as DNA methylation, histone modifications, and the presence of histone variants (Jenuwein 2002). Specific combinations of these marks are thought to determine the local chromatin structure that affects transcription and genome stability (Jenuwein 2002). In plants and mammals, DNA methylation is the best-studied epigenetic modification. Its faithful propagation is not only critical for proper development but also silences transposable elements (Finnegan 1996; Miura et al. 2001; Singer et al. 2001; Kato et al. 2003; Chan et al. 2005). Thus, apart from a role in development, DNA methylation protects genome integrity.
Methylation modifies cytosines preceding guanines (mCG) and in plants METHYLTRANSFERASE 1 (MET1), a homolog of the mammalian Dnmt1, is required for propagating mCG patterns during DNA replication (Finnegan 1996). Loss of MET1 leads to almost a complete erasure of CG methylation and indirect losses of plant-specific non-CG methylation (Saze et al. 2003). Loss of MET1 also results in the suppression of DNA demethylation activities, altered RNA directed de novo methylation, and the redistribution of other repressive marks such as histone H3 dimethylation in Lys 9 and trimethylation in Lys 27 (Soppe et al. 2002; Tariq et al. 2003; Mathieu et al. 2005, 2007), creating further epigenetic variation. Thus, transgenerational inheritance of the epigenome in plants is coordinated by the faithful replication of mCG patterns (Mathieu et al. 2007). Notably, loss of mCG results in hypomethylated epi-alleles that, at some loci, may be stably inherited over many generations (Kakutani et al. 1996; Mathieu et al. 2007; Vaughn et al. 2007). This is similar to inheritance of epigenetic states of ribosomal DNA following intercrossed Arabidopsis accessions (Riddle and Richards 2002, 2005; Woo and Richards 2008). Despite considerable efforts to assemble epigenomic maps for several eukaryotes, including humans, the extent of stable transgenerational epi-allelic inheritance across the genome is not known.
We created a population of Arabidopsis thaliana inbred lines with random distribution of epi-alleles derived from the wild-type and a met1-3-null mutant (Saze et al. 2003) to assess the feasibility of integrating phenotypic and epigenomic data into a quantitative genetics framework, as recently proposed (Johannes et al. 2008). met1-3 was crossed to the isogenic wild type to create an F2 of unique epigenetic variants. To immediately restore stable transgenerational inheritance of segregating DNA methylation polymorphisms, only MET1 homozygous F2 plants were propagated by self-pollination and single-seed descent. Inbreeding of these lines resulted in a population of epigenetic recombinant inbred lines referred to as “epiRILs.” The stability and inheritance of epi-alleles was analyzed in these epiRILs, as well as the impact of the induced epigenetic variation on phenotypic variation and genome stability.
Results
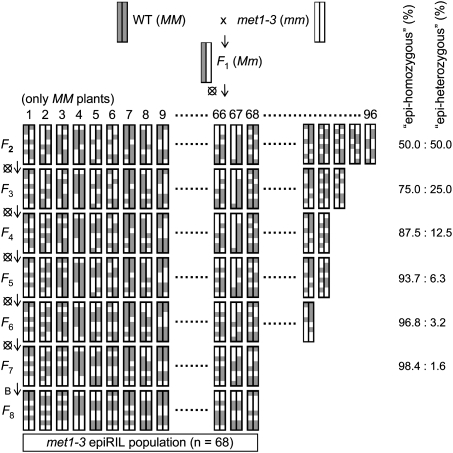
Generation of the met1-3 epiRIL population
F2 individuals from a selfed F1 hybrid of a wild type and a homozygous met1-3 mutant (both Col-0 ecotype) were expected to inherit various patterns of wild-type- and met1-3-derived epi-alleles that recombined during F1 meiosis (Fig. 1). A group of 96 MET1 F2 plants was propagated further to allow transgenerational inheritance of the novel epi-allele combinations. Each selected F2 plant was selfed and one random individual from 100 F3 plants was forwarded to the next generation. Single-seed descent was applied until the seventh generation, when seeds of nine random F7 plants were bulk harvested to give the F8 for phenotyping and DNA analysis (Fig. 1). For parental epi-alleles (referred to as wild-type-like or met-like) not subjected to selection and inherited in a Mendelian fashion, the frequency of heterozygosity (referred to as “epi-heterozygosity”) should decline by 50% in each subsequent epiRIL generation, resulting in <2% probability of epi-heterozygosity at any chosen locus by the F7 (Fig. 1). Due to loss of fitness, only 68 of the 96 epiRIL lines were advanced to the F8, which may reflect selection against demethylated epi-alleles at loci influencing plant fitness.
Figure 1.
Construction of the epiRIL population, illustrated by one pair of chromosomes. Parental chromosomes and their segments are marked wild type (gray, WT, MM) and met1-3 (white, met1-3, mm). Eight generations of inbreeding (vertical) are marked as F2 to F8 (crossed circles at the left mark single-seed descent). A bulk harvest of individuals at F7 is marked by B. Predicted levels of “epi-homozygosity” and “epi-heterozygosity” at each generation are indicated on the right.
Phenotypic variation within the epiRIL population
Developmental variation within and between epiRILs was observed during inbreeding; however, altered phenotypes often arose stochastically and/or were unstable (Supplemental Fig. 1). Moreover, such abnormalities were generally difficult to stabilize by phenotypic selection. Therefore, only traits that appeared stable in particular epiRILs, including flowering time, plant growth, and stress responses, were examined.
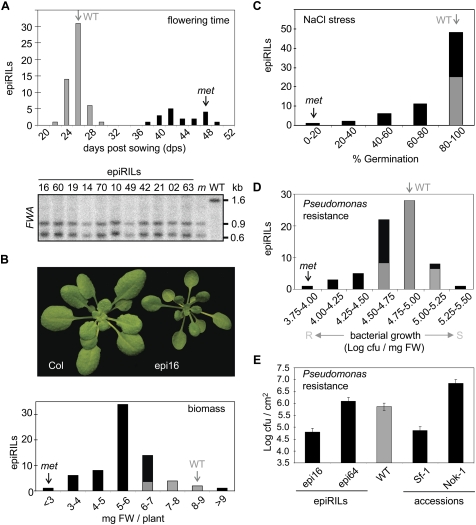
There was a bimodal distribution in flowering time, with 26% of epiRILs exhibiting delayed flowering (Fig. 2A). Since SINE elements residing within the promoter of the flowering repressor FWA (FLOWERING WAGENINGEN) are methylated in wild-type plants (Fujimoto et al. 2008) and demethylated in met1-3 (Saze et al. 2003), causing ectopic FWA transcription and delayed flowering (Soppe et al. 2000), FWA methylation was examined in 11 late-flowering lines (Fig. 2A). Each line had a met-like fwa epi-allele, consistent with stable inheritance of this met1-3-derived trait. However, the proportion of epiRILs with a late-flowering phenotype deviated from the 50% expected for inheritance not subjected to selection pressure; the wild-type early-flowering phenotype predominated (74%). To avoid phenotypic selection for early flowering, seeds of all plants were allowed to mature fully. Thus, the predominance of the wild-type-like phenotype suggests selection against demethylated loci, possibly linked to FWA.
Figure 2.
Phenotypic variation within the epiRILs (see the Supplemental Material for details). All graphs (except E) display trait variation (X-axis) and the number of epiRILs per trait interval (Y-axis). The proportions of epiRILs statistically equal to or different from wild type (95% confidence level) are marked by gray and black areas, respectively. The wild-type and met1-3 averages are indicated by marked gray and black arrows, respectively. (dps) Days post-sowing; (cfu) colony-forming units; (FW) fresh weight. (A) Flowering time (top panel) and Southern blot analysis at the FWA locus using CfoI-digested DNA (bottom panel). Late flowering epiRILs with numbers indicated above each lane were compared with the parental lines. (WT) Wild type; (m) met1-3. The FWA probe was designed as described previously (Soppe et al. 2000). (B) Biomass. Representative wild-type and epi16 plants at 35 dps are shown above the graph. (C) Salt-stress response. Germination rates of seeds (% Germination) in the presence of 150 mM NaCl at 4 dps. (D) Pseudomonas resistance. Resistant (R) and susceptible (S) responses are indicated (X-axis). (E) The resistance to Pseudomonas of epiRILs and selected Arabidopsis accessions. The average bacterial titer levels at 3 d post-infection (dpi) in Log cfu per square centimeter (error bars, SD; Fisher's LSD test; 95% confidence limit).
For other characteristics, the phenotypic variation had the continuous distribution observed for complex traits, suggesting multiple epi-alleles with partial, quantitative effects or with incomplete penetrance. The phenotypic distributions were quantified (Supplemental Table 1). The biomasses of 21-d-old plants showed significant differences (P < 0.05), with the growth of 85% of epiRILs being retarded compared with the wild type; e.g., epi16 (Fig. 2B). Repeated testing of a subset of lines showing large growth differences supported the initial results (Supplemental Fig. 2).
The epiRIL population was examined for resistance to selected abiotic and biotic stresses. Although most epiRIL seeds germinated normally under standard conditions (Supplemental Fig. 3), germination of 60% of the epiRILs was delayed significantly compared with the wild type under elevated salinity (Fig. 2C; Supplemental Fig. 3A–D). Salt-sensitive lines forwarded to the F9 exhibited similar responses (Supplemental Fig. 3E–H), confirming stable inheritance of this trait.
In a screen for resistance to the biotrophic bacterial pathogen Pseudomonas syringae pv. tomato (Pst), 34% and 4% of epiRILs showed increased resistance or susceptibility, respectively (Fig. 2D). Heritability was confirmed in the F9 and F10 for one susceptible and four resistant lines (Supplemental Fig. 4A,B), with epi16 and epi64 being the most resistant and susceptible lines, respectively. To compare the range of Pst resistance within the epiRILs to that of Arabidopsis accessions, the most resistant and susceptible accessions reported amongst 127 accessions (Perchepied et al. 2006; Fan et al. 2008) were screened. Sf-1 and Nok-1 were the most resistant and susceptible, respectively (Supplemental Fig. 4C). Sf-1 and Nok-1 and selected Arabidopsis mutants with altered pathogen resistance were then examined together with epi16 and epi64 (Fig. 2E; Supplemental Fig. 4B). The differences between Sf-1 and Nok-1 (2.3 Logs of cfu/cm2) and epi16 and epi64 (1.3 Logs of cfu/cm2) suggested that the range of epiRIL disease resistance covers ∼58% of the phenotypic variation within 127 Arabidopsis accessions.
Inheritance of DNA methylation during epiRIL inbreeding
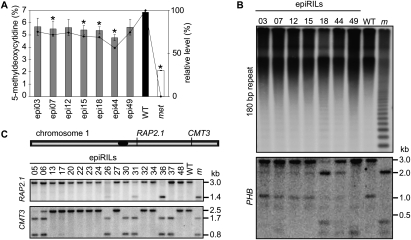
In general, examination of the epiRIL phenotypes revealed induction of variability that was stably heritable for some traits and, therefore, the heritability of parental DNA methylation during epiRIL inbreeding was examined. Stable propagation in the absence of selection of all epi-alleles inherited from the parental lines within the epiRIL population should result in a mean methylation level for the epiRIL population equal to the mid-parent level (or slightly above, due to MET1 selection in the F2). The total 5-methyldeoxycytidine (mC) levels of the parental lines were 6.8% and 2.2% for wild type and met1-3, respectively (Fig. 3A), similar to recently published values (Rozhon et al. 2008). Comparison of seven epiRILs at the F8 generation to the parental lines revealed a bias toward wild-type levels, averaging 80% of the mC content of the wild type (Fig. 3A). This may reflect either preferential transmission of methylated chromosomal regions or de novo methylation, as observed previously in met1-3 mutants (Zhang et al. 2006; Mathieu et al. 2007; Cokus et al. 2008; Lister et al. 2008; Reinders et al. 2008). Given that mC is abundant at centromeric and pericentromeric repeats (Pruitt and Meyerowitz 1986), preferential inheritance of methylation at these regions in the epiRILs could explain the high mC levels. Southern blots with HpaII-digested DNA that assayed methylation of either of the cytosines in the CCGG recognition motif probed with centromeric 180-base-pair (bp) repeats revealed near wild-type mC levels (Fig. 3B). This suggests favored transmission of wild-type-derived centromeres or the efficient remethylation of met1-3 centromeres during inbreeding.
Figure 3.
Inheritance of DNA methylation. (A) mC content in DNA of epiRILs and parental lines. (Left) mC relative to total C. (Right) mC relative to the parental lines (wild type, 100%; met1-3 level, 0%; error bars, SD) (B) Southern blots of HpaII-digested DNA hybridized to probes specific to the centromeric 180-bp repeat (top blot) or the PHB locus (bottom blot). (C) DNA methylation patterns at RAP2.1 and CMT3 loci. Southern blots of HpaII-digested DNA from 17 epiRILs (numbers above the lanes) and parental controls. (WT) Wild type; (m) met1-3. The loci positions are shown on the chromosome above the blot (centromere marked black). Restriction maps corresponding to the fragment sizes (shown at right) are shown in Supplemental Figure 5.
Given such high levels of centromeric methylation, the maintenance of epigenetic variation within euchromatin of these epiRILs was investigated. HpaII sites that are methylated in the wild type at the PHABULOSA (PHB) locus (Bao et al. 2004) were examined (Fig. 3B; Supplemental Table 2 for all primers; Supplemental Fig. 5 for restriction maps). Epi-allele segregation at PHB revealed wild-type-like patterns (epi03, epi07, epi12, and epi15) and hypomethylation (epi18 and epi44), indicating persistence of hypomethylation in euchromatic regions despite highly methylated centromeres (Fig. 3B). These results are consistent with active remethylation of centromeric DNA rather than preferential transmission of wild-type-like chromosomes. Noticeably, the epi49 methylation pattern did not correspond to either parent (Fig. 3B), indicating that hypermethylation can form transgressive epi-alleles.
DNA methylation was also analyzed on chromosome 1 at the RAP2.1 promoter, which was shown previously to be hypomethylated in met1-3 plants (Saze et al. 2003). Here, 14 of 17 epiRILs (82%) had wild-type methylation patterns and only one line had a met-like epi-allele (epi36) (6%) (Fig. 3C). Moreover, epi26 and epi31 appeared to carry both parental epi-alleles, suggesting epi-heterozygosity at an unexpectedly high frequency (12%) (Fig. 3C).
The locus CMT3 was found previously to be differentially methylated during met1-3 inbreeding (Mathieu et al. 2007). As this locus resides on the same chromosomal arm as RAP2.1 (Fig. 3C), possible coinheritance of parental epi-alleles was examined at these two loci. In mCG analysis at HpaII sites, 10 (59%) and 6 (35%) epiRILs were scored as wild-type-like or met-like, respectively; epi05 (met-like) and epi17 (wild-type-like) had faint wild-type and met1-3 signals, respectively (Fig. 3C). These signals may originate from a low epi-allele frequency within the 10 bulked individuals examined (see the Materials and Methods) or somatic epi-allelic instability. A methylation pattern consistent with epi-heterozygosity (epi06) was also detected at the CMT3 locus.
Together, these results confirm that parental epi-alleles can be maintained within the epiRILs for eight generations and support independent assortment of epi-alleles at these two loci within 6 lines (epi05, epi06, epi26, epi30, epi31, epi37) and the possible linkage of wild-type-like epi-alleles in 10 lines (epi13, epi17, epi20, epi22, epi23, epi24, epi27, epi32, epi34, epi48) and met-like epi-alleles in one line (epi36). However, the relative proportions of parental epi-alleles were not 1:1 due to overrepresentation of wild-type-like epi-alleles (82% and 59% for RAP2.1 and CMT3, respectively). There was also a surprising persistence of epi-heterozygotes.
Dynamic epi-allelic interactions within recombined parental chromosomal segments
For any heritable phenotypic variation exhibited by the epiRILs, the underlying genomic regions affecting the phenotype should be associated with a particular epi-allele (e.g., fwa in Fig. 2A) or a combination of epi-alleles. However, given the identical DNA sequence of the parental lines, classical genetic mapping of traits cannot be performed and only DNA methylation-based mapping is appropriate. Therefore genome-wide DNA methylation profiling was performed to gain more information about genomic methylation distribution in the epiRILs. We examined the DNA methylation profiles of three random epiRILs using bisulfite conversion and hybridization to tiling microarrays as previously described (Reinders et al. 2008). Briefly, bisulfite modification of genomic DNA results in cytosine-to-thymine transitions following PCR, but methylated cytosines remain cytosines. Decreased signal intensity of a specific 25-mer array feature upon hybridization of the labeled PCR product to an Affymetrix tiling array reflects an increase in mismatches. Therefore, only highly methylated DNA will conserve sequence complimentarity to the array and produce strong hybridization signals (Reinders et al. 2008). Importantly, such DNA methylation profiling does not differentiate CG from non-CG methylation due to the 25-bp resolution, corresponding to the probe length on an array feature. Therefore, the analysis only used the subset of features described below that clearly distinguished parental epi-alleles.
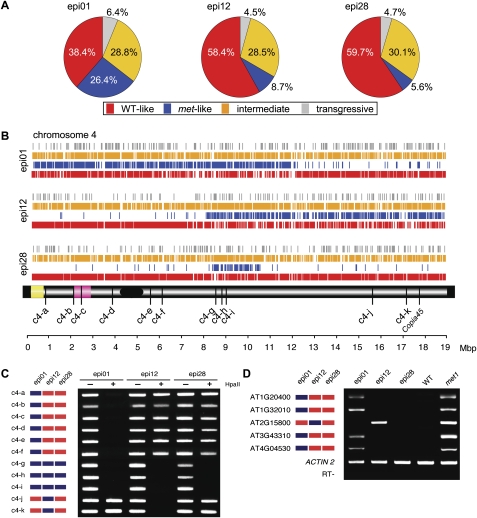
Removal of nonspecific sequences from the >3.2 million array features (Zhang et al. 2008) restricted the analysis to >1.68 million features containing 8,531,303 cytosines. Methylation of ∼5.26% of these cytosines (Lister et al. 2008) distributed in proportion to the other nucleotides within each 25-mer feature sequence would give differential signals due to methylation changes with a maximum of 71,800 features, representing the “methylome.” The results showed that 33,828 (47%), 37,327 (51%), and 39,775 (55%) of the expected features had signals in epi01, epi12, and epi28, respectively, distinguishing the epiRIL from one or both parents (see the Materials and Methods; Supplemental Table 3). On average, only 3% of 6554 control features without cytosines were significantly different from the parental lines for any epiRIL, compared with an expected error rate of 5%. Thus, ∼50% of the methylome represented on the array was detected as differentially methylated in these epiRILs.
For signal differences to be informative with respect to the parental origin of chromosomal regions, it is necessary to assign them confidently to one of the parents. Approximately 33% of the tiles (average of 12,450 tiles) (Supplemental Table 3) detecting differences among the epiRILs showed no differences between the parents and were discarded; only the remaining 67% tiles with clear parental origin were considered (average of 24,526 tiles per epiRIL) (Supplemental Table 3). Within this subset, >65% could be assigned confidently to one of the parents (averaging 16,109 tiles) (Supplemental Table 3). Wild-type-like signals predominated, varying from 38.4% to 59.7% and averaging 52.2% (12,773 tiles) (Fig. 4A; Supplemental Table 3), whereas the proportion of met-like varied from 26.4% to 5.6% between different epiRILs (averaging 13.5%; 3336 of 24,526 total tiles) (Fig. 4A; Supplemental Table 3). Given the appearance of novel, transgressive methylation patterns and the unexpectedly high frequency of epi-heterozygotes detected by Southern blot analysis (Fig. 3B,C), it was anticipated that some signals would not be assignable to their parental origins. Indeed, the fraction of nonparental hybridization signals (intermediate and transgressive) (see the Materials and Methods for the class description) was significant (averaging 34.3%; 12,773 tiles) and relatively constant among the three epiRILs (Fig. 4A; Supplemental Table 3).
Figure 4.
Epigenomic mapping of DNA methylation polymorphisms. (A) Genome-wide DNA methylation polymorphism composition (%) per epiRIL (for more detail, see Supplemental Table 3). (B) Distribution of signals along chromosome 4 for each of three epiRILs and a simplified chromosome 4 map below indicating the centromere (black), the heterochromatic knob (pink), and the nucleolus organizer region (yellow) (distances in Mbp shown at the bottom). The hybridization signals are color-coded as in A. (C) Validation of parental methylation polymorphisms (color-coded as in A and displayed on the left for each locus). PCR amplification was performed with undigested (−) and HpaII-digested (+) DNA. (D) EpiRIL transcription patterns. Methylation profiling patterns are shown on the left (as in C). The epiRIL transcripts were detected by RT–PCR and compared with the parental expression patterns. (WT) wild type; (met1) met1-3. Actin2 and RT− are displayed for each locus (RT− reactions without reverse transcriptase using primers for centromeric repeats as described before in Mathieu et al. (2007).
The methylation polymorphisms along the chromosomes were aligned for each epiRIL. The distribution patterns of nonparental signals appeared random for all chromosomes of each epiRIL, suggesting that nonparental epi-alleles were formed and inherited independently of the parental origin (Fig. 4B; Supplemental Fig. 6). In contrast, wild-type-like and met-like signals appeared in reciprocal blocks, consistent with the visualization of parental regions defined by meiotic recombination break points. Interestingly, the parental chromosomal origins were more clearly visualized by blocks of met-like signals than the wild-type-like signals (blue and red, respectively, in Fig. 4B; Supplemental Fig. 6).
Since progressive remethylation via the RNA-directed DNA methylation (RdDM) pathway (Mette et al. 2000; Wassenegger 2000) may restore wild-type methylation levels at selective targets during inbreeding (Teixeira et al. 2009), the contribution to this process of small RNAs (sRNAs) derived from each parent (Lister et al. 2008) was examined. The distribution of parental sRNAs in a met1-derived region of all three profiled epiRILs (chromosome 4, 8.1–10.1 Mbp) (Fig. 4) across loci prone to methylation changes was compared with random loci. We observed preferential association of especially wild-type sRNAs with epi-alleles prone to methylation changes (Supplemental Fig. 7A) supporting previous observations (Lister et al. 2008). This tendency, consistent with the involvement of sRNAs in remethylation of hypomethylated epi-alleles during inbreeding (Teixeira et al. 2009), was confirmed across the entire chromosome 4 (Supplemental Fig. 7B).
Methylation-sensitive PCR assays directed toward targets on chromosome 4 were used to validate the methylation profiling data. Eleven random loci encoding HpaII sites were examined, for which profiling data suggested hypomethylation in met1-3 relative to the wild type. The results were consistent with the methylation profiling data for all 11 loci across the entire chromosome (Fig. 4C). Associations between inherited epi-allele states and transcript levels were investigated by comparing epiRIL methylation profiling data with previously reported up-regulated expression levels in met1-3 (Lister et al. 2008). Within the three profiled epiRILs, the expression levels for the five examined loci within the observed chromosomal segments corresponded to their parental origin (Fig. 4D).
The overlapping distributions of methylation polymorphisms when a parental segment was shared between the profiled epiRILs were also assessed. For example, the met-like chromosome 4 segments (Supplemental Fig. 8A) shared 16%, 33%, 29%, and 34% of the transgressive, intermediate, met-like, and wild-type-like tiles, respectively (Supplemental Fig. 8A). Very similar patterns were detected within the met-like chromosome 2 region (Supplemental Fig. 8A). Analysis within wild-type-derived segments indicated high levels (>60%) of the expected wild-type-like signals (Supplemental Fig. 8B), low levels (average of 10%) of met-like signals, but surprisingly similar levels of transgressive and intermediate patterns compared with the met-derived segments (25% and 7%, respectively). Since the levels of intermediate and transgressive signals seemed to be independent of parental origin (Supplemental Fig. 8C), these were probably established or maintained independently of linkage-based inheritance of parental epi-alleles.
To further examine whether intermediate methylation levels reflect reduced levels of methylation due to epi-heterozygous states, as indicated above (Fig. 3C), or are associated with novel methylation patterns, Southern blot analysis was performed at a locus encoding a putative Copia45 retro-element (AT4G37705) for which the array data indicated wild-type methylation levels in epi01 and epi28 and an intermediate level in epi12 (Fig. 5A). Given that the informative array feature containing an HpaII site was adjacent to the 3′ LTR of the element, the hybridization probe included the LTR (Fig. 5B). Due to 5′ LTR cross-hybridization, this probe revealed a 9.4-kb DNA fragment in wild type, epi01, and epi28, and a 4.8-kb fragment in met1-3 and epi12 (Fig. 5C). Probe hybridization in the 3′ LTR region of Copia45 revealed a range of HpaII fragments forming patterns reflecting the levels and distribution of DNA methylation in this region (Fig. 5C). These patterns were used as molecular indicators to examine the inheritance of DNA methylation in epi12 at this locus. Analysis of 10 individuals of epi12 at F8 revealed variation in methylation patterns that persisted during the inbreeding process (Fig. 5D). F9 progeny of four random F8 individuals were analyzed to determine how this variability is inherited. Surprisingly, the inheritance of epi-allelic methylation patterns at this locus appeared unstable, revealing a significant degree of stochastic variability in F9 progenies (Fig. 5E). This variability and the novel methylation patterns were not consistent with the epi-heterozygosity. For example, in the F9 progeny of epi12 #09, siblings 3 and 5 regained methylation at the target HpaII site, whereas sibling 4 exhibited novel methylation patterns due to further demethylation. These results are rather unexpected and illustrate the rapid formation of novel methylation patterns.
Figure 5.
Inheritance of DNA methylation patterns at the Copia45 (AT4G37705) locus. (A) Hybridization signals of methylation profiling at Copia45 region for wild type, met1-3, epi01, epi12, and epi28. The parental assignment of informative tiles is provided below each signal graph of epiRILs (color-coded as in Fig. 4A; purple hybridization signal intensity). (B) HpaII restriction map of the Copia45 locus. The diagnostic HpaII site is marked by an asterisk in A and B. (C) Validation of the Copia45 methylation profiles by Southern blot analyses of F8 generation using HpaII-digested DNA isolated from 10 siblings per sample. (D) Southern blot revealing variation of DNA methylation patterns among F8 siblings. (E) Inheritance of DNA methylation patterns to F9 progeny. Four to five progeny (marked 1 to 5) were analyzed for each F8 progenitor plant (marked above the blot).
In contrast to the presence of wild-type-like signals within met-like regions, which can be explained by RdDM, met-like signals interspaced within wild-type-derived regions (Supplemental Fig. 8B) were somewhat surprising, and implied that met-derived demethylated regions impinge on wild-type epigenetic information during the formation and/or inbreeding of mosaic epigenomes. Given the selection of the MET1 allele in the F2 generation, wild-type-derived regions were expected flanking the MET1 locus within each epiRIL and were apparent in the profiling analysis (Fig. 6A). Importantly, the region adjacent to the MET1 gene provided a unique opportunity to test the methylation status at loci that were epi-heterozygous only during the F1 generation.
Figure 6.
Inheritance of DNA methylation within wild-type-derived chromosome 5 region. (A) DNA methylation polymorphism distributions along chromosome 5 (color-coded as in Fig. 4A). The shared wild-type-derived chromosome 5 segments are marked by the red box (15.5–22.2 Mbp) (see also Supplemental Fig. 8B). Physical and genetic distances to the MET1 gene (Liu et al. 1996) for analyzed flanking loci are provided below. (B) Methylation polymorphisms in epi01, epi12, epi28, and an additional 17 epiRILs. Southern blots of HpaII-digested DNA hybridized with probes for loci c5-a to c5-c (details of probe design are given in Supplemental Table 2). Asterisks color-coded as for the methylation profiling indicate methylation patterns differing from wild type. (C) Transgenerational Southern blot analysis of DNA methylation patterns in epi12 (F3 to F9). (D) Southern blot methylation analysis of 10 individual plants of epi12 at F8.
Bisulfite sequencing 10 kb downstream from the MET1 gene, where the array data indicated that all three epiRILs had intermediate patterns, supported non-wild-type methylation in this region (Supplemental Fig. 9). Furthermore, we performed Southern blot analyses with HpaII-digested DNA hybridized with additional probes located in regions linked to MET1, which indicated array signal deviated from wild-type values (Fig. 6B–D). Although the results for a further 20 epiRILs showed predominant wild-type-like patterns (85%–65%), met-like epi-heterozygous and transgressive patterns were also detected in this region (Fig. 6B). In particular, at loci c5-a and c5-b, but not c5-c, transgressive patterns were detected that cannot be explained by inheritance from either parental chromosome. Together, these results strongly suggest that epigenetic variation observed within the MET1 region that was genetically selected at the F2 generation originates from the interaction of met1-3 and wild-type chromosomes in the F1 hybrid.
To examine how these methylation patterns were subsequently inherited, a transgenerational analysis was performed using HpaII-digested pooled DNA from 10 siblings of the F3 to F9 generations for epi12 that appeared wild-type-like and epi-heterozygous in the F8 at c5-b and c5-c, respectively (Fig. 6C). Intriguingly, novel methylation patterns (indicated by two bands at 7.8 and 9.1 kb) were detected at the c5-b locus in early generations (Fig. 6C). At the c5-c locus, epi12 maintained epi-heterozygosity in all generations tested, with variable stochiometry between met-like and wild-type-like patterns in the F7 to F9 (Fig. 6C). Further analysis of DNA from 10 individual epi12 siblings of F8 showed unexpected epi-allelic variation between the plants. Six wild-type-like individuals were detected at the c5-b locus and three siblings had intermediate methylation patterns (Fig. 6D). Moreover, five wild-type-like, two epi-heterozygous and three met-like patterns were detected at the c5-c locus (Fig. 6D). Taken together, these results reveal astonishingly high levels of epigenetic variation inherited both across the population and within individual epiRILs that appear also at the wild-type-derived chromosomal segments.
Genetic instability within the epiRIL population
Since the parental lines were genetically identical, except at the MET1 locus, it was assumed that genetic polymorphisms could be neglected unless genomic instability or transposon movements were induced during epiRIL population development. For example, the DNA transposon CACTA was reported to transpose in ddm1, cmt3-7, and cmt3-7/met1-1 double mutants (Miura et al. 2001; Singer et al. 2001; Kato et al. 2003) but not in met1 single mutants. Nevertheless, the parental lines were examined for possible CACTA transposition.
Even though the parental met1-3 plant used to create the epiRILs was a first-generation met1-3 homozygote, given the previously reported transgenerational epigenetic instabilities in met1-3 (Mathieu et al. 2007), a fourth-generation met1-3 was also included as an additional control for the inbreeding process. Southern blots of DNA from the parental lines showed no transposition of CAC1 or CAC2 (Fig. 7A), the two active CACTA elements previously reported (Miura et al. 2001). Unexpectedly, CACTA transposition was observed in a significant proportion of epiRILs (28%) (exemplified in Fig. 7A; Supplemental Fig. 10). Additional Southern blot analysis of progressive generations of epi07 revealed accumulation of CACTA movements over the generations (Fig. 7B). Random F4 plants of epi07 were examined to test whether the induction of CACTA transposition was gradual in all progeny or was activated in random siblings. Southern blot analysis revealed stochastic transposition of CACTA within specific individuals (Fig. 7C).
Figure 7.
Activation of CACTA transposition in epiRILs revealed by Southern blot analyses of EcoRV-digested DNA and probed as reported previously (Miura et al. 2001). (A) The parental lines wild type, second, and fourth generation met1-3 (WT, m2, and m4, respectively) (left panel), and four random epiRILs (right panel). (B) Transgenerational analysis (F3 to F9) of epi07 (see also Supplemental Fig. 10). (C) Stochastic onset of CACTA transposition in epi07 siblings at the F4 generation.
A simple explanation for the observed transposon activity is that transcription of a known epigenetic regulator was altered in CACTA-active epiRILs, which allowed its movement. However, there was no correlation between mRNA levels of epigenetic regulators and CACTA activity (Supplemental Fig. 11). Thus, the direct involvement of these regulators in CACTA transposition is doubtful and an alternative transposition regulatory mechanism must be sought.
Discussion
Transgenerational heritability of epigenetic marks in plants relies on the faithful propagation of DNA methylation at CG sites (Soppe et al. 2002; Tariq et al. 2003; Mathieu et al. 2005, 2007) and mutants defective in this process accumulate severe developmental abnormalities during inbreeding (Kakutani et al. 1996; Mathieu et al. 2007). Therefore, it has not been possible so far to examine epigenetic variation generated by interfering with maintenance of CG methylation at the population level or to determine stability of its inheritance in many generations. Here we report a strategy of “epigenetic inbreeding” that aims to stabilize Arabidopsis lines with mosaic epigenomes consisting of chromosomal segments derived from the wild-type and mCG-depleted chromosomes.
For some traits, such as flowering time and stress responses, phenotypic classes were apparent and/or quantitative differences were statistically significant. Furthermore, some of these traits appeared to be inherited from the met1-3 parent. These analyses provide the first insight into the extent of phenotypic variability generated by randomly combining epi-alleles and their interactions. The phenotypic variability of the epiRILs was similar to the range of variation among natural Arabidopsis ecotypes occurring in response to Pseudomonas syringae infection, suggesting that it is possible to implement epigenetic breeding techniques to native alleles to expose phenotypic variation buffered by epigenetic mechanisms. This could be especially valuable for an organism with narrow genetic diversity.
However, CACTA transposition was observed in a significant proportion of epiRILs, despite the fact that CACTA is immobile in both parental strains. Therefore, a fraction of these traits may reflect genetic, transposon-induced variation. Yet, the frequencies of phenotypic alterations in CACTA-active and CACTA-silent lines are similar (Supplemental Table 1), and thus, CACTA activity per se does not explain the high degree of phenotypic variation observed within this population.
Although stable inheritance of parental methylation patterns was detected, reflected best by the parental chromosomal origins visualized within the methylation profiles, epigenetic inheritance in the epiRILs at several loci did not follow Mendelian predictions for random segregation of alleles. Genome-wide methylation was 30% higher than the midparent value and overrepresentation of wild type over met-derived epi-alleles was observed (Fig. 3). This could be the result of selection for methylation maintenance at loci contributing to plant fitness and/or active de novo methylation and suppression of the demethylation activities operating within the epiRIL epigenomes, as reported previously for met1 mutants (Huettel et al. 2006; Mathieu et al. 2007). The latter two mechanisms are also likely to be involved in establishing and maintaining the novel methylation patterns detected at several loci (Figs. 3B, 5D,E, 6B–D).
The high level of epigenetic variability and epi-heterozygosity that endured the inbreeding process is most intriguing. Since this also applies to loci in the vicinity of the MET1 gene selected at the F2, “epigenetic conversions” of the wild-type loci to met-like alleles seem to be initiated already in the F1 hybrid. Since passive loss of DNA methylation due to MET1 haplo-insufficiency was excluded previously (Saze et al. 2003), the mechanism could involve active methylation/demethylation resulting from sRNA activities (Vazquez 2006; Mosher et al. 2008; Zheng et al. 2008). Alternatively, a paramutation-like activity (Chandler and Alleman 2008), possibly activated by epi-heterozygosity of the F1, could lead to in trans interactions of epi-alleles on homologous chromosomes, resulting in altered methylation patterns. Yet, these two processes do not adequately explain the surprising persistence of epi-heterozygosity during inbreeding by single-seed descent. We postulate that such metastable epi-alleles either continue interacting, possibly through newly acquired chromatin states or altered epigenetic regulation (Nakayama et al. 2000; Hall et al. 2002), or they retain the intrinsic ability to alternate between epi-allelic states. Similar activities may be involved in the formation of transgressive methylation patterns. Transmission of such a propensity to the progeny would then explain the persistence of apparent epi-heterozygosity and variation in nonparental methylation patterns. Regardless of the molecular mechanisms, likely involving well-established Arabidopsis epigenetic regulators acting downstream from mCGs, our results demonstrate that epi-allele inheritance in epiRILs can significantly deviate from Mendelian expectations.
One of the important questions remaining is whether integrating epigenomic data into a quantitative genetics framework could facilitate mapping of epigenetic quantitative trait loci (epi-QTL). Although epi-QTL mapping is an exciting prospect, the frequent occurrence of dynamic epi-alleles within parental chromosomal segments that do not correspond to that parent may complicate this process. This implies that linkage-based epi-QTL mapping may not be the most suitable, and that associative studies may be more suited for this purpose. A better understanding of the dynamic nature of epigenomes is needed to resolve this issue.
The results also raise interesting questions about the rate of epigenetic evolution. Recent studies addressing the natural variation of epigenetic diversity suggested high stability of epi-alleles on an evolutionary scale, especially for transposable elements (Vaughn et al. 2007; Zhai et al. 2008; Zhang et al. 2008). Here, we show that this stability is fragile and can be lost rapidly when patterns of CG methylation are altered. The implication is that the destabilization of an organism's epigenome, perhaps by a spontaneous or induced down-regulation of DNA methylation (e.g., by aza-cytidine), will rapidly induce novel epigenetic diversity. Since the MET1 gene has a documented gametophytic function and no functional redundancy (Kankel et al. 2003; Saze et al. 2003), its suppression or deficiency (also as a heterozygote), which may occur in nature, would significantly contribute to natural epigenetic variation. This could confer adaptive benefits, as previously suggested (Finnegan 2002; Kakutani 2002; Richards 2006, 2008), but could also be harmful due to loss of fitness. Thus, there is still much to learn about the epigenetic and phenotypic consequences once an epigenome has been “unlocked.”
Materials and methods
EpiRIL population development
The F2 population was derived from one self-fertilized F1 plant resulting from the cross of a wild-type ecotype Col-0 and a met1-3-null mutant (Col-0 background) as the maternal and paternal parents, respectively. The F2 seeds were stratified (2 d) and grown under long-day (LD) conditions (16 h light, 8 h dark, 22°C). F2 plants (n = 396) were genotyped at the MET1 locus, as previously described (Saze et al. 2003). Individuals (n = 96) homozygous for MET1 alleles were selected, grown to maturity, and seeds of each plant were harvested. Aliquots of 100 seeds per plant were sown and one random plant was transplanted after 10 d and advanced to the next generation. This method of single-seed descent was used up to the F7 generation. At the F7, nine random plants per line were advanced and their seeds were bulk harvested to create the F8 seed stock used for analyses.
DNA extraction and methylation analysis
For the HPLC and Southern blot analyses, DNA was extracted from leaf tissue using the Qiagen MaxiPrep kit (Qiagen), with the following modifications. After the column was washed with ethanol, it was spun again (15 min), dried at 37°C (45 min), and eluted. Southern blots were performed as described previously (Mathieu et al. 2007). Total mC was determined as described previously (Rozhon et al. 2008). DNA was extracted from two biological replicates (10 siblings per replicate) and quantified using high-performance cation-exchange chromatography. The biological replicates for each epiRIL were then compared with the wild type using a two-sample t-test (α = 0.05).
Bisulfite methylation profiling and microarray data analysis
Briefly, three biologically replicated DNA samples, each from leaf tissue of 10 individuals, were profiled as described previously (Reinders et al. 2008) with 200 ng of bisulfite-converted DNA subjected to genome-wide amplification. The data were processed as described previously (Zhang et al. 2008). With this approach, 1,683,620 unique perfect match features were spatially corrected and quantile normalized with all arrays together. Log2-transformed signal intensity ratios were made between each epiRIL to each parent using the previously reported parental data (Reinders et al. 2008). Using the limma package (http://www.bioconductor.org) within R (http://www.R-project.org), the empirical Bayes method was used to calculate F-test statistics and was corrected for multiple comparisons using the Benjamini Hochberg method. The number of statistically significant comparisons at the 95% confidence level was determined for each entry.
For signal differences to be scored as wild-type-like, the epiRIL value had to be equal to the wild type and significantly different from met1-3. Conversely, for the epiRIL signal to be scored as met-like, the epiRIL value had to be significantly different from the wild type but equal to met1-3. If the epiRIL signal value was between significantly different parental values, the epiRIL was scored as intermediate. If the epiRIL signal value was significantly greater than or lower than both parental values, the epiRIL value was scored as transgressive.
The data described here have been submitted to the Gene Expression Omnibus (GEO) at NCBI under accession number GSE13438.
Methylation-sensitive cleavage amplification polymorphism assays
After measuring DNA concentrations using a Nanodrop 1000 spectrophotometer (Thermo Scientific), 500 ng of genomic DNA was digested in the presence or absence of methylation-sensitive enzyme (20 μL final volume) according to the manufacturer's instructions. Digestions were incubated overnight at 37°C, followed by a denaturing treatment (15 min at 65°C). Reaction volumes were diluted to 100 μL with TE and equal DNA amounts were used for each PCR assay, followed by electrophoresis in a 1% agarose gel stained with ethidium bromide and visualized under UV light.
RT–PCR analysis
RNA isolation and RT–PCR were as described previously (Mathieu et al. 2007).
Acknowledgments
We thank all members of the Paszkowski laboratory, but especially Dr. Sally Adams, Larissa Broger, Dr. Olivier Mathieu, and Dr. Isabelle Vaillant. We thank Dr. Patrick Descombes for DNA methylation profiling assistance and Pat King for editing the manuscript. The computations using R (http://www.R-project.org) were performed at the Vital-IT (http://www.vital-it.ch) Center for high-performance computing of the Swiss Institute of Bioinformatics. This work was supported by grants from the Swiss National Science Foundation (3100A0-102107), the European Commission through The Epigenome (LSHG-CT-2004-503433), and TAGIP (018785). B.W. was supported by the Human Frontier Science Program.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.524609.
Supplemental material is available at http://www.genesdev.org.
References
- Bao N., Lye K.W., Barton M.K. MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Dev. Cell. 2004;7:653–662. doi: 10.1016/j.devcel.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Chan S.W., Henderson I.R., Jacobsen S.E. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat. Rev. Genet. 2005;6:351–360. doi: 10.1038/nrg1601. [DOI] [PubMed] [Google Scholar]
- Chandler V., Alleman M. Paramutation: Epigenetic instructions passed across generations. Genetics. 2008;178:1839–1844. doi: 10.1093/genetics/178.4.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus S.J., Feng S., Zhang X., Chen Z., Merriman B., Haudenschild C.D., Pradhan S., Nelson S.F., Pellegrini M., Jacobsen S.E. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J., Crooks C., Lamb C. High-throughput quantitative luminescence assay of the growth in planta of Pseudomonas syringae chromosomally tagged with Photorhabdus luminescens luxCDABE. Plant J. 2008;53:393–399. doi: 10.1111/j.1365-313X.2007.03303.x. [DOI] [PubMed] [Google Scholar]
- Finnegan E.J. The role of DNA methylation in plant development. In: Russo V., et al., editors. Epigenetic mechanisms of gene regulation. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1996. pp. 127–140. [Google Scholar]
- Finnegan E.J. Epialleles—A source of random variation in times of stress. Curr. Opin. Plant Biol. 2002;5:101–106. doi: 10.1016/s1369-5266(02)00233-9. [DOI] [PubMed] [Google Scholar]
- Fujimoto R., Kinoshita Y., Kawabe A., Kinoshita T., Takashima K., Nordborg M., Nasrallah M.E., Shimizu K.K., Kudoh H., Kakutani T. Evolution and control of imprinted FWA genes in the genus Arabidopsis. PLoS Genet. 2008;4:e1000048. doi: 10.1731/journal.pgen.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall I.M., Shankaranarayana G.D., Noma K.-i., Ayoub N., Cohen A., Grewal S.I.S. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- Huettel B., Kanno T., Daxinger L., Aufsatz W., Matzke A.J., Matzke M. Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. EMBO J. 2006;25:2828–2836. doi: 10.1038/sj.emboj.7601150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T. Molecular biology. An RNA-guided pathway for the epigenome. Science. 2002;297:2215–2218. doi: 10.1126/science.1077903. [DOI] [PubMed] [Google Scholar]
- Johannes F., Colot V., Jansen R.C. Epigenome dynamics: A quantitative genetics perspective. Nat. Rev. Genet. 2008;9:883–890. doi: 10.1038/nrg2467. [DOI] [PubMed] [Google Scholar]
- Kakutani T. Epi-alleles in plants: Inheritance of epigenetic information over generations. Plant Cell Physiol. 2002;43:1106–1111. doi: 10.1093/pcp/pcf131. [DOI] [PubMed] [Google Scholar]
- Kakutani T., Jeddeloh J.A., Flowers S.K., Munakata K., Richards E.J. Developmental abnormalities and epimutations associated with DNA hypomethylation mutations. Proc. Natl. Acad. Sci. 1996;93:12406–12411. doi: 10.1073/pnas.93.22.12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankel M.W., Ramsey D.E., Stokes T.L., Flowers S.K., Haag J.R., Jeddeloh J.A., Riddle N.C., Verbsky M.L., Richards E.J. Arabidopsis MET1 cytosine methyltransferase mutants. Genetics. 2003;163:1109–1122. doi: 10.1093/genetics/163.3.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., Miura A., Bender J., Jacobsen S.E., Kakutani T. Role of CG and non-CG methylation in immobilization of transposons in Arabidopsis. Curr. Biol. 2003;13:421–426. doi: 10.1016/s0960-9822(03)00106-4. [DOI] [PubMed] [Google Scholar]
- Lister R., O'Malley R.C., Tonti-Filippini J., Gregory B.D., Berry C.C., Millar A.H., Ecker J.R. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.G., Mitsukawa N., Lister C., Dean C., Whittier R.F. Isolation and mapping of a new set of 129 RFLP markers in Arabidopsis thaliana using recombinant inbred lines. Plant J. 1996;10:733–736. doi: 10.1046/j.1365-313x.1996.10040733.x. [DOI] [PubMed] [Google Scholar]
- Mathieu O., Probst A., Paszkowski J. Distinct regulation of histone H3 methylation at lysines 27 and 9 by CpG methylation in Arabidopsis. EMBO J. 2005;24:2783–2791. doi: 10.1038/sj.emboj.7600743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu O., Reinders J., Caikovski M., Smathajitt C., Paszkowski J. Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell. 2007;130:851–862. doi: 10.1016/j.cell.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Mette M.F., Aufsatz W., van der Winden J., Matzke M.A., Matzke A.J. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 2000;19:5194–5201. doi: 10.1093/emboj/19.19.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura A., Yonebayashi S., Watanabe K., Toyama T., Shimada H., Kakutani T. Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature. 2001;411:212–214. doi: 10.1038/35075612. [DOI] [PubMed] [Google Scholar]
- Mosher R.A., Schwach F., Studholme D., Baulcombe D.C. PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proc. Natl. Acad. Sci. 2008;105:3145–3150. doi: 10.1073/pnas.0709632105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J.-i., Klar A.J.S., Grewal S.I.S. A chromodomain protein, Swi6, performs imprinting functions in fission yeast during mitosis and meiosis. Cell. 2000;101:307–317. doi: 10.1016/s0092-8674(00)80840-5. [DOI] [PubMed] [Google Scholar]
- Perchepied L., Kroj T., Tronchet M., Loudet O., Roby D. Natural variation in partial resistance to Pseudomonas syringae is controlled by two major QTLs in Arabidopsis thaliana. PLoS One. 2006;1:e123. doi: 10.1371/journal.pone.0000123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt R.E., Meyerowitz E.M. Characterization of the genome of Arabidopsis thaliana. J. Mol. Biol. 1986;187:169–183. doi: 10.1016/0022-2836(86)90226-3. [DOI] [PubMed] [Google Scholar]
- Reinders J., Delucinge Vivier C., Theiler G., Chollet D., Descombes P., Paszkowski J. Genome-wide, high-resolution DNA methylation profiling using bisulfite-mediated cytosine conversion. Genome Res. 2008;18:469–476. doi: 10.1101/gr.7073008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards E.J. Inherited epigenetic variation—Revisiting soft inheritance. Nat. Rev. Genet. 2006;7:395–401. doi: 10.1038/nrg1834. [DOI] [PubMed] [Google Scholar]
- Richards E.J. Population epigenetics. Curr. Opin. Genet. Dev. 2008;18:221–226. doi: 10.1016/j.gde.2008.01.014. [DOI] [PubMed] [Google Scholar]
- Riddle N.C., Richards E.J. The control of natural variation in cytosine methylation in Arabidopsis. Genetics. 2002;162:355–363. doi: 10.1093/genetics/162.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle N.C., Richards E.J. Genetic variation in epigenetic inheritance of ribosomal RNA gene methylation in Arabidopsis. Plant J. 2005;41:524–532. doi: 10.1111/j.1365-313X.2004.02317.x. [DOI] [PubMed] [Google Scholar]
- Rozhon W., Baubec T., Mayerhofer J., Mittelsten Scheid O., Jonak C. Rapid quantification of global DNA methylation by isocratic cation exchange high-performance liquid chromatography. Anal. Biochem. 2008;375:354–360. doi: 10.1016/j.ab.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Saze H., Mittelsten Scheid O., Paszkowski J. Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat. Genet. 2003;34:65–69. doi: 10.1038/ng1138. [DOI] [PubMed] [Google Scholar]
- Singer T., Yordan C., Martienssen R.A. Robertson's mutator transposons in A. thaliana are regulated by the chromatin-remodeling gene decrease in DNA Methylation (DDM1) Genes & Dev. 2001;15:591–602. doi: 10.1101/gad.193701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppe W.J., Jacobsen S.E., Alonso-Blanco C., Jackson J.P., Kakutani T., Koornneef M., Peeters A.J. The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol. Cell. 2000;6:791–802. doi: 10.1016/s1097-2765(05)00090-0. [DOI] [PubMed] [Google Scholar]
- Soppe W.J., Jasencakova Z., Houben A., Kakutani T., Meister A., Huang M.S., Jacobsen S.E., Schubert I., Fransz P.F. DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J. 2002;21:6549–6559. doi: 10.1093/emboj/cdf657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tariq M., Saze H., Probst A.V., Lichota J., Habu Y., Paszkowski J. Erasure of CpG methylation in Arabidopsis alters patterns of histone H3 methylation in heterochromatin. Proc. Natl. Acad. Sci. 2003;100:8823–8827. doi: 10.1073/pnas.1432939100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira F.K., Heredia F., Sarazin A., Roudier F., Boccara M., Ciaudo C., Cruaud C., Poulain J., Berdasco M., Fraga M.F., et al. A role for RNAi in the selective correction of DNA methylation defects. Science. 2009;323:1600–1604. doi: 10.1126/science.1165313. [DOI] [PubMed] [Google Scholar]
- Vaughn M.W., Tanurd Ic M., Lippman Z., Jiang H., Carrasquillo R., Rabinowicz P.D., Dedhia N., McCombie W.R., Agier N., Bulski A., et al. Epigenetic natural variation in Arabidopsis thaliana. PLoS Biol. 2007;5:e174. doi: 10.1471/journal.pbio.00500174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F. Arabidopsis endogenous small RNAs: Highways and byways. Trends Plant Sci. 2006;11:460–468. doi: 10.1016/j.tplants.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Wassenegger M. RNA-directed DNA methylation. Plant Mol. Biol. 2000;43:203–220. doi: 10.1023/a:1006479327881. [DOI] [PubMed] [Google Scholar]
- Woo H., Richards E. Natural variation in DNA methylation in ribosomal RNA genes of Arabidopsis thaliana. BMC Plant Biol. 2008;8:92. doi: 10.1186/1471-2229-8-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J., Liu J., Liu B., Li P., Meyers B.C., Chen X., Cao X. Small RNA-directed epigenetic natural variation in Arabidopsis thaliana. PLoS Genet. 2008;4:e1000056. doi: 10.1371/journal.pgen.1000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Yazaki J., Sundaresan A., Cokus S., Chan S.W., Chen H., Henderson I.R., Shinn P., Pellegrini M., Jacobsen S.E., et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Zhang X., Shiu S., Cal A., Borevitz J.O. Global analysis of genetic, epigenetic and transcriptional polymorphisms in Arabidopsis thaliana using whole genome tiling arrays. PLoS Genet. 2008;4:e1000032. doi: 10.1371/journals.pgen.1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Pontes O., Zhu J., Miki D., Zhang F., Li W.-X., Iida K., Kapoor A., Pikaard C.S., Zhu J.-K. ROS3 is an RNA-binding protein required for DNA demethylation in Arabidopsis. Nature. 2008;455:1259–1262. doi: 10.1038/nature07305. [DOI] [PMC free article] [PubMed] [Google Scholar]