Abstract
Quality control mechanisms operate in various steps of ribosomal biogenesis to ensure the production of functional ribosome particles. It was reported previously that mature ribosome particles containing nonfunctional mutant rRNAs are also recognized and selectively removed by a cellular quality control system (nonfunctional rRNA decay [NRD]). Here, we show that the NRD of 25S rRNA requires a ubiquitin E3 ligase component Rtt101p and its associated protein Mms1p, identified previously as factors involved in DNA repair. We revealed that a group of proteins associated with nonfunctional ribosome particles are ubiquitinated in a Rtt101–Mms1-dependent manner. 25S NRD was disrupted when ubiquitination was inhibited by the overexpression of modified ubiquitin molecules, demonstrating a direct role for ubiquitin in this pathway. These results uncovered an unexpected connection between DNA repair and the quality control of rRNAs. Our findings support a model in which responses to DNA and rRNA damages are triggered by a common ubiquitin ligase complex during genotoxic stress harmful to both molecules.
Keywords: Ubiquitin, ribosome, genotoxic stress, quality control, rRNA
Gene mutations often result in the production of nonfunctional RNA molecules. In addition, RNA itself is continuously damaged by endogenous and exogenous stress, including ionizing radiation, exposure to certain chemical compounds, and the intracellular generation of reactive oxygen species (Bregeon and Sarasin 2005). Rare but measurable errors in transcription also produce mutant RNAs that do not properly fulfill their roles and aims. In order to avoid a breakdown of cellular order, it is important for cells to detect and selectively dismantle such irregular RNA molecules continuously. It is well documented that various types of aberrant RNAs are selectively removed in eukaryotic cells (Doma and Parker 2007). Three pathways requiring distinct factors degrade different classes of aberrant mRNAs, including mRNAs with a nonsense mutation in their ORFs (nonsense-mediated mRNA decay) (Isken and Maquat 2007), mRNAs with no termination codon (nonstop mRNA decay) (Frischmeyer et al. 2002; van Hoof et al. 2002), and mRNAs with a highly stable structure that prevents ribosomal progression (no-go mRNA decay) (Doma and Parker 2006). Recently, it has been reported that tRNAs with hypomodifications are also selectively degraded in vivo, indicating that stable RNAs are monitored by cellular quality control systems (Kadaba et al. 2004; Chernyakov et al. 2008). However, it is not clear how the quality control of ribosomal RNAs (rRNAs), another species of stable RNAs, is achieved, although rRNAs are highly abundant and essential for life.
The eukaryotic ribosome is a massive ribonucleoprotein (RNP) complex that consists of four rRNAs and about 80 ribosomal proteins (Venema and Tollervey 1999). The precursor 35S rRNA transcribed by RNA polymerase (Pol) I is processed into three parts; 18S, 5.8S, and 25S rRNA. 5S rRNA is synthesized independently by Pol III. The 40S subunit is formed with 18S rRNA and 33 proteins, whereas the 60S subunit is formed with 25S, 5.8S, and 5S rRNA and 46 proteins (46 in budding yeast, 47 in mammalian cells). Each subunit is assembled in the nucleolus and then exported separately through the nuclear pores. In the cytoplasm, the two subunits eventually join to form an 80S particle on the initiation codon of mRNA. The amount and production rate of the ribosomes are strictly regulated by the nutritional condition of the cell, since the biosynthesis of the ribosome is a major energy-utilizing process in the cell (Warner 1999).
Over the past few years, several studies have revealed that the quality of the ribosome is also controlled in various steps of ribosomal production in eukaryotes. It was shown that mutations that impair a specific step in ribosomal biogenesis do not result in the corresponding pre-rRNAs being accumulated in large amounts, suggesting that quality control mechanisms detect and degrade irregular pre-rRNAs that cannot be properly assembled (Hage and Tollervey 2004). Recently, Dez et al. (2006) reported that inactivation of Sda1, a component of pre-60S particles, caused the nuclear retention and polyadenylation of 25S rRNA and 27S pre-rRNA by the TRAMP complex, followed by degradation by the exosome complex. Pre-60S, the TRAMP complex, and the exosome accumulate in a distinct nucleolar focus termed No-body, shortly after Sda1 is inactivated. This structure is considered as the site where RNA components of unsuccessful preribosomes are degraded. Interestingly, mature 40S and 60S are also monitored by quality control system(s) (LaRiviere et al. 2006). LaRiviere et al. (2006) showed that nonfunctional 18S rRNA with a point mutation in its decoding center and 25S rRNA with a mutation in its peptidyl-transferase center (PTC) are both rapidly degraded after their assembly into 40S or 60S particles. Such selective degradation of nonfunctional rRNAs was termed nonfunctional rRNA decay or NRD, but the molecular mechanism of this pathway remained to be elucidated.
In contrast to the degradation of aberrant mRNAs and tRNAs, the degradation of nonfunctional rRNAs may face more difficulties due to their specific spatial arrangement. Because rRNAs are surrounded by 80 ribosomal proteins constituting a very stable RNP complex, the dissociation of these proteins and the exposure of naked RNAs might be required before RNases can access the rRNAs to be digested. However, little is known about the molecular mechanism of RNP degradation.
In this study, we performed a screen for genetic requirements of NRD, using a collection of budding yeast knockout (YKO) strains (Winzeler et al. 1999). We showed that functional 25S NRD requires at least two factors, Mms1 and Rtt101, which are components of an E3 ubiquitin ligase complex involved in DNA repair. We also found that additional ubiquitinated proteins emerged in the ribosomal fractions when nonfunctional 25S rRNAs were expressed. These ubiquitinated proteins were not observed in mms1Δ and rtt101Δ strains, suggesting that nonfunctional ribosomes are ubiquitinated by these two factors. Furthermore, the overexpression of Myc-tagged ubiquitin, which prevents the degradation of target proteins (Hochstrasser et al. 1991), interfered with the degradation of nonfunctional rRNAs, demonstrating the direct role of ubiquitin molecules in NRD in vivo. These results revealed a novel role for ubiquitin in 25S NRD and an unexpected link between DNA repair and the quality control of rRNAs. The biological significance of this link will be discussed.
Results
Nonfunctional mutant rRNAs (A2451U and C2452G) are selectively degraded in yeast
The aim of this study was to reveal the molecular framework of NRD by identifying factors involved in this pathway. To this end, we took advantage of the budding yeast Saccharomyces cerevisiae. This organism was used previously for the demonstration of NRD (LaRiviere et al. 2006).
It was reported that a mutant strain, NOY401, with a temperature-sensitive mutation of RNA Pol I, can survive at nonpermissive temperatures when harboring a plasmid pNOY102 that produces 35S rRNA from the GAL7 promoter (Nogi et al. 1991). On a galactose plate, rRNAs transcribed from pNOY102 by Pol II can compensate for the loss of chromosomal rDNA transcription at a restrictive temperature in NOY401. It was also reported that an 18-nucleotide (nt) tag sequence can be inserted into a nonessential stem–loop of 25S rRNA without any detectable interference with the activity (Musters et al. 1989). A combination of these two technologies, introducing the 18-nt tag into pNOY102, enabled us to monitor the function and to track the fate of mutant 25S rRNAs in vivo. We experimentally confirmed that the resultant plasmid pWT1 can indeed suppress the temperature sensitivity of NOY401 (Supplemental Fig. S1). Essentially the same system was used for the first demonstration of NRD (LaRiviere et al. 2006).
The atomic structure of the 50S ribosome of Haloarcula marismortui suggested A2451 as a putative active site nucleotide of the PTC (Ban et al. 2000). We introduced the transversion mutation A2451U or C2452G into pWT1 (named pA2451U and pC2452G, respectively). These mutations are located in the same positions as the transition mutations A2451G and C2452U previously characterized by LaRiviere et al. (2006), who showed that A2451G mutant rRNA formed nonfunctional 60S subunits and was rapidly degraded, while C2452U formed functional and stable 60S subunits. Recently, another report showed that C2452U is functional in vivo but C2452G is not (Rakauskaite and Dinman 2008). If the NRD machinery recognizes mutant rRNAs by their loss of function, then the nonfunctional C2452G should be recognized and degraded selectively, in contrast to the functional C2452U. If the machinery requires a specific structural change at A2451, C2452G might be spared from NRD.
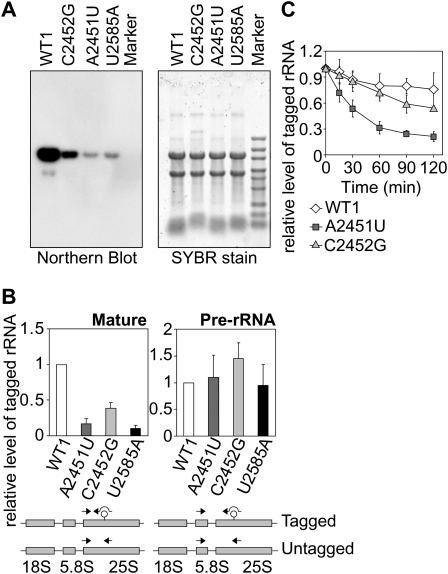
We first confirmed that our mutants (A2451U and C2452G) produce nonfunctional rRNAs, using a NOY401 complementation assay (Supplemental Fig. S1). Then we tested whether these inactive mutant rRNAs are less abundant than the wild-type rRNAs in yeast. As was the case with NRD substrates, the inactive mutant rRNAs created here were less abundant when analyzed by Northern blotting (Fig. 1A) and by quantitative real-time RT–PCR (qRT–PCR; ∼15% of pWT1-derived rRNA for the A2451U mutant, ∼40% for the C2452G mutant) (Fig. 1B). In contrast, the quantity of pre-rRNAs containing the mutation in PTC did not differ significantly from the wild-type counterpart (Fig. 1B). Finally, we also evaluated the stability of the tagged rRNAs and found that the inactive mutants, A2451U and C2452G, are highly unstable in vivo (Fig. 1C). Taken together, we concluded that A2451U and C2452G are novel NRD substrates. The recognition by the NRD machinery here appears to correspond to the loss of function of the mutant rRNAs, rather than to the specific change in the A2451 residue.
Figure 1.
The A2451U and C2452G mutants are selectively degraded in the wild-type yeast strain. (A) Strains expressing tagged rRNAs were cultured in SD-galactose medium to mid-log phase and harvested. Equal amounts of total RNA isolated from each strain were separated by electrophoresis, and the mutant rRNAs were probed by Northern blotting using a probe for the 18-nt tag sequence. The image of the gel stained with SYBR gold shows equal loading in each lane. (B) The mutant rRNAs were quantified by qRT–PCR using the indicated primer sets. The amount of tagged rRNAs was normalized by the amount of untagged rRNAs. (C) Cells at mid-log phase grown in SD-galactose medium were collected and resuspended in SD-glucose medium to shut-off the expression from the GAL7 promoter. The amounts of tagged rRNAs were evaluated by qRT–PCR as B at the indicated time points after transcriptional shut-off.
Mms1 is required for the selective degradation of various 25S NRD substrates
To identify the factor(s) involved in NRD, we started our initial screening with the examination of known RNA degradation pathways by using the newly identified NRD substrates. We tested 18 mutant strains with gene disruptions and one growth condition that prevents RTD (rapid tRNA decay) (Chernyakov et al. 2008). The results clearly showed that the selective degradation of nonfunctional rRNAs was dependent on none of these factors (Supplemental Fig. S2), suggesting that NRD is a novel pathway with previously unidentified factors, although the possibility that multiple pathways are required simultaneously for NRD cannot be excluded. In any case, we decided to screen the YKO collection (Winzeler et al. 1999) for mutants in which nonfunctional rRNAs are stabilized.
Because genes involved in the quality control mechanism for other classes of RNA are in many cases nonessential for yeast growth, we expected that at least some genes involved in NRD might be also nonessential. Each YKO strain contains a null deletion of a single ORF. A deletion of a NRD-related gene would result in accumulation of nonfunctional rRNAs, which can be specifically detected using the 18-nt tag sequence. The yeast colony Northern technique (Stepien and Butow 1990) was used to screen colonies that may accumulate nonfunctional rRNA, and qRT–PCR was employed to subsequently confirm the results.
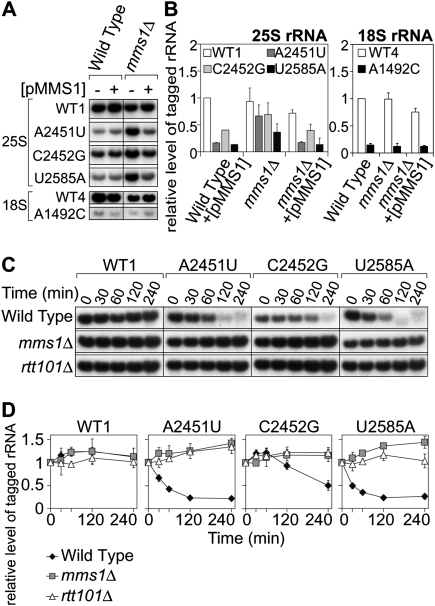
As shown in Figure 2A, in a YKO strain, mms1Δ, the cellular abundance of A2451U rRNAs was comparable with that of pWT1-derived rRNAs. When a plasmid with the MMS1 gene was introduced into these strains, the amount of A2451U rRNA was again reduced, whereas the amount of wild-type tagged rRNA was unaffected (Fig. 2A,B).
Figure 2.
The mutant rRNAs were stabilized in strain mms1Δ. (A) Quantification of the mutant rRNAs in the wild-type and mms1Δ strains. The wild-type strain and mms1Δ with various tagged rRNA plasmids and pMMS1 (+) or an empty vector (−) were grown in SD-galactose medium. Total RNA was isolated, resolved by electrophoresis, and analyzed by Northern blotting. For 25S mutants, a probe for the 18-nt tag in 25S rRNA was used. For the 18S mutant and a control plasmid pWT4, another probe for the 16-nt tag in 18S rRNA was used. (B) The same RNAs were used for the quantification by qRT–PCR. (C) Time-course experiments with tagged 25S rRNA mutants. The indicated yeast strains with various tagged rRNA plasmids were grown in SD-galactose medium, and the medium was replaced by SD-glucose medium at mid-log phase to shut-off transcription from the GAL7 promoter. Cells were harvested and analyzed by Northern blotting at the indicated time points after transcriptional shut-off. (D) The signals in C were quantified.
We also tested whether other NRD substrates could be accumulated in the mms1Δ strain. Besides pWT1 and pA2451U, three plasmids, pC2452G, pU2585A, and pA1492C:18S, were separately introduced into mms1Δ. U2585 neighbors A2451 in the three-dimensional structure, and the mutant U2585A was shown previously to be a NRD substrate (LaRiviere et al. 2006). pA1492C:18S contains a mutation in the decoding center of 18S rRNA. The mutant A1492C:18S was also shown to be another NRD substrate (LaRiviere et al. 2006). We compared the abundance of tagged rRNAs between the wild type and mms1Δ for each mutant rRNA. We found that C2452G rRNA also increased in abundance in mms1Δ to a level comparable with that of wild-type tagged rRNA (Fig. 2A,B). The accumulation of C2452G rRNA in mms1Δ was abolished when a plasmid with the MMS1 gene was cointroduced. Similarly, the reduction of U2585A rRNA was at least partially suppressed in mms1Δ. The half-lives of all 25S NRD substrates tested were prolonged in mms1Δ (Fig. 2C,D). This suggests that Mms1 is involved in the degradation of 25S NRD substrates. Also, no significant difference was observed for the pre-rRNA levels of A2451U, C2452G, and wild type in mms1Δ (Supplemental Fig. S3), indicating the effect of Mms1's deletion on the production of these NRD substrates to be negligible. In contrast, the pre-rRNA level of U2585A in mms1Δ was somewhat lower than the levels of other pre-rRNA species, suggesting that an additional pathway may be involved in the clearance of U2585A (see the Discussion). These results demonstrate that Mms1 is not specific to A2451U, but rather is a more general factor involved in multiple NRD substrates. Interestingly, another NRD substrate, A1492C:18S, which contains a critical mutation in 18S rRNA, was not accumulated in the mms1Δ strain, suggesting that the NRD of the 40S subunit differs from that of the 60S subunit.
Nonfunctional 60S particles are accumulated in the cytoplasm of mms1Δ strain
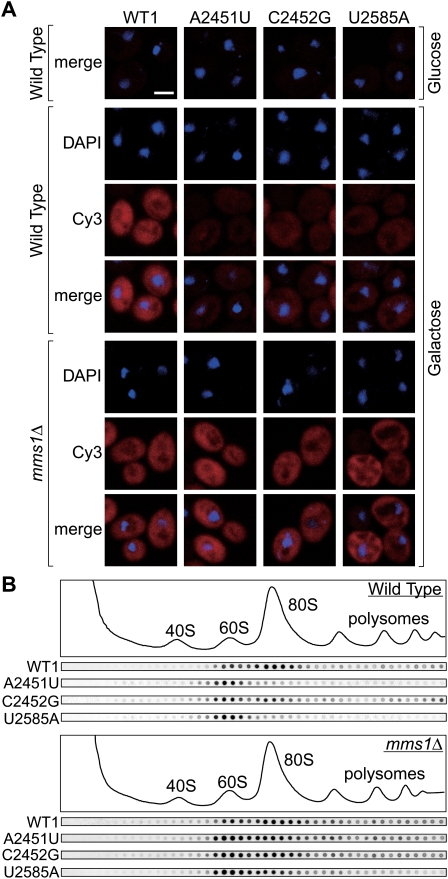
To understand the precise role of Mms1 in NRD, we examined the effect of Mms1's disruption on the subcellular localization of NRD substrates, using in situ hybridization with a probe complementary to the 18-nt tag sequence (Fig. 3A). In the wild-type strain, NRD substrates were observed in both the nucleus and cytoplasm, suggesting that they are exported to the cytoplasm at least to some extent. When inspected more carefully, we noticed that some cells accumulate nonfunctional rRNAs in spots juxtaposed to the nucleus (Supplemental Fig. S4A; see the Discussion). When Mms1 was deleted, the cytoplasmic signals for these substrates increased overall in the mms1Δ strain (Fig. 3A; Supplemental Fig. S4B), indicating that Mms1 is involved in the cytoplasmic clearance of NRD substrates.
Figure 3.
60S particles containing nonfunctional rRNAs are accumulated in the cytoplasm of mms1Δ. (A) Wild-type tagged rRNAs or nonfunctional mutant rRNAs in the wild type and mms1Δ were visualized by in situ hybridization. Cells grown in SD-glucose medium or in SD-galactose medium were used. Bar, 2 μm. (B) The wild type or strain mms1Δ with various tagged mutant rRNAs was grown, and cleared lysate was resolved by a 10%–40% sucrose gradient sedimentation assay. Fractions were monitored by A260 and the amount of tagged rRNAs was visualized by Northern hybridization.
It was reported that the NRD substrates A2451G and U2585A were found in 60S particles prior to their degradation (LaRiviere et al. 2006). However, it was not clear whether our newly constructed NRD substrates, A2451U and C2452G, were incorporated into the 60S or 80S particle. Also, we had no information about the form of the NRD substrates accumulated in the cytoplasm of mms1Δ cells. To address these issues, we performed a sucrose density sedimentation assay, followed by Northern blotting with a probe for the 18-nt tag.
In the wild-type strain, signals for all the NRD substrates tested were found in the 60S fraction, with very few, if any, signals in the lighter fractions (Fig. 3B), indicating that these mutant rRNAs are incorporated into the 60S subunit. This is consistent with the cytoplasmic signals of these rRNAs shown in Figure 3A, since the nuclear export of rRNAs occurs only after the assembly of the 40S/60S subunits. In sharp contrast to the 60S signals, the signals from the 80S fraction and polysomal fractions of each NRD substrate differed significantly (Fig. 3B).
In strain mms1Δ, the patterns of NRD substrates obtained by sucrose density sedimentation differed greatly in several respects. In particular, the signals for 80S particles were significantly intensified for all substrates tested (Fig. 3B; for quantification, see Supplemental Fig. S5). The signals from 60S particles and polysomal fractions were also increased in mms1Δ compared with the wild-type strain, especially for A2451U mutant rRNA (Supplemental Fig. S5). These results of in situ hybridization and sucrose density sedimentation show that 60S- and 80S-containing NRD substrates are ubiquitously accumulated in the cytoplasm of mms1Δ cells. We conclude that Mms1 is required to prevent accumulation of nonfunctional 80S particles in the cytoplasm of wild-type cells.
NRD substrates constitutes noncatalytic ribosomes
Originally, nonfunctional rRNA was identified by showing a deficiency of the complementation of defective rRNA synthesis in the host strain by the mutant rRNA being examined (LaRiviere et al. 2006; Rakauskaite and Dinman 2008). These nonfunctional rRNAs were analyzed and exceptionally short half-lives were observed for all, providing the basis for the concept of NRD. However, we now realize that these results do not necessarily exclude an alternative interpretation. Because the NRD substrates were unstable and mostly absent in the cells used in the complementation assay, we cannot conclude that those mutant rRNAs are catalytically inactive. Instead, it is possible that we occasionally selected highly unstable rRNAs that cannot constitute enough ribosomes. Therefore, whether catalytically nonfunctional rRNAs are recognized and selectively degraded in eukaryotic cells is still an open question.
To determine whether a mutant rRNA is catalytically functional or nonfunctional in vivo, we took advantage of strain mms1Δ, which accumulates several candidates for NRD substrates in the cytoplasm (Fig. 3A,B). We examined if these mutant rRNAs can complement the defective rRNA synthesis in a host strain with a disrupted MMS1. Although we initially employed the temperature-sensitive strain NOY401, the effect of Mms1's deletion was not fully observed at a higher temperature. A2451U was not entirely stable at 37°C even in mms1Δ without the Pol I ts mutation (data not shown) for unknown reasons. Thus we used a conditional knockout strain (Hughes et al. 2000) in which the expression of a subunit of Pol I ceased after doxycycline (Dox) was added to the culture medium.
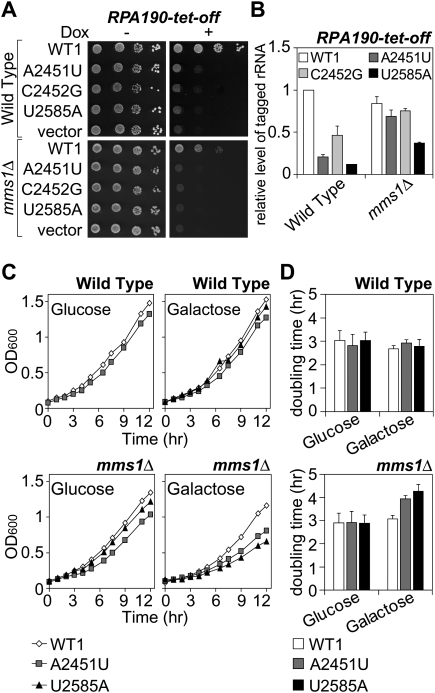
The results are shown in Figure 4A. When Dox was added, the Pol I tet-off strain did not form colonies. Such growth repression by Dox was abrogated by the introduction of pWT1 into the strain. On the other hand, introduction of pA2451U, pC2452G, or pU2585A did not rescue the growth of the Pol I tet-off strain in the presence of Dox. Interestingly, the pattern of complementation by the series of plasmids was exactly the same when the Pol I tet-off-mms1Δ strain was used instead (Fig. 4A). Figure 4B shows that the rRNAs from pA2451U and pC2452G are almost fully stabilized in this mutant, indicating that a sufficient amount of mutant rRNAs from pA2451U or pC2452G cannot compensate for the defective transcription of chromosomal rDNA. Therefore, we concluded that rRNAs with the mutation A2451U or C2452G are not catalytically active enough to support the growth of the yeast strains. Unfortunately, these findings do not lead to any conclusion for U2585A, since the corresponding rRNAs were not sufficiently stabilized by the MMS1 deletion (Figs. 2B, 4B). These results clearly show that a series of catalytically inactive rRNAs is recognized and degraded in eukaryotic cells, as was predicted by LaRiviere et al. (2006) This is the first time that substrates subject to selective degradation have been defined as functionally deficient rRNAs.
Figure 4.
(A,B) The degradation substrates, ribosomes containing A2451U or C2452G, are catalytically inactive. (A) Complementation assay of the Pol I-Tet-off strain and Pol I-Tet-off-mms1Δ strain with various plasmids. Transformants were spotted on SD-galactose ± Dox plates. (B) The amounts of tagged rRNAs from the strains used in A were measured by qRT–PCR assay. (C,D) Accumulation of nonfunctional rRNAs in mms1Δ interferes with cell growth. The wild-type strain and strain mms1Δ with various mutant rRNA plasmids were grown in glucose or in galactose medium. (C) OD600 was monitored and recorded at the indicated time points. (D) The doubling time was calculated for each strain.
Accumulation of nonfunctional rRNAs in mms1Δ is harmful to cell growth
In spite of the apparent importance of the elimination of nonfunctional rRNAs from the cytoplasm, its biological significance has not been assessed. Although it was reported that the growth rate of wild-type S. cerevisiae was not affected by the expression of nonfunctional rRNAs (LaRiviere et al. 2006), it is still unclear if the accumulation of nonfunctional rRNAs in the strain mms1Δ is harmful to the cells. To examine the effect of such an accumulation, we measured the growth rate of mms1Δ in the presence or absence of nonfunctional rRNAs.
As presented in Figure 4, C and D, the growth of the mutant strains with a defect in the Mms1 pathway was significantly slowed by the expression of nonfunctional rRNAs. Moreover, no reduction in the rate of growth was observed when functional (tagged wild-type) rRNAs were expressed. When growth rates were measured in glucose-containing medium that represses the expression of plasmids coding for rRNAs, no differences were found among the transformants.
We also confirmed that no reduction in growth rates on the expression of nonfunctional rRNAs occurred in the wild-type strain (Fig. 4C,D), in which nonfunctional rRNAs are rapidly removed by the Mms1 pathway. When we evaluated the effect of the overexpression of nonfunctional rRNAs in the wild-type strain with a microarray analysis, we could not detect any significant differences in the transcriptome before and after the expression (data not shown). This result also supports the idea that eukaryotic cells are resistant to the occurrence of nonfunctional rRNAs when the Mms1 pathway is intact.
Taken together, these results demonstrate the biological significance of the clearance of nonfunctional rRNAs in eukaryotic cells. This is in contrast to bacteria, which lack an apparent Mms1 homolog. It was reported that expression of nonfunctional mutant rRNAs repressed the growth of Escherichia coli in a dominant-negative fashion (Thompson et al. 2001).
Rtt101 is also required for functional NRD
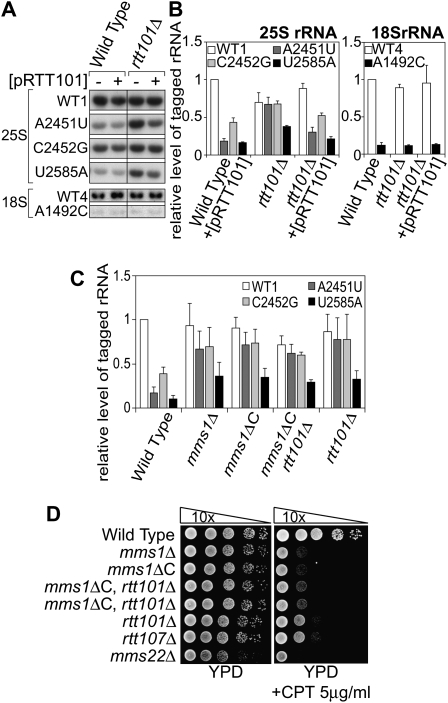
Mms1 was originally isolated as a factor involved in the recovery of DNA damaged by an alkylating agent, methyl methanesulfonate (MMS) (Prakash and Prakash 1977; Hryciw et al. 2002). Also, Mms1 was independently identified as a factor required for the suppression of the Ty1 retrotransposon element (Scholes et al. 2001). According to functional genomic analyses, Mms1 belongs to the Mms22 module (Pan et al. 2006), members of which (Mms22, Rtt101, Rtt107, and Mms1) form an epistasis group for MMS sensitivity. Physical interactions between the members are also reported (Krogan et al. 2006). Given that our screening of YKO was not perfect, we could not exclude the possibility that other members of the Mms22 module are involved in NRD together with Mms1. Thus we performed a qRT–PCR assay using mutant strains with deletions of each of the genes of the module. We found that nonfunctional rRNAs from pA2451U accumulated in strain rtt101Δ as well as strain mms1Δ (Fig. 5A). It seemed that other factors are not involved in NRD (Supplemental Fig. S6).
Figure 5.
Rtt101 is another factor involved in the Mms1 pathway of NRD. (A) The wild type and strain rtt101Δ with various tagged rRNA plasmids and pRTT101 (+) or an empty vector (−) were analyzed as in Figure 2A. (B) The same RNAs were analyzed by qRT–PCR as in Figure 2B. (C) The amounts of various NRD substrates in different backgrounds were evaluated by qRT–PCR. (D) The mutant strains used in this study and related mutants were spotted onto a CPT-containing plate after a series of 10-fold dilutions.
To understand the role of Rtt101 in NRD, we examined whether other NRD substrates are accumulated in rtt101Δ. The results are shown in Figure 5B. The rate for the accumulation of each substrate in rtt101Δ was quite similar to that in mms1Δ. The amounts of A2451U and C2452G were comparable with the amount of control rRNA from pWT1 in rtt101Δ. In contrast, the degradation of U2585A was only partially suppressed in this mutant, as observed in mms1Δ. These effects were abolished when a plasmid with the RTT101 gene was introduced into the host strain, proving that Rtt101 is required for the breakdown of NRD substrates.
As was done in strain mms1Δ, the stability of each NRD substrate was examined in strain rtt101Δ. Figure 2, C and D, shows the time course of the change in amounts of NRD substrates in rtt101Δ, measured by Northern blotting at various time points after transcriptional shut-off. As was observed in mms1Δ, all NRD substrates tested were more stable in rtt101Δ than in the wild-type strain. These results led us to conclude that Rtt101 is another factor required for the degradation of NRD substrates.
In analogy to the DNA repair pathway, Rtt101 and Mms1 may work together in NRD. To examine this issue, we next performed a genetic analysis of these two genes.
We showed that A2451U and C2452G are stabilized in strain mms1Δ or rtt101Δ (Fig. 2C,D). However, there was always a small reduction (10%–15%) in the amounts of A2451U and C2452G even in these mutant strains (Figs. 2B, 5B). In the case of U2585A, the amount of mutant rRNA recovered in mms1Δ and rtt101Δ only partially. These observations were used as indicators for the next epistatic analysis. If Mms1 and Rtt101 work separately in different pathways, the double mutant will accumulate more NRD substrates than the single mutant of each gene. We used a null allele of MMS1 (mms1ΔC) for this assay. mms1ΔC is a chromosomal mutation that lacks the C-terminal half of the MMS1 ORF.
The results are shown in Figure 5C. Clearly, the recovery of U2585A in the mms1ΔC rtt101Δ double mutant was in the same range as that in the single mutants. Also, it seemed that the small reduction of A2451U and C2452G observed in the single mutants remained. The same effect was observed for the DNA repair pathway, when assessed based on resistance to camptothecin (CPT, a topoisomerase I inhibitor) (Fig. 5D). Taken together, we concluded that Mms1 and Rtt101 function in the same pathway of NRD. This conclusion is consistent with the physical interaction between Mms1p and Rtt101p reported recently (Suter et al. 2007).
Mms1–Rtt101-dependent ubiquitination of the ribosomal fraction is induced when nonfunctional rRNAs are expressed
Rtt101p belongs to a family of proteins called cullins, scaffold components of SCF (Skp1–Cullin–F-box protein) ubiquitin E3 ligases (Bosu and Kipreos 2008). It was reported that Rtt101p physically binds to a ring finger-containing subunit Hrt1p and the E2-conjugating enzyme Cdc34p, resulting in the formation of an active ubiquitin ligase complex (Michel et al. 2003). Also, as cited above, Rtt101p physically binds to Mms1p. These results led us to hypothesize that a Mms1p–Rtt101p-containing complex functions in NRD by ubiquitinating certain proteins. The simplest model is that nonfunctional ribosomal particles are ubiquitinated before degradation, similar to the ubiquitination of damaged proteins before proteolysis.
To examine this model, we searched for ubiquitinated proteins in the ribosomal fraction in the presence or absence of nonfunctional rRNAs. Myc-tagged ubiquitin and various NRD substrates (including 18-nt tagged wild-type rRNA as a control) are expressed in a strain that carries Flag-tagged ribosomal protein L28 (RPL28-Flag). The ribosome fraction was affinity purified with RPL28-Flag, and proteins in the fraction were analyzed by SDS-PAGE followed by immunoblotting probed with anti-Myc antibody for ubiquitinated proteins (Fig. 6A). Several intensified signals for ubiquitinated proteins emerged in the presence of nonfunctional rRNAs (Fig. 6A, lanes 2–4). Intensified bands corresponding to 40, 50, and 60 kDa were observed when any three nonfunctional rRNAs were expressed, whereas very faint bands were observed in these areas when wild-type tagged rRNAs were expressed (Fig. 6A, lane 1). These ubiquitinated proteins were not observed when RPL28-Flag was replaced with intact RPL28, showing that the ubiquitination occurred in the ribosome-associated fraction (Fig. 6A, lane 6). The bands observed in the presence of wild-type tagged rRNAs (Fig. 6A, lane 1) were also observed even when the tagged rRNAs were not expressed (data not shown), suggesting that a low level of ribosomal ubiquitination occurs on natural ribosomes in a Mms1–Rtt101-dependent manner.
Figure 6.
Ribosomal fractions containing nonfunctional rRNAs are ubiquitinated in a Mms1–Rtt101-dependent manner. (A) Immunoblotting of ribosomal fractions purified from the wild-type strain expressing various tagged rRNAs, pMyc-Ubi, and RPL28-Flag. Ubiquitinated proteins were probed by anti-Myc polyclonal antibody. For lane 5, an empty vector was used instead of pMyc-Ubi. For lane 6, the wild-type strain with untagged RPL28 was used. (B) In addition to the wild-type strain (lanes 7,8), strain mms1Δ (lanes 9,10) and strain rtt101Δ (lanes 11,12) were used. Both deletion strains carry the Flag-tagged L28 gene. (C) The efficiency of immunoprecipitation with anti-Myc antibody for the wild-type strain expressing wild-type tagged rRNAs or A2451U. Efficiency was calculated based on the amounts of tagged and untagged rRNAs measured by qRT–PCR. The tagged rRNAs from strain A2451U were nonfunctional. All other rRNAs (tagged and untagged rRNAs from strain WT1, and untagged rRNAs from strain A2451U) are functional rRNAs. Myc-Ubi(−) columns indicate the background of anti-Myc. (D) The effect of overexpression of Myc-Ubi on NRD. The wild-type strain with pWT1 or pA2451U containing pMyc-Ubi, pUbi, or an empty vector was grown, and Myc-tagged or untagged ubiquitin was overexpressed. The tagged rRNAs at indicated time points were detected as in Figure 2C. The intensity of the bands was quantified and is summarized in E.
When mms1Δ or rtt101Δ was used as a host strain for the expression of Myc-Ubi and NRD substrates, these ubiquitinated proteins were mostly (if not completely) eliminated from the ribosomal particles precipitated (Fig. 6B, lanes 10,12). This result led us to conclude that ribosomal or ribosome-associated proteins are ubiquitinated in a Mms1–Rtt101-dependent manner when nonfunctional rRNAs exist.
We also confirmed that ubiquitinated ribosomes contain nonfunctional rRNAs. When the ubiquitinated ribosomes were immunoprecipitated by the anti-Myc antibody from the ribosomes eluted by 3× Flag peptides, nonfunctional mutant rRNAs were always precipitated more efficiently than functional rRNAs (Fig. 6C). This is in contrast to the immunoprecipitation by anti-Flag antibody, which is totally unrelated to the functionality of rRNAs (data not shown).
We showed that both Mms1 and Rtt101 are required for the ubiquitination of ribosomes containing nonfunctional rRNAs and the subsequent degradation of those rRNAs. We next examined if the ubiquitination itself is required for NRD.
It was reported that the overexpression of a N-terminal Myc-tagged ubiquitin stabilized ubiquitinated and nonubiquitinated forms of target proteins (Hochstrasser et al. 1991), presumably by interfering with the proteasomes and ubiquitin-binding proteins. If ubiquitin molecules, besides the ubiquitin ligase complex with Mms1p and Rtt101p, play a role in NRD, the overexpression of Myc-tagged ubiquitin might interfere with the degradation of nonfunctional rRNAs. To explore this possibility, we transiently overproduced Myc-Ubi with the CUP1 promoter by adding CuSO4 (Supplemental Fig. S7A,B) and tracked the fate of nonfunctional rRNAs after the transcriptional shut-off of the GAL7 promoter. By Northern blotting of tagged rRNAs, we showed that a nonfunctional rRNA of A2451U was significantly stabilized when Myc-Ubi was overproduced (Fig. 6D, for quantification, see E). When a comparable amount of untagged ubiquitin was overproduced from a control plasmid (Supplemental Fig. S7A,B), no interference in the degradation of A2451U was observed. These results indicate that ubiquitin molecules are directly involved in the clearance of nonfunctional rRNAs.
Discussion
Identification of a complex involved in NRD
In this study, we searched for the factors involved in the degradation of nonfunctional 25S rRNA, and revealed that Mms1 and Rtt101 are both required for this pathway. Also, based on a report that Rtt101 constitutes a ubiquitin E3 ligase complex (Michel et al. 2003), we explored the role of ubiquitin molecules in the degradation of nonfunctional rRNAs. The results presented here clearly showed that ubiquitin molecules are conjugated to certain protein(s) in the ribosomal fractions in a Mms1–Rtt101-dependent manner when nonfunctional rRNAs are expressed. The experiment in which Myc-tagged ubiquitin was overexpressed revealed the direct involvement of ubiquitin molecules in the NRD pathway.
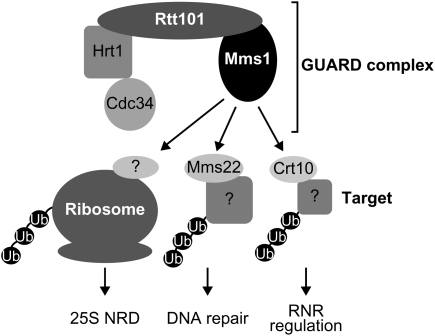
Mms1 and Rtt101 were originally isolated as factors involved in the repair of DNA (Prakash and Prakash 1977; Hryciw et al. 2002) and in the suppression of Ty1 retrotransposable elements (Scholes et al. 2001). Rtt101 is a member of the cullin family, which constitutes ubiquitin E3 ligase complexes as a scaffold (Bosu and Kipreos 2008). It was shown biochemically that Rtt101 binds to Mms1, Mms22, an E2 conjugating enzyme (Cdc34), and a ring finger protein (Hrt1) (Krogan et al. 2006; Suter et al. 2007). Among these proteins, Rtt101, Mms1, and Mms22 form an epistasis group (Pan et al. 2006), suggesting that they function together in the same complex. In this study, we examined whether Mms22 is also involved in NRD, but found no such evidence. These results lead us to propose the following hypothesis. The core ubiquitin ligase complex may consist of Mms1p, Rtt101p, Cdc34p, and Hrt1p, and Mms22p may be an adaptor protein that recruits factors involved in DNA repair to the core complex for their ubiquitination. Recently, it was reported that Mms22p and Crt10p (a transcriptional regulator of subunits of the ribonucleotide reductase) physically bind to Rtt101p in a Mms1p-dependent manner (Zaidi et al. 2008). Crt10p may be another adaptor protein for the regulation of the ribonucleotide reductase. In the case of ubiquitination for NRD, another unidentified adaptor protein will replace these two factors to recruit ribosomes containing nonfunctional rRNAs to the core complex. Since this complex is involved in both DNA repair and NRD, we hereinafter term the core complex the GUARD complex, for genotoxic stress-related ubiquitin ligase associated with RNA and DNA damage.
Recognition of NRD substrates by GUARD complex
In the present and previous studies, it was shown that various nonfunctional rRNAs are selectively degraded in vivo. If loss of enzymatic activity is the only criterion for the degradation, and if nonfunctional rRNAs all follow the same pathway for degradation, they should have similar half-lives. However, we and others observed various half-lives for different nonfunctional rRNA species. Based on the behavior of U2585A, we hypothesize that at least two pathways exist for the degradation of nonfunctional rRNAs. Unlike A2451U and C2452G, U2585A might be a good substrate in the alternative pathway, because the degradation of U2585A is only partially suppressed in strain mms1Δ. We observed that the pre-rRNA level of U2585A in mms1Δ was somewhat lower than other pre-rRNA species (Supplemental Fig. S3). This reduction of pre-rRNA should at least partly contribute to the incompleteness of the recovery of mature U2585A rRNA in mms1Δ. It is possible that unidentified quality control mechanism(s) remove some portion of U2585A pre-rRNA in the nucleus. Since the pre-rRNA level of U2585A in wild-type strain is not lower than that of other pre-rRNA species (Fig. 1B; Supplemental Fig. S3), this alternative (Mms1-independent) pathway might be triggered only when the Mms1 pathway is inactivated. Further experiments will be necessary to address this point.
On careful inspection, we observed about 10% degradation of A2451U and C2452G even in strains mms1Δ (Fig. 2B) and rtt101Δ (Fig. 5B), suggesting that an unidentified alternative pathway is also involved in the degradation of A2451U and C2452G. It is possible that the combination of two parallel pathways with different reaction rates and different preferences for mutants determines the rate of decay, depending on the mutation. Although we focused on the PTC mutants in this study, it would be worth examining whether other types of mutant rRNAs are recognized and degraded selectively and whether the GUARD complex is involved in the degradation.
We showed in this study that the amount of nonfunctional rRNA incorporated into 80S particles increased in the mms1Δ strain (Fig. 3B; Supplemental Fig. S5). This result prompted us to build two models for the substrate recognition mechanism of the GUARD complex. In one model, the complex recognizes nonfunctional 60S subunits and conjugates ubiquitin onto them, inhibiting the incorporation of nonfunctional 60S subunits into 80S particles. When Mms1 is missing, the ubiquitination of nonfunctional 60S subunits is eliminated, resulting in the accumulation of nonfunctional and harmful 80S particles in the cytoplasm. In the second model, the GUARD complex recognizes nonfunctional 80S particles and conjugates ubiquitins to them, leading to the degradation of the nonfunctional rRNAs contained in the particles. When Mms1 is absent, 80S particles, the target of the ubiquitination, are accumulated. At present, we do not know which model is correct; however, we prefer the latter, because we cannot assume that there is a simple way to distinguish functional and nonfunctional 60S subunits. It would be extremely difficult to identify nonfunctional 60S subunits based on structural changes. On the other hand, it might be easier to distinguish functional and nonfunctional 80S particles, since a “functional test” of the particles is theoretically possible for 80S particles. For instance, long-term retention of a certain elongation factor on the nonfunctional 80S could become a good signal for the ribosomes with various kinds of defects. In this study, we showed that C2452G forms nonfunctional 60S subunits and is degraded by the Mms1 pathway, although in sharp contrast another substitution of the same nucleotide, C2452U, constitutes functional and stable 60S subunits (LaRiviere et al. 2006). This supports the second model that the enzymatic function of ribosomes rather than structural change at the active site is important for NRD. Further analysis and identification of the interacting partners of the GUARD complex will be required to clarify which model is correct.
The fate of ubiquitinated ribosomes
Although we observed at least three ubiquitinated protein bands in a GUARD complex-dependent manner (Fig. 6B), the relevant targets of GUARD complex ubiquitination are still unknown. Far more details will be revealed when the target proteins are identified from among the 79 ribosomal proteins and more than 180 proteins associated with ribosomes (Fleischer et al. 2006).
The ubiquitinated ribosomes might be disassembled in close proximity to the nucleus, considering the results shown in Figure 3A and Supplemental Figure S4A. When inspected carefully, the nonfunctional rRNAs were found to be relatively concentrated in characteristic structures juxtaposed to the nuclei in some cells (Supplemental Fig. S4A). These structures are missing in strain mms1Δ, suggesting that Mms1 is involved in the hypothetical relocalization of nonfunctional rRNA during NRD.
In a recent report by Kaganovich et al. (2008), it was shown that a specific class of misfolded proteins are collected in a distinct structure (termed JUNQ, juxtanuclear quality control compartment) next to the nucleus in a ubiquitination-dependent fashion, where they are degraded by enriched proteasomes. It is worth examining whether the structure observed in this study (Supplemental Fig. S4A) is related to JUNQ. Since we now know that ubiquitination is also necessary for the clearance of nonfunctional ribosomes, it is possible that the same or a very similar system is used in both cases.
Recently, it was reported that mature ribosomes are selectively digested by a specific type of autophagy (termed ribophagy) under conditions of nitrogen starvation (Kraft et al. 2008). That study also showed that the deubiquitination enzymes Bre5 and Ubp3 are required for the degradation of ribosomes. From these observations and the results presented here, an overall picture of ribosomal turnover gradually emerges. Ribosomes in a steady state contain a variety of ubiquitinated proteins even when nonfunctional rRNAs are absent (Fig. 6A, lane 1). Also, a comprehensive study of ubiquitinated proteins showed that most ribosomal proteins and ribosome-associating proteins can be ubiquitinated to some extent (Peng et al. 2003). Nevertheless, when focusing on each protein in the ribosomal complex, we estimate that only a small fraction of each protein is ubiquitinated in the steady state, because the intensity of each ubiquitinated protein in Figure 6A is relatively low (lane 1, cf. lanes 2–4; note that only a small portion of rRNAs are nonfunctional in lanes 2–4, although distinct ubiquitination signals are clearly observed). When the function of the ribosome is disrupted by RNA damage, the GUARD complex ubiquitinates a large fraction of certain protein(s) in the ribosome, leading to the selective degradation of nonfunctional rRNAs. On the other hand, under conditions of nitrogen starvation, ribosomes have to be digested to supply nutrition to the cell, regardless of their functionality. Such conditions should require a different type of ribosomal modification; that is, deubiquitination by Bre5 or Ubp3 (Kraft et al. 2008). Thus the stability of ribosome particles greatly depends on a delicate balance of ubiquitination among the components. One of our next aims will be to find out how cells maintain the fine balance of ribosomal ubiquitination and how changes in this balance can trigger downstream events.
Coordination of responses to nucleic acid damages by GUARD complex
We revealed that the GUARD complex is required for the clearance of nonfunctional rRNAs, as well as being involved in DNA repair. It would be reasonable for cells to place these distinct pathways under the control of the same complex, considering that these pathways have to be activated at the same time. Under genotoxic conditions harmful to genomic DNA, rRNAs, which are the predominant cytoplasmic RNAs, will also be damaged. In such a situation, the GUARD complex should (1) repair DNA damage by recruiting Mms22p, (2) optimize dNTP concentrations with Crt10p through the transcriptional regulation of ribonucleotide reductases, and (3) remove damaged and nonfunctional harmful rRNAs with currently unidentified adaptors (Fig. 7). Then, why is the GUARD complex also required for the suppression of Ty1? Curcio et al. (2007) showed that the transposition of Ty1 was enhanced by the treatment of cells with hydroxyurea (HU). HU treatment is a typical genotoxic stress known to down-regulate cellular dNTPs and induce replication fork arrest (Koc et al. 2004). This result suggests that cells under genotoxic stress face a crisis of genomic destruction caused by insertions of Ty1, in addition to DNA damage and RNA damage. Therefore, it would also make sense for the GUARD complex to suppress the transposition of Ty1. The GUARD complex with various recruited adaptors might be a molecular guardian that deals with a variety of problems caused by genotoxic stress. In the future, we need to clarify how the expression of this complex is regulated, how the adaptors are exchanged, and what the targets of ubiquitination are, under stressful conditions.
Figure 7.
A model for various functions of GUARD complex. For the NRD pathway, the adaptor molecule is unidentified. For DNA repair and RNR regulation, adaptors (Mms22 and Crt10, respectively) have been identified but the substrate of ubiquitination is unknown.
Materials and methods
Plasmids
pNOY102 was a gift from Dr. M. Nomura (University of California at Irvine). pWT1 was made by inserting an 18-nt sequence into pNOY102, as described previously (Musters et al. 1989). pA2451U, pC2452G, and pU2585A were established by introducing a single point mutation using overlap extension PCR with pWT1 as the templates. A 16-nt tag sequence was inserted into pWT1 to produce pWT4. pA1492C:18S was created by introducing a point mutation into pWT4. The numbering system for E. coli rRNA was used to assign the corresponding nucleotides in yeast, as was used by LaRiviere et al. (2006).
pUbi was made by the cloning of a PCR fragment containing a CUP1 promoter-Ubiquitin-CYC terminator cassette from YEp96 (kindly provided by Dr. D. Finley) into pYO323. pMyc-Ubi was created by inserting the Myc tag sequence at the start codon of the ubiquitin gene of pUbi.
For pMms1 and pRtt101, each ORF was amplified by PCR with the upstream and downstream 1 kb. Those PCR fragments were cloned into YCplac111.
Yeast strains and growth conditions
The YKO collection and yTHC collection were purchased from Openbiosystems. NOY401 was provided by Dr. M. Nomura. To delete the MMS1 gene in a RPA190-Tet-off strain in the yTHC collection, a hygromycin resistance cassette from pFA6 (gift from Dr. Y. Murakami) was used for selection after targeting. The mms1ΔC strain was made by transforming a PCR cassette with the LEU2 gene into the wild-type strain. The fragment between the 401st codon and the stop codon in the MMS1 gene in the genome was replaced by a short Flag peptide sequence (the C-terminal 1007 amino acids are lost). Strain mms1ΔC rtt101Δ was made similarly to strain rtt101Δ in the YKO collection.
The Cyh2-Flag strain was established as follows. First, the mutation Q38E (a cyh2 mutation, conferring resistance to cycloheximide) was introduced into a PCR fragment of the RPL28(YGL103W) gene containing its promoter, its ORF and a terminator. A Flag tag was then added to the C terminus of this gene by overlap extension. The resultant PCR fragment was directly transformed into BY20693 and cycloheximide-resistant transformants were selected (at 1 μg/mL). About a half of the resistant colonies tested expressed Flag-tagged Cyh2 protein.
The genomic DNA of all the strains modified was checked by two sets of PCR to confirm the presence of the modified allele and absence of the intact allele.
For the expression from the GAL7 promoter, we used SD medium supplemented with 2% galactose. For the expression from the CUP1 promoter, 0.1 mM CuSO4 was added to the culture 2 h before using the cells for the downstream experiments. To terminate the transcription from the GAL7 promoter, cells were harvested and resuspended into prewarmed SD medium with 2% glucose.
Yeast colony Northern blotting
All the strains in the YKO collection (slightly less than 5000) were transformed with pA2451U as described (Akada et al. 2000). The cells were then plated on SD-Gal plates and incubated for 3 d. Colonies were transferred onto a membrane (Hybond-N+, GE Healthcare) and grown overnight. The colonies were treated with Zymolyase and alkaline lysis and washed on the membrane, according to the yeast colony Northern protocol described by Stepien and Butow (1990). The Northern hybridization to detect tagged rRNAs was performed with the probe MK253 labeled with 32P by kination. We used Perfecthyb Plus buffer purchased from Sigma and followed the manufacturer's instructions for the Northern blotting experiments.
RNA and polysome analysis
To extract total RNA from yeast cells, we used the MasterPure Yeast RNA Purification Kit (Epicentre Biotechnologies). Sepasol I Super (Nacalai tesque) was employed for the extraction of RNA from lysates or purified ribosomes. For the polysome analysis, 100 OD600 unit cells were lysed by the glass beads method, and cleared lysate was used for centrifugation with a 10%–40% sucrose gradient. To make the sucrose gradient and to fractionate the samples, Gradient Station model 153 (BioComp Instruments, Inc.) was used. Dot blotting of the fractions was performed by spotting 5 μL of solution from 200-μL fractions onto a membrane. After UV cross-linking for 3 min, the membrane was probed as described above. In the Northern analyses of stability, each lane contained total RNAs from 0.5 ODU of yeast cells.
The qRT–PCR was done using the SuperScript III Platinum SYBR Green One-Step qRT–PCR Kit (Invitrogen) with the Applied Biosystems 7500 Real-time PCR System by the standard curve method. For the quantification of plasmid-derived tagged rRNA, the primers MK251 and MK253 were used. The quantity of chromosome-derived untagged rRNA, measured with the primers Kota030 and Kota031, was used for normalization of the input RNA amount. The sequences of all the primers used are listed in the Supplemental Table.
In situ hybridization
Cells were grown to log phase (OD600 0.8–1.0) and collected by centrifugation, resuspended in 0.1 M K-PO4 (pH 6.0)/4% formaldehyde, incubated for 4 h at 30°C, washed twice with 0.1 M K-PO4 (pH 6.0), and washed once with Sorbitol buffer (0.1 M K-PO4 at pH 6.0/1.2 M sorbitol). The pellet was resuspended in Sorbitol buffer plus 10 mM DTT and spheroplasted with 100T Zymolyase (ICN Biochemials) at a concentration of 1 mg/mL for 30 min at 30°C on slides coated with 0.1% polyethyleneimine (Sigma). The slides were plunged in 70%, 80%, and 90% ethanol for 5 min each and dried. Cells were incubated in hybridization buffer (4× SSC, 5× Denhardt's solution, 1 mg/mL yeast tRNA) for 1 h at room temperature and incubated with 0.4 μM Cy3-labeled oligo DNA probe (MK253) in hybridization buffer overnight at 37°C. Cells washed twice with 4× SSC and twice with 2× SSC for 15 min at 42°C. Nuclei were stained with 125 μg/mL of DAPI. The slides were washed with PBS, mounted, and examined under a Leica TCS SP2 AOBS confocal microscope.
Immunoprecipitation and imunoblotting
The purification of ribosomes by immunoprecipitation was performed using anti-Flag M2 agarose beads from Sigma. The cells were crushed by the glass beads method. IPP150 (10 mM Tris-HCl at pH 8.0, 150 mM NaCl, 0.1% NP40) supplemented with Complete protease inhibitor cocktail (Roche); RNase inhibitor RNasin (Promega) and 20 mM N-ethylmaleimide were used for lysis, antibody–antigen binding, and washing. To elute the ribosomes, the same buffer with 3× Flag peptide (Sigma) at a concentration of 0.1 mg/mL was used. For the immunoprecipitation of ubiquitinated ribosomes, 3× Flag eluates were filtered and precipitated by anti-Myc polyclonal antibody (Sigma) conjugated to rProtein A-Sepharose (GE Healthcare).
For the immunoblotting of ubiquitinated ribosomes, equal amounts of ribosomes in the 3× Flag eluates (determined from rRNA content) were subjected to 12.5% SDS-PAGE. After transfer of the proteins to a membrane, the membrane was probed by anti-Myc polyclonal antibody (1:300; Sigma) or anti-Flag polyclonal antibody (1:300; Sigma). ECL (GE Healthcare) was used for visualization.
Acknowledgments
We thank Drs. Masayasu Nomura, Yasuhisa Nogi, Yota Murakami, and Daniel Finley for generously providing plasmids and yeast strains, and Drs. Katsura Asano, Toru Yoshihisa, Toshifumi Inada, Naoyuki Kataoka, and Ichiro Taniguchi for technical advice and discussions. This work was supported by PRESTO, JST, to M.K.; CREST, JST, to M.O.; and grants from MEXT (Ministry of Education, Culture, Sports, Science and Technology of Japan) to M.K. and M.O. K.F. and A.M. were supported by the 21st Century COE Program of MEXT. A.M. is a JSPS (Japan Society for the Promotion of Science) Research Fellow.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1775609.
Supplemental material is available at http://www.genesdev.org.
References
- Akada R., Kawahata M., Nishizawa Y. Elevated temperature greatly improves transformation of fresh and frozen competent cells in yeast. Biotechniques. 2000;28:854–856. [PubMed] [Google Scholar]
- Ban N., Nissen P., Hansen J., Moore P.B., Steitz T.A. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- Bosu D.R., Kipreos E.T. Cullin–RING ubiquitin ligases: Global regulation and activation cycles. Cell Div. 2008;3:7. doi: 10.1186/1747-1028-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bregeon D., Sarasin A. Hypothetical role of RNA damage avoidance in preventing human disease. Mutat. Res. 2005;577:293–302. doi: 10.1016/j.mrfmmm.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Chernyakov I., Whipple J.M., Kotelawala L., Grayhack E.J., Phizicky E.M. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5′-3′ exonucleases Rat1 and Xrn1. Genes & Dev. 2008;22:1369–1380. doi: 10.1101/gad.1654308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio M.J., Kenny A.E., Moore S., Garfinkel D.J., Weintraub M., Gamache E.R., Scholes D.T. S-phase checkpoint pathways stimulate the mobility of the retrovirus-like transposon Ty1. Mol. Cell. Biol. 2007;27:8874–8885. doi: 10.1128/MCB.01095-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dez C., Houseley J., Tollervey D. Surveillance of nuclear-restricted pre-ribosomes within a subnucleolar region of Saccharomyces cerevisiae. EMBO J. 2006;25:1534–1546. doi: 10.1038/sj.emboj.7601035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma M.K., Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma M.K., Parker R. RNA quality control in eukaryotes. Cell. 2007;131:660–668. doi: 10.1016/j.cell.2007.10.041. [DOI] [PubMed] [Google Scholar]
- Fleischer T.C., Weaver C.M., McAfee K.J., Jennings J.L., Link A.J. Systematic identification and functional screens of uncharacterized proteins associated with eukaryotic ribosomal complexes. Genes & Dev. 2006;20:1294–1307. doi: 10.1101/gad.1422006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischmeyer P.A., van Hoof A., O'Donnell K., Guerrerio A.L., Parker R., Dietz H.C. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science. 2002;295:2258–2261. doi: 10.1126/science.1067338. [DOI] [PubMed] [Google Scholar]
- Hage A.E., Tollervey D. A surfeit of factors: Why is ribosome assembly so much more complicated in eukaryotes than bacteria? RNA Biol. 2004;1:10–15. [PubMed] [Google Scholar]
- Hochstrasser M., Ellison M.J., Chau V., Varshavsky A. The short-lived MATα2 transcriptional regulator is ubiquitinated in vivo. Proc. Natl. Acad. Sci. 1991;88:4606–4610. doi: 10.1073/pnas.88.11.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hryciw T., Tang M., Fontanie T., Xiao W. MMS1 protects against replication-dependent DNA damage in Saccharomyces cerevisiae. Mol. Genet. Genomics. 2002;266:848–857. doi: 10.1007/s00438-001-0605-x. [DOI] [PubMed] [Google Scholar]
- Hughes T.R., Marton M.J., Jones A.R., Roberts C.J., Stoughton R., Armour C.D., Bennett H.A., Coffey E., Dai H., He Y.D., et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- Isken O., Maquat L.E. Quality control of eukaryotic mRNA: Safeguarding cells from abnormal mRNA function. Genes & Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- Kadaba S., Krueger A., Trice T., Krecic A.M., Hinnebusch A.G., Anderson J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes & Dev. 2004;18:1227–1240. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaganovich D., Kopito R., Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koc A., Wheeler L.J., Mathews C.K., Merrill G.F. Hydroxyurea arrests DNA replication by a mechanism that preserves basal dNTP pools. J. Biol. Chem. 2004;279:223–230. doi: 10.1074/jbc.M303952200. [DOI] [PubMed] [Google Scholar]
- Kraft C., Deplazes A., Sohrmann M., Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat. Cell Biol. 2008;10:602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- Krogan N.J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A.P., et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- LaRiviere F.J., Cole S.E., Ferullo D.J., Moore M.J. A late-acting quality control process for mature eukaryotic rRNAs. Mol. Cell. 2006;24:619–626. doi: 10.1016/j.molcel.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Michel J.J., McCarville J.F., Xiong Y. A role for Saccharomyces cerevisiae Cul8 ubiquitin ligase in proper anaphase progression. J. Biol. Chem. 2003;278:22828–22837. doi: 10.1074/jbc.M210358200. [DOI] [PubMed] [Google Scholar]
- Musters W., Venema J., van der Linden G., van Heerikhuizen H., Klootwijk J., Planta R.J. A system for the analysis of yeast ribosomal DNA mutations. Mol. Cell. Biol. 1989;9:551–559. doi: 10.1128/mcb.9.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi Y., Yano R., Nomura M. Synthesis of large rRNAs by RNA polymerase II in mutants of Saccharomyces cerevisiae defective in RNA polymerase I. Proc. Natl. Acad. Sci. 1991;88:3962–3966. doi: 10.1073/pnas.88.9.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Ye P., Yuan D.S., Wang X., Bader J.S., Boeke J.D. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell. 2006;124:1069–1081. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- Peng J., Schwartz D., Elias J.E., Thoreen C.C., Cheng D., Marsischky G., Roelofs J., Finley D., Gygi S.P. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- Prakash L., Prakash S. Isolation and characterization of MMS-sensitive mutants of Saccharomyces cerevisiae. Genetics. 1977;86:33–55. doi: 10.1093/genetics/86.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakauskaite R., Dinman J.D. rRNA mutants in the yeast peptidyltransferase center reveal allosteric information networks and mechanisms of drug resistance. Nucleic Acids Res. 2008;36:1497–1507. doi: 10.1093/nar/gkm1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholes D.T., Banerjee M., Bowen B., Curcio M.J. Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics. 2001;159:1449–1465. doi: 10.1093/genetics/159.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepien P.P., Butow R.A. Yeast colony northern: A fast method for detection of transcripts by colony hybridization. Nucleic Acids Res. 1990;18:380. doi: 10.1093/nar/18.2.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter B., Fetchko M.J., Imhof R., Graham C.I., Stoffel-Studer I., Zbinden C., Raghavan M., Lopez L., Beneti L., Hort J., et al. Examining protein–protein interactions using endogenously tagged yeast arrays: The cross-and-capture system. Genome Res. 2007;17:1774–1782. doi: 10.1101/gr.6667007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J., Kim D.F., O'Connor M., Lieberman K.R., Bayfield M.A., Gregory S.T., Green R., Noller H.F., Dahlberg A.E. Analysis of mutations at residues A2451 and G2447 of 23S rRNA in the peptidyltransferase active site of the 50S ribosomal subunit. Proc. Natl. Acad. Sci. 2001;98:9002–9007. doi: 10.1073/pnas.151257098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A., Frischmeyer P.A., Dietz H.C., Parker R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002;295:2262–2264. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- Venema J., Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- Warner J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- Winzeler E.A., Shoemaker D.D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J.D., Bussey H., et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Zaidi I.W., Rabut G., Poveda A., Scheel H., Malmstrom J., Ulrich H., Hofmann K., Pasero P., Peter M., Luke B. Rtt101 and Mms1 in budding yeast form a CUL4(DDB1)-like ubiquitin ligase that promotes replication through damaged DNA. EMBO Rep. 2008;9:1034–1040. doi: 10.1038/embor.2008.155. [DOI] [PMC free article] [PubMed] [Google Scholar]