Abstract
We report DNA and predicted protein sequence similarities, implying homology, among genes of double-stranded DNA (dsDNA) bacteriophages and prophages spanning a broad phylogenetic range of host bacteria. The sequence matches reported here establish genetic connections, not always direct, among the lambdoid phages of Escherichia coli, phage φC31 of Streptomyces, phages of Mycobacterium, a previously unrecognized cryptic prophage, φflu, in the Haemophilus influenzae genome, and two small prophage-like elements, φRv1 and φRv2, in the genome of Mycobacterium tuberculosis. The results imply that these phage genes, and very possibly all of the dsDNA tailed phages, share common ancestry. We propose a model for the genetic structure and dynamics of the global phage population in which all dsDNA phage genomes are mosaics with access, by horizontal exchange, to a large common genetic pool but in which access to the gene pool is not uniform for all phage.
The double-stranded DNA (dsDNA)-containing bacteriophages are very likely the most numerically abundant group of similar organisms in the biosphere, and nearly 4,500 different dsDNA phages capable of infecting a large diversity of bacterial hosts have been described (1). However, these phages have proven difficult to classify, in part because of the breadth of their genetic variation. For example, phages with similar morphologies, modes of replication, and overall genomic architectures may be completely unrelated at the nucleotide level. By comparing the genomes of several newly characterized phages and cryptic prophages, it appears that the vast majority of dsDNA tailed phages have common ancestry and that they undergo profuse exchange of functional genetic elements drawn from a large shared pool.
The classification of bacteriophages by their host range, morphology, or available life-cycles has led to conflicting conclusions regarding their origins and evolution (1). Phages can be found in virtually all places where their bacterial hosts exist, although only a small number have been investigated in detail (2, 3). As phages with near-identical genomes are rarely isolated from independent sources in nature, the term “species” is of limited use in describing relationships among phages (4). Groups of phages related to each other by common gene organization and some degree of sequence similarity clearly do exist (for example, the “lambdoid” or λ-like group), and evidence for horizontal transfer among tail fiber genes has been reported (5–8). However, it has been unclear whether, and in what ways, different groups of phages—particularly phage groups with phylogenetically distant hosts—are related to each other.
The complete genomic sequences of several phages closely related to phage λ have been determined and shown to have similar genomic organizations (R. Juhala, M.E.F., R. L. Duda, A. Youlton, G.F.H., and R.W.H., unpublished work). However, the genomes are clearly mosaic in nature, with regions of obvious sequence similarity interspersed with segments that are apparently unrelated (refs. 9–12; R. Juhala, M.E.F., R. L. Duda, A. Youlton, G.F.H., and R.W.H., unpublished work). This argues for the existence of extensive horizontal genetic exchange among members of this group of phages. Other bacteriophages—such as mycobacteriophage L5—have no clear sequence similarity with these phages but do possess a similar genomic architecture, raising the possibility that they share common ancestry with the λ-like phages (13, 14).
Recently, researchers determined the genome sequences of mycobacteriophages D29 (a relative of L5; ref. 15) and TM4 (16), coliphages HK97 and HK022 (closely related members of the lambdoid group; R. Juhala, M.E.F., R. L. Duda, A. Youlton, G.F.H., and R.W.H., unpublished work), and actinophage φC31 (host: Streptomyces spp.; R.N.B. and M.C.M.S., unpublished work). Although none of the mycobacteriophages have any recognizable sequence similarity to HK97 or HK022 (nor to other λ-like phage genomes), φC31 provides a clear bridge between these two apparently distant groups. On the one hand, the head protein genes of φC31 have a very similar organization to those of HK97 and HK022, and some of these are clearly homologous on the basis of sequence similarity. Thus, the putative terminase, portal, protease, and capsid genes from HK97 and φC31 are colinear and share 28, 29, 28, and 20% amino acid sequence identity, respectively (17, 18). A similar mode of capsid subunit processing buttresses the proposed relationship between these phages (Fig. 1). On the other hand, several φC31 genes match genes in the mycobacteriophages. For example, ORFs 9 and 10 of φC31 (19) [now known to be a single gene called 9a (R.N.B. and M.C.M.S., unpublished work)] encode a protein related to gp70 of mycobacteriophage TM4 (16); φC31 gp11 is a DNA polymerase with greatest similarity to gp44 of L5 (43% identity); φC31 gp16 is related to gp48 of L5 (53% identity); and gp20 of φC31 is a dCMP deaminase closely related to gp36.1 of mycobacteriophage D29 (55% identity). Although these are all early proteins, we note that the putative terminase of φC31 (gp33) shares similarity with the putative terminase (gp13) of L5 (27% identity), as it also does with the HK97 terminase, gp2, noted above. A pairwise alignment of the HK97 gp2 and L5 gp13 terminase sequences does not alone make a convincing case that they are related, but their mutual similarity to the φC31 protein argues that all three terminases are homologues. Taken together, the observations that these genes match across the phylogenetic chasms separating their hosts argue strongly that they—and, perhaps, most phage genes—are derived from a shared pool. Although the modest levels of similarity that we see between the groups of phages infecting phylogenetically distant hosts argues against direct horizontal exchange of DNA in recent evolutionary time, we believe the observations are consistent with an ongoing pattern of exchanges among extensive chains of intermediates connecting the particular genomes that we have examined.
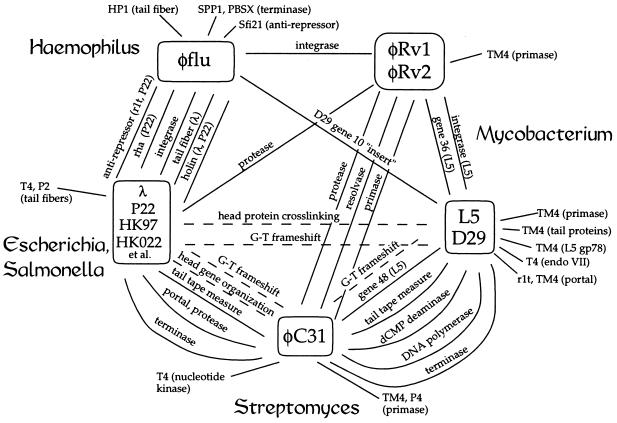
Figure 1.
Sequence connections among phages and prophages. The relationships among phage and prophage sequences are indicated, with the solid lines representing sequence similarities and the dotted lines corresponding to commonalities of gene organization or gene function. Closely related phages are shown in boxes, and bacterial hosts are shown at the perimeter of the web. Sequence comparisons were performed by using blast and gapped blast programs available at the National Center for Biotechnology Information web site (http://www.ncbi.nlm.nih.gov/) and the fasta, tfasta, and bestfit programs within the Genetics Computer Group (Madison, WI) package. Protein sequences were considered to be related (that is, probable homologues) if they could be aligned over a substantial portion of their lengths with 20% or greater amino acid identities or if the blast output reported a high probability of relatedness. A table of the protein similarities is available at http://www.pitt.edu/∼gfh/table.html). Accession numbers for phage genome sequences are HK97, AF069529; HK022, AF069308; φC31, AJ006589; L5, Z18946; D29, AF022214; and TM4, AF068845.
A somewhat different set of examples of sequence relationships among “distant” phages comes from analysis of the genome sequence of mycobacteriophage D29, a close relative of phage L5 (15). The left arms of the L5 and D29 genomes are very similar to each other (≈80% sequence identity) and contain closely related genes in colinear positions. A notable departure occurs in gene 10 of D29, which, while sharing sequence similarity with L5 gene 10, is 603 bp larger (1,481 bp vs. 878 bp) and encodes a putative protein product (gp10) that is 201 amino acids bigger than its L5 counterpart (Fig. 2). Alignment of the coding sequences shows that D29 gp10 contains a contiguous block of 194 residues between codons 173 and 174 that is absent in L5.
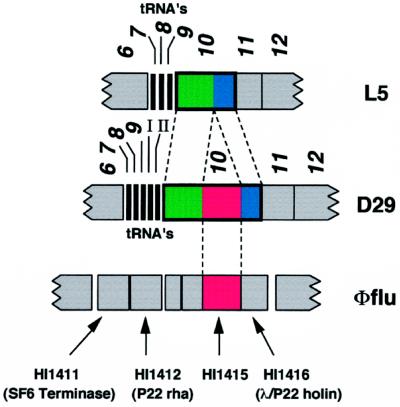
Figure 2.
Relationship between mycobacteriophages L5 and D29 and H. influenzae gene HI1415. Segments of the genomes of the closely related mycobacteriophages L5 and D29 are shown, with genes 6–12 represented as boxes (L5 genes 7–9 and D29 genes 7–9; I and II encode tRNAs as indicated). Below is shown a segment of the H. influenzae genome from within the putative cryptic prophage φflu (shown in full in Fig. 3). Gene 10 of D29 is larger than L5 gene 10 because of an additional 600 bp within the coding region that results in a gene product (gp10) that is 200 amino acids larger than L5 gp10. The upstream parts of gp10 (green) are significantly more similar than the downstream segments (blue), having 81 and 50% amino acid identity, respectively. The “insert” in D29 gene 10 (red) encodes a protein sequence that has 34% identity with the product of H. influenzae HI1415.
Comparison of L5 gp10 with the protein databases fails to identify any close relatives (13). However, database searches with D29 gp10 reveal similarity to the putative 200-aa product of the HI1415 gene of Haemophilus influenzae, whose function is unknown (20). The similarity is restricted to the region of D29 gp10 that is absent from L5 gp10, and this can be aligned with the HI1415 product with 34% identity (Fig. 2). The evolutionary events that account for the relationship between HI1415 and gene 10 of L5 and D29 are not obvious, although it seems unlikely that an intein is involved because the sequence features of known inteins (21) are not present.
A closer examination of the H. influenzae genome surrounding HI1415 suggests that this gene is part of a previously unidentified prophage that we propose to call φflu (Fig. 3). In particular, we note that several other genes in this region have sequence similarity to known phage genes; these are HI1403, HI1410, HI1411, HI1412, HI1415, HI1416, HI1418, HI1422, and HI1424 (Fig. 3). The phages whose genes they match are associated with a wide diversity of bacterial hosts (Fig. 3). Other noteworthy features are an integrase gene (HI1424) with a tRNA gene immediately upstream, a gene (HI1407) encoding a product related to traN of plasmid RP4, and the genes (HI1392 and HI1393) encoding the HindIII restriction/modification system (Fig. 3). There are several ORFs encoding proteins that do not match existing database entries. Because the only phage particles reported to be released from the sequenced strain H. influenzae Rd are those of a Mu-like prophage (20), it seems likely that φflu is cryptic and unable to produce infectious viral particles.
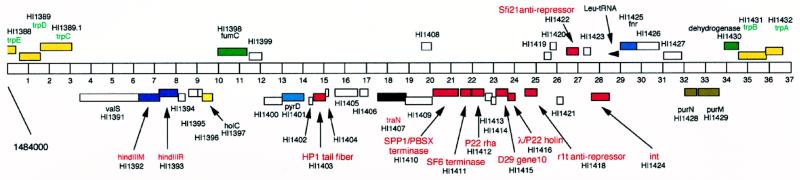
Figure 3.
Organization of φflu, a cryptic prophage of H. influenzae. A 37-kilobase segment of the H. influenzae genome is shown corresponding to coordinates 1484000–1521000 (20). Genes and their putative functions are shown according to the published annotation (20) and are shown either above (leftward) or below (rightward) depending on the direction of transcription. Genes with sequences similar to known phage genes are shown in red. White boxes represent genes that do not give a significant database match. The genetic constitution of the φflu prophage suggests that it is unlikely to form infectious particles, although we cannot rule out the possibility that it can do so under certain conditions.
The phage-related genes lie within an ≈31.5-kilobase segment of the H. influenzae genome that is flanked by the trpB and trpA genes (HI1431 and HI1432, respectively) on one side and trpE, trpD, and trpC (HI1388, HI1389, and HI1389.1, respectively) on the other (Fig. 3). This entire region contains ≈40 genes, including several that are not obviously phage-related (e.g., HI1391, valS; HI1398, fumC; HI1401, pyrD; HI1425, fnr; HI1428, purN; and HI1429, purM). A simple explanation accounting for this genome structure is that a defective transducing particle—generated by aberrant excision—integrated into the trp operon through illegitimate recombination.
The φflu cryptic prophage is remarkable in that the combination of identifiable phage genes has not been seen in genomes of infectious bacteriophages. Moreover, the phages that carry genes homologous to φflu genes (i.e., HP1, SF6, P22, D29, λ, Sfi21, and r1t) infect a wide range of bacterial hosts (Haemophilus, Bacillus, Salmonella, Mycobacterium, Escherichia, Streptococcus, and Lactococcus) and have little or no sequence similarity with each other. As with the phage examples described above, these observations argue that there is substantial genetic exchange of phage genes across host phylogenetic boundaries.
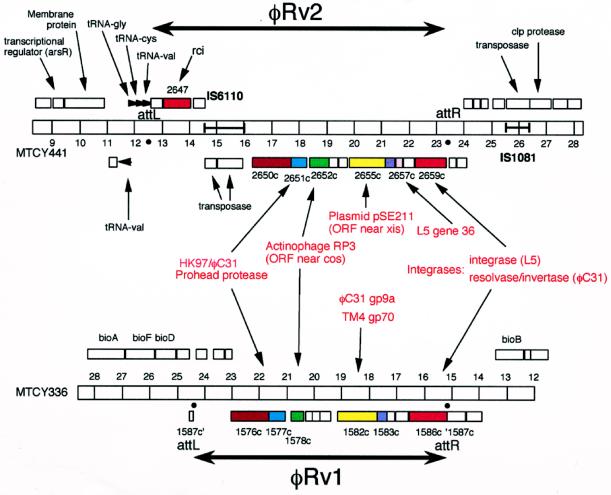
The unusual combination of phage genes in φflu is not peculiar to this particular cryptic prophage but appears to be a feature of other, unrelated prophages. For example, the genome of Mycobacterium tuberculosis H37Rv (22, 23) contains at least two apparent cryptic phages (Fig. 4). Both of these are relatively small (≈10 kilobases) but contain several phage-related genes. One of these prophages (φRv2) has at least two homologues of mycobacteriophage genes: integrase (Rv2659c, a member of the phage integrase-related family of recombinases but most closely related to integrases of phages L5 and D29) and a relative of L5 gene 36 (Rv2657c). In addition, it contains a second recombinase (Rv2647, related to the plasmid-encoded recombinase rci), a homologue of the prohead protease genes of phages HK97 and φC31, and a homologue of a gene product of actinophage RP3 of unknown function (Fig. 4). The second cryptic prophage (φRv1) is rather similar in overall structure to φRv2; it also contains homologues of the HK97/φC31 prohead protease and the actinophage RP3 gene. It has a gene similar to gene 70 of mycobacteriophage TM4 (16) and to gene 9a of Streptomyces phage φC31, encoding a probable DNA primase (R.N.B. and M.C.M.S., unpublished work). Finally, it has a recombinase (Rv1586c) that—like the integrase of φC31 (24)—is related to the transposon resolvase/DNA invertase family of site-specific recombinases (25). This cryptic prophage is flanked by two 12-bp direct repeats, which may represent recognition sites for this recombinase (26). Mahairas et al. (26) noted that this region is only present in 16% of M. tuberculosis clinical isolates and is absent from bacille Calmette-Guérin—an avirulent derivative of the M. tuberculosis complex of bacteria—which has only a single copy of the 12-bp repeat. Presumably, the recombination functions associated with this prophage are active and mediate integration and/or excision of this DNA segment. The φRv2 element also may be recombinationally active because the attachment junctions (attL and attR) are present and appear to derive from a phage attachment site (attP) that is structurally similar to L5 attP (27); the excisionase is provided by the homologue of L5 gene 36 (J. Lewis and G.F.H., unpublished observations). Although the two M. tuberculosis prophages appear too small and too deficient in virion structural genes to encode a complete virion, they could in fact be “complete” satellite phages in the manner of coliphage P4, which uses the structural genes of another phage to package its genome (28). In addition, they could be analogs of some other form of integrating plasmid, such as the Streptomyces integrating plasmid pSE211 (29), which has a gene with sequence similarity to a gene in φRv2 (Fig. 4) as well as an indirect connection by sequence similarity through φC31 gene 9a to the DNA primase gene of phage P4 (R.N.B. and M.C.M.S., unpublished work).
Figure 4.
Cryptic prophages of M. tuberculosis H37Rv. Portions of two segments of the M. tuberculosis H37Rv genome present in cosmids MYCY441 (φRv2) and MTCY336 (φRv1) are shown (accession nos. Z80225 and Z95586, respectively). Horizontal bars represent the genomes with markers at 1-kilobase intervals; numbers correspond to the cosmid coordinates. Genes are shown either above or below the bar, depending on the direction of transcription, with genes below transcribed leftward. Related genes are shown in similar colors. Putative attachment junctions, attL and attR, delineate the boundaries of the cryptic prophages φRv2 and φRv1. The location of genes and their putative functions are from annotations by the Sanger Center, Cambridge, U.K.
Regardless of the provenance and current function of these cryptic prophages, their unusual combinations of genes demonstrate that phages with apparently unrelated genomes (e.g., mycobacteriophage L5 and coliphage HK97) do not exist in genetic isolation. The bridge that φC31 provides between the L5 and HK97 sequences and the diverse combination of phage genes in φflu lead to similar conclusions, and we expect that many more such links will become apparent as new phage and prophage sequences become available. These connections are also consistent with previous observations that phages frequently have specific functional properties in common [for example, the crosslinked head proteins of L5 and HK97 (18) and ribosomal frameshifting in tail genes (4)] even when sequence similarity is not evident (Fig. 1). The combination of these functional, organizational, and now sequence similarities suggests that a significant number of dsDNA bacteriophages are in fact related, having had—and continuing to have—access to a large pool of functional genetic elements. The network of connections shown in Fig. 1 is presumed to be representative of a vastly larger network of relationships among phage sequences that exists in nature. The sequence connections shown in the figure are unlikely to be the result of direct genetic exchanges between the indicated phage genomes; rather, we believe they are the visible end products of long chains of genetic interactions. We anticipate that these processes not only fuel the generation of new bacteriophages but that similar events are involved in the evolution of novel viruses of eukaryotic and archael hosts.
DISCUSSION
It has been estimated that there are 4–6 × 1030 prokaryotic cells in the biosphere (30), and direct counts on environmental samples typically show ≈10-fold more tailed phage particles than cells (3). Thus, the total number of extant phages is enormous. We have no way of calibrating the age of phages as a group, but we presume them to be ancient—possibly comparable in age to their bacterial hosts. Against this very large and probably very old population, there are ≈30 complete DNA sequences of contemporary phage genomes available, including those reported here, plus several prophage sequences contained in bacterial genome sequences. Here, we discuss what our observations of this sample imply about the genetic structure and dynamics of the phage genome population as a whole and more explicitly present our view of the most plausible interpretation of these data.
We take our observations of sequence similarities among genes found in phages and prophages of diverse hosts to imply common ancestry for those genes. We expect that the same is true for those genes for which sequence similarities are not yet apparent in the currently available data but that have similar functions, although this remains to be proven. Thus, we suggest, on the basis of the sequence similarities, that many and probably most of the genes of contemporary phages derive from a common ancestral pool of genes. The view of common ancestry gains support as well from many striking similarities of phage gene organization and function, often in the absence of any remaining sequence similarity (refs. 14–16 and 31 and our unpublished observations).
In comparing phage genomes, it is clear that only some pairs of genes show significant sequence similarities. We believe that this argues strongly for horizontal exchange of sequences among the ancestors of the contemporary phage. Thus, the juxtaposition of those genes that match with substantial sequence similarity between L5 and φC31 together with those that have no detectable similarity seems most plausibly explained by past horizontal exchange of sequences. The same is true for the similarity of gene organization—and, in some cases, sequence similarity—between the late genes of HK97 and φC31, which are joined to the differently organized and sequence-dissimilar early genes of these phages. The most extreme case, however, is in φflu, where genes with unequivocal sequence similarities to genes from a large group of phylogenetically diverse hosts are found but, for each homologous pair, the adjacent genes have no sequence or functional resemblance. We find it very difficult to understand how such a set of relationships could have arisen without multiple horizontal exchanges. An alternative model that these phages have derived from a common ancestor in the absence of horizontal exchange cannot be ruled out formally. In this scenario, differences in the degree of sequence similarity for different genes could be attributable to gene-specific differences in the rate of the mutational clock, but it is difficult to understand why, within different pairwise comparisons of genomes, different sets of genes would have slower mutational clocks than others.
If we allow that horizontal exchange has occurred, then we can ask when it happened. An extreme model is that most of the horizontal exchange giving rise to the current diversity occurred early in the history of phages and since some point early in evolutionary history—for example, the beginnings of bacterial speciation—has been passed down to contemporary phages largely by vertical transmission. If the phage gene pool was sufficiently diverse, the observed sequence similarities over large (host) phylogenetic distances between some phage genes, but not others, simply could reflect the assortment of the various combinations of genes at this early stage. Such a model might allow horizontal exchange to continue among phages infecting a single bacterial species. A less extreme model, and one that we strongly prefer, is one in which horizontal exchange has continued up to the present. This is in part because we doubt that the degree of sequence similarity we see in some genes could have persisted over a period of time when other genes—for example, the virion structural genes, which are arguably homologous on the basis of shared gene organization and function—often have diverged past the point of recognizable similarity. Of more importance, a model with widespread contemporary horizontal exchange is more consistent with our knowledge of the biology of phages and their hosts.
It has been clear for some time that there is vigorous and ongoing horizontal exchange among the well studied lambdoid phages of Escherichia coli and Salmonella—originally so named because they are able to form viable genetic hybrids with the prototype phage λ (ref. 11; R. Juhala, M.E.F., R. L. Duda, A. Youlton, G.F.H., and R.W.H., unpublished work). Thus, for example, we can find pairs of genes in two lambdoid phages (such as HK97 and P22) that have nearly identical sequences juxtaposed with genes with little or no sequence similarity, implying horizontal exchange in the very recent evolutionary past. This exchange presumably happens most often when two phage genomes find themselves in the same cell, either as two coinfecting phages or, perhaps of more importance, as a single phage infecting a cell that carries one or more prophages. Sequence comparisons by us and others (15, 16, 31–34) of genomes of phages that infect other single or closely related groups of host species are beginning to make a case for similar clusters of intensely exchanging phages centered around these hosts. However, we believe it is very unlikely that horizontal exchange is confined to these species-related groups of phages. Phage host ranges often are not confined to a single bacterial species, and phages with overlapping host ranges should be able to exchange sequences whether or not their host ranges are otherwise similar. Other mechanisms exist for transfer of bacterial DNA sequences, potentially including phage or prophage sequences, between bacterial species, and it has been estimated that E. coli replaces ≈16 kbp of its genome by horizontal exchange from outside sources every million years (35). Thus, we believe that there are ample opportunities for phage DNA sequences to travel among host species in the local phylogenetic neighborhood. However, our data and similar observations place an important constraint on how this process happens on a larger scale. Thus, all of the sequence similarities we see between phages or prophages associated with phylogenetically distant hosts are of only a modest level of sequence identity; this argues against direct horizontal exchange between these phage in recent evolutionary time. This most likely means that there is no mechanism available for direct exchange between two such phages—for example, that there is no common host for coliphage HK97 and actinophage φC31—but it also might mean that a sequence adapted to function in one host might find itself at a disadvantage after leaping directly into a very different host. In either case, this argues that for a phage sequence to travel to a phylogenetically distant host requires a journey of many steps—a sort of random walk through phylogenetic space.
What, then, is our view of the genetic structure of the global phage population? We favor a model in which all of the dsDNA phage and prophage genomes are mosaics with access by horizontal exchange to a large common gene pool. However, access is clearly not uniform. There are phylogenetically local areas of free and intense exchange of genetic information, as, for example, the lambdoid phages of enteric hosts or the phage L5 family of the mycobacteria. In addition, there is exchange beyond the confines of the local neighborhood, but only with reduced frequency. Thus, we imagine that any given phage has access to all of the sequences in the global pool but that the frequency of that access depends strongly on the number of barriers (e.g., host ranges) between any particular sequence and that phage and, therefore, how many individual steps of genetic exchange are required to bring them together. The veracity of this view of bacteriophage population genetics and evolution, and the quantitative nature of the relationships implied, will only be determined, we believe, after substantially more data are determined of sequences and genetic organization of phages and their hosts.
Acknowledgments
We thank members of our laboratories and Jeffrey Lawrence for useful discussions. This work was supported in part by National Institutes of Health Grant GM51975 to R.W.H. and G.F.H. and grants from the Medical Research Council (G9301410MB) and Royal Society to M.C.M.S. and in part by a grant from the Pittsburgh Supercomputing Center through the National Institutes of Health National Center for Research Resources resource Grant 2 P41 RR06009.
ABBREVIATION
- dsDNA
double-stranded DNA
Footnotes
References
- 1.Coetzee J. In: Phage Ecology. Goval S M, Gerba C, Bitton G, editors. New York: Wiley; 1987. pp. 45–85. [Google Scholar]
- 2.Ackermann H-W. Arch Virol. 1996;141:209–218. doi: 10.1007/BF01718394. [DOI] [PubMed] [Google Scholar]
- 3.Bergh Ø, Børsheim Y, Bratbak G, Heldal M. Nature (London) 1989;340:467–468. doi: 10.1038/340467a0. [DOI] [PubMed] [Google Scholar]
- 4.Casjens S R, Hatfull G F, Hendrix R W. Semin Virol. 1992;3:383–397. [Google Scholar]
- 5.Haggard-Ljungquist E, Halling C, Calendar R. J Bacteriol. 1992;174:1462–1477. doi: 10.1128/jb.174.5.1462-1477.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sandmeier H, Iida S, Arber W. J Bacteriol. 1992;174:3936–3944. doi: 10.1128/jb.174.12.3936-3944.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tetart F, Repoila F, Monod C, Krisch H M. J Mol Biol. 1996;258:726–731. doi: 10.1006/jmbi.1996.0281. [DOI] [PubMed] [Google Scholar]
- 8.Monod C, Repoila F, Kutateladze M, Tetart F, Krisch H M. J Mol Biol. 1997;267:237–249. doi: 10.1006/jmbi.1996.0867. [DOI] [PubMed] [Google Scholar]
- 9.Campbell A, Botstein D. In: The Bacteriophages. Calendar R, editor. I. New York: Plenum; 1988. pp. 1–14. [Google Scholar]
- 10.Highton P, Chang Y, Myers R. Mol Microbiol. 1990;4:1329–1340. doi: 10.1111/j.1365-2958.1990.tb00712.x. [DOI] [PubMed] [Google Scholar]
- 11.Campbell A. Annu Rev Microbiol. 1994;48:193–222. doi: 10.1146/annurev.mi.48.100194.001205. [DOI] [PubMed] [Google Scholar]
- 12.Simon M N, Davis R W, Davidson N. In: The Bacteriophage Lambda. Hershey A D, editor. Plainview, New York: Cold Spring Harbor Lab. Press; 1971. pp. 313–328. [Google Scholar]
- 13.Hatfull G F, Sarkis G. Mol Microbiol. 1993;7:395–405. doi: 10.1111/j.1365-2958.1993.tb01131.x. [DOI] [PubMed] [Google Scholar]
- 14.Hatfull G F, Jacobs J R., Jr . In: Tuberculosis: Pathogenesis, Protection and Control. Bloom B R, editor. Washington, DC: Am. Soc. Microbiol.; 1994. pp. 165–183. [Google Scholar]
- 15.Ford M E, Sarkis G J, Belanger A E, Hendrix R W, Hatfull G F. J Mol Biol. 1998;279:143–164. doi: 10.1006/jmbi.1997.1610. [DOI] [PubMed] [Google Scholar]
- 16.Ford M E, Hendrix R W, Hatfull G F. Tubercle Lung Dis. 1998;79:63–73. doi: 10.1054/tuld.1998.0007. [DOI] [PubMed] [Google Scholar]
- 17.Popa M P, McKelvey T A, Hempel J, Hendrix R W. J Virol. 1991;65:3227–3237. doi: 10.1128/jvi.65.6.3227-3237.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duda R L, Martincic K, Xie Z, Hendrix R W. FEMS Microbiol Rev. 1995;17:41–46. doi: 10.1111/j.1574-6976.1995.tb00186.x. [DOI] [PubMed] [Google Scholar]
- 19.Hartley N M, Murphy G O, Bruton C J, Chater K F. Gene. 1994;147:29–40. doi: 10.1016/0378-1119(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 20.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, et al. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 21.Perler F B, Olsen G J, Adam E. Nucleic Acids Res. 1997;25:1087–1093. doi: 10.1093/nar/25.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Philipp W J, Poulet S, Eiglmeier K, Pascopella L, Balasubramanian V, Heym B, Bergh S, Bloom B R, Jacobs W R, Jr, Cole S T. Proc Natl Acad Sci USA. 1996;93:3132–3137. doi: 10.1073/pnas.93.7.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cole S T. Nature (London) 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 24.Kuhstoss S, Rao N J. J Mol Biol. 1991;222:897–908. doi: 10.1016/0022-2836(91)90584-s. [DOI] [PubMed] [Google Scholar]
- 25.Stark W M, Boocock M R, Sherratt D J. Trends Genet. 1992;12:432–439. [PubMed] [Google Scholar]
- 26.Mahairas G G, Sabo P J, Hickey M J, Singh D C, Stover C K. J Bacteriol. 1996;178:1274–1282. doi: 10.1128/jb.178.5.1274-1282.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peña C E A, Lee M H, Pedulla M L, Hatfull G F. J Mol Biol. 1997;266:76–92. doi: 10.1006/jmbi.1996.0774. [DOI] [PubMed] [Google Scholar]
- 28.Lindqvist B H, Deho G, Calendar R. Microbiol Rev. 1993;57:683–702. doi: 10.1128/mr.57.3.683-702.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown D P, Idler K B, Katz L. J Bacteriol. 1990;172:1877–1888. doi: 10.1128/jb.172.4.1877-1888.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whitman W B, Coleman D C, Wiebe W J. Proc Natl Acad Sci USA. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lucchini S, Desiere F, Brüssow H. Virology. 1998;246:63–73. doi: 10.1006/viro.1998.9190. [DOI] [PubMed] [Google Scholar]
- 32.Desiere F, Lucchini S, Brüssow H. Virology. 1998;241:345–356. doi: 10.1006/viro.1997.8959. [DOI] [PubMed] [Google Scholar]
- 33.Neve H, Zenz K I, Desiere F, Koch A, Heller K J, Brüssow H. Virology. 1998;241:61–72. doi: 10.1006/viro.1997.8960. [DOI] [PubMed] [Google Scholar]
- 34.Brussow H, Bruttin A, Desiere F, Lucchini S, Foley S. Virus Genes. 1998;16:95–109. doi: 10.1023/a:1007957911848. [DOI] [PubMed] [Google Scholar]
- 35.Lawrence J G, Ochman H. Proc Natl Acad Sci USA. 1998;95:9413–9417. doi: 10.1073/pnas.95.16.9413. [DOI] [PMC free article] [PubMed] [Google Scholar]