Abstract
DNA double-strand breaks (DSBs) are among the most deleterious forms of DNA lesions in cells. Here we induced site-specific DSBs in yeast cells and monitored chromatin dynamics surrounding the DSB using Chromosome Conformation Capture (3C). We find that formation of a DSB within G1 cells is not sufficient to alter chromosome dynamics. However, DSBs formed within an asynchronous cell population result in large decreases in both intra- and interchromosomal interactions. Using live cell microscopy, we find that changes in chromosome dynamics correlate with relocalization of the DSB to the nuclear periphery. Sequestration to the periphery requires the nuclear envelope protein, Mps3p, and Mps3p-dependent tethering delays recombinational repair of a DSB and enhances gross chromosomal rearrangements. Furthermore, we show that components of the telomerase machinery are recruited to a DSB and that telomerase recruitment is required for its peripheral localization. Based on these findings, we propose that sequestration of unrepaired or slowly repaired DSBs to the nuclear periphery reflects a competition between alternative repair pathways.
Keywords: Chromatin, DNA double-strand break, DNA repair, Mps3p, Rad52, 3C
DNA double-strand breaks (DSBs) are among the most deleterious and ubiquitous forms of genomic damage. A single human cell needs to detect and repair tens of DSBs every day (Vilenchik and Knudson 2003). The fact that most cells survive and proliferate despite this high level of genomic damage suggests that they have developed a sophisticated DSB detection and repair system. Much of the damage that occurs in a cell is endogenous in nature. A major source of endogenous DSBs is the result of replication fork collapse at single-strand lesions (mispairing, abasic sites, nicks, etc.) during S phase. Induced DSBs are less common and occur as a result of exposure to chemical mutagens and ionizing radiation. However, not all DSBs are damaging in nature. There are specialized mechanisms for DSB production that operate during meiosis and during immune cell differentiation to generate distinct DNA recombinants (Bassing and Alt 2004; Keeney and Neale 2006 and references therein).
DSBs are repaired by two major pathways—homologous recombination (HR) and nonhomologous end joining (NHEJ), both of which are well-conserved from yeast to humans (Paques and Haber 1999). HR is an error-free process in which a homologous donor sequence is used as a template to repair the DSB. This repair event occurs primarily in the G2 phase of the cell cycle when the homologous sister chromatid is available as the donor (Sung et al. 2003). In contrast, NHEJ involves error-prone end-to-end ligation of the DSB ends, and this pathway is most prevalent in G1 phase of both yeast and human cells (Riha et al. 2006). The choice of DSB repair by the HR or NHEJ pathway is dictated in part by the presence or absence of 5′-to-3′ resection of the DNA ends. This resection has been shown to be controlled by CDK and cyclin activity that is cell cycle-specific (Aylon et al. 2004; Ira et al. 2004). Processing of the DNA ends generates long 3′ single-stranded tails that provide a substrate for Rad52p-mediated formation of a Rad51p nucleoprotein filament. This nucleoprotein filament performs the homology search, followed by strand invasion to form a joint molecule that is a prerequisite for the subsequent recombination events (Paques and Haber 1999).
In addition to HR and NHEJ, cells can use alternative repair pathways to heal the chromosome, often leading to gross chromosomal rearrangements (GCRs). Spontaneous GCRs are most likely due to stalling and collapse of replication forks that lead to DSBs. GCRs include nonreciprocal translocations, chromosome fusions, and isoduplications (Pennaneach et al. 2006). Interestingly these rearrangements reflect much of the genomic instability seen in cancer cells. The NHEJ machinery appears to facilitate formation of spontaneous translocation events, as inactivation of the Lig4p component suppresses chromosome translocations due to inactivation of Rad52p, Mre11p, or Rfa1p (Myung et al. 2001; Myung and Kolodner 2003). Typically, however, translocations constitute a minor percentage of GCR events, whereas most GCRs are a result of deletion and loss of genetic material distal to the DSB and subsequent de novo telomere addition. These events probably occur in the S–G2 phase of the cell cycle when telomerase activity is the highest (Diede and Gottschling 1999). It is unclear whether de novo telomere addition is an aberrant pathway resulting from telomerase recruitment to a DSB that is masquerading as a chromosome end or whether it represents a bona fide repair pathway. This question is further complicated by the fact that much of the DNA damage/checkpoint response consists of proteins that are also implicated in telomere homeostasis (Viscardi et al. 2005).
Several studies have defined mechanisms for recruitment of telomerase to a telomere DNA end during S phase (Diede and Gottschling 1999, 2001; Bianchi et al. 2004; Bianchi and Shore 2008). The yeast telomerase complex consists of Est1p, Est2p (the catalytic component), Est3p, Sm proteins, and the TLC1 RNA (Seto et al. 1999). At natural telomeric ends, long tracts of single-stranded, TG1–3 telomeric repeats provide high-affinity binding sites for Cdc13p, a ssDNA-binding protein that recruits the telomerase machinery by direct interactions with the Est1p subunit (Bianchi et al. 2004). A second pathway for telomerase recruitment also exists that involves interactions between TLC1, Est2p, and the Ku70/80 heterodimer (Stellwagen et al. 2003; Chan et al. 2008) and may involve an independent role for RPA (Schramke et al. 2004). This latter mechanism appears to play a key role in de novo telomere formation at DSBs that lack extensive TG repeats (Stellwagen et al. 2003; Negrini et al. 2007).
Spontaneous GCR events are suppressed by numerous mechanisms, including intact HR and NHEJ pathways (e.g., Rad52p, Lig4p), inhibitors of telomerase (e.g., Pif1p), and a functional DNA damage checkpoint response (e.g., Mre11p, Mec1p) (Myung et al. 2001). Several studies have shown that a SUMO-dependent ubiquitin ligase complex, Slx5p/Slx8p, also suppresses GCR events (Zhang et al. 2006; Nagai et al. 2008). Slx5p/Slx8p are associated with the Nup84 nuclear pore subcomplex, and they can be cross-linked to chromatin surrounding an unrepaired DSB (Nagai et al. 2008). Gasser and colleagues (Nagai et al. 2008) have proposed that Slx5p/Slx8p may target unrepaired DSBs for a SUMO-dependent ubiquitinylation event at the nuclear periphery that activates an alternative repair pathway and suppresses GCR events.
Chromatin structure also plays a central role in the orchestration of the DNA damage response. Immediately following DSB formation, histone H2AX is phosphorylated by the ATM or ATR checkpoint kinases within a large chromatin domain surrounding the DSB (Downs et al. 2000). In addition, several ATP-dependent chromatin remodeling enzymes are recruited to the DSB irrespective of DSB processing (Shroff et al. 2004; Shim et al. 2007). Several groups have suggested that DSB formation may lead to large-scale chromatin changes including decondensation or “relaxation,” and that such changes in chromatin higher-order structure may play a role in subsequent checkpoint activation as well as directly facilitate repair of the lesion (Bakkenist and Kastan 2003; Aten et al. 2004; Kruhlak et al. 2006; Ziv et al. 2006). However, previous studies examining chromatin structure in response to DNA damage involved induction of DNA damage by irradiation that results in many DSBs throughout the genome. Thus it is not clear whether a single DSB also results in dramatic changes in chromosome structure, although it is sufficient to activate the DNA damage checkpoint (Paques and Haber 1999).
To investigate possible changes in chromosome dynamics in response to a single, site-specific DSB in a yeast chromosome, we applied Chromosome Conformation Capture (3C). 3C is a relatively new method that probes the frequency with which two chromosomal regions interact within intact cells (Dekker et al. 2002). This technique has proved invaluable for detecting looping interactions between gene regulatory sequences (Tolhuis et al. 2002; Vakoc et al. 2005), for studying global chromosome dynamics along an entire yeast chromosome (Dekker et al. 2002) and for developing a large-scale interaction map of the nucleus (Dostie et al. 2006; Simonis et al. 2006; Zhao et al. 2006). We find that formation of a DSB within asynchronous yeast cells leads to decreased 3C interactions between chromatin fragments harboring the DSB and all other fragments. We find that changes in 3C interactions correlate with sequestration of the DSB to the nuclear periphery, consistent with a previous study (Nagai et al. 2008). We demonstrate that changes in chromosome conformation and peripheral localization require the nuclear envelope protein Mps3p, and they correlate with recruitment of the telomerase machinery. Furthermore, we present evidence that suggests that tethering of a DSB to Mps3p at the nuclear periphery stimulates GCR events that are antagonized by the Pif1p telomerase inhibitor and Slx5p/Slx8p. Our data suggest that sequestration of unrepaired or slowly repaired DSBs to the nuclear periphery reflects a competition between alternative pathways for healing chromosome breaks, some of which contribute to GCRs.
Results
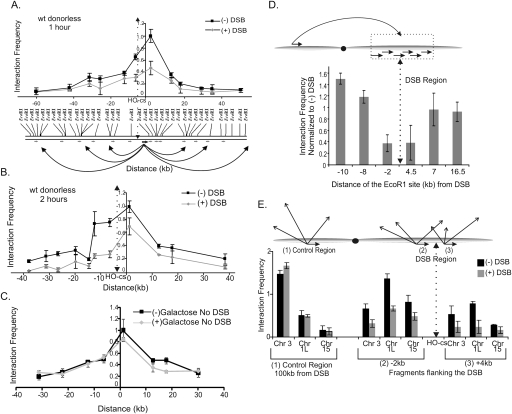
A single DSB was created within the MAT locus on yeast chromosome III by expression of the HO endonuclease from the galactose-inducible GAL10 promoter (Paques and Haber 1999). This DSB is normally repaired using homologous sequences located at two unexpressed HM donor loci (HMLα and HMRa) on each end of the same chromosome, with MATa and MATα strains preferentially using information from HML and HMR, respectively (Paques and Haber 1999). However, in order to eliminate possible complications in the 3C analysis due to recombinational repair, we induced a DSB in a yeast strain where the HML and HMR donor loci have been deleted (JKM179) (Lee et al. 1998). Formaldehyde was added to intact cells to capture chromosome interactions, 1 or 2 h after DSB induction by addition of galactose. Cells were then lysed, and cross-linked chromatin was digested to completion with EcoRI and religated under dilute conditions to generate novel EcoRI junctions. Chromosome interactions were detected by PCR, using a common “anchor” primer located at the DSB region in combination with primers that monitor each of the EcoRI fragments within an ∼100-kb region that surrounds the DSB (Fig. 1A, schematic). 3C analysis in this “donorless” strain showed a dramatic decrease in the interaction frequencies between the fragment bearing the DSB and the surrounding fragments (Fig. 1A,B). These DSB-induced changes in interaction frequencies were not unique to the MAT locus, as similar changes were observed in a strain where an HO-induced DSB was located on chromosome V (Supplemental Fig. S1). Importantly, no changes in interaction frequencies were detected on a chromosome that lacked the DSB (chromosome VI), nor were changes detected in a yeast strain that lacked the HO recognition sequence at MAT (Supplemental Fig. S2 and Fig. 1C, respectively).
Figure 1.
A single DNA DSB causes a decrease in chromosome interaction frequencies. (A,B) 3C interactions of the DSB region with ∼100 kb of surrounding region in the absence of a DSB and at 1 h (A) and 2 h (B) after galactose addition. The DSB (HO-cs) is located on an ∼6-kb EcoRI restriction fragment (see Supplemental Fig. S4). Using a primer at the 3′ end of the DSB-containing EcoRI fragment, we determined its interactions with the surrounding region by PCR using specific primers for each fragment. Primers are shown as small arrows below the schematic. Interactions were normalized to a region on chromosome VI to adjust for experimental variation. (C) The decrease in 3C interactions is not a property of the MAT sequences and does not occur in the absence of a DSB. A DSB was introduced at the LEU2 locus on chromosome III in the strain YMV45 in which the HO recognition site at MAT and the donors are deleted. 3C interactions were determined using the same primer pairs as in A albeit in the absence of a DSB at MAT. (D) Formation of a DSB leads to chromosome-wide decreases in interaction frequencies. Using the same 3C template as in B, 3C analysis was performed using an EcoRI fragment that is ∼106 kb from the DSB. Interactions of this fragment were determined with EcoRI fragments close to the DSB by PCR with primers specific for each fragment. The interaction frequencies were normalized to frequencies in the absence of a DSB. The primer at the 4.5-kb EcoRI site measures the interactions with the fragment containing the DSB. The primer at the −2-kb EcoRI site measures the interactions of the fragment immediately adjacent to the DSB site that is ∼4 kb in size. Hence, this interaction represents the interactions of the region from −2 kb to −6 kb from the DSB. (E) Interchromosomal 3C interactions with the DSB region also decrease. 3C interactions of the EcoRI fragments next to and bearing the DSB—(2) and (3), respectively—were determined with EcoRI fragments from the chromosome III subtelomeric region, the left arm of chromosome I, and the right arm of chromosome XV. The control primer (1) is the same one used in D. Interactions were normalized to a region on chromosome VI to control for experimental variation. In all the panels, error bars represent one standard deviation from the mean for three separate PCRs. Results were confirmed with at least two independent 3C templates.
To further determine the extent of the decrease in chromosome interactions, we chose an EcoRI fragment ∼100 kb distal from the DSB and found that its interactions were also decreased with the fragments next to and bearing the DSB (Fig. 1D). In addition, to determine if the decrease extends to interchromosome interactions, we monitored interactions between the two fragments flanking the DSB with regions on other chromosomes (Fig. 1E). In each case, we detected decreased interaction frequencies, and thus the DSB region appears to be generally sequestered from chromosome interactions located in cis and in trans.
An HO-induced DSB is processed for recombinational repair by exonucleolytic removal of the 5′ to 3′ strands (Paques and Haber 1999). To determine if processing of a DSB is required for the observed changes in chromatin interactions, we induced HO expression and performed 3C analyses in G1-arrested, donorless cells in which exonucleolytic processing of a DSB does not occur (Ira et al. 2004). To our surprise, induction of a DSB in G1 cells did not lead to changes in interaction frequencies (Fig. 2A). Given that DSB formation (Supplemental Fig. S3), detection, checkpoint activation, phosphorylation of histone H2AX, and recruitment of various chromatin remodeling enzymes occur normally at DSB sites in G1-arrested cells (Ira et al. 2004; Shroff et al. 2004; Shim et al. 2007), our results suggest that these processes are likely not sufficient for the changes in 3C interactions detected in cycling cells. Interestingly, we also do not observe a specific loss of interactions between the two EcoRI fragments that span the HO recognition site; these data are consistent with previous studies indicating that the two ends of an HO-induced DSB do not dissociate (Kaye et al. 2004; Lobachev et al. 2004).
Figure 2.
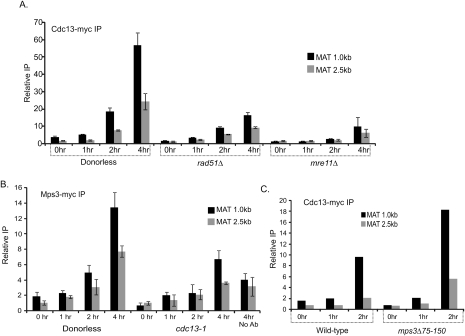
The DSB-dependent decreases in 3C interaction frequencies requires RAD51, RAD52, and MPS3. (A) 3C interactions in G1-arrested cells. CY1276 cells (MATa “donorless,” isogenic to JKM179) were arrested in the G1 phase of the cell cycle by treatment with 4 μM α-factor prior to DSB induction with galactose. 3C analysis was carried out as in Figure 1B using a primer on the DSB-containing fragment and various primers specific for the neighboring fragments. G1 arrest was monitored by flow cytometry, and DSB formation was confirmed by Southern blotting (Supplemental Fig. S3). (B,C) Recombination proteins are required for the decrease in interactions of the DSB seen by 3C. 3C interactions in asynchronous rad51Δ (B) and rad52Δ (C) strains were analyzed as in Figure 1B. (D) The Mps3p N-terminal domain is required for DSB-induced changes in 3C interaction frequencies. 3C interactions in asynchronous wild-type or mps3Δ75-150 cells were analyzed as in Figure 1B. For each panel, representative experiments of at least three independent data sets are shown. Error bars represent one standard deviation from the mean for three separate PCRs. DSB formation was shown to occur normally in each of these mutants (Supplemental Fig. S8).
How can processing of the DSB lead to changes in chromosome interaction frequencies? One trivial possibility is that processing of the DSB results in the destruction of the EcoRI sites that are located ∼4 kb and 2.5 kb from the DSB. However, we find that 90% of cells still harbor the EcoRI site that is 4 kb from the DSB even 2 h after induction of HO (Supplemental Fig. S4A). In addition, we isolated DNA from 3C preparations prior to the ligation step and analyzed the efficiency of EcoRI cleavage by semiquantitative PCR analysis. As shown in Supplemental Figure S4B, the EcoRI site adjacent to the DSB is cleaved as efficiently as a control site on Chromosome V, even 2 h after DSB induction. Thus, the accessibility of the EcoRI site is unaltered after induction of a DSB and considerable end-processing. Furthermore, we found that the efficiency of EcoRI cleavage in 3C samples provides a qualitative measurement of the extent of formaldehyde cross-linking, and thus, the nearly equivalent levels of EcoRI cleavage in samples that contain or lack a DSB indicate that processing of the DSB does not alter cross-linking efficiency (Dekker 2007).
Since processing of the DSB is essential for repair by HR, we tested whether components of the recombinational repair machinery might be required for the observed changes in chromosome interaction frequencies. Deletion of the gene that encodes Rad51p or its loading factor, Rad52p, eliminated the DSB-induced changes in interaction frequencies in the donorless strain (Fig. 2B,C). Importantly, neither of these mutations alters the rate or efficiency of DSB processing (Lee et al. 2003), providing further confirmation that processing is not sufficient to alter 3C interactions. In contrast, removal of either Rad54p or Rad55p, neither of which is essential for full recruitment of Rad51p, did not alter DSB-induced changes in interaction frequencies (Supplemental Fig. S5). These data suggest that the decrease in chromosome interactions seen in the vicinity of a DSB depends on the Rad52-dependent loading of Rad51p onto a processed DNA DSB.
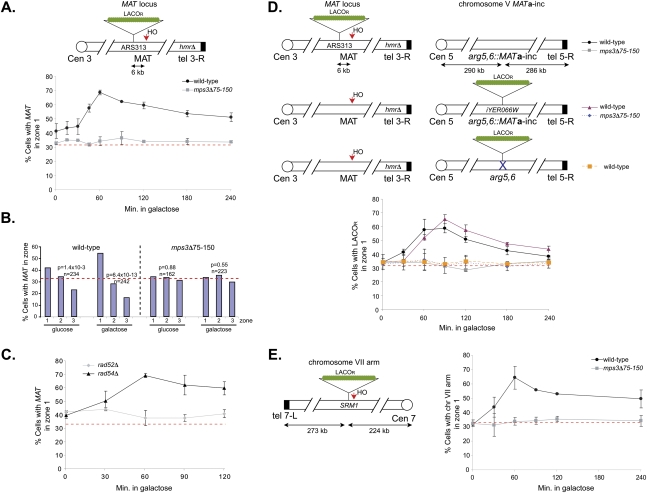
Relocalization of an unrepairable DSB to the nuclear periphery
Our 3C studies indicate that chromatin surrounding the DSB becomes partially sequestered from other chromosomal interactions. This might occur if the DSB becomes compartmentalized within the nucleus, restricting its ability to interact with other chromatin regions in cis and/or in trans. To directly visualize the position of DSBs in the nucleus, we created two donorless yeast strains that contained integrated arrays of lac operators (LacOR) adjacent to an HO recognition site at either the MAT locus on chromosome III or on the arm of chromosome VII (Fig. 3A,E; Belmont and Straight 1998). The expression of a LacI-GFP fusion protein along with the nucleoporin Nup49 fused to GFP allowed us to visualize the LacOR in interphase cells and determine its position relative to the nuclear periphery. In the absence of a DSB, the LacOR was found at the nuclear periphery in ∼30%–40% of cells (Fig. 3, zone 1 represents the outer 33% cross-sectional area of one focal plane of the nucleus and thus a value of 33% represents random distribution; Hediger et al. 2002). However, following formation of the HO-induced DSB, we observe an increase in the percentage of LacOR foci localized to the nuclear periphery for both DSB locations, consistent with a recent study (Nagai et al. 2008). Enhanced peripheral localization was detected within 30 min of galactose addition, indicating that it occurs soon after DSB formation. DSB-induced peripheral localization increased with time, peaking at 60 min following DSB induction and persisting for at least 4 h. Recruitment of LacOR to the nuclear membrane in the presence of a DSB was not due to changes in cell cycle position since it was also observed in cells arrested in metaphase prior to HO induction (Fig. 3B). Importantly, a LacOR located on chromosome VI did not relocalize to the nuclear periphery in response to an HO-induced DSB on chromosome III (Supplemental Fig. S6B; Supplemental Table S1). Furthermore, as was the case for changes in chromosome interactions, the DSB-induced changes in nuclear localization require Rad52p but not Rad54p (Fig. 3C).
Figure 3.
DSBs are sequestered at the nuclear periphery by Mps3p. (A) Schematic of ∼256 LacOR sites integrated at ARS313, which is located on the arm of chromosome III ∼6 kb from the HO cleavage site at MAT. Following induction of the DSB with 2% galactose, the percentage of wild-type and mps3Δ75-150 cells showing localization of this chromosome to zone 1, the outermost region of the nucleus, was determined by epifluorescence microscopy. Localization to other zones is presented in Supplemental Table S1. The red horizontal bar at 33% corresponds to a random distribution. (B) Cells were arrested in mitosis using 15 μg/mL nocodazole and 30 μg/mL benomyl for 3 h at 25°C prior to induction or repression of HO for 2 h with 2% galactose or 2% glucose, respectively. The distribution of ARS313 spots in zones 1, 2, and 3 are indicated. Confidence values (p) for the χ2 test were calculated for each data set between random and test distributions. In addition, a χ2 test was also used to compare the distributions obtained between samples grown in glucose (expected) and galactose (observed); for wild type P = 1.9 × 10−4 and for mps3Δ75-150 P = 7.8 × 10−6. The number of cells examined in each data set is n. (C) rad52Δ and rad54Δ cells containing the HO cleavage site at MAT and LacOR at ARS313 were grown and analyzed as in A. (D) Addition of 2% galactose induced an HO-cleavage at MAT on chromosome III in wild-type and mps3Δ75-150 cells containing the MATa-inc donor sequence at ARG5,6 on chromosome V (top two schematics). Localization was monitored as in A using LacOR sites integrated at ARS313 (top schematic) or iYER066W (middle schematic), respectively, as indicated in the schematic. (Bottom schematic) The “donorless” strain (SLJ2826) contains ∼256 LacOR sites on chromosome V but lacks MATa-inc. (E) Wild-type and mps3Δ75-150 cells containing ∼256 LacOR sites and an HO cleavage site integrated on the arm of chromosome VII were analyzed as in A.
Mps3p is an essential inner nuclear envelope protein required for spindle pole body duplication and telomere positioning at the nuclear periphery (Jaspersen et al. 2002; Nishikawa et al. 2003; Antoniacci et al. 2007; Bupp et al. 2007). Deletions of the acidic N-terminal domain of Mps3p (mps3Δ75-150), which is predicted to extend into the nucleoplasmic space, results in increased sensitivity to DNA damaging agents compared with wild-type cells (Supplemental Fig. S7). The Mps3p N-terminal domain is not essential for spindle-pole body duplication, nor is it required for localization of Mps3p to the nuclear envelope (Bupp et al. 2007). Strikingly, deletion of the Mps3p N-terminal acidic domain eliminates the DSB-induced changes in chromatin interaction frequencies detected by 3C (Fig. 2D), as well as the localization of DSBs to the nuclear periphery (Fig. 3; Supplemental Fig. S6A). The efficiency of DSB formation and recruitment of Rad51p were not affected in mps3Δ75-150 mutants, suggesting that these changes are not simply due to delayed formation of the presynaptic filament that localizes to the nuclear periphery (Supplemental Figs. S8, S9). Instead, chromatin immunoprecipitation (ChIP) analyses indicate that DSB formation leads to the recruitment of Mps3p to adjacent chromatin in a manner that depends on the Mps3p N terminus as well as Rad51p (Fig. 4A), consistent with relocalization of the DSB region to the nuclear periphery. Mps3p recruitment was detected at the DSB by 1 h after galactose addition, which is similar to the kinetics of peripheral localization detected by live cell imaging. However, cross-linking of Mps3p to DSB chromatin was much higher at 2 h, even though the peripheral localization detected by live cell imaging was beginning to decrease at this later time point. A likely explanation is that the DSBs that remain at the nuclear periphery at later time points continue to recruit additional Mps3p protein, leading to an enhanced ChIP signal. Consistent with this view, the DSB region was observed at the nuclear periphery even 4 h after galactose addition in a significant fraction of cells (Fig. 3A,E). Taken together, the combination of 3C, live cell imaging, and ChIP assays indicate that a HO-induced DSB is recruited to the nuclear periphery by interactions with the nuclear envelope protein Mps3p.
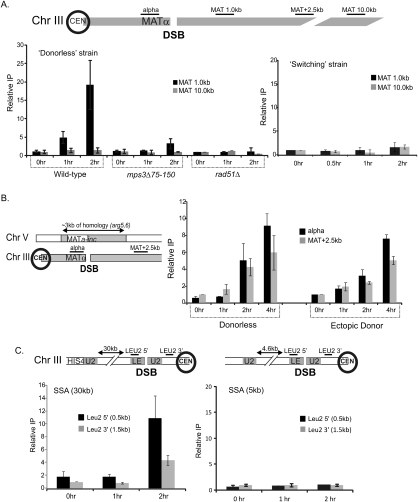
Figure 4.
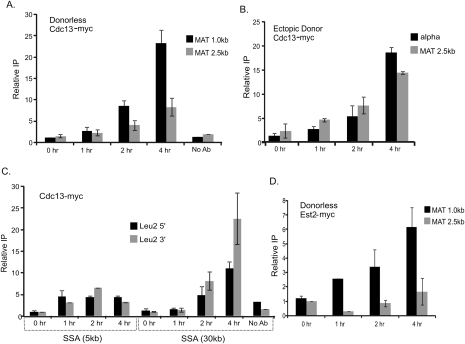
Mps3p interacts with chromatin surrounding a DSB. (A) ChIP analysis of Mps3p was conducted in MPS3-13Myc, mps3Δ75-150-13Myc, and rad51Δ MPS3-13Myc strains using polyclonal anti-myc antiserum (9E10; Santa Cruz Biotechnologies). DSBs were induced in these derivatives of the MATα donorless strain (CY915) as well as the isogenic MATa “switching” strain (CY924), and samples were collected before and 1 and 2 h after induction of a DSB. Immunoprecipitated (IP) DNAs were amplified by real-time PCR using primer pairs for regions either 1 kb or 10 kb distal to the HO recognition site. The percent immunoprecipitated (IP/Input) values were normalized to the percent immunoprecipitated for the control ACT1 ORF. To compare between each of the two independent experiments shown, the percent immunoprecipitated values were normalized to the time 0 samples to yield the fold IP values plotted on the Y-axis. Primers used for ChIP analysis in this and subsequent figures are represented in the schematics accompanying each panel. The “alpha” represents a primer set ∼0.5 kb to the left of the DSB (in the Yalpha region), and MAT 1.0, 2.5, 10.0 represent primers 1.0, 2.5, and 10.0 kb from the DSB. Primer sequences are available on request. (B) Mps3p is recruited to a DSB in the presence of a donor. ChIP analysis was carried out as above for Mps3p in the Ectopic Donor strain (yJK17). yJK17 contains an HO recognition site at MATα on chromosome III, deletions of HML and HMR, and an incleavable MATa-inc locus integrated at arg5,6 on chromosome V that can act as a donor. For ChIP detection, we used amplicons in the unique Yalpha region next to the DSB and a region 2.5 kb from MAT that are not common with the donor. Primer sequences are available on request. (C) Mps3p is recruited to a DSB at a location other than MAT. DSB was induced in the SSA strains YMV2 and YMV45 where the HO-cs is present within the LEU2 gene on chromosome III. The DSB is repaired by SSA after ∼30 kb and ∼5 kb of resection, respectively. Unique primers were designed next to the DSB, and ChIP was carried out and analyzed as above. All ChIPs plotted on the same panel were always carried out simultaneously. Error bars represent one standard error of the mean for three independent experiments.
Localization of repairable DSBs to the nuclear periphery
One possibility is that only a DSB that cannot be repaired is sequestered to the nuclear periphery (e.g., in strains that lack a homologous donor). To test this idea, we used ChIP to monitor recruitment of Mps3p to a DSB that is induced in yeast strains that harbor a homologous donor sequence either at HMLα on the same chromosome (a “switching” strain, JKM154) or at an ectopic location on chromosome V (an ectopic donor strain, yJK17). Consistent with previous live cell imaging (Bystricky et al. 2008; Nagai et al. 2008), Mps3p was not recruited to the DSB when the donor was present on the same chromosome (Fig. 4A, “switching strain”). However, Mps3p was recruited to the DSB when the donor was located at the ectopic location (Fig. 4B). In the latter case, the kinetics and magnitude of Mps3p recruitment were similar to that observed for recruitment to the unrepairable DSB in the donorless strain (Fig. 4B). To confirm that Mps3p recruitment in the ectopic donor strain reflects localization to the nuclear periphery, we introduced a LacOR adjacent to either the HO recognition site on chromosome III or adjacent to the Mata-inc donor sequence on chromosome V and analyzed chromosome position within the nucleus by live cell imaging (Fig. 3D). Consistent with the ChIP results, formation of the DSB led to the Mps3p-dependent relocalization of the LacOR adjacent to MAT on chromosome III to the nuclear periphery. Surprisingly, the LacOR adjacent to the donor sequence on chromosome V also relocalized to the nuclear periphery, but with delayed kinetics compared with the DSB (Fig. 3D). These results indicate that even DSBs that are actively undergoing recombinational repair can be relocalized to the nuclear periphery, and furthermore, the data indicate that completion of the homology search step of HR occurs either during or after this relocalization. This result is quite surprising given that peripheral localization of the DSB leads to a general decrease in 3C interaction frequencies.
To further test whether Mps3p recruitment is a hallmark of unrepaired or slowly repaired DSBs, an HO-induced DSB was created in strains that contain two defective copies of the LEU2 gene that are separated by either 5 kb or 30 kb of intervening DNA (see Fig. 4C for schematic; Vaze et al. 2002). In these strains, the DSB within one copy of LEU2 is repaired by a single-strand annealing (SSA) pathway of HR in which resection of the intervening DNA uncovers homology within the second copy of LEU2. The rate of resection determines the kinetics of DSB repair, with successful repair of the 5-kb reporter occurring within 1–2 h and repair of the 30-kb reporter requiring >5 h. Interestingly, Mps3p was not recruited to the DSB region of the rapidly repaired, 5-kb SSA reporter, whereas robust recruitment of Mps3p was detected with the slowly repaired, 30-kb SSA reporter (Fig. 4C). These data indicate that DSBs that are unrepairable or slowly repaired are competent for relocalization to the nuclear periphery and consequently are bound by Mps3p.
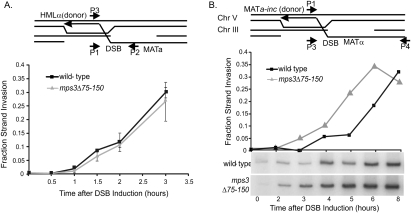
To test whether the MPS3-dependent localization of a DSB has functional consequences for repair, we monitored an early step of HR, strand invasion and extension, in mps3Δ75-150 strains that contained a donor at HMLα (switching strain) or at the ectopic location on chromosome V (see schematic in Fig. 5A). When repair used the donor at HMLα, deletion of the Mps3p N terminus had no significant effect on the rate of strand invasion/extension (Fig. 5A) or on the overall efficiency of repair (data not shown). These results are consistent with the fact that this DSB does not relocalize to the periphery as assayed by Mps3p recruitment (Fig. 4A) and live cell imaging (Bystricky et al. 2008; Nagai et al. 2008). However, when the donor locus was present at the ectopic location, deletion of the Mps3p N-terminal domain enhanced the rate of strand invasion/extension (Fig. 5B). Whereas detection of significant levels of strand invasion/extension product required at least 4 h in the wild-type strain, we consistently observe high levels of product by 2–3 h (Fig. 5B). This result suggests that localization of a DSB to the nuclear periphery inhibits the rate of recombinational repair.
Figure 5.
The N-terminal region of Mps3p impairs ectopic recombination. (A) Mps3p does not influence the rate of recombinational repair between a DSB at MAT and HMLα. Cells (JKM154) were grown in raffinose to OD 0.5, a DSB was induced at MAT by addition of 2% galactose for 1 h (t = 0), and then HO expression was repressed by addition of 2% glucose (t = 1). Strand invasion and subsequent extension of the joint at HMLα was monitored at each time point after glucose addition by real-time PCR analysis with the DNA primers P2 and P3 that are depicted in the schematic. Values were normalized to the ACT1 ORF. Fraction strand invasion value was calculated using the level of the P1–P2 amplicon prior to DSB induction as a value of 1.0. (B) Mps3p inhibits the rate of recombinational repair between a DSB at MAT and an ectopic donor. A DSB at MAT was induced in the Ectopic Donor bearing strain yJK17, and strand invasion was monitored by PCR using primers P1 and P4. (Bottom panel) Primer P4 is located in a region of heterology outside the MAT locus, and due to the large size of the amplicon, PCR with radioactive nucleotides had to be used. Products were quantified by PhosphorImager analysis using ImageQuant ver1.3 for Mac and values were normalized to the levels of ACT1 ORF determined by real-time PCR analysis. Fraction strand invasion was calculated as in A. The experiment shown is representative of four independent experiments. In all experiments, DSB formation was equivalent between wild-type and mps3Δ75-150 strains.
Role of the telomerase machinery in DSB relocalization
Why are unrepairable or slowly repaired DSBs localized to the nuclear periphery? We considered the possibility that peripheral localization might reflect the activation of an alternative pathway for DSB repair. Specifically, we investigated whether peripheral localization is linked to de novo telomere formation, especially in light of the fact that previous studies have demonstrated that the Mps3p N-terminal domain is required for efficient tethering of telomeres to the nuclear periphery (Bupp et al. 2007) and that Mps3p interacts with at least one component of the telomerase complex (Antoniacci et al. 2007). To test whether components of the telomerase machinery are recruited to HO-induced DSBs, we monitored recruitment of Cdc13p and the telomerase catalytic subunit, Est2p, by ChIP. We find robust recruitment of both Cdc13p and Est2p to an unrepairable HO-induced DSB with kinetics that parallel the recruitment of Mps3p (Fig. 6A,D, also cf. Figs. 4A and 7B). In addition, Cdc13p was recruited to the slowly repaired DSBs within the ectopic donor or 30-kb SSA reporter strains, but much less Cdc13p was recruited to the DSB at the rapidly repaired 5-kb SSA reporter (Fig. 6B,C), similar to Mps3p.
Figure 6.
The telomerase machinery is recruited to an unrepaired or slowly repaired DSB. (A) Cdc13p is recruited to a DSB in the donorless strain. ChIP analysis of Cdc13p recruitment was conducted in a “donorless” CDC13-13Myc strain as described in Figure 4. (B,C) Cdc13p is recruited to a DSB that is slowly repaired. Cdc13-myc recruitment was studied in the strain with the Ectopic Donor yJK17 (B) and in the SSA strain YMV2 (C) in a manner similar to A. (D) Telomerase is recruited to a DSB. ChIPs for Est2p-13Myc show that it is recruited to the DSB locus in a donorless cell with kinetics that mirror those of Cdc13p. All ChIPs plotted on the same panel were always carried out simultaneously. Error bars represent one standard error of the mean for at least three independent experiments.
Figure 7.
Recruitment of Mps3p to a DSB requires Cdc13p (A) Recruitment of Cdc13p requires Rad51p and Mre11p. ChIPs for Cdc13-13myc were carried out in isogenic donorless rad51Δ and mre11Δ strains. DSB induction was shown to be equivalent by real-time PCR analysis of DNA extracted in parallel at each time point (data not shown). (B) Optimal recruitment of Mps3p to a DSB is dependent on Cdc13p. ChIP was carried out for Mps3-13myc in a cdc13-1 strain that carries a ts allele of CDC13. Cells were maintained at 22°C throughout their lifetime. ChIP experiments were carried out by inducing a DSB 1 h after shifting cells to 37°C in a water bath. Equal DSB induction was confirmed by real-time PCR. (C) Cdc13p recruitment to a DSB is not dependent on the recruitment of the DSB to the periphery. CDC13-13Myc ChIP was carried out in a donorless strain lacking the N-terminal region of Mps3p. All ChIPs plotted on the same panel were always carried out simultaneously. Error bars represent one standard error of the mean for at least three independent experiments.
Furthermore, we find that Cdc13p recruitment to the DSB requires the Mre11p processing enzyme, and it is largely RAD51-dependent (Fig. 7A). Strikingly, recruitment of Mps3p to an unrepairable DSB was substantially decreased in a cdc13-1 mutant (Fig. 7B), but recruitment of Cdc13p was not disrupted in the mps3Δ75-150 mutant (Fig. 7C). These data are consistent with a model in which loading of the telomerase machinery onto an unrepairable or slowly repaired DSB plays a major role in recruiting the DSB to the nuclear periphery.
Mps3p promotes spontaneous gross chromosomal rearrangements
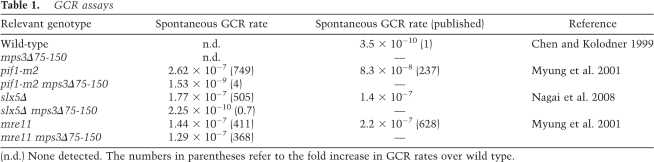
One key question is whether Mps3p also plays a role in the peripheral localization and repair of spontaneous DSBs. In particular, we tested whether Mps3p regulates the occurrence of GCR events. In wild-type yeast, spontaneous GCRs occur at a very low frequency (<3.5 × 10−10) (Chen and Kolodner 1999; Myung et al. 2001), and deletion of the N-terminal domain of Mps3p does not yield detectable GCR events (Table 1). In contrast, inactivation of Slx5p or Mre11p increases GCR events ∼400–500-fold (Table 1). Likewise, GCR events are increased 750-fold in a pif1-m2 mutant that abrogates negative regulation of telomerase activity by the Pif1p helicase (Table 1). In this case, nearly 100% of the GCR events are due to de novo telomere additions (Myung et al. 2001). Strikingly, deletion of the N-terminal domain of Mps3p eliminates GCR events due to inactivation of Slx5p and decreases GCR events in the pif1-m2 mutant by nearly 200-fold (Table 1). In contrast, enhanced GCRs due to inactivation of Mre11p are not significantly decreased by mps3Δ75-150 (Table 1). Thus, the N-terminal domain of Mps3p is not generally required for high rates of GCR, but our data suggest that Mps3p is in the same pathway(s) as Slx5p and Pif1p. Furthermore, these data suggest that Mps3p-dependent tethering of spontaneous DSBs to the nuclear periphery is required for GCR events in the absence of Slx5p and that peripheral localization is required for efficient de novo telomere formation in the absence of Pif1p.
Table 1.
GCR assays
(n.d.) None detected. The numbers in parentheses refer to the fold increase in GCR rates over wild type.
Discussion
Here we used 3C to monitor the frequency of intrachromosomal interactions before and after formation of a single, site-specific DSB within a yeast chromosome. Surprisingly, we found that an unprocessed DSB within G1-arrested cells did not alter 3C interactions, suggesting that a single DSB is not sufficient to alter large-scale chromosome dynamics. In contrast, 3C interactions were significantly decreased when a DSB was processed for HR in asynchronous cells. Decreased 3C interactions required the nuclear envelope protein Mps3p, and they correlated with binding of Mps3p to DSB chromatin and relocalization of the DSB to the nuclear periphery. Furthermore, we found that efficient recruitment of Mps3p to a DSB required Cdc13p, a factor that regulates recruitment of the telomerase machinery to DSBs. Our data suggest a model in which the recruitment of the telomerase machinery to a DSB leads to its relocalization to the nuclear periphery by direct interactions between Mps3p and components of the telomerase complex. Given our finding that peripheral localization of a DSB inhibits the rate of HR and stimulates GCR events, the nuclear periphery may represent an environment where multiple, alternative repair pathways can compete for repair of a DSB.
The 3C method probes the frequency with which two chromosomal regions interact, and when applied along the length of a chromosome, it can provide a measurement of conformational flexibility. Typically, the strength of 3C interactions along a chromosome is proportional to the distance between the probed chromatin fragments. In contrast, here we find that localization of a DSB to the nuclear periphery decreases interaction frequencies between chromatin fragments that surround a DSB and all other chromatin fragments tested. Surprisingly, these changes in 3C interactions do not seem to preclude the ability of donor sequences to capture homology, as our live cell imaging results indicate that a donor sequence located on chromosome V is able to find the homologous sequences on chromosome III, even when these chromatin fragments are localized to the nuclear periphery. Our results are consistent with recent work showing that the general interaction frequencies at telomeres, which are also localized to the periphery, are lower than that of nontelomeric loci (A. Miele and J. Dekker, in prep.).
Role of Mps3p in DSB relocalization
Mps3p is the yeast member of the conserved SUN family of inner nuclear envelope proteins (Jaspersen et al. 2006). Although these proteins have divergent N-terminal domains, multiple family members from yeast to humans bind to telomeres during meiosis and many play critical roles in meiotic recombination (de La Roche Saint-Andre 2008). Indeed, it has been proposed in mammalian cells that SUN family proteins Sun1p and Sun2p may act to couple the nuclear matrix to the cytoplasm (Crisp et al. 2006; Haque et al. 2006; Ding et al. 2007). Together with the data we presented, we anticipate that other SUN proteins will also play a role in mitotic growth in recruitment and repair of DSBs based on their ability to sequester proteins or regions of the genome at the nuclear periphery following DNA damage. A previous study indicated that the NUP84 nuclear pore subcomplex is involved in tethering yeast telomeres to the nuclear membrane and in the repair of DSBs located close to chromosome ends (Therizols et al. 2006). Additionally, the yeast NUP84 complex interacts with Slx5p/Slx8p, a SUMO-dependent ubiquitin ligase, and both the Nup84 and Slx5p/Slx8p complexes are recruited to an unrepaired DSB (Nagai et al. 2008). However, we find that loss of the Nup84p or Nup133p components of the NUP84 subcomplex does not substantially alter DSB-induced changes in chromosome interaction frequencies that are detected by 3C (Supplemental Fig. S10), nor do these mutations alter repair of DSBs that are located distal from the telomere (Therizols et al. 2006). Thus, recruitment of DSBs to the nuclear periphery via SUN family members may represent a more general mechanism for compartmentalization of a DSB at the nuclear periphery.
Role of the telomerase machinery in DSB relocalization
How does Mps3p promote relocalization of a DSB to the nuclear periphery? The simplest model posits that the N-terminal domain of Mps3p directly interacts with one or more proteins that are bound to a processed DSB. We were unable to detect a physical interaction between Mps3p and a Rad51p nucleoprotein filament in vitro (P. Oza and C.L. Peterson, unpubl.), and Mps3p does not interact with Rad52p as assayed by a coimmunoprecipitation assay from cell extracts (S.L. Jaspersen, unpubl.). Previous two-hybrid studies have shown that Mps3p interacts with the conserved acetyltransferase Eco1p/Ctf7p, which is required for establishment of chromosome cohesion, and Est1p, a component of the telomerase machinery (Antoniacci et al. 2004, 2007). Mps3p does play a role in chromosome cohesion, and there may be a role for Eco1p in cohesion establishment as well as DSB repair (Unal et al. 2007; Ben-Shahar et al. 2008). However, given our result that Cdc13p is required for optimal recruitment of Mps3p to a DSB, we favor a model in which components of the telomerase machinery direct the DSB to Mps3p at the periphery. Given that Mps3p binds to telomere proteins during meiosis, this is a particularly attractive possibility (Conrad et al. 2007). This model is also consistent with recent results demonstrating that a LexA–Est2p fusion protein can recruit a chromosomal locus to Mps3p at the nuclear periphery (Schober et al. 2009).
Examination of spontaneous Rad52p foci suggests that most HR occurs within the nuclear lumen (Lisby et al. 2003). Likewise, we and others (Fig. 4A; Bystricky et al. 2008; Nagai et al. 2008) have found that DSBs that are rapidly repaired by either HR or SSA were not sequestered to the periphery. One possibility is that the periphery represents a “disposal” compartment for DSBs that cannot be repaired. Indeed, both our data shown here, as well as a previous study demonstrate that a persistent, unrepairable DSB is localized to the nuclear periphery (Nagai et al. 2008). In this latter case, an unrepairable DSB at the MAT locus appeared to colocalize with nuclear pores, and components of the NUP84 subcomplex could be formaldehyde cross-linked to chromatin surrounding a DSB. However, it is not clear if the NUP84 subcomplex is required for peripheral localization of a DSB or whether the DSB simply interacts with several components of the nuclear periphery. Based on these studies, there may exist several pathways for recruitment of DSBs to the periphery (e.g., nuclear pores or Mps3p) or, alternatively, DSBs are first localized to the periphery via Mps3p, and they then interact with the nuclear pores.
We did find two cases in which repairable DSBs were also localized to the nuclear periphery. In one case, a DSB was repaired by HR using a donor located on a different chromosome, and in the other case, a DSB was repaired by SSA when two defective copies of LEU2 were separated by 30 kb. In both cases, the repair event occurred with very slow kinetics, and consequently the DNA damage checkpoint was activated (Vaze et al. 2002; Keogh et al. 2006). One possibility is that checkpoint signaling is required for the recruitment of a slowly repaired DSB to the nuclear periphery. This would be consistent with a recent report showing that the Mec1p and Tel1p checkpoint kinases (orthologs of mammalian ATM and ATR, respectively) are required for relocalization of an unrepairable DSB to the nuclear periphery (Nagai et al. 2008). A likely target for checkpoint signaling would be one or more components of the telomerase machinery; indeed, previous studies have suggested that checkpoint kinases may influence telomerase recruitment by Cdc13p. Cdc13p is phosphorylated in vitro by the Tel1 and Mec1 checkpoint kinases on several serine residues, two of which are required for telomere maintenance in vivo (Tseng et al. 2006). Interestingly, both amino acids are contained within a small domain of the protein responsible for telomerase recruitment (Pennock et al. 2001; Bianchi et al. 2004). It is thus tempting to speculate that persistent activation of the DNA damage checkpoint regulates both telomere addition and DSB relocalization through Cdc13p phosphorylation. A requirement for persistent checkpoint activation may provide an explanation for why a previous study did not observe peripheral localization of a DSB that was repaired from an ectopic donor (Nagai et al. 2008).
At natural telomeric ends, long tracts of single-stranded TG1–3 repeats provide high-affinity binding sites for Cdc13p that can recruit the telomerase machinery by direct interactions with the Est1p subunit. We were surprised to find robust recruitment of Cdc13p to an HO-induced DSB at the MAT locus, since this region lacks extensive TG repeats and has no putative telomere seed sequences (Mangahas et al. 2001). Cdc13p does have a relatively high affinity for non-TG ssDNA, much like the ssDNA-binding protein RPA (Hughes et al. 2000; Lin et al. 2001). Indeed, Cdc13p and RPA may compete for binding to ssDNA at an HO-induced DSB. We propose that this competition may be influenced by formation of the Rad51p filament, given that RAD51 is required for optimal recruitment of Cdc13p to an HO-induced DSB. The RAD51 dependence of Cdc13p binding provides a molecular explanation for the RAD51 dependence of DSB relocalization to the nuclear periphery. Alternatively, mammalian Rad51p interacts with the nuclear matrix, and this interaction may contribute to DSB localization (Mladenov et al. 2006).
Functional connections among Mps3, Slx5p, and Pif1p in spontaneous GCRs
Repair of chromosomal DNA DSBs is essential for cell survival and genome stability. Consequently, cells have devised numerous mechanisms to heal this type of chromosomal damage. HR and NHEJ are the most prevalent repair mechanisms as they provide the best opportunities for error-free repair. However, if these two pathways are inactive or operate too slowly, alternative pathways may also be activated that promote chromosome healing. Our results suggest that slowly repaired DSBs are recognized by components of the telomerase machinery that recruit these DSBs to the nuclear periphery. We propose that the nuclear periphery provides an environment that is permissive for de novo telomere additions as well as other alternative repair pathways that may result in GCRs. Indeed, our results suggest that the N-terminal domain of Mps3p greatly stimulates de novo telomere additions that occur at spontaneous DSBs in a pif1-m2 mutant. Our data are also consistent with a key role for Slx5p/Slx8p in preventing GCR events that occur at the nuclear periphery, as deletion of the Mps3p N-terminal domain eliminates GCR events due to inactivation of Slx5p.
How the Slx5p/Slx8p SUMO-dependent ubiquitin ligase prevents GCR events is not clear. It is likely that sumoylation plays a role in the removal of the replication machinery from collapsed forks. Sumolyation of the replication factor PCNA facilitates the loading of trans-lesion polymerases at collapsed forks and recruits the anti-recombinogenic factor Srs2p (Pfander et al. 2005; Moldovan et al. 2007). In the absence of PCNA sumolyation, DSB generation and telomere addition may become more common events. Given our genetic interactions between MPS3, PIF1, and SLX5, it is also possible that the Slx5p/Slx8p machinery directly antagonizes GCRs by blocking the action of telomerase at DSBs. Since Mps3p is required for much if not all of the GCR events that occur in the absence of Slx5p or Pif1p, our data suggest that localization of DSBs to the nuclear periphery may promote chromosome healing at the expense of increased genomic instability.
Materials and methods
Strain construction
All strains are based on the JKM series (Moore and Haber 1996), and strains used are isogenic to strain JKM179 (CY915) that has the relevant genotype of MATα Δho Δhml∷ADE1 Δhmr∷ADE1 ade1-110 leu2,3-112 lys5 trp1∷hisG ura3-52 ade3∷GAL10:HO (Table 2). The “switching” strain (CY924) is based on JKM154, which is isogenic to JKM179, except that it is MATa HMLα HMRa. Strain CY1276 is isogenic to CY915, except that it is MATa. Strain CY1392 (yJK17) has the relevant genotype of MATα hmlΔ hmrΔ arg5,6∷MATainc ade3∷GAL-HO. CY1442 was generated by the introduction of the HO endonuclease recognition site on chromosome V as follows. The 36-base-pair (bp) HO-cs recognition site (Sugawara and Haber 2006) was cloned into the KpnI–XhoI fragment of pRS406. The plasmid was linearized with Pst1 and integrated at the URA3 locus on chromosome V. The mps3Δ75-150 and MPS3-13Myc alleles were introduced by one-step replacement of MPS3 using PCR. The r\ad51Δ, rad52Δ, rad54Δ, and rad55Δ strains were made in CY915 by one-step gene deletion by PCR. The SSA strains used in the study were YMV45, which has 5 kb of DNA between two defective LEU2 repeats, and YMV2, which is isogenic except that it has 30 kb of intervening DNA (Vaze et al. 2002).
Table 2.
Strain list
Strains to visualize the DSB at MAT are derivatives of CY915 and were made by one-step PCR-mediated gene replacement of NUP49 with NUP49-GFP-NATMX followed by integration of a GFP-LACI fusion construct into the URA3 locus (Kaye et al. 2004). Next, a 662-bp fragment containing ARS313 was cloned into the XhoI–SacI sites of AFS52 (Hediger et al. 2002) to create pSJ728. Following digestion with StyI, integration of ∼256 LacOR and TRP1 into the ARS313 locus was verified by Southern blotting, and the resulting strain designated SLJ2698 (MATα hmlΔ∷ADE1 hmrΔ∷ADE1 ade1-110 leu2-3,112 trp1∷hisG ura3∷CUP1-GFP-LACI-URA3 nup49Δ∷NUP49-GFP-NATMX ARS313∷LACOR-TRP1 ade3∷GAL-HO lys5). Versions of this strain containing Mps3Δ∷mps3Δ75-150-HYGMX (SLJ2699), rad52Δ∷KANMX (SLJ2749), and rad54Δ∷KANMX (SLJ2750) were created by replacement or disruption of the endogenous gene by PCR. To visualize DSBs on chromosome VII, NUP49-GFP-NATMX was introduced into yJK38.2 (Kaye et al. 2004) to create SLJ2744 (MatΔ hmrΔ hmlΔ nup49Δ∷NUP49-GFP-NATMX can1 lys5 ade2 ade3∷GALHO trp1 his3 ura3 leu2 armVII∷TRP-HOcs ura3∷CUP1-LACI-GFP-URA3 lys5∷LACOR-LYS5). SLJ2745 was derived by one-step gene replacement and contains Mps3Δ∷mps3Δ75-150-HYGMX; it displays a partial increase in ploidy. To visualize the position of MATa-inc, which is integrated at ARG5,6 on chromosome V, a 700-bp fragment containing the intergenic region of YER066W was cloned into the XhoI–SacI sites of AFS52 (Hediger et al. 2002) to create pSJ776. Following digestion with NcoI, this plasmid was integrated into versions of CY915 or CY1392 containing NUP49-GFP and GFP-LacI to create SLJ2826 (MATα hmlΔ∷ADE1 hmrΔ∷ADE1 ade1-110 leu2-3,112 trp1∷hisG ura3∷CUP1-GFP-LACI-URA3 nup49Δ∷NUP49-GFP-NATMX YER066Wi∷LACOR-TRP1 ade3∷GAL-HO lys5) and SLJ2821 (MATα hmlΔ∷ADE1 hmrΔ∷ADE1 ade1-110 leu2-3,112 trp1∷hisG ura3∷CUP1-GFP-LACI-URA3 nup49Δ∷NUP49-GFP-NATMX YER066Wi∷LACOR-TRP1 arg5,6∷MATa-inc-HYGMX ade3∷GAL-HO lys5). Mps3Δ∷mps3Δ75-150-KANMX (SLJ2823) was created by PCR-mediated gene replacement in SLJ2821.
DSB induction
A single DNA DSB was generated by adding 2% galactose to mid-log-phase cells growing in YEP with 2% raffinose to induce expression of the HO endonuclease. Cells were collected at various time points after DSB induction and analyzed by 3C, Southern blotting, ChIP, PCR, or microscopy.
3C template generation
3C template was generated from log-phase yeast cells as previously published (Dekker et al. 2002). Briefly, cells at an OD600 of 0.5–1.0 were cross-linked with 1% formaldehyde in YEP media to ensure immediate capture of any dynamic events. Cross-linked cells were resuspended in EcoRI restriction enzyme buffer (Buffer #2; New England Biolabs) and flash-frozen in liquid nitrogen. These cells were then ground while still in liquid nitrogen using a porcelain mortar and pestle that was kept cold on dry ice. The cells were checked microscopically for ∼70% breakage before collecting the lysate. The OD600 of the lysate was adjusted to 10.0 with more buffer to ensure equal DNA in all samples. The lysate was then taken through the 3C procedure. Control template was generated from CY915 using non-cross-linked cells as published previously (Dekker et al. 2002) and used at equal concentrations throughout the study.
3C interaction mapping
Specific primers were designed for the 3′ end of each EcoRI restriction fragment tested (primer sequences available on request). The linear range for PCR, ethidium bromide staining, and quantification was determined by titration of each 3C template with two or more primers (∼20–40 kb away) over a large range of concentrations. A concentration at the lower end of this range was then used in all subsequent experiments with the same template to ensure that all quantifications were carried out within the linear range of the assay. PCR products were electrophoresed on 1.5% ethidium bromide–stained gels and quantified using LabWorks version 4.5 software (UVP LLC). PCRs with the control template were performed in parallel with the 3C template reactions, and products were electrophoresed on the same gel. After quantitation, the interaction frequency for each fragment was normalized to the control (non-cross-linked) template interactions for the very same primers to control for amplification efficiency of each primer pair. In addition, the peak of interactions for the templates without DSB induction was set to 1, and all the other interactions were normalized accordingly. The experiment in Figure 1C was also repeated using SybrGreen based quantitative PCR (Applied Biosystems) and a set of redesigned primers with similar results (data not shown).
Southern blotting
Total genomic DNA prepared by glass bead lysis (Wolner and Peterson 2005) was loaded onto 1% agarose gels and electrophoresed for 3 h at 120 V before capillary transfer to a Nylon membrane (Stratagene). Probes were made by PCR amplification of the region of interest and labeled by random primer labeling. Denaturing gels were electrophoresed in 0.1 N NaOH. Blots were quantified using ImageJ (NIH) and were normalized for loading using a probe for the ENA1 ORF.
ChIP assay
ChIP was carried out essentially as described previously (Wolner and Peterson 2005). Real-time SyBrGreen-based PCR analysis was carried out on an ABI 7300 machine to quantify the DNAs. Primer pairs were checked and optimized to ensure linear amplification. At the time of PCR, Ct values and melting curves were used to ensure uniform and specific amplification of all samples. Care was taken to ensure all Ct values were in the linear range. All ChIPs were repeated at least thrice and PCRs repeated a similar number of times to arrive at final values. Error bars represent ±SEM of three or more separate experiments. All ChIPs shown on the same panel were always conducted simultaneously.
Strand invasion assays
In strain JKM154, strand invasion was monitored by real-time PCR of genomic DNA using primers P3 and P2. Formation of an amplification product requires strand invasion at HMLα and extension of the joint by at least 18 nucleotides so that primer P3 can anneal. In the strain with the ectopic donor (yJK17), the region of homology between MATa-inc and MATα is much larger, and consequently the first unique product of strand invasion and joint extension that can be detected by PCR is >2 kb in size. Hence, PCR with a radioactive nucleotide was used to detect formation of this product as described (Wolner and Peterson 2005). While the same P1 primer was used to ensure similar conditions for amplification, the primer P2 was replaced with P4, which is >2 kb from P3. Equal loading was ensured by parallel real-time PCR to detect the amplification of ACT1. Primers are available on request.
Cytological techniques
The position of GFP spots was determined as previously described (Hediger et al. 2002; Bupp et al. 2007). Briefly, a Zeiss AxioImager with a 100× Zeiss α-Plan-Fluar objective (NA = 1.45) and a Hamamatsu Orca-ER digital camera was used to capture 19-image stacks of 150-nm step-size through nuclei of log-phase cells at room temperature. The spot-to-periphery distance and the nuclear diameter were determined in a single Z-stack image, where the spot was most concentrated using Axiovision 4.6.3 (Zeiss). By dividing the spot-to-periphery distance by the diameter, each spot fell into one of three zones of equal surface. Zone 1 has a width of 0.184 × the nuclear radius (r), zone 2 has a width of 0.184r to 0.422r, and zone 3 has a width of 0.422r. At least two independent transformants of each genotype were analyzed in three independent experiments.
GCR assay
GCR assays were carried out in YEPD or in YEPD with 0.1% MMS for 2 h as described (Myung and Kolodner 2003; Banerjee and Myung 2004). Colonies were counted on 5-FOA/canavanine plates and GCR rate determined using the method of the median (Foster 2006; Schmidt et al. 2006).
Acknowledgments
We are grateful to D. Toczyski, K. Myung, and J. Haber for strains, and to J. Haber, J. Schwartz, J. Gerton, M. Lisby, M. Papamichos-chronakis, S. Hawley, and members of the Peterson and Jaspersen laboratories for valuable discussions and comments on the manuscript. We thank Susan Gasser for communicating results prior to publication. S.L.J. is supported by the Stowers Institute for Medical Research and a Basil O'Connor Award from the March of Dimes. C.L.P. is supported by grants from the NIH. J.D. is supported by grants from the NIH, the Keck foundation, and the Cystic Fibrosis Foundation.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1782209.
Supplemental material is available at http://www.genesdev.org.
References
- Antoniacci L.M., Kenna M.A., Uetz P., Fields S., Skibbens R.V. The spindle pole body assembly component Mps3p/Nep98p functions in sister chromatid cohesion. J. Biol. Chem. 2004;279:49542–49550. doi: 10.1074/jbc.M404324200. [DOI] [PubMed] [Google Scholar]
- Antoniacci L.M., Kenna M.A., Skibbens R.V. The nuclear envelope and spindle pole body-associated Mps3 protein bind telomere regulators and function in telomere clustering. Cell Cycle. 2007;6:75–79. doi: 10.4161/cc.6.1.3647. [DOI] [PubMed] [Google Scholar]
- Aten J.A., Stap J., Krawczyk P.M., van Oven C.H., Hoebe R.A., Essers J., Kanaar R. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science. 2004;303:92–95. doi: 10.1126/science.1088845. [DOI] [PubMed] [Google Scholar]
- Aylon Y., Liefshitz B., Kupiec M. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 2004;23:4868–4875. doi: 10.1038/sj.emboj.7600469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkenist C.J., Kastan M.B. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- Banerjee S., Myung K. Increased genome instability and telomere length in the elg1-deficient Saccharomyces cerevisiae mutant are regulated by S-phase checkpoints. Eukaryot. Cell. 2004;3:1557–1566. doi: 10.1128/EC.3.6.1557-1566.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassing C.H., Alt F.W. The cellular response to general and programmed DNA double strand breaks. DNA Repair (Amst.) 2004;3:781–796. doi: 10.1016/j.dnarep.2004.06.001. [DOI] [PubMed] [Google Scholar]
- Belmont A.S., Straight A.F. In vivo visualization of chromosomes using lac operator–repressor binding. Trends Cell Biol. 1998;8:121–124. doi: 10.1016/s0962-8924(97)01211-7. [DOI] [PubMed] [Google Scholar]
- Ben-Shahar T.R., Heeger S., Lehane C., East P., Flynn H., Skehel M., Uhlmann F. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science. 2008;321:563–566. doi: 10.1126/science.1157774. [DOI] [PubMed] [Google Scholar]
- Bianchi A., Shore D. How telomerase reaches its end: Mechanism of telomerase regulation by the telomeric complex. Mol Cell. 2008;31:153–165. doi: 10.1016/j.molcel.2008.06.013. [DOI] [PubMed] [Google Scholar]
- Bianchi A., Negrini S., Shore D. Delivery of yeast telomerase to a DNA break depends on the recruitment functions of Cdc13 and Est1. Mol. Cell. 2004;16:139–146. doi: 10.1016/j.molcel.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Bupp J.M., Martin A.E., Stensrud E.S., Jaspersen S.L. Telomere anchoring at the nuclear periphery requires the budding yeast Sad1-UNC-84 domain protein Mps3. J. Cell Biol. 2007;179:845–854. doi: 10.1083/jcb.200706040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystricky K., Van Attikum H., Montiel M.-D., Dion V., Gehlen L., Gasser S.M. Regulation of nuclear positioning and dynamics of the silent mating type loci by the yeast Ku70/Ku80 complex. Mol. Cell. Biol. 2008;29:835–848. doi: 10.1128/MCB.01009-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A., Boule J.-B., Zakian V.A. Two pathways recruit telomerase to Saccharomyces cerevisiae telomeres. PLoS Genet. 2008;4:e1000236. doi: 10.1371/journal.pgen.1000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Kolodner R.D. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat. Genet. 1999;23:81–85. doi: 10.1038/12687. [DOI] [PubMed] [Google Scholar]
- Conrad M.N., Lee C.-Y., Wilkerson J.L., Dresser M.E. MPS3 mediates meiotic bouquet formation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 2007;104:8863–8868. doi: 10.1073/pnas.0606165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp M., Liu Q., Roux K., Rattner J.B., Shanahan C., Burke B., Stahl P.D., Hodzic D. Coupling of the nucleus and cytoplasm: Role of the LINC complex. J. Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J. GC- and AT-rich chromatin domains differ in conformation and histone modification status and are differentially modulated by Rpd3p. Genome Biol. 2007;8:R116. doi: 10.1186/gb-2007-8-6-r116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J., Rippe K., Dekker M., Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- de La Roche Saint-Andre C. Alternative ends: Telomeres and meiosis. Biochimie. 2008;90:181–189. doi: 10.1016/j.biochi.2007.08.010. [DOI] [PubMed] [Google Scholar]
- Diede S.J., Gottschling D.E. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases α and δ. Cell. 1999;99:723–733. doi: 10.1016/s0092-8674(00)81670-0. [DOI] [PubMed] [Google Scholar]
- Diede S.J., Gottschling D.E. Exonuclease activity is required for sequence addition and Cdc13p loading at a de novo telomere. Curr. Biol. 2001;11:1336–1340. doi: 10.1016/s0960-9822(01)00400-6. [DOI] [PubMed] [Google Scholar]
- Ding X., Xu R., Yu J., Xu T., Zhuang Y., Han M. SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev. Cell. 2007;12:863–872. doi: 10.1016/j.devcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- Dostie J., Richmond T.A., Arnaout R.A., Selzer R.R., Lee W.L., Honan T.A., Rubio E.D., Krumm A., Lamb J., Nusbaum C., et al. Chromosome Conformation Capture Carbon Copy (5C): A massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs J.A., Lowndes N.F., Jackson S.P. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature. 2000;408:1001–1004. doi: 10.1038/35050000. [DOI] [PubMed] [Google Scholar]
- Foster P.L. Methods for determining spontaneous mutation rates. Methods Enzymol. 2006;409:195–213. doi: 10.1016/S0076-6879(05)09012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque F., Lloyd D.J., Smallwood D.T., Dent C.L., Shanahan C.M., Fry A.M., Trembath R.C., Shackleton S. SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol. Cell. Biol. 2006;26:3738–3751. doi: 10.1128/MCB.26.10.3738-3751.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hediger F., Neumann F.R., Van Houwe G., Dubrana K., Gasser S.M. Live imaging of telomeres: yKu and Sir proteins define redundant telomere-anchoring pathways in yeast. Curr. Biol. 2002;12:2076–2089. doi: 10.1016/s0960-9822(02)01338-6. [DOI] [PubMed] [Google Scholar]
- Hughes T.R., Weilbaecher R.G., Walterscheid M., Lundblad V. Identification of the single-strand telomeric DNA binding domain of the Saccharomyces cerevisiae Cdc13 protein. Proc. Natl. Acad. Sci. 2000;97:6457–6462. doi: 10.1073/pnas.97.12.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G., Pellicioli A., Balijja A., Wang X., Fiorani S., Carotenuto W., Liberi G., Bressan D., Wan L., Hollingsworth N.M., et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen S.L., Giddings T.H., Jr, Winey M. Mps3p is a novel component of the yeast spindle pole body that interacts with the yeast centrin homologue Cdc31p. J. Cell Biol. 2002;159:945–956. doi: 10.1083/jcb.200208169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen S.L., Martin A.E., Glazko G., Giddings T.H., Jr, Morgan G., Mushegian A., Winey M. The Sad1–UNC-84 homology domain in Mps3 interacts with Mps2 to connect the spindle pole body with the nuclear envelope. J. Cell Biol. 2006;174:665–675. doi: 10.1083/jcb.200601062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye J.A., Melo J.A., Cheung S.K., Vaze M.B., Haber J.E., Toczyski D.P. DNA breaks promote genomic instability by impeding proper chromosome segregation. Curr. Biol. 2004;14:2096–2106. doi: 10.1016/j.cub.2004.10.051. [DOI] [PubMed] [Google Scholar]
- Keeney S., Neale M.J. Initiation of meiotic recombination by formation of DNA double-strand breaks: Mechanism and regulation. Biochem. Soc. Trans. 2006;34:523–525. doi: 10.1042/BST0340523. [DOI] [PubMed] [Google Scholar]
- Keogh M.-C., Kim J.-A., Downey M., Fillingham J., Chowdhury D., Harrison J.C., Onishi M., Datta N., Galicia S., Emili A., et al. A phosphatase complex that dephosphorylates γH2AX regulates DNA damage checkpoint recovery. Nature. 2006;439:497–501. doi: 10.1038/nature04384. [DOI] [PubMed] [Google Scholar]
- Kruhlak M.J., Celeste A., Dellaire G., Fernandez-Capetillo O., Muller W.G., McNally J.G., Bazett-Jones D.P., Nussenzweig A. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J. Cell Biol. 2006;172:823–834. doi: 10.1083/jcb.200510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.E., Moore J.K., Holmes A., Umezu K., Kolodner R.D., Haber J.E. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- Lee S.E., Pellicioli A., Vaze M.B., Sugawara N., Malkova A., Foiani M., Haber J.E. Yeast Rad52 and Rad51 recombination proteins define a second pathway of DNA damage assessment in response to a single double-strand break. Mol. Cell. Biol. 2003;23:8913–8923. doi: 10.1128/MCB.23.23.8913-8923.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.-C., Hsu C.-L., Shih J.-W., Lin J.-J. Specific binding of single-stranded telomeric DNA by Cdc13p of Saccharomyces cerevisiae. J. Biol. Chem. 2001;276:24588–24593. doi: 10.1074/jbc.M101642200. [DOI] [PubMed] [Google Scholar]
- Lisby M., Mortensen U.H., Rothstein R. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat. Cell Biol. 2003;5:572–577. doi: 10.1038/ncb997. [DOI] [PubMed] [Google Scholar]
- Lobachev K., Vitriol E., Stemple J., Resnick M.A., Bloom K. Chromosome fragmentation after induction of a double-strand break is an active process prevented by the RMX repair complex. Curr. Biol. 2004;14:2107–2112. doi: 10.1016/j.cub.2004.11.051. [DOI] [PubMed] [Google Scholar]
- Mangahas J.L., Alexander M.K., Sandell L.L., Zakian V.A. Repair of chromosome ends after telomere loss in Saccharomyces. Mol. Biol. Cell. 2001;12:4078–4089. doi: 10.1091/mbc.12.12.4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mladenov E., Anachkov B., Tsaneva I. Sub-nuclear localization of Rad51 in response to DNA damage. Genes Cells. 2006;11:513–524. doi: 10.1111/j.1365-2443.2006.00958.x. [DOI] [PubMed] [Google Scholar]
- Moldovan G.L., Pfander B., Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Moore J.K., Haber J.E. Capture of retrotransposon DNA at the sites of chromosomal double-strand breaks. Nature. 1996;383:644–646. doi: 10.1038/383644a0. [DOI] [PubMed] [Google Scholar]
- Myung K., Kolodner R.D. Induction of genome instability by DNA damage in Saccharomyces cerevisiae. DNA Repair (Amst.) 2003;2:243–258. doi: 10.1016/s1568-7864(02)00216-1. [DOI] [PubMed] [Google Scholar]
- Myung K., Chen C., Kolodner R.D. Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature. 2001;411:1073–1076. doi: 10.1038/35082608. [DOI] [PubMed] [Google Scholar]
- Nagai S., Dubrana K., Tsai-Pflugfelder M., Davidson M.B., Roberts T.M., Brown G.W., Varela E., Hediger F., Gasser S.M., Krogan N.J. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science. 2008;322:597–602. doi: 10.1126/science.1162790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini S., Ribaud V., Bianchi A., Shore D. DNA breaks are masked by multiple Rap1 binding in yeast: Implications for telomere capping and telomerase regulation. Genes & Dev. 2007;21:292–302. doi: 10.1101/gad.400907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa S., Terazawa Y., Nakayama T., Hirata A., Makio T., Endo T. Nep98p is a component of the yeast spindle pole body and essential for nuclear division and fusion. J. Biol. Chem. 2003;278:9938–9943. doi: 10.1074/jbc.M210934200. [DOI] [PubMed] [Google Scholar]
- Paques F., Haber J.E. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennaneach V., Putnam C.D., Kolodner R.D. Chromosome healing by de novo telomere addition in Saccharomyces cerevisiae. Mol. Microbiol. 2006;59:1357–1368. doi: 10.1111/j.1365-2958.2006.05026.x. [DOI] [PubMed] [Google Scholar]
- Pennock E., Buckley K., Lundblad V. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell. 2001;104:387–396. doi: 10.1016/s0092-8674(01)00226-4. [DOI] [PubMed] [Google Scholar]
- Pfander B., Moldovan G.L., Sacher M., Hoege C., Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- Riha K., Heacock M.L., Shippen D.E. The role of the nonhomologous end-joining DNA double-strand break repair pathway in telomere biology. Annu. Rev. Genet. 2006;40:237–277. doi: 10.1146/annurev.genet.39.110304.095755. [DOI] [PubMed] [Google Scholar]
- Schmidt K.H., Pennaneach V., Putnam C.D., Kolodner R.D., Judith L.C., Paul M. Analysis of gross chromosomal rearrangements in Saccharomyces cerevisiae. Methods Enzymol. 2006;409:462–476. doi: 10.1016/S0076-6879(05)09027-0. [DOI] [PubMed] [Google Scholar]
- Schober H., Ferreira H., Kalck V., Gehlen L.R., Gasser S.M. Telomerase and SUN domain protein Mps3 anchor and protect telomeres in budding yeast. Genes & Dev. 2009 doi: 10.1101/gad.1787509. (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramke V., Luciano P., Brevet V., Guillot S., Corda Y., Longhese M.P., Gilson E., Geli V. RPA regulates telomerase action by providing Est1p access to chromosome ends. Nat. Genet. 2004;36:46–54. doi: 10.1038/ng1284. [DOI] [PubMed] [Google Scholar]
- Seto A.G., Zaug A.J., Sobel S.G., Wolin S.L., Cech T.R. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature. 1999;401:177–180. doi: 10.1038/43694. [DOI] [PubMed] [Google Scholar]
- Shim E.Y., Hong S.J., Oum J.H., Yanez Y., Zhang Y., Lee S.E. RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin. Mol. Cell. Biol. 2007;27:1602–1613. doi: 10.1128/MCB.01956-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff R., Arbel-Eden A., Pilch D., Ira G., Bonner W.M., Petrini J.H., Haber J.E., Lichten M. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr. Biol. 2004;14:1703–1711. doi: 10.1016/j.cub.2004.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis M., Klous P., Splinter E., Moshkin Y., Willemsen R., de Wit E., van Steensel B., de Laat W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat. Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- Stellwagen A.E., Haimberger Z.W., Veatch J.R., Gottschling D.E. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes & Dev. 2003;17:2384–2395. doi: 10.1101/gad.1125903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N., Haber J.E. Repair of DNA double strand breaks: In vivo biochemistry. Methods Enzymol. 2006;408:416–429. doi: 10.1016/S0076-6879(06)08026-8. [DOI] [PubMed] [Google Scholar]
- Sung P., Krejci L., Van Komen S., Sehorn M.G. Rad51 recombinase and recombination mediators. J. Biol. Chem. 2003;278:42729–42732. doi: 10.1074/jbc.R300027200. [DOI] [PubMed] [Google Scholar]
- Therizols P., Fairhead C., Cabal G.G., Genovesio A., Olivo-Marin J.C., Dujon B., Fabre E. Telomere tethering at the nuclear periphery is essential for efficient DNA double strand break repair in subtelomeric region. J. Cell Biol. 2006;172:189–199. doi: 10.1083/jcb.200505159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B., Palstra R.-J., Splinter E., Grosveld F., de Laat W. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol. Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- Tseng S.-F., Lin J.-J., Teng S.-C. The telomerase-recruitment domain of the telomere binding protein Cdc13 is regulated by Mec1p/Tel1p-dependent phosphorylation. Nucleic Acids Res. 2006;34:6327–6336. doi: 10.1093/nar/gkl786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unal E., Heidinger-Pauli J.M., Koshland D. DNA double-strand breaks trigger genome-wide sister-chromatid cohesion through Eco1 (Ctf7) Science. 2007;317:245–248. doi: 10.1126/science.1140637. [DOI] [PubMed] [Google Scholar]
- Vakoc C.R., Letting D.L., Gheldof N., Sawado T., Bender M.A., Groudine M., Weiss M.J., Dekker J., Blobel G.A. Proximity among distant regulatory elements at the β-globin locus requires GATA-1 and FOG-1. Mol. Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Vaze M.B., Pellicioli A., Lee S.E., Ira G., Liberi G., Arbel-Eden A., Foiani M., Haber J.E. Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol. Cell. 2002;10:373–385. doi: 10.1016/s1097-2765(02)00593-2. [DOI] [PubMed] [Google Scholar]
- Vilenchik M.M., Knudson A.G. Endogenous DNA double-strand breaks: Production, fidelity of repair, and induction of cancer. Proc. Natl. Acad. Sci. 2003;100:12871–12876. doi: 10.1073/pnas.2135498100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viscardi V., Clerici M., Cartagena-Lirola H., Longhese M.P. Telomeres and DNA damage checkpoints. Biochimie. 2005;87:613–624. doi: 10.1016/j.biochi.2004.10.022. [DOI] [PubMed] [Google Scholar]
- Wolner B., Peterson C.L. ATP-dependent and ATP-independent roles for the Rad54 chromatin remodeling enzyme during recombinational repair of a DNA double strand break. J. Biol. Chem. 2005;280:10855–10860. doi: 10.1074/jbc.M414388200. [DOI] [PubMed] [Google Scholar]
- Wolner B., van Komen S., Sung P., Peterson C.L. Recruitment of the recombinational repair machinery to a DNA double-strand break in yeast. Mol. Cell. 2003;12:221–232. doi: 10.1016/s1097-2765(03)00242-9. [DOI] [PubMed] [Google Scholar]
- Zhang C., Roberts T.M., Yang J., Desai R., Brown G.W. Suppression of genomic instability by SLX5 and SLX8 in Saccharomyces cerevisiae. DNA Repair (Amst.) 2006;5:336–346. doi: 10.1016/j.dnarep.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Zhao Z., Tavoosidana G., Sjolinder M., Gondor A., Mariano P., Wang S., Kanduri C., Lezcano M., Sandhu K.S., Singh U., et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat. Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- Ziv Y., Bielopolski D., Galanty Y., Lukas C., Taya Y., Schultz D.C., Lukas J., Bekker-Jensen S., Bartek J., Shiloh Y. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat. Cell Biol. 2006;8:870–876. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]