Abstract
Assembly of quantitative models of large complex networks brings about several challenges. One of them is combinatorial complexity, where relatively few signaling molecules can combine to form thousands or millions of distinct chemical species. A receptor that has several separate phosphorylation sites can exist in hundreds of different states, many of which must be accounted for individually when simulating the time course of signaling. When assembly of protein complexes is being included, the number of distinct molecular species can easily increase by a few orders of magnitude. Validation, visualization, and understanding the network can become intractable. Another challenge appears when the modeler needs to recast or grow a model. Keeping track of changes and adding new elements present a significant difficulty. We describe an approach to solve these challenges within the Virtual Cell (VCell). Using (i) automatic extraction from pathway databases of model components, and (ii) rules of interactions that serve as reaction network generators, we provide a way for semi-automatic generation of quantitative mathematical models that also facilitates the reuse of model elements. In this approach, kinetic models of large, complex networks can be assembled from separately constructed modules, either directly or via rules. To implement this approach, we have combined the strength of several related technologies: the BioPAX ontology, the BioNetGen rule-based description of molecular interactions, and the VCell modeling and simulation framework.
1. Introduction
For biologists, modularity usually refers to the concept that physiological and cell biological regulatory mechanisms can be described as being composed of more or less well-defined functional modules, with sparse connectivity across the boundaries of such modules [1]. We generalize this approach to address the combinatorial complexity that often arises when detailed quantitative models of intracellular networks and pathways are being sought. Similar to describing metabolism as modules that can be reused in different pathways [2], one can view proteins that are composed of multiple domains as functional modules composed of many elements - e.g., Src homology 2 (SH2) binding sites and tyrosines phosphorylation sites [3]. This is a typical situation that generates combinatorial complexity in signaling pathways. For example, in the case of Epidermal Growth Factor Receptor, EGFR [4], a receptor with 10 tyrosine phospho-sites can exist in 210=1024 different phosphoforms, and dimerization and interaction with other molecules then leads to millions of possible distinct complexes. At present, kinetics models accounting for dozens of different molecular species are a norm [5], and models accounting for hundreds of species and reactions are no longer rare [6, 7]. Visualization of such networks is difficult at best, and manually specifying the list of species and reactions becomes error-prone and slow. A solution for this challenge can be provided by the modular approach, in the form of (i) defining smaller reusable model components for quantitative models (modeling modules), and (ii) specifying rules of interaction, be it at the protein/molecular complex level, or arbitrary functional level (e.g. kinetic of ligand-receptor binding is independent of receptor phosphoforms). Quantitative models of complex networks can then be assembled from separately constructed and validated components, either directly or via rules.
To implement this strategy, we have combined the use of the Biological Pathways Exchange (BioPAX) ontology ([8], http://biopax.org), and of the BioNetGen rule-based description of molecular interaction ([9, 10], http://bionetgen.lanl.gov) within the Virtual Cell (VCell) modeling and simulation software framework ([11, 12], http://vcell.org), using the Systems Biology Markup Language (SBML) as a vehicle for interchanging models in simulation-ready format ([13], http://sbml.org/). VCell uses a biophysically and mathematically consistent description of kinetic models that are being stored in a relational database and can be easily shared and re-used at various levels of granularity. BioPAX is a pathway exchange format that aims to facilitate sharing of pathway information between databases and users. Each element of a BioPAX file is linked to an originating biological database, providing for a well-documented biological identification for each element of the model. Any groups of species and reactions annotated with BioPAX can be easily encapsulated in reusable modeling modules. Two approaches are used to generate models without manual specification of each and every species and reactions. The first is using BioPAX data imported from the BioPAX-compatible databases, e.g. Reactome [14]. A BioPAX@VCell application automatically generates an SBML file that can be simulated after kinetic parameters are added by the modeler. Moreover, it also allows for better visualization of the model (Figure 1). The second approach is to specify a model in the form of molecular interaction rules that generate (parts of) the reaction network [9]. This approach, developed originally into a general-purpose software, BioNetGen [10], has been implemented as a BioNetGen@VCell application. The modeler uses his or her knowledge of the system to specify classes of molecules and their interacting and modification modules, (such as tyrosines and SH2 domains), and rules of activities and interactions among modules and molecules (Figure 2). This information is then used by the software to automatically generate a model comprised of all possible distinct chemical species that can arise in the reaction network, as well as all transitions among these species.
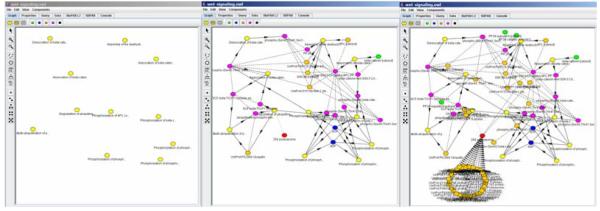
Figure 1.
The screenshot of BioPAX@VCell representation. The file with BioPAX file describing Signaling by Wnt [Homo Sapiens] was loaded from Reactome database. The pathway describes 11 interactions, but SBML file generated by Reactome includes about 100 species, to account for all proteins that constitute complexes in the reaction network. Visualization of this file in any simulation tool is unreadable. BioPAX@VCell provides a better visualization using two key features: (1) different coloring and symbols for different types of species and reactions; and (2) different level of granularity in representation, where the user can view, for example, reactions only (the first panel), reactions with reaction participants (second panel), or expansion of all complexes to view their components (the third panel). The user is able to collapse selected portions of the network into a single super-node, thus significantly improving network readability.
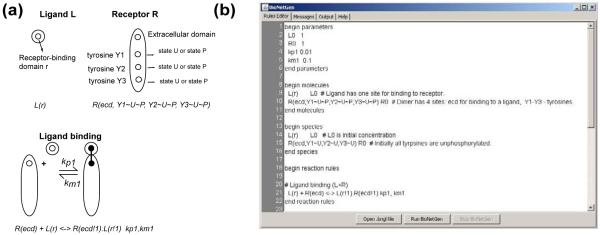
Figure 2.
(a) Rules are based on the knowledge of modular structure of proteins, such as interaction of ligand with extracellular domain of the receptor is independent from the state of intracellular tyrosines. Here, a receptor consists of 4 elements (domains): extracellular ecd and 3 tyrosines that can be in two states: either unphosphorylated (U) or phosphorylated (P). Thus, the total number of potential phosphoforms of this receptor is 23=8. However, the rule does not specify a state of any of phosphosites, which implies that the rule is applied to all potential phosphoforms, thus corresponding to at least 8 bidirectional reactions parameterized by the same on and off rates. Lines in italic demonstrate encoding of this description in BioNetGen Language (BNGL). (b) The screenshot of rules representation in the BNG@VCell application. The model illustrates BNGL file that includes specification of parameters, molecules, initial species, and rules.
2. Introducing reusable components in VCell
Several repositories of mathematical models exist that provide validated, annotated models of cellular processes. The BioModels database [15] is one such fast growing repository, providing several hundred of models in simulation-ready SBML format, which is the emerging community standard for kinetic modeling [13]. VCell has a public repository of models in a proprietary VCML format (a richer format than SBML, that includes among others, spatial descriptions for models), which can be exported into the SBML format. Most software tools now provide a facility to translate into SBML working models, or models from their own repository, if they have one. However, often the translation has a significant flaw, that all elements (species and reactions) can not be easily compared and exchanged between the models without manual intervention. Even the well-annotated models stored in BioModels repository that are compliant with MIRIAM (Minimum information requested in the annotation of biochemical models) standard [16] present challenges. For example, currently there is no standard way to distinguish between different states of the same molecules. Thus, all phosphoforms of the same receptor will be annotated with the same reference identifier (GO term, UniProt key, etc.). This means that there still will be often the case that it is impossible to automatically tell whether species X of model A and species Y of model B, which have the same reference identifier, are indeed identical and should be mapped into the same species in a merged model. We address this by adding compatibility with the BioPAX standard, which has an advantage of a fine grained unique identification of all elements of interactions and signaling pathways across multiple databases.
Currently, several online resources provide information about pathways in BioPAX format, for example Reactome ([14], http://www.reactome.org/), Pathway Interaction Database (http://pid.nci.nih.gov/), BioCyc collection of Pathway/Genome databases ([17], http://biocyc.org), Integrating Network Objects with Hierarchies (INOH) database (http://www.inoh.org/).
The data may range from complete signaling pathways to the biomolecules participating in pathways and individual interactions. To make use of this data, we have designed a BioPAX@VCell modeling framework intended to obtain, store and merge data in BioPAX, to facilitate generating of kinetic models expressed in SBML ([18], http://ccam.uchc.edu/mblinov/biopax). The BioPAX model lacks simulation-related information (such as concentrations, kinetic laws etc) and has auxiliary information which is not essential for simulations, but valuable for understanding and reusing the modes (such as organisms, different names, linking species to a variety of databases, etc). A BioPAX model can be easily visualized, by giving different BioPAX object classes (proteins, small molecules, complexes etc) different representations (e.g. colors), and each object is linked to biological information from public databases. Several such BioPAX models can then be easily merged into a larger model. After adding information to convert a BioPAX model into a computable kinetic model, it can be exported into the VCell software framework for kinetic simulations, or converted into SBML for use with other simulation software. Remarkably, the resulting kinetic model remembers all BioPAX information. Therefore, a merged model can be compactly visualized as a set of modules, where all elements of the same BioPAX model are compressed into a single container node. The BioPAX model provides a “model skeleton” where each species is being automatically assigned a unique identifier which allows semi-automatic merging of VCell models.
To facilitate the organization of the data and to make selections, the BioPAX@VCell framework provides a graphical interface that can handle any OWL model but is specifically developed to support the BioPAX ontology. Ontologies provide a great deal of flexibility in data representation, analysis and visualization, for example allowing the user to select which resources are visible and which are hidden, similar to Cytoscape visualization framework ([19], http://cytoscape.org). Arbitrary sets of objects can be collapsed and expanded again. The user can decide which kinds of property relationships are represented by graph edges. As the framework is based on the existing VCell software, it displays a graph consisting of interactions among BioPAX physical entities in the VCell style as a bipartite graph in its fully flattened form (with nodes for both physical entities and interactions - see Figure 1). For each class of physical entities and interactions the framework provides a separate symbol. Each p-interaction is connected by an edge with the p-entities participating in it. Complexes are displayed in a way that alludes to their components. All other objects are hidden, until they are properties of an object which becomes selected.
3. Using rules to generate models in VCell
The rule-based approach to define models [9, 10] has the advantage of inherently handling combinatorial complexity. It allows one to account comprehensively and precisely for the possible molecular species implied by the specified interactions, activities, and modifications of the molecules in a system. The model is built by defining the rules that govern how molecules interact (to form complexes, modify internal states, or degrade), as illustrated in Figure 2a. These rules can thus specify how new molecular species can be produced and are used to automatically generate the reaction network, freeing the user from the intense bookkeeping that would be required to enumerate such a network by hand. A general-purpose software package, BioNetGen ([9, 10]), was developed that implements this approach. This tool was used to build several models, including the models of receptor-mediated signaling events stimulated by antigen [20] and epidermal growth factor [4], and the function of Shp2 phosphotase in intracellular signal transduction [21]. The model is usually based on assumptions about modular interactions of proteins and the mechanistic details of protein-protein interactions [3, 22]. BioNetGen can also solve the resulting system of ordinary differential equations, simulating the time courses for each species. The software can also determine time courses using the Gillespie algorithm, which may become important when the concentrations are low. Moreover, species and reactions in a network can be generated only as needed during the course of a network simulation [23, 24], which is important when the number of potential chemical species is too big for any reasonable computations (e.g. ∼108 species in the example of independent proteins binding to EGFR tyrosines considered above). Since many of the quantities measured in experiments are properties of ensembles, rather than distinct microscopic species, BioNetGen allows the modeler to calculate functions of microscopic ensembles. For example, the level of phosphorylation of a protein is a function of the concentrations of all microscopic species containing the protein. The appropriate sums of species concentrations that correspond to a defined measurable property of the system are computed automatically.
We have implemented BioNetGen as a stand-alone application invoked by VCell (http://vcell.org/bionetgen; see Figure 2b for a screenshot). A set of precompiled BioNetGen executables has been created for different platforms (total size is less than 3 MB). These are placed in a resource folder for VCell and downloaded when it is launched. In the VCell user interface, BioNetGen appears as a menu item. Users can upload a BioNetGen model, manually edit it in the Rule Editor tab, see the progress of automatic network generation in the Messages tab, visualize and save BioNetGen outputs (.cdat and .gdat files with timecourses for concentrations of all species and observables) using regular VCell graphical capabilities. The Help tab is provided with links to various help topics. Network generation, time courses simulation and visualization are performed within BioNetGen service. No jobs are sent to the VCell server and no model is uploaded to the VCell Database Server unless specifically requested by the user. To do so, a VCell BioModel can be automatically created from an SBML file exported by BioNetGen. The user can thus create a biochemical reaction network in BioNetGen and then use the VCell capabilities for enhancing the model, such as adding multiple compartments and resolved spatial geometries, running multiple simulations and parameter scans, etc.
4. Bridging BioPAX and BioNetGen
The BioPAX@VCell framework provides reading of pathway data from BioPAX-supported databases with explicit specification of each and every species and reactions. BioNetGen@VCell provides generation of VCell models from user-specified rules. Both approaches can be used synergistically. Indeed, the data may allow multiple interpretations. For example, a given interaction can be applied to several phosphoforms of a protein, thus providing a rule instead of a single interaction. These efforts should be concurrent with the development of the next level of BioPax ontology (that will better describe protein modifications), as well as the SBML level 3 extension (that will allow description of multi-component multi-state species).
A serious problem is mapping BioPAX semantics to BioNetGen. BioNetGen-generated models, as well as BioPAX-based models, can be translated into SBML, so they can be shared with other tools using the SBML format [9]. However, this does not supplant exchange of the actual rule-based models or models generated from BioPAX data, as essential features that were used to generate the reaction networks (e.g. components of molecules and rules for rule-based models or protein ids for BioPAX data) can be lost. The SBML development of Level 3 extensions [25] should allow inclusion of all BioNetGen language features into SBML. Additionally, SBML provides for several mechanisms to include such information from different formats and languages in separate namespaces. However, even with the currently planned SBML extensions, not everything that can be expressed in BioNetGen will be able to be expressed in BioPAX, and transforming BioPAX to BioNetGen may not always be unambiguous. To make exact correspondence between two formats,, it is necessary to create and store knowledge about a variety of relationships between SBML (and its extensions) and BioPAX objects. To represent such knowledge, we need a framework that includes terms compatible to all relevant SBML and BioPAX terms. To describe such a framework, we are currently devloping an ontology that will act as a glue and which will include terms compatible with all relevant SBML or BioPAX terms. The goal is for this ontology to effectively be used as an extension or annotation scheme, allowing the target languages to support the representation of all relevant information, and thus,exchange between SBML and BioPAX can occur without semantic losses.
5. Conclusions and future directions
A highly desirable feature for modeling tools is the automated data retrieval and verification using external web resources. After the user picks elements to be included in a model, an intelligent framework should try to infer additional elements (interactions, modifications, kinetic constants) to make a kinetic model complete, and then request this information from available public resources. Qualitative information, such as reaction catalysts, can be requested from databases like Reactome that provide an API for querying and retrieving BioPax data over the web. Quantitative information, such as kinetic constants, can be requested from emerging databases of reaction kinetics, such as SABIO-RK (http://sabio.villa-bosch.de/SABIORK/). When fully implemented, such capabilities will provide a powerful data-driven modeling environment.
Another issue that needs to be addressed in parallel with tool development is standardization. Both BioPAX and SBML are established exchange formats for different communities: quantitative models for simulations are often published or exchanged via SBML, while pathway databases often store information in BioPAX. Extensive joint efforts of both communities are needed to standardize such integration which can eventually lead to an accurate and effective exchange of experimental and theoretical biological information on multiple levels: data, models and visualization.
Acknowledgments
We would like to thank James R. Faeder (Pittsburgh University), William S. Hlavacek (Los Alamos National Lab), Michael Hucka (Caltech), and James C. Schaff (UCHC) for many ideas and helpful discussions regarding this project. The project was supported in part by NIH R01 GM076570 grant (MLB) and NIH U54 RR022232 grant (OR, IIM).
References
- [1].Moraru II, Loew LM. Intracellular signaling: Spatial and temporal control. Physiology. 2005;20:169–179. doi: 10.1152/physiol.00052.2004. [DOI] [PubMed] [Google Scholar]
- [2].Ravasz E, Somera AL, Mongru DA, Oltvai ZN, Barabasi AL. Hierarchical organization of modularity in metabolic networks. Science. 2002;297:1551–1555. doi: 10.1126/science.1073374. [DOI] [PubMed] [Google Scholar]
- [3].Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–52. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- [4].Blinov ML, Faeder JR, Goldstein B, Hlavacek WS. A network model of early events in epidermal growth factor receptor signaling that accounts for combinatorial complexity. Biosystems. 2006;83:136–51. doi: 10.1016/j.biosystems.2005.06.014. [DOI] [PubMed] [Google Scholar]
- [5].Kholodenko BN, Demin OV, Moehren G, Hoek JB. Quantification of short term signaling by the epidermal growth factor receptor. J Biol Chem. 1999;274(42):30169–81. doi: 10.1074/jbc.274.42.30169. [DOI] [PubMed] [Google Scholar]
- [6].Li QJ, Dinner AR, Qi S, Irvine DJ, Huppa JB, Davis MM, Chakraborty AK. CD4 enhances T cell sensitivity to antigen by coordinating Lck accumulation at the immunological synapse. Nat Immunol. 2004;5(8):791–9. doi: 10.1038/ni1095. [DOI] [PubMed] [Google Scholar]
- [7].Schoeberl B, Eichler-Jonsson C, Gilles ED, Müller G. Computational modeling of the dynamics of the MAP kinase cascade activated by surface and internalized EGF receptors. Nat Biotechnol. 2002;20(4):370–5. doi: 10.1038/nbt0402-370. [DOI] [PubMed] [Google Scholar]
- [8].Luciano JS. PAX of mind for pathway researchers. Drug Discov Today. 2005;10:937–42. doi: 10.1016/S1359-6446(05)03501-4. [DOI] [PubMed] [Google Scholar]
- [9].Hlavacek WS, Faeder JR, Blinov ML, Posner RG, Hucka M, Fontana W. Rules for modeling signal-transduction systems. Sci STKE. 2006;2006(344):re6. doi: 10.1126/stke.3442006re6. [DOI] [PubMed] [Google Scholar]
- [10].Blinov ML, Faeder JR, Goldstein B, Hlavacek WS. BioNetGen: software for rule-based modeling of signal transduction based on the interactions of molecular domains. Bioinformatics. 2004;20:3289–91. doi: 10.1093/bioinformatics/bth378. [DOI] [PubMed] [Google Scholar]
- [11].Loew LM, Schaff JC. The Virtual Cell: A software environment for computational cell biology. Trends in Biotechnology. 2001;19:401–406. doi: 10.1016/S0167-7799(01)01740-1. [DOI] [PubMed] [Google Scholar]
- [12].Moraru II, Schaff JC, Loew LM. In: Perrone LF, Wieland FP, Liu J, Lawson BG, Nicol DM, Fujimoto RM, editors. Think simulation - think experiment: the Virtual Cell paradigm; Proceedings of the 2006 Winter Simulation Conference; 2006.pp. 1713–1719. [Google Scholar]
- [13].Hucka M, et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–31. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- [14].Vastrik I, D’Eustachio P, Schmidt E, Joshi-Tope G, Gopinath G, Croft D, de Bono B, Gillespie M, Jassal B, Lewis S, Matthews L, Wu G, Birney E, Stein L. Reactome: a knowledge base of biologic pathways and processes. Genome Biol. 2007;8(3):R39. doi: 10.1186/gb-2007-8-3-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Le Novère N, Bornstein B, Broicher A, Courtot M, Donizelli M, Dharuri H, Li L, Sauro H, Schilstra M, Shapiro B, Snoep JL, Hucka M. BioModels Database: a free, centralized database of curated, published, quantitative kinetic models of biochemical and cellular systems. Nucleic Acids Res. 2006;34:D689–91. doi: 10.1093/nar/gkj092. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Laibe C, Le Novere N. MIRIAM Resources: tools to generate and resolve robust cross-references in Systems Biology. BMC Syst Biol. 2007;1(1):58. doi: 10.1186/1752-0509-1-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Karp PD, Ouzounis CA, Moore-Kochlacs C, Goldovsky L, Kaipa P, Ahrén D, Tsoka S, Darzentas N, Kunin V, López-Bigas N. Expansion of the BioCyc collection of pathway/genome databases to 160 genomes. Nucleic Acids Res. 2005;33(19):6083–9. doi: 10.1093/nar/gki892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ruebenacker O, Moraru II, Schaff JC, Blinov ML. Kinetic Modeling Using BioPAX Ontology; Proceedings of the 2007 IEEE International Conference on Bioinformatics and Biomedicine; 2007; pp. 339–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Faeder JR, Hlavacek WS, Reischl I, Blinov ML, Metzger H, Redondo A, Wofsy C, Goldstein B. Investigation of early events in Fc epsilon RI-mediated signaling using a detailed mathematical model. J Immunol. 2003;170(7):3769–81. doi: 10.4049/jimmunol.170.7.3769. [DOI] [PubMed] [Google Scholar]
- [21].Barua D, Faeder JR, Haugh JM. Structure-based kinetic models of modular signaling protein function: focus on Shp2. Biophys J. 2007;92(7):2290–30. doi: 10.1529/biophysj.106.093484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bhattacharyya RP, Reményi A, Yeh BJ, Lim WA. Domains, motifs, and scaffolds: the role of modular interactions in the evolution and wiring of cell signaling circuits. Annu Rev Biochem. 2006;75:655–80. doi: 10.1146/annurev.biochem.75.103004.142710. [DOI] [PubMed] [Google Scholar]
- [23].Lok L, Brent R. Automatic generation of cellular reaction networks with Moleculizer 1.0. Nat Biotechnol. 2005;23(1):131–6. doi: 10.1038/nbt1054. [DOI] [PubMed] [Google Scholar]
- [24].Faeder JR, Blinov ML, Hlavacek WS. Rule-based modeling of biochemical networks. Complexity. 2005;10:22–41. [Google Scholar]
- [25].Blinov ML, Moraru II. XML Encoding of Features Describing Rule-Based Modeling of Reaction Networks with Multi-Component Molecular Complexes; Proceedings of the 7th IEEE International Conference on Bioinformatics and Bioengineering; 2007; pp. 987–994. [DOI] [PMC free article] [PubMed] [Google Scholar]