Abstract
Cholesterol is essential for proper function and regulation of eukaryotic membranes, and significant amounts of metabolic energy are dedicated to controlling cellular cholesterol levels. Oxidation products of cholesterol, the oxysterols, are enzymatically produced molecules that play a major role in mediating cholesterol homeostasis through mechanisms which have not yet been fully elucidated. Certain oxysterols are known to have direct effects on membrane permeability and structure; effects that are strikingly different from that of cholesterol. We use molecular dynamics simulations of these oxysterols in 1-palmitoyl 2-oleoyl phosphatidylcholine (POPC) bilayers to explain the structural origins for the differing effects of cholesterol and 25-hydroxycholesterol on bilayer properties. In particular, we demonstrate that the source for these differing perturbations is the much wider range of molecular orientations accessible to 25-hydroxycholesterol when compared to cholesterol. This study shows that direct membrane perturbation by side-chain oxysterols is significant, and suggests that these membrane perturbations may play a role in the oxysterol regulation of cholesterol homeostasis.
1 Introduction
While the major components of cellular membranes are phospholipids, sterols are essential for membrane function.1 Cholesterol is the most prevalent sterol in mammalian cells, where it is distributed unevenly among mammalian membranes with the plasma membrane containing significantly more cholesterol than the mitochondrial or ER membranes.1, 2 Cholesterol is required by all mammalian cells, and can either be produced endogenously or taken up from plasma lipoproteins.1, 2 Its functions in the cell include binding to sterol-sensing domains to regulate protein function,3, 4 participating in the formation of lipid rafts,5, 6 and serving as a precursor for steroid hormone and bile acid synthesis.7 Homeostasis of cholesterol levels is maintained through regulation of de novo synthesis, cholesterol uptake, and cholesterol efflux.8, 9 Cholesterol influences cellular behavior both directly and indirectly. Conserved sterol-sensing domains (SSDs) are found in many different membrane proteins and respond to concentrated levels of sterols in the local membrane by changing their binding affinities and enzymatic activities, allowing sterols to signal in a number of pathways through these proteins.4 In particular, both the cholesterol synthesis and intracellular cholesterol transport pathways contain proteins with SSDs, indicating a role for the SSD in sterol homeostasis.4 Cholesterol also alters membrane structure, increasing membrane thickness, bending modulus, and lipid order while decreasing membrane uidity.10–15 These physical changes can affect membrane proteins as demonstrated in membrane protein sorting,16 cellular signaling,6 and changes in ion channel properties.17
Oxysterols are also known to exhibit a variety of biological activities. Of particular interest is their effect on cholesterol synthesis through feedback inhibition.18 Transcriptional regulation of cholesterol synthesis is mediated by sterol regulatory-element binding proteins (SREBPs), a family of membrane-bound transcription factors.8, 18 SREBPs form a complex with the SREBP cleavage-activating protein (SCAP). When this complex moves to the Golgi apparatus, SREBP is cleaved by Golgi-resident proteases, and the transcription factor domain of SREBP is released to activate transcription of cholesterol biosynthetic enzymes.8, 18 High levels of membrane sterols also induce binding of SCAP to ER-resident Insig proteins, retaining the SREBP-SCAP complex in the ER and blocking upregulation of cholesterol synthesis.8, 18 While cholesterol alone is sufficient to induce this feedback inhibition, it has been known for many years that oxysterols, including 25-hydroxycholesterol, are greater than 50 times more effective at suppressing the expression of sterol synthetic enzymes such as HMGCoA reductase.18, 19 In conjunction with the discovery of oxysterol-synthesizing proteins and oxysterol-binding proteins, this has led to speculation that cholesterol’s feedback inhibition may be partially mediated through oxysterols.20 Like cholesterol, the 25-hydroxycholesterol oxysterol can act through both specific ligand-protein interactions20, 21 and by altering the structural properties of membranes. Specifically, 25-hydroxycholesterol has been shown to increase membrane permeability and monolayer per-lipid area.22–24
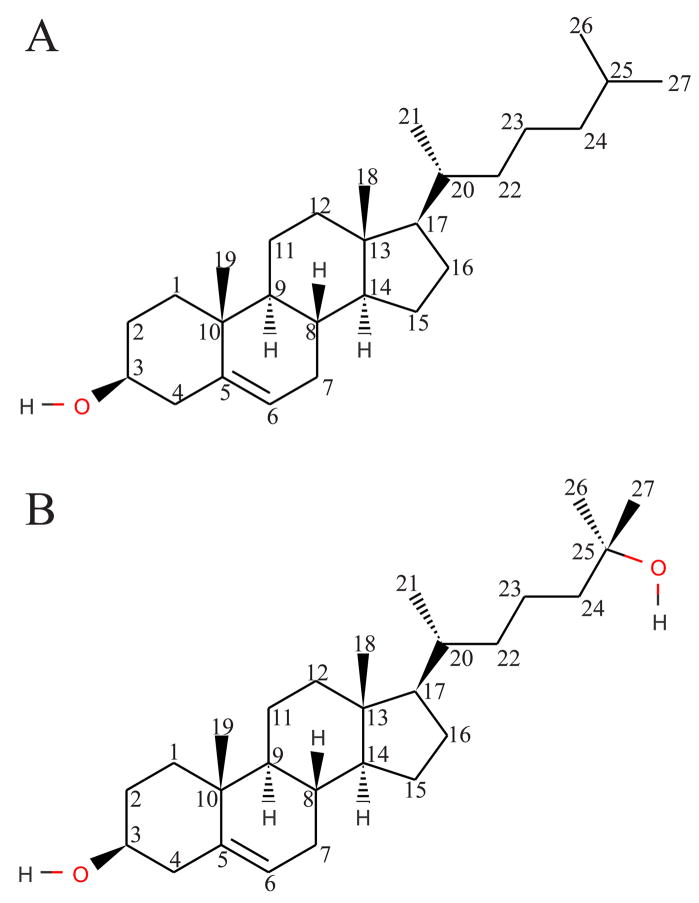
The molecular structure of cholesterol (Fig. 1A) is based around four fused rings in a trans configuration, making the ring structure planar and rigid. This rigid ring structure contains two methyl groups protruding out of one face of the planar ring and is connected to a flexible iso-octyl hydrocarbon chain at carbon 17. Due to the hydroxyl group located at carbon 3, cholesterol is an amphiphile. In the membrane, this tends to orient cholesterol with the hydroxyl group facing water and the polar regions of nearby phospholipids to maximize hydrogen-bonding interactions.1 Another important aspect of cholesterol is that the 3-hydroxyl group, the iso-octyl hydrocarbon chain, and the two methyl groups are all attached to the same face of the planar ring, giving it “smooth” and “rough” faces that influence its interactions with other molecules.1 While cholesterol is the dominant sterol within mammalian cells, oxidation products of cholesterol are formed at low levels by both reactive oxygen species and enzymatic action on cholesterol.25, 26 The oxysterol 25-hydroxycholesterol (Figure 1B) is one such oxidation product which contains an additional hydroxyl group at the end of the iso-octyl tail, on the other side of the molecule from the 3-hydroxyl group. Addition of this 25-hydroxyl group to a nonpolar region of the cholesterol structure significantly alters the amphiphilic characteristics of this molecule.
Figure 1.
Cholesterol (A) and 25-Hydroxycholesterol (B), with standard carbon numbering schemes.
The molecular-level interactions of cholesterol and 25-hydroxycholesterol with phospholipid membranes are essential to understanding their dramatically different effects on mammalian cells. Both of these sterols can perturb the bulk properties of membranes1, 22, 23 with significant effects on the behavior of membrane-bound proteins.6, 16, 17 In this study, we use molecular dynamics simulations of mixed sterol/phospholipid membranes to examine the influence of chemical differences in sterol structure on membrane interactions. This technique, while limited to very fine details of interactions and simple membrane structures, allows us to gather atomic-level information about how these small molecules interact with membranes and the mechanisms by which they can perturb membrane behavior.
2 Methods
2.1 Parameters and Structures
The initial united atom structure and GROMACS topology for cholesterol modeling were taken from Höltje et al.27 An additional hydroxyl group was added to both the structure and topology to produce 25- hydroxycholesterol (Fig. 1B).
Atomic charges for both cholesterol and 25-hydroxycholesterol molecules were calculated using quantum mechanical/molecular mechanical (QM/MM) methods. Our starting structures for these calculations were taken from short MD simulations of a single sterol (cholesterol or 25-hydroxycholesterol) solvated in SPC water with imprecise atom charges, taken from the original Höltje topology.27 QM/MM minimization was performed with the QSite program.28 The QM/MM methodology has been described elsewhere.29, 30 A solvation sphere of 15 Å around the sterol was retained, and the outer 3 Å of waters were constrained in place. Water molecules were modeled using MM methods, using the OPLS-AA force field.31 The sterol molecule was converted to an all-atom structure and modeled using density functional QM methods with the B3LYP functional32, 33 in combination with the LACVP* basis set.34, 35
Sterol atom charges were obtained from the minimized QM/MM systems by fitting the molecular electrostatic potential (ESP) surface to atomic point charges.36–38 Charges for non-hydroxyl hydrogen atoms were added onto their attached heavy atom’s charge to prepare united atom charges. These charges were then adjusted slightly to create net-neutral charge groups for MD simulation and are shown in Supporting Information.
1-palmitoyl-2-oleoyl-phosphatidylcholine (POPC) lipids were simulated using the united atom parameters of Berger and Lindahl,39 along with SPC water40 and Straatsma-Berendsen potassium and chloride ion parameters.41 Sterol bonded and non-bonded parameters were taken from the GROMOS force-field native to GROMACS.42 This combination of force-field parameters has been successfully used in a number of previous studies39, 43–45 and yields good agreement with experimental observables such as area per lipid headgroup and tail order parameters. While good results have also been obtained using all-atom models for POPC,46, 47 united atom models allow us to use longer timesteps and simulate larger systems while still retaining a useful level of molecular detail. An initial bilayer structure of 128 POPC molecules was obtained from Tieleman and coworkers.44 This structure was replicated in the bilayer plane and trimmed to produce a larger, 256 lipid bilayer, with approximate dimensions of 10 nm × 10 nm in the bilayer plane.
To prepare low-concentration mixed sterol/POPC bilayers, the 256 POPC bilayer was solvated with 14260 SPC water molecules along with 30 K+ and Cl− ions for a nominal KCl concentration of 110 mM. A single molecule of cholesterol or 25-hydroxycholesterol was placed 1–2 nm from the surface of each monolayer at the start of the low-concentration simulation. This system was then simulated for 40 ns to allow the sterols to associate with the POPC bilayer. Both cholesterol and 25-hydroxycholesterol inserted themselves into the bilayer within 10 ns. High-concentration sterol/POPC structures were prepared from the converged portions of these low-concentration simulations as follows. As illustrated in Supporting Information, 7 sterol and 16 POPC molecules were extracted from the converged low-concentration simulation and arranged in a 5 × 5 array to form an oriented monolayer. This monolayer was then stacked on an inverted copy of itself to produce a 14 sterol, 32 POPC bilayer. These very small bilayers were simulated for 10 to 15 ns to allow them to relax. The relaxed structure was then copied 3 × 3 in the plane of the bilayer and 16 POPC and 7 sterols removed from each monolayer of the resulting structure. This process produced final structures consisting of 256 POPC and 112 sterols, or bilayers of about 30 mole percent of sterols. These structures were solvated with 17541 (cholesterol) or 17325 (oxysterol) SPC water molecules and 36 K+ and Cl− ions for an approximate molar concentration of 110mM KCl.
2.2 Simulations
All molecular dynamics (MD) simulations were performed using GROMACS version 3.3.1.42, 48 All simulations followed the same molecular dynamics protocol. Conjugate gradient energy minimization was first performed on the initial structures to relax any unfavorable contacts between molecules. The system was then gradually warmed with a series of 30 ps constant temperature, constant pressure MD simulations from 0 to 300 K in 15 K increments, with 2 fs time steps. Production simulations were then run for 207 (cholesterol) or 208 (25-hydroxycholesterol) ns. Anisotropic pressure coupling was applied at 1 atm using the Parrinello- Rahman method with a time constant of 1 ps.49 Temperature coupling was applied independently to lipids and solvent using the Nosé-Hoover algorithm with a time constant of 0.2 ps.50 Electrostatic interactions were calculated using the particle-mesh Ewald method (PME), with both the direct space PME cutoff and the Lennard-Jones cutoffs set to 1 nm.51 Constraints were applied to all bonds52, 53 using the LINCS algorithm incorporated in GROMACS to allow 2 fs timesteps.54
2.3 Analytical Methods
2.3.1 Bootstrap Errors
Some time-independent observables, such as bulk membrane properties or hydrogen bonding probability distributions, are calculated as cumulative properties of an entire stationary trajectory. For such observables, errors can be estimated using a bootstrap method.55, 56 In the following description, we assume we are analyzing a trajectory containing N frames of data f1, f2, …, fN and are interested in calculating an observable property P(f1, f2, …, fN) which depends on multiple frames of this trajectory. Furthermore, we use the statistical inefficiency method described in Supporting Information to estimate the number N′ of trajectory frames that are statistically independent. The main step in the bootstrap method is the generation of a synthetic trajectory dataset with N′ frames of data {f1′, f2′, …, fN′}. Generation of this synthetic dataset proceeds by drawing N′ frames randomly with replacement from the real trajectory. The observable of interest is then calculated from this synthetic trajectory as P({f1′, f2′, …, fN′}). A distribution of observable values is produced by repeating this main resampling step over many randomly-generated synthetic trajectories. The distribution of these synthetic observable values can then be used to estimate errors in the calculated value by measuring the standard deviation (or other error metrics) from the bootstrapped distribution of observable values.
2.3.2 SASA Calculations
Solvent-accessible surface area (SASA) calculations for our systems were performed with APBS version 1.0.0, using a 1.4 Å radius solvent probe.57 For these calculations, the systems were were stripped of solvent and ion molecules and replicated 3 × 3 times in the xy plane to reduce edge effects. Only the central image of this replicated bilayer was used for analysis. The SASA of each atom in the central structure’s lipids in the replicated system is calculated with APBS, and the contributions from each lipid’s atoms summed to obtain a molecular surface area for each lipid.
3 Results
3.1 Equilibration
In order to determine whether a lipid simulation is sufficiently equilibrated (e.g., with observables sampling a stationary distribution), we would like to examine those properties of the system which are slow to converge to a steady-state value. For bilayer systems, the total cross-sectional area Atot of the simulation is a useful metric; it generally drifts more slowly than other properties of the system. Furthermore, the area offers useful information about the bilayer structure that can be compared with experimental values.
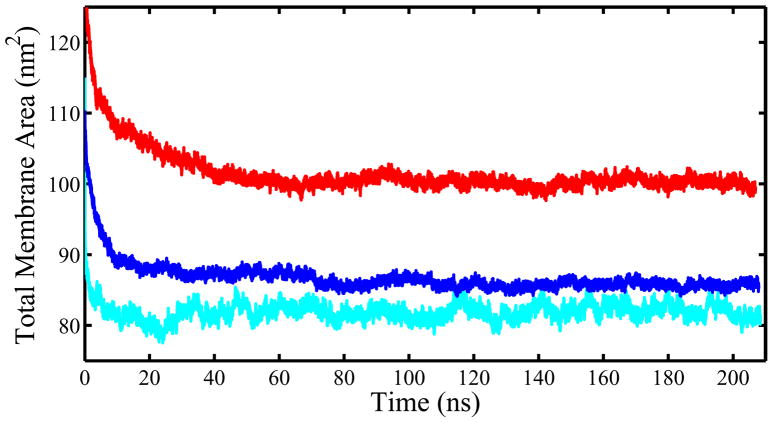
The total system areas of our three simulations are shown in Figure 2. For equilibration purposes, it is necessary to make an estimation of when the initial drift in the simulations has vanished. The length of this initial drift phase varies somewhat for each system. For the pure POPC membrane, this drift phase is relatively short, reaching steady-state values in under 5–10 ns. The cholesterol system relaxes slightly more slowly, approaching steady-state values after approximately 15 ns, although with a slight change at 70 ns. Finally, the 25-hydroxycholesterol system converges much more slowly, not approaching steady state values until after 40 ns. Based on observation of these areas, as well as additional metrics described in Supporting Information, the first 80 ns of all simulations were removed as an equilibration phase. Subsequent “production” analyses were performed using only data from the final 128 ns of the 208 ns trajectories.
Figure 2.
The total projected membrane area of the three membrane simulations — pure POPC (cyan), 30% mole fraction cholesterol (blue), and 30% mole fraction 25-hydroxycholesterol (red).
Statistical inefficiency tests were applied to the “production” 128 ns of these area plots to determine the relaxation time of our systems (see Supporting Information). Approximate area relaxation times of 3.0–4.0 ns were obtained. Thus for all later statistical analyses, each trajectory was conservatively treated as containing 32 independent bilayer conformations. For analysis of individual molecules, each lipid was treated independently for each independent frame.
Similar analyses assessing the equilibration and relaxation of other simulation observables are shown in Supporting Information.
3.2 Areas and Volumes
In simple bilayers consisting of only one type of lipid, the total area can be converted to an area per lipid A by simply dividing the total area by N, the number of lipids per monolayer. For more complex bilayers composed of a mixture of different lipids, the mean area per lipid will differ for each component, and the computation of the mean area of each component is not straightforward. Numerous techniques have been proposed for partitioning the area of a phospholipid/sterol bilayer: allocating all area evenly between the two components, allocating all area to the phospholipid, assuming a constant mean area for the sterol equal to that seen in a sterol monolayer,58 assuming a constant mean volume for the sterol equal to that observed in the crystal structure,59 partitioning the area of a slice through the membrane using VdW radii of the component atoms,53 among others.52, 60, 61 Partial molecular areas have also been used to examine the effects of cholesterol on membranes.60, 62 However this type of analysis requires multiple simulations with different molar concentrations of the additional molecule and is beyond the scope of our current focus (or resources). One may also estimate phospholipid area by structural analysis based on volume and thickness information.59, 63, 64
After removing the equilibration portion of our simulations, we calculated the average area per POPC by simply dividing the total system area by the number of POPC lipids in a monolayer, resulting in 64.0 ± 0.8 Å2 per POPC for the pure POPC simulation, 67.1 ± 0.4 Å2 for the cholesterol/POPC simulation, and 78.4 ± 0.5 Å2 for the 25-hydroxycholesterol/POPC simulation. The pure POPC results agree with simulation results of Róg et al. who obtained 63.5 ± 0.5 Å2, while experimental data shows a somewhat larger area of 68.3 ± 1.5 Å2.58, 65 Róg et al. also performed simulations of POPC/cholesterol mixtures, and the per-POPC area for a 22 mole percent cholesterol simulation can be calculated from their data as 70.2 ± 0.5 Å2.58
The change in per-phospholipid area with the addition of small molecules can be partitioned into a direct increase of area due to additional molecules in the system and an indirect effect of the small molecules on nearby phospholipids. Cholesterol is known to have an area-decreasing effect on nearby phospholipids, but in simulations of relatively high cholesterol concentration, the direct increase in system area from the additional volume of the cholesterol molecules dominates and creates a slight increase in total area per phospholipid.12, 53, 59 We observe a 3.1 Å2 increase in area per phospholipid between the 0 and 30 mole percent cholesterol simulations. This change is consistent with previous results and shows a larger direct than indirect influence of cholesterol on membrane area. 25-hydroxycholesterol, however, shows a much larger increase of 14.4 Å2 in per-POPC area between 0 and 30 mole percent 25-hydroxycholesterol simulations. Together with the solvent-accessible area data (below), this observation suggests that 25-hydroxycholesterol takes up much more area in the membrane than cholesterol.
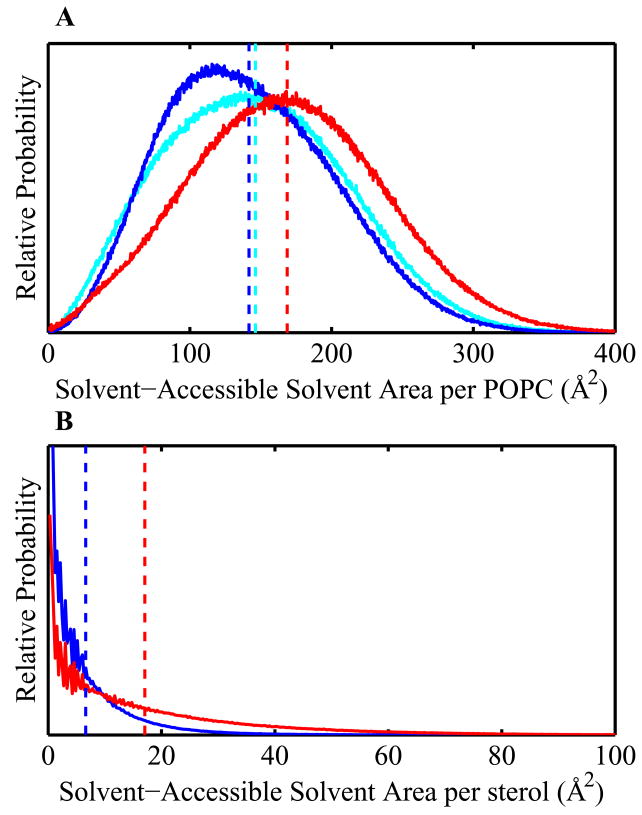
In order to decompose the area-altering effect of cholesterol and 25-hydroxycholesterol on system area into direct and indirect contributions, the solvent-accessible surface area (SASA) of each snapshot from the three trajectories was calculated and partitioned into molecular SASAs. This decomposition gave distributions of SASA for each type of molecule, shown in Figure 3. This allows assessment of the indirect effect of sterols on system area through their perturbation of individual POPC area from the direct increase of area due to the additional molecules. Differences in distributions are compared using the Kolmogorov-Smirnov test, described in Supporting Information. In the cholesterol-containing simulation, the distribution of POPC molecular SASAs clearly shifts to smaller values by depletion of large area contributions. Conversely, in the 25-hydroxycholesterol-containing simulation, the distribution of POPC surface areas shifts to much larger areas. Even larger differences between distributions are seen for the cholesterol and 25-hydroxycholesterol molecule SASAs, where the mean area of cholesterol is much smaller than that of 25-hydroxycholesterol and the distribution of cholesterol SASA is much more tightly peaked.
Figure 3.
Membrane component solvent-accessible areas. (A) Solid colored lines show the probability distributions of per-POPC solvent-accessible surface area (SASA) in simulations of pure POPC (cyan), 30% mole fraction cholesterol (blue), and 30% mole fraction 25-hydroxycholesterol (red). The mean SASAs for these distributions are 150 ± 60, 140 ± 60, and 170 ± 60 Å2, respectively. (B) Solid colored lines show the probability distributions and means of per-sterol SASA, cholesterol in blue and 25-hydroxycholesterol in red. Dashed vertical lines show the mean SASA for the whole distribution. Mean SASAs for these distributions are 10 ± 20 and 17 ± 20 Å2, respectively. p-values for distribution differences are calculated using the Kolmogorov-Smirnov test (see Supporting Information), and are all less than 0.1%.
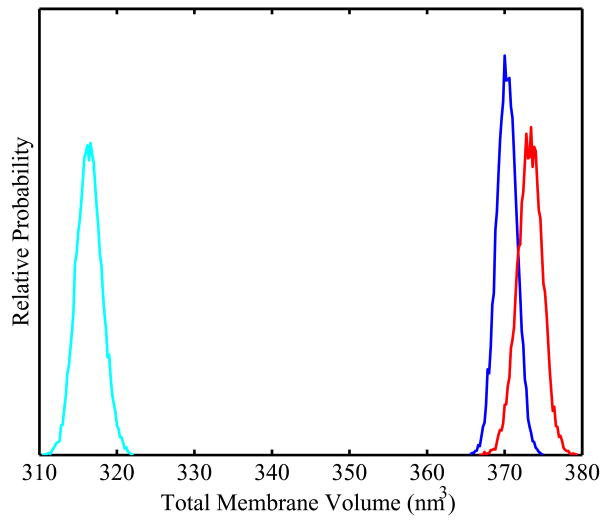
The volumes of the membranes were calculated as the product of the system area with the thickness of the membrane, estimated as the mean phosphate-to-phosphate distance.58 Volume distributions over the course of the three simulations are shown in Figure 4. We observed a large increase in membrane volume with the addition of 30% of either sterol, with a slightly larger increase in volume for 25-hydroxycholesterol as compared to cholesterol. This suggests that the perturbation of total volume by sterols is primarily due to the additional volume from the sterols themselves and that the differential effects of cholesterol and 25-hydroxycholesterol on total membrane area are compensated by changes in membrane thickness.
Figure 4.
The distributions of total membrane volume for a pure POPC membrane (cyan), 30% mole fraction cholesterol (blue), and 30% mole fraction 25-hydroxycholesterol (red). Mean membrane volumes are 316 ± 2, 370 ± 1, and 373 ± 2 nm3, respectively. p-values for distribution differences are calculated using the Kolmogorov-Smirnov test (see Supporting Information), with the p-values for all distribution differences less than 0.1%.
3.3 Densities
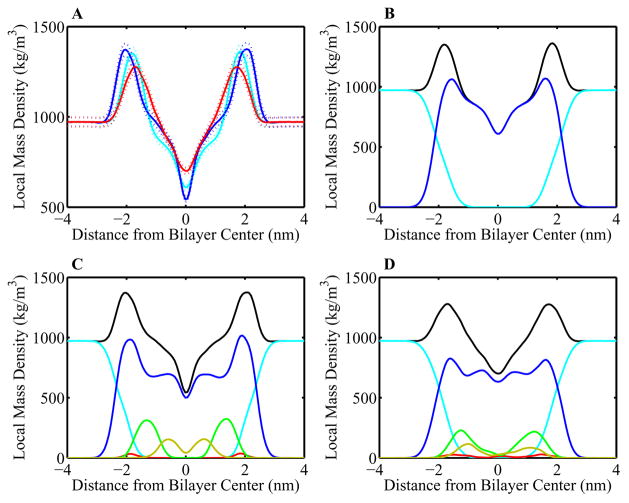
The cross-sectional mass densities of the simulations were calculated by taking each frame and dividing the system into slabs approximately 2 Å thick and calculating the mass density of each slab. These densities are averaged over the 128 ns of equilibrated simulation time. The total mass densities for all three simulations calculated this way are shown in Figure 5A. The general density profile is similar for all simulations: a symmetric profile where uniform bulk water density gives way to a strong peak, corresponding to the heavy phospholipid phosphate group, which in turn decays to a value lower than bulk water in the less-populated hydrophobic interior of the bilayer. The major effect of sterols on the total mass density profiles is seen in the peak locations. In particular the peak-to-peak distance between the two largest peaks in the total mass density is 37 Å in the pure POPC bilayer, 41 Å in the cholesterol/POPC system, and 34 Å in the oxysterol/POPC system. The increased thickness of the cholesterol-containing bilayer is similar to that seen experimentally;12 this increased and the decreased thickness of the oxysterol-containing bilayer are consistent with our observation of compensatory area and volume changes resulting in small overall volume differences between the two sterol-containing systems.
Figure 5.
Mass density profiles of POPC bilayers. (A) Total electron density profiles for 256 POPC bilayer systems with 0% sterol (cyan), 30% mole fraction cholesterol (blue), and 30% mole fraction 25-hydroxycholesterol (red). Errors, calculated using a bootstrap sampling method (Sec. 2.3.1), are shown as dotted lines. (B), (C), and (D) Component densities for 0% sterol, 30% cholesterol, and 30% 25-hydroxycholesterol simulations, respectively. Water and ions (cyan), POPC (blue), sterol ring (green), sterol tail (yellow), and sterol hydroxyl groups (red).
We also evaluated the contributions of the membrane components to the total mass density, examining the mass density profiles of water, POPC, and the sterol ring, tail and hydroxyls (Figures 5B, 5C, and 5D). Of particular interest is the correlation between the location of sterol density and its effect on POPC density. While the total density profiles are largely similar (Figure 5A), the component profiles change quite substantially, with shift of POPC density towards the bilayer center when sterols are present. This phenomenon is pronounced in the oxysterol simulation, and implies a displacement effect of sterols; by integrating themselves into the bilayer, they shift POPC density into other regions. Information about sterol organization can also be obtained from these component densities: the cholesterol density is quite ordered, with hydroxyl density furthest out, followed by ring, then tail density. Oxysterol density, on the other hand, shows significant overlap and spread, suggesting less ordered orientations.
3.4 Membrane Mechanical Properties
Mechanical properties of the membrane such as bending modulus, area compressibility, and volume compressibility, were also calculated. Bending moduli are evaluated as previously established.66–68 Briefly, the height of each lipid is defined by the position of the first phospholipid glycerol carbon. The height of an evenly spaced grid of ≈5 Å was then fit based on the lipid positions. The undulatory motion of the system was estimated by taking the average of the top and bottom leaflet heights. The square of the 2-dimensional Fourier transform of the undulatory height grid was calculated, giving the spectral intensity at a given x-axis wavenumber m and y-axis wavenumber n. The 1-dimensional undulation mode spectrum was calculated by reducing m and n into a single wavenumber q:
| (1) |
Under conditions where the thickness of a bilayer is significantly smaller than the area, membrane mechanics can be modeled as an elastic sheet. At the zero surface tension conditions of our simulations, the relationship between spectral intensity and wavenumber can be described as
| (2) |
where u2(q) is the spectral intensity at a given wavenumber q, β is the inverse thermal energy 1/kBT, and Abox the system area.66–68 By performing a linear regression of log(〈u2(q)〉) and log(〈q〉) over the trajectories, we can confirm that our system behaves quartically in 〈q〉 and thus that the elastic sheet model is valid over the scales we simulate. As shown in Table 1, these regressions give slopes not significantly different than −4.00, showing that the undulation intensity scales as expected to ~ q−4. The bending modulus can be determined from the total spectral intensity as follows:66–68
Table 1.
The results of linear regression on the logarithm of spectral intensity versus the logarithm of wavenumber (Eq. 2). Errors are calculated from the linear regression to the data using standard techniques.56
| System | Slope | Intercept | Pearson’s R2 |
|---|---|---|---|
| No sterol | −4.06±0.09 | −2.81±0.07 | 98.5% |
| Cholesterol | −4.09±0.13 | −2.85±0.09 | 97.2% |
| Oxysterol | −4.09±0.14 | −2.78±0.09 | 97.1% |
| (3) |
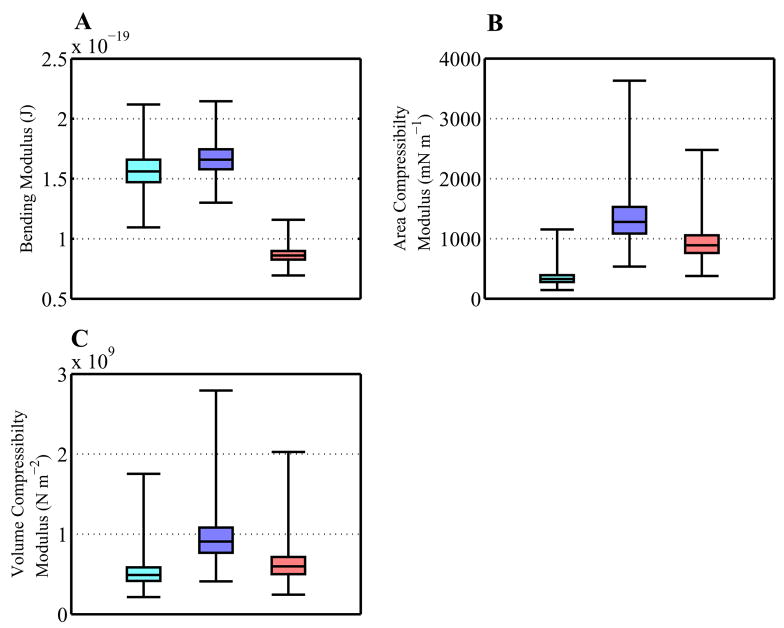
The results of these calculations are shown in Figure 6A. The experimentally determined bending modulus for pure POPC membranes is 8.5 × 10−20 J,69 thus our calculated bending moduli are within a factor of 2 of the experimental values, similar to that seen in previous lipid simulations.67 We see a small but significant increase in bending modulus upon the addition of cholesterol to the membrane, consistent with experimental results.14, 70 25-hydroxycholesterol, however, has the opposite effect of lowering the bending modulus These changes in bending modulus mean that 25-hydroxycholesterol-containing bilayers are slightly easier to bend than pure POPC bilayers, while cholesterol makes the membrane more resistant to bending.
Figure 6.
Calculated bulk membrane properties of our pure POPC (cyan), mixed POPC/cholesterol (blue), and mixed POPC/oxysterol (red) membranes. All errors were calculated using the bootstrap method described in Sec. 2.3.1. Box-and-whiskers plots are shown for bootstrapped distributions of calculated bulk membrane properties. The central box shows the interquartile range and median of the distribution, while the whiskers show the full range of calculated values. Membrane-to-membrane comparison of calculated properties are significant with p-values < 0.1% for all properties and membrane comparisons. (A) The bending modulus of the bilayers, calculated from the total power in the undulation spectrum as described in the text. (B) The area compressibility of the bilayers, calculated from the size of fluctuations in total system area. (C) The volume compressibility of the bilayer, calculated from the size of fluctuations in total system volume.
The area and volume compressibilities of a system can be calculated from the fluctuations in the total system area or volume by:67,71,72
| (4) |
| (5) |
Using the projected area of the system (Figure 2A), the area compressibilities of our systems (Figure 6B), are calculated to be 320 ± 100, 1250 ± 370, and 870 ± 210 mN m−1 for the sterol-free, cholesterol, and oxysterol simulations, respectively. The value for the sterol-free system is reasonably close to the experimental value for pure POPC membranes of 278 mN m−1.69 Cholesterol has been previously shown to increase the area compressibility modulus73 in a manner consistent with our results. Compared to cholesterol, 25- hydroxycholesterol has a smaller but significant effect on the area compressibility. Volume compressibility moduli can be calculated the same way, from fluctuations in the total system volume as shown in Figure 4. Results are shown in Figure 6C, and behave similarly to the area compressibility moduli, with sterols increasing the compressibility modulus and 25-hydroxycholesterol having a smaller effect than cholesterol.
3.5 Lipid Structure
The tail order of membrane component lipids can be used to measure the ordering of the lipid acyl chains in the bilayer and provide a convenient comparison with experimental carbon deuterium order parameters measured by NMR spectroscopy.74 Lipid order is normally characterized by the order parameter tensor for each tail atom i::67, 75, 76
| (6) |
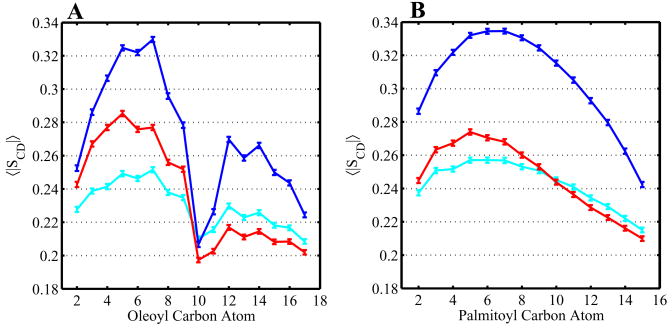
where α, β = x, y, z and θα(i) is the angle between the z-axis and the αth axis of the ith atom. In particular, θz(i) is estimated by the angle between the vector pointing from the (i+1)th atom to the (i − 1)th atom and the z-axis. The experimentally relevant deuterium order parameter is related by and we present |SCD| for closer comparison with experiment. Larger numbers indicate a more ordered orientation, with increased alignment of the chain at that position along the bilayer normal axis, while smaller numbers indicate a more disordered orientation. This analysis was applied to the atoms of the oleoyl unsaturated and palmitoyl saturated chains of POPC in the three simulations. The average results across all lipids in our simulations are shown in Figure 7. We see, as has been observed in other simulations, an increase in POPC tail order in the mixed cholesterol/POPC simulation as compared with the pure POPC simulation for all tail atoms except those directly around the double bond in the unsaturated oleoyl tail.77 This has been attributed to packing of the phospholipid tails around the rigid cholesterol ring structure.78 The effect of 25-hydroxycholesterol on POPC tail order is more ambiguous, with an moderate ordering of tail atoms near the head group region transitioning to a slight disordering of tail atoms away from the head groups.
Figure 7.
The mean tail order parameters for atoms in the oleoyl unsaturated chain of POPC (A) and the palmitoyl saturated chain of POPC (B). Order parameters for the pure POPC bilayer shown in cyan, the mixed POPC/cholesterol bilayer in blue, and the mixed POPC/oxysterol bilayer in red. Smaller atom indices are closer to the POPC headgroup. Errors calculated using a bootstrap method.
Hydrogen bonding patterns among lipids were analyzed as well. Hydrogen bonds were defined based on geometric criteria among all potential acceptor (A), donor (D), and hydrogen (H) atoms: the acceptor- donor distance rAD, the acceptor-hydrogen distance rAH, and the angle formed by the hydrogen, donor, and acceptor atoms θHDA. When rAD < 3.4 Å, rAH < 2.425 Å, and θHDA < 30.0°, the interaction is defined as a hydrogen bond.79, 80 Of the lipids, the only potential hydrogen bond donors are the sterol hydroxyl groups, while both the sterol hydroxyl groups as well as POPC phosphate and glycerol groups can accept hydrogen bonds.
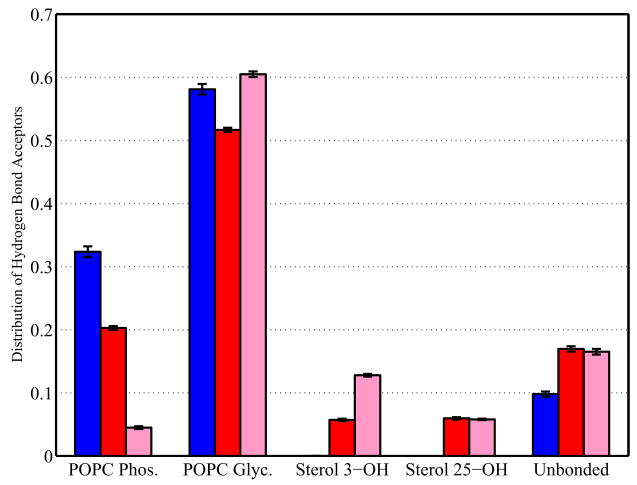
Each sterol hydroxyl group was analyzed to determine which of these potential partners it is bonded with, and total distributions calculated. The equilibrium distributions for the sterol simulations are shown in Figure 8. Hydrogen bonds between membrane components and waters were not considered; thus, molecules which hydrogen bond only to water are counted in the “unbonded” category. The cholesterol hydroxyl group is mostly bound to POPC, and about twice as likely to be bound to a POPC glycerol oxygen as to a POPC phosphate oxygen. Only about 10% of cholesterols were unbound or bound to only water. A negligible fraction of cholesterol hydroxyl groups hydrogen bonded to other cholesterol molecules. The oxycholesterol 3-hydroxyl group shows reduced propensity for POPC bonding. This compound has an increased likelihood to remain unbound from lipid or to form sterol-sterol hydrogen bonds. The 25-hydroxyl group at the end of the tail shows similar sterol-sterol hydrogen bond formation. An examination of the bilayer structures reveals that these sterol-sterol hydrogen bonds are often found as clusters of 2 to 4 sterols in the hydrophobic center of the bilayer. These clusters may have relevance for the increase in membrane permeability to polar and charged molecules observed upon addition of 25-hydroxycholesterol.22, 23
Figure 8.
Hydrogen bonding distributions for sterol hydroxyl groups, shown as a fraction of sterol hydroxyls bonded to specific hydrogen bond acceptors. Hydrogen bond distributions of cholesterol 3-hydroxyl are shown in blue, oxycholesterol 3-hydroxyl in red, and oxycholesterol 25-hydroxyl in pink. The non-bonded category includes hydroxyl groups with no hydrogen bonds at all as well as those only bound to water. Errors are calculated using the bootstrap method described in Section 2.3.1.
We also performed radial distribution function calculations between the C9 atom of the POPC oleoyl chain and the C18 β-methyl atom of the sterols in a manner similar to Pandit et al. for cholesterol.62 Plots of these distribution functions are shown in Supporting Information. There are two distinct peaks corresponding to packing of the oleoyl chain around the rough and smooth faces of the sterols. The oleoyl chains of POPC appear to pack evenly around both faces of cholesterol, while there is a slight bias towards packing of the unsaturated chains around the smooth rather than the rough face of 25-hydroxycholesterol.
3.6 Sterol Orientation and Organization
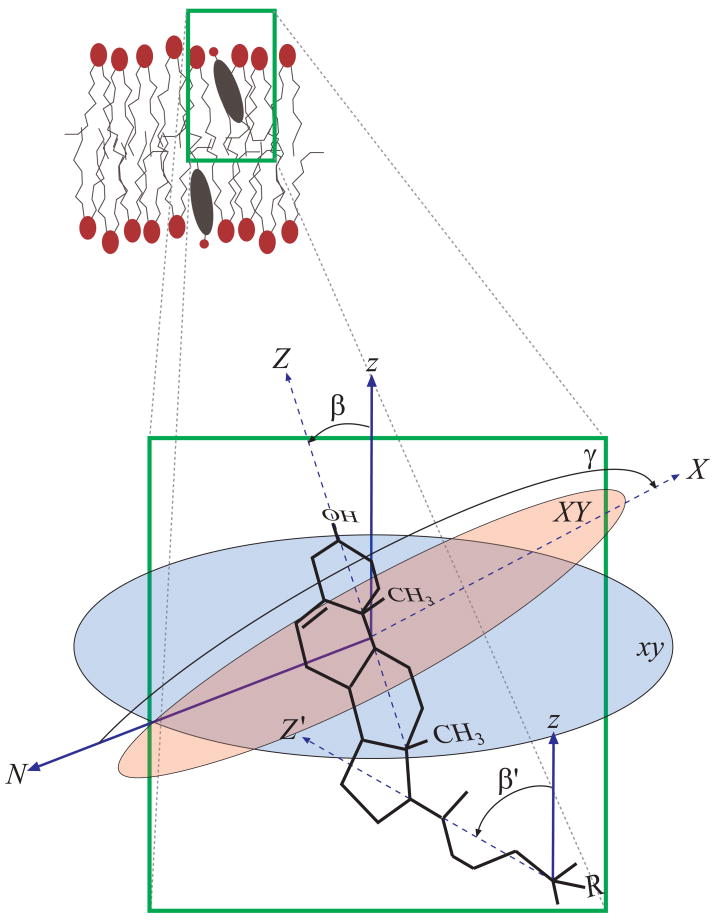
In order to investigate whether the distributions of sterol orientations within the membrane bilayer are related to the differential effects of sterols on membrane and lipid properties, we require a method to define sterol orientation. This is done with a series of Euler angles,81 shown in Figure 9. First, molecular axes are defined. The ring Z axis is defined from carbon 13 to carbon 3, pointing towards the head of the sterol along the ring structure. The ring X axis is defined from carbon 10 to 19, pointing outwards along the protruding methyl groups on one face of the ring. Finally, the tail Y axis is defined from carbon 25 to 17, from the end of the tail to the beginning.
Figure 9.
A diagram showing the methods by which individual sterol orientation is defined using Euler angles relating molecular axes of the sterol to reference axes of the bilayer as a whole. The reference z axis is defined along the membrane normal, while the reference xy is the plane of the bilayer. A molecular ring axis Z is defined from carbon 13 to 3 along the length of the ring, and a molecular tail axis Z′ is defined from carbon 25 to 17 along the length of the tail. Finally, a ring normal axis X is defined from carbon 10 to 19, outward through the protruding methyl groups. Three Euler angles are then calculated. The sterol ring tilt β is defined as the angle between the reference z-axis and sterol ring Z-axis, the sterol ring twist γ is defined as the angle between the intersection N of the reference xy plane and sterol ring XY plane and the sterol ring X axis, and the sterol tail tilt β′ is defined as the angle between the reference z-axis and sterol tail Z′-axis.
Euler angles defining the orientation of the sterol can be calculated using these molecular axes and their relationship to the reference bilayer axes (the box axes in the top leaflet, the negative of the box axes in the bottom leaflet). The ring tilt β is defined as the angle between the reference z axis and the sterol ring Z axis, and varies from 0 to 180 degrees, where 0 degrees indicates a parallel orientation, with the ring aligned so that the head points out of the bilayer and the tail inside, and 180 degrees indicates an antiparallel orientation, with the head pointing inside the bilayer. The ring twist γ is defined as the angle between the sterol ring X axis and the reference xy plane, measured along the ring XY plane. This twist varies from −180 to 180 degrees, where 90 degrees indicates the sterol lying with the rough face facing out of the bilayer and −90 degrees indicates the sterol lying with the rough face facing into the bilayer. Finally, the sterol tail tiltβ; is defined as the angle between the reference z axis and the sterol tail Z′-axis, varying, as with the sterol ring tilt, from 0 to 180 degrees. In addition to these angles, we also calculated an average ring height, as the mean distance of carbon 13 and 3 from the bilayer center.
The orientation of each sterol in each frame of the equilibrated region of our systems was analyzed and combined into a multidimensional orientation distribution for both cholesterol and 25-hydroxycholesterol. Contour plots of 2-dimensional slices through these distributions are shown for cholesterol in Figure 10 and for 25-hydroxycholesterol in Figure 11.
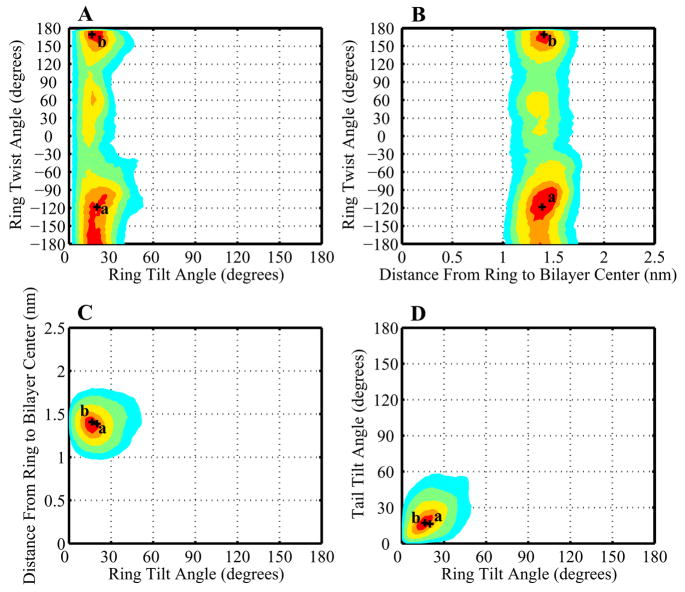
Figure 10.
A contour plot showing the distribution of cholesterol orientations in mixed POPC/cholesterol bilayers. Regions containing the densest 10%, 25%, 50%, 75%, and 90% of the total probability density are shown by areas of red, orange, yellow, green, and cyan respectively. (A) Ring tilt β vs. ring twist γ. (B) Ring tilt β vs. ring height, calculated as the average distance of carbons 3 and 13 from the bilayer center. (C) Ring height vs. ring twist γ (D) Ring tilt β vs. tail tilt β′. Labelled crosses show the orientations of the specific sterol molecules depicted in Figure 12.
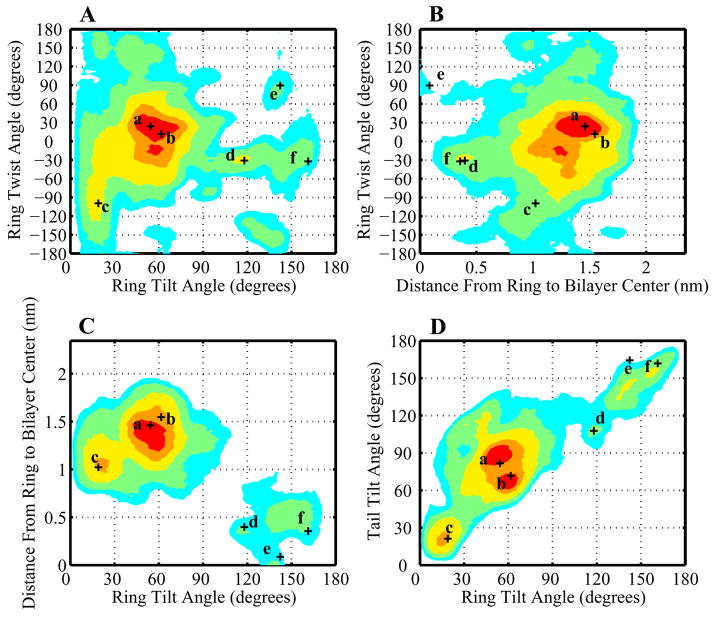
Figure 11.
A contour plot as in Figure 10 showing the distribution of 25-hydroxycholesterol orientations in mixed POPC/oxysterol bilayers. Regions containing the densest 10%, 25%, 50%, 75%, and 90% of the total probability density are shown by areas of red, orange, yellow, green, and cyan respectively. (A) Ring tilt β vs. ring twist γ (B) Ring tilt β vs. ring height, calculated as the average distance of carbons 3 and 13 from the bilayer center. (C) Ring height vs. ring twist γ (D) Ring tilt β vs. tail tilt β′. Labelled crosses show the orientations of the specific sterol molecules depicted in Figure 13.
The cholesterol distribution contours show a relatively tight distribution. All parameters other than the ring twist are sharply peaked: ring tilt at ≈ 20 degrees, ring height at ≈ 1.4 Å, and tail tilt at ≈ 20 degrees. Ring twist does show a wider distribution, though with a definite peak at −120 degrees. The main peak of the distribution corresponds to a sterol fully extended in the bilayer with the tail stretched out along the same angle as the rigid ring (Figure 12A).
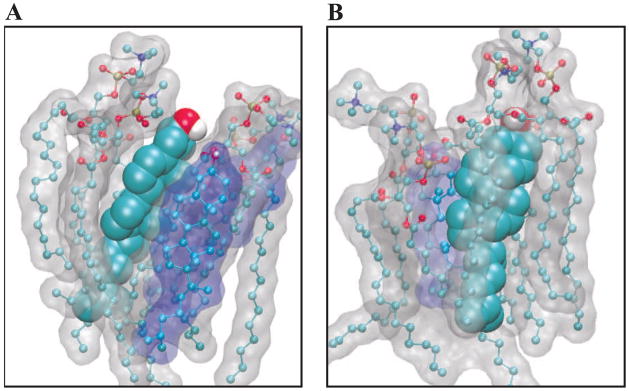
Figure 12.
Depiction of cholesterol molecules from our molecular dynamics simulations. The cholesterol of interest is shown as a space-filling model, while nearby POPC molecules as ball-and-stick models with a gray molecular surface and nearby cholesterol molecules shown as ball-and-stick models with a blue molecular surface. Each subfigure shows a section from a single leaflet of the bilayer, with the lipid/water interface at the top of the figure. The sterol labels (A) through (B) correspond to the marked orientations shown in Figure 10.
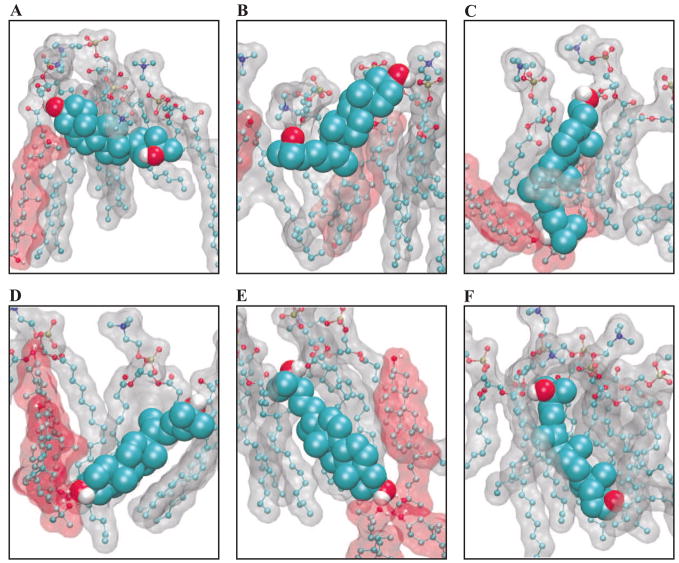
The distribution of 25-hydroxycholesterol orientation is much more complicated. To aid with visualization, a variety of representative oxysterol orientations are shown in Figure 13. The orientations of oxysterols depicted in Figure 13 are marked on the distribution shown in Figure 11. Figure 11C plots height versus ring tilt and shows two distinct populations of molecules, one with ring tilts of about 20 to 70 degrees and large ring heights of ≈ 1.0 – 1.5 Å, and another smaller population of ring tilts ≈ 150 degrees and heights of ≈ 0 – 0.5 Å. The first population corresponds to normally-oriented sterols with rings tilted towards the bilayer interface and tails in the bilayer interior. The second population corresponds to inverted sterols with ringd in the bilayer interior and the tails oriented towards the bilayer interface. Figure 11D, shows the relative orientation of the tail with respect to the ring. This figure includes the same inverted (in the top right corner) and normal (in the bottom left corner) orientations, but also indicates that the normally-oriented sterol distribution has a generally-larger tail than the ring tilt. We observe that many of the sterols adopt an orientation in which one of their hydroxyl groups is buried in the bilayer interior form hydrogen bond interactions with other such buried sterol hydroxyl groups, forming clusters of polar hydroxyl groups in the hydrophobic interior of the membrane.
Figure 13.
Depiction of 25-hydroxycholesterol molecules from our molecular dynamics simulations. The 25-hydroxycholesterol of interest is shown as a space-filling model, while nearby POPC molecules as ball-and- stick models with a gray molecular surface and nearby 25-hydroxycholesterol molecules shown as ball-and- stick models with a red molecular surface. Each subfigure shows a section from a single leaflet of the bilayer, with the lipid/water interface at the top of the figure. The sterol labels (A) through (F) correspond to the marked orientations shown in Figure 11.
4 Discussion
We observe significant changes in the bulk properties of POPC membranes when relatively high concentrations of cholesterol or 25-hydroxycholesterol are added. Most obviously, the membrane volumes and areas increase upon the addition of these sterols. However, while the volume increase is similar in both sterol systems, the area increase is much larger for membranes containing 25-hydroxycholesterol as compared to cholesterol. We attribute the observed volume increases to the direct effects of additional sterol volume, which is largely the same between the two sterols. The large difference in area effects by these sterols suggest that they have altered interaction with the POPC lipids, and that indirect effects of sterols on POPC structure contribute to these changes. Examination of membrane density profiles shows that the cholesterol system has thickened with respect to the sterol-free system, while the 25-hydroxycholesterol system has thinned. Cholesterol, as has been previously observed both in simulation and experiment,11–13, 53, 59 condenses and thickens membranes. On the other hand, we show 25-hydroxycholesterol to have an opposing effect, expanding the membrane laterally and thinning it. In addition to changes in gross membrane dimensions, the membrane’s mechanical properties are altered. Sterol-containing systems show less resistance to bending, with a corresponding increase in membrane undulations. These systems also show increased volume and area compressibility modulus, making them more difficult to compress and with smaller area and volume fluctuations. In all of these mechanical properties, oxysterol-containing membranes show a larger deviation from pure phospholipid membranes than cholesterol-containing membranes. Supporting Information includes a table with a complete summary of the membrane structural changes and comparison with available experimental data.
The bulk membrane changes induced by cholesterol and 25-hydroxycholesterol necessarily have their roots in the altered molecular structure of the membranes, both through perturbations in POPC structure as well as addition of new molecules forming sterol-POPC interactions. Solvent-accessible surface area calculations for POPC show that POPC has a smaller surface area in cholesterol-containing membranes, while POPC in 25-hydroxycholesterol-containing membranes has a much larger surface area. These differences suggest a compression of POPC by cholesterol and an expansion by 25-hydroxycholesterol. Calculated POPC order parameters are consistent with this observation: cholesterol orders POPC acyl tails while 25-hydroxycholesterol has both ordering and disordering effects, with ordering dominant near the head group, and disordering dominant near the end of the tails. Looking at differences in sterol structure, we examined hydrogen bonding patterns for the sterol hydroxyl groups. Cholesterol engages in almost no cholesterol-cholesterol hydrogen bonding, preferring to hydrogen bond to the POPC headgroup. However, 25-hydroxycholesterol shows significant levels of sterol-sterol hydrogen bonding, with reduced sterol-POPC bonding. These hydrogen bonding patterns are rooted in the different orientations of sterols within the membrane bilayer.
We defined these sterol orientations with respect to the membrane normal axis and found that, while cholesterol was overwhelmingly found in a single orientation roughly parallel to the phospholipid tails, 25-hydroxycholesterol was found in a very wide range of orientations. Cholesterol adopts an orientation with its steroid ring at a slight tilt to the membrane normal axis and its iso-octyl tail fully extended into the center of the bilayer (Fig. 10a and b). While this orientation is accessible to 25-hydroxycholesterol (Fig. 11c), only a small fraction of 25-hydroxycholesterol molecules are found in that orientation. This cholesterol-like orientation is presumably disfavored for the oxysterol because the 25-hydroxyl group would then be buried in the nonpolar center of the bilayer with few hydrogen bonding partners. The preferred orientation for 25-hydroxycholesterol is, instead, with a relatively deep ring tilt of about 60° and the iso-octyl tail with its hydroxyl group bent up towards the hydrophilic interfacial layer (Fig. 11a and b). This orientation allows both hydroxyl groups to avoid the hydrophobic bilayer interior. Also accessible are inverted orientations, with the steroid rings buried in the bilayer interior and the iso-octyl tails extending into the interfacial layer (Fig. 11d, e, and f).
The orientations of cholesterol and 25-hydroxycholesterol are relevant to the effects observed on bulk membrane and phospholipid properties. The parallel alignment of the rigid cholesterol ring in bilayer has been thought to order nearby phospholipid tails by inducing them to stretch out along its rigid structure.78 This effect can not only produce the increased POPC order parameters we observe but also changes the thickness, density, and melting point of cholesterol-containing bilayers.11–13 25-hydroxycholesterol however, does not align neatly in the bilayer, but instead adopts a diverse array of orientations which cause it to have a more chaotic, disordering effect.
Given the varied responses of cells to oxysterols it seems appropriate to consider the possible consequences for oxysterol addition to membranes on membrane proteins. The change in membrane thickness and the disorder of the aliphatic chains are prominent effects observed in our simulations. Hydrophobic matching is a major mechanism of protein-membrane interaction and this has been proposed as one important role for cholesterols in biological membranes.82, 83 The difference we see between the cholesterol membrane and the oxysterol membrane represents a dramatic change in the hydrophobic matching region for many proteins.84, 85 In addition, oxysterol-related changes in aliphatic chain order would be expected to influence the mobility of transmembrane portions of membrane transporters, channels and transmembrane receptors/signaling complexes.86, 87
5 Conclusions
The change in cholesterol from an amphiphilic to bisamphiphilic character by the addition of the 25-hydroxyl group changes the energy landscape of orientational space. Specific regions of the space are disfavored, including the aligned cholesterol orientation, while other regions are favored, shifting the distribution of molecular orientations. We predict that the occupancy of these different regions of orientational space is what drives the biophysical effects observed on phospholipid membranes. In particular, the 25-hydroxyl group preferentially biases the molecule towards regions of orientational space that increase bilayer area and affect lipid order.
We can make predictions about how these sterols may interact with membranes of different lipid composition as well. Saturated lipids such as DPPC have more ordered lipid chains and thus the presence of these lipids should promote more ordered and extended orientations of sterols, similar to those preferred by cholesterol. Unsaturated lipids or polyunsaturated lipids such as DOPC, however, have more disordered tail chains that will disfavor ordered sterol orientations and shift the equilibrium towards the tilted and clustered orientations observed in our 25-hydroxycholesterol simulation. These expected changes in the energies of various orientations would then show their effects in the perturbation of membrane structure by sterols, with cholesterol having stronger condensing effects on unsaturated lipids and 25-hydroxycholesterol conversely showing stronger expansive effects on saturated lipids. These different lipid sensitivities have been observed in a recent study examining the area expansive effects of 25-hydroxycholesterol on lipid bilayers.88 Unsaturated DOPC membranes showed a larger area increase than POPC membranes upon the addition of 25-hydroxycholesterol, while DPPC membranes showed relatively little change.
25-hydroxycholesterol has previously been shown to increase the permeability of membranes to ions and small osmolytes.22–24 Our understanding of how this sterol perturbs membrane structure suggests two potential mechanisms for this permeability. Firstly, the oxysterol directly thins the membrane. Secondly, the orientations of these sterols produces clusters of hydrogen-bonded hydroxyl groups in the hydrophobic interior of the bilayer. These hydroxyl clusters would allow limited hydrogen bonding interactions with polar molecules traversing the membrane, lowering the energy barrier to permeation. This mechanism is currently being tested in ongoing simulations.
These structural effects of 25-hydroxycholesterol on lipid membranes have biological relevance. A recent study has shown that the effects of 25-hydroxycholesterol on cellular cholesterol homeostasis are not enantioselective; the enantiomer of 25-hydroxycholesterol has identical effects on membrane biophysical properties and suppression of the cholesterol synthetic pathway.88 This lack of stereospecificity suggests that 25-hydroxycholesterol does not act in this pathway through protein binding but rather that the signal is transduced through perturbation of the membrane, potentially through some of the mechanisms we have observed. This result reveals the role of the membrane in mediating signal cascades, and suggests that the biological effects of not only 25-hydroxycholesterol but potentially other signaling molecules as well may be effected through their perturbation of membrane structure.
Supplementary Material
Acknowledgments
The authors wish to thank Daniel Ory and Doug Covey for encouragement of this work in its early stages, as well as Daniel Ory, Doug Covey, David Holland, Agata Bielska, and Katherine Henzler-Wildman for critical reading of the manuscript. The authors are grateful to Victor Guallar for assistance with QM/MM calculations. We would also like to thank the anonymous reviewers for their helpful suggestions. Computational resources were provided by the Texas Advanced Computing Center through Teragrid grants TG-MCB060053 and TG-MCA08X003 to NAB. Brett Olsen was supported by the Cellular and Molecular Biology Training Grant T32 GM007067.
Footnotes
Supporting information available
Additional material is provided that discusses statistical methods, details of system preparation, molecular parameters, and system analyses. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Ohvo-Rekila H, Ramstedt B, Leppimaki P, Slotte JP. Progress in Lipid Research. 2002;41:66–97. doi: 10.1016/s0163-7827(01)00020-0. [DOI] [PubMed] [Google Scholar]
- 2.Liscum L, Munn NJ. Biochimica et Biophysica Acta - Molecular and Cellular Biology of Lipids. 1999;1438:19–37. doi: 10.1016/s1388-1981(99)00043-8. [DOI] [PubMed] [Google Scholar]
- 3.Epand RM. Progress in Lipid Research. 2006;45:279–294. doi: 10.1016/j.plipres.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Kuwabara PE, Labouesse M. Trends in Genetics. 2002;18:193–201. doi: 10.1016/s0168-9525(02)02640-9. [DOI] [PubMed] [Google Scholar]
- 5.Incardona JP, Eaton S. Current Opinion in Cell Biology. 2000;12:193–203. doi: 10.1016/s0955-0674(99)00076-9. [DOI] [PubMed] [Google Scholar]
- 6.Edidin M. Annual Review of Biophysics and Biomolecular Structure. 2003;32:257–283. doi: 10.1146/annurev.biophys.32.110601.142439. [DOI] [PubMed] [Google Scholar]
- 7.Chiang JYL. Journal of Hepatology. 2004;40:539–551. doi: 10.1016/j.jhep.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 8.Horton JD, Goldstein JL, Brown MS. Journal of Clinical Investigation. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ory DS. Circulation Research. 2004;95:660–670. doi: 10.1161/01.RES.0000143422.83209.be. [DOI] [PubMed] [Google Scholar]
- 10.Nezil FA, Bloom M. Biophysical Journal. 1992;61:1176–1183. doi: 10.1016/S0006-3495(92)81926-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Purdy P, Fox M, Graham J. Cryobiology. 2005;51:102–112. doi: 10.1016/j.cryobiol.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 12.Hung W, Lee M, Chen F, Huang HW. Biophysical Journal. 2007;92:3960–3967. doi: 10.1529/biophysj.106.099234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warschawski D, Devaux P. European Biophysics Journal. 2005;34:987–996. doi: 10.1007/s00249-005-0482-z. [DOI] [PubMed] [Google Scholar]
- 14.Martinez GV, Dykstra EM, Lope-Piedrafita S, Job C, Brown MF. Physical Review E. 2002;66:050902. doi: 10.1103/PhysRevE.66.050902. [DOI] [PubMed] [Google Scholar]
- 15.Endress E, Heller H, Casalta H, Brown MF, Bayer TM. Biochemistry. 2002;41:13078–13086. doi: 10.1021/bi0201670. [DOI] [PubMed] [Google Scholar]
- 16.Lundbaek JA, Andersen OS, Werge T, Nielsen C. Biophysical Journal. 2003;84:2080–2089. doi: 10.1016/S0006-3495(03)75015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McIntosh TJ, Simon SA. Annual Review of Biophysics and Biomolecular Structure. 2006;35:177–198. doi: 10.1146/annurev.biophys.35.040405.102022. [DOI] [PubMed] [Google Scholar]
- 18.Goldstein JL, Rawson RB, Brown MS. Archives of Biochemistry and Biophysics. 2000;397:139–148. doi: 10.1006/abbi.2001.2615. [DOI] [PubMed] [Google Scholar]
- 19.Humphries GMK, McConnell HM. Journal of Immunology. 1979;122:121–126. [PubMed] [Google Scholar]
- 20.Gill S, Chow R, Brown AJ. Progress in Lipid Research. 2008;47:391–404. doi: 10.1016/j.plipres.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 21.Radhakrishnan A, Ikeda Y, Kwon HJ, Brown MS, Goldstein JL. Proceedings of the National Academy of Sciences. 2007;104:6511–6518. doi: 10.1073/pnas.0700899104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Theunissen JJH, Jackson RL, Kempen HJM, Demel RA. Biochimica et Biophysica Acta. 1986;860:66–74. doi: 10.1016/0005-2736(86)90499-2. [DOI] [PubMed] [Google Scholar]
- 23.Holmes RP, Yoss NL. Biochimica et Biophysica Acta. 1984;770:15–21. doi: 10.1016/0005-2736(84)90067-1. [DOI] [PubMed] [Google Scholar]
- 24.Kauffman JM, Westerman PW, Carey MC. Journal of Lipid Research. 2000;41:991–1003. [PubMed] [Google Scholar]
- 25.Russell DW. Biochimica et Biophysica Acta - Molecular and Cell Biology of Lipids. 2000;1529:126–135. doi: 10.1016/s1388-1981(00)00142-6. [DOI] [PubMed] [Google Scholar]
- 26.Smith LL. Chemistry and Physics of Lipids. 1987;44:87–125. doi: 10.1016/0009-3084(87)90046-6. [DOI] [PubMed] [Google Scholar]
- 27.Höltje M, Fórster T, Brandt B, Engels T, von Rybinski W, Höltje H. Biochimica et Biophysica Acta - Biomembranes. 2001;1511:156–167. doi: 10.1016/s0005-2736(01)00270-x. [DOI] [PubMed] [Google Scholar]
- 28.QSite, 2001.
- 29.Philipp DM, Friesner RA. Journal of Computational Chemistry. 1999;20:1468–1494. [Google Scholar]
- 30.Murphy RB, Philipp DM, Friesner RA. Journal of Computational Chemistry. 2000;21:1442–1457. [Google Scholar]
- 31.Kaminski G, Friesner RA, Tirado-Rives J, Jorgensen WL. Journal of Physical Chemistry B. 2001;105:6474–6487. [Google Scholar]
- 32.Becke AD. Journal of Chemical Physics. 1993;98:5648–5652. [Google Scholar]
- 33.Stephens PJ, Devlin FJ, Chabalowski CF, Frisch MJ. Journal of Physical Chemistry. 1994;98:11623–11627. [Google Scholar]
- 34.Hay PJ, Wadt WR. Journal of Chemical Physics. 1985;82:299–310. [Google Scholar]
- 35.Friesner RA, Murphy RB, Beachy MD, Ringnalda MN, Pollard WT, Dunietz BD, Cao Y. Journal of Physical Chemistry A. 1999;103:1913–1928. [Google Scholar]
- 36.Breneman CM, Wiberg KB. Journal of Computational Chemistry. 1990;11:361–373. [Google Scholar]
- 37.Woods RJ, Khalil M, Pell W, Moffat SH, Jr, VHS Journal of Computational Chemistry. 1990;11:297–310. [Google Scholar]
- 38.Chirlian LE, Francl MM. Journal of Computational Chemistry. 1987;8:894–905. [Google Scholar]
- 39.Berger O, Edholm O, Jahnig F. Biophysical Journal. 1997;72:2002–2013. doi: 10.1016/S0006-3495(97)78845-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berendsen HJC, Grigera JR, Straatsma TP. Journal of Physical Chemistry. 1987;91:6269–6271. [Google Scholar]
- 41.Straatsma TP, Berendsen HJC. Journal of Chemical Physics. 1988;89:5876–5886. [Google Scholar]
- 42.Lindahl E, Hess B, van der Spoel D. Journal of Molecular Modeling. 2001;7:306–317. [Google Scholar]
- 43.Bockmann RA, Hac A, Heimburg T, Grubmuller H. Biophysical Journal. 2003;85:1647–1655. doi: 10.1016/S0006-3495(03)74594-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tieleman DP, Forrest LR, Sanson MSP, Berendsen HJC. Biochemistry. 1998;37:17554–17561. doi: 10.1021/bi981802y. [DOI] [PubMed] [Google Scholar]
- 45.Tieleman DP, Berendsen HJC. Journal of Chemical Physics. 1996;105:4871–4880. [Google Scholar]
- 46.Huber T, Rajamoorthi K, Kurze VF, Beyer K, Brown MF. Journal of the American Chemical Society. 2002;124:298–309. doi: 10.1021/ja011383j. [DOI] [PubMed] [Google Scholar]
- 47.Feller SE, Jr, ADM Journal of Physical Chemistry B. 2000;104:7510–7515. [Google Scholar]
- 48.Berendsen H, van der Spoel D, van Drunen R. Computer Physics Communications. 1995;91:43–56. [Google Scholar]
- 49.Parrinello M, Rahman A. Journal of Applied Physics. 1981;52:7182–7190. [Google Scholar]
- 50.Hoover WG. Physical Review A. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 51.Darden T, York D, Pedersen LG. Journal of Chemical Physics. 1993;98:10089–10092. [Google Scholar]
- 52.Pandit SA, Vasudevan S, Chiu SW, Mashl RJ, Jakobsson E, Scott HL. Biophysical Journal. 2004;87:1092–1100. doi: 10.1529/biophysj.104.041939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Falck E, Patra M, Karttunen M, Hyvönen MT, Vattulainen I. Biophysical Journal. 2004;87:1076–1091. doi: 10.1529/biophysj.104.041368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. Journal of Computational Chemistry. 1997;18:1463–1472. [Google Scholar]
- 55.Efron B, Tibshirani RJ. An Introduction to the Bootstrap. 1. Chapman & Hall/CRC; 1994. [Google Scholar]
- 56.Press WH, Teukolsky SA, Vetterling WT, Flannery BP. Numerical Recipes in C: The Art of Scientific Computing. 2. Cambridge University Press; 1992. [Google Scholar]
- 57.Baker NA, Sept D, Holst MJ, McCammon JA. Proceedings of the National Academy of Sciences. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Róg T, Pasenkiewicz-Gierula M. Biochimie. 2006;88:449–460. doi: 10.1016/j.biochi.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 59.Hofsäβ C, Lindahl E, Edholm O. Biophysical Journal. 2003;84:2192–2206. doi: 10.1016/S0006-3495(03)75025-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Edholm O, Nagle JF. Biophysical Journal. 2005;89:1827–1832. doi: 10.1529/biophysj.105.064329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chiu SW, Jakobsson E, Mashl RJ, Scott HL. Biophysical Journal. 2002;83:1842–1853. doi: 10.1016/S0006-3495(02)73949-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pandit SA, Chiu S, Jakobsson E, Grama A, Scott HL. Langmuir. 2008;24:6858–6865. doi: 10.1021/la8004135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Petrache HI, Dodd SW, Brown MF. Biophysical Journal. 2000;79:3172–3192. doi: 10.1016/S0006-3495(00)76551-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rajamoorthi K, Petrache HI, McIntosh TJ, Brown MF. Journal of the American Chemical Society. 2005;127:1576–1588. doi: 10.1021/ja046453b. [DOI] [PubMed] [Google Scholar]
- 65.Kučerka N, Tristram-Nagle S, Nagle JF. Journal of Membrane Biology. 2006;203:193–202. doi: 10.1007/s00232-005-7006-8. [DOI] [PubMed] [Google Scholar]
- 66.Lindahl E, Edholm O. Biophysics Journal. 2000;79:426–433. doi: 10.1016/S0006-3495(00)76304-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Song Y, Guallar V, Baker N. Biochemistry. 2000;44:13425–13438. doi: 10.1021/bi0506829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Edholm O. Current Topics in Membranes. 2008;60:91–110. [Google Scholar]
- 69.Mathai JC, Tristram-Nagle S, Nagle JF, Zeidel ML. The Journal of General Physiology. 2007;131:69–76. doi: 10.1085/jgp.200709848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Martinez GV, Dykstra EM, Lope-Piedrafita S, Brown MF. Langmuir. 2004;20:1043–1046. doi: 10.1021/la036063n. [DOI] [PubMed] [Google Scholar]
- 71.Feller SE, Pastor RW. Journal of Chemical Physics. 1999;111:1281–1287. [Google Scholar]
- 72.Hansen J-P, McDonald IR. Theory of Simple Liquids. Vol. 2. Academic Press; San Diego: 2000. [Google Scholar]
- 73.Needham D, Nunn RS. Biophysical Journal. 1990;58:997–1009. doi: 10.1016/S0006-3495(90)82444-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Seelig A, Seelig J. Biochemistry. 1974;13:4839–4485. doi: 10.1021/bi00720a024. [DOI] [PubMed] [Google Scholar]
- 75.Egberts E, Berendsen HJC. Journal of Chemical Physics. 1988;89:3718–3732. [Google Scholar]
- 76.Patra M, Salonen E, Terama E, Vattulainen I, Faller R, Lee BW, Holopainen J, Karttunen M. Biophysical Journal. 2006;90:1121–1135. doi: 10.1529/biophysj.105.062364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pandit SA, Chiu S, Jakobsson E, Grama A, Scott HL. Biophysical Journal. 2007;92:920–927. doi: 10.1529/biophysj.106.095034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Greenwood AI, Tristram-Nagle S, Nagle JF. Chemistry and Physics of Lipids. 2006;143:1–10. doi: 10.1016/j.chemphyslip.2006.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Luzar A. The Journal of Chemical Physics. 2000;113:10663–10675. [Google Scholar]
- 80.Guardia E, Marti J, Garcia-Tarres L, Laria D. Journal of Molecular Liquids. 2005;117:63–67. [Google Scholar]
- 81.Landau LD, Lifschitz EM. Mechanics. 3. Pergamon Press; Oxford, England: 1976. [Google Scholar]
- 82.Mouritsen OG, Bloom M. Biophysical Journal. 1984;46:141–153. doi: 10.1016/S0006-3495(84)84007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Marsh D. Biophysical Journal. 2008;94:3996–4013. doi: 10.1529/biophysj.107.121475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McIntosh TJ. Chemistry and Physics of Lipids. 2004;130:83–98. doi: 10.1016/j.chemphyslip.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 85.Marsh D. Biochimica et Biophysica Acta - Biomembranes. 2008;778:1545–1575. doi: 10.1016/j.bbamem.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 86.Yuan C, O’Connell RJ, Jacob RF, Mason RP, Treistman SN. Journal of Biological Chemistry. 2007;282:7276–7286. doi: 10.1074/jbc.M607593200. [DOI] [PubMed] [Google Scholar]
- 87.Lee KJB. Physical Review E. 2005;72:031917. doi: 10.1103/PhysRevE.72.031917. [DOI] [PubMed] [Google Scholar]
- 88.Gale SE, Westover EJ, Dudley N, Krishnan K, Merlin S, Scherrer DE, Han X, Zhai X, Brockman HL, Brown RE, Covey DF, Schaffer JE, Schlesinger P, Ory DS. Journal of Biological Chemistry. 2009;284:1755–1764. doi: 10.1074/jbc.M807210200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.