Abstract
Estrogen modulates gene expression through interactions with estrogen receptors (ERs) that bind chromosomal target genes. Recent studies have suggested an interaction between the cytoskeletal system and estrogen signaling; these have implicated a role of cytoplasmic microtubules in scaffolding ERα and enhancing nongenomic function; in addition, other experiments demonstrate that dynein light chain 1 may chaperone ERα to the nucleus, indirectly increasing transcriptional potency. Actin/myosin and dynein light chain 1 are also required for estrogen-mediated chromosomal movement that is required for transcriptional up-regulation of ERα targets. We present evidence that the dynactin component, p150/glued, directly influences the potency of nuclear ER function. Increasing the stoichiometric ratio of p150/glued and ERα by overexpression enhances estrogen responses. ERα enhancement by p150/glued does not appear to be influenced by shifts in subcellular localization because microtubule disruption fails to increase nuclear ERα. Rather, we find that modest amounts of p150/glued reside in the nucleus of cells, suggesting that it plays a direct role in nuclear transcription. Notably, p150/glued is recruited to the pS2 promoter in the presence of hormone, and, in MCF-7 cells, knockdown of p150/glued levels reduces estrogen-dependent transcription. Our results suggest that p150/glued modulates estrogen sensitivity in cells through nuclear mechanisms.
The p150/glued subunit of dynactin was shown to bind to and enhance the function of ER(α), further linking the microtubule system to nuclear hormone function.
Estrogen receptor-α (ERα) is a ligand-activated nuclear hormone receptor that regulates gene expression. The classical mechanism of ERα action remains the best characterized and requires ligand-dependent transformation of the receptor, enabling dimerization and recognition of DNA-regulatory elements on chromatin [estrogen response elements (EREs), reviewed in Ref. 1]. Bound ERα then recruits coregulator proteins (reviewed in Ref. 2) that influence a series of complex but choreographed steps, including histone acetylation, recruitment of RNA polymerase II and associated proteins, and phosphorylation of RNA polymerase C-terminal domain (3,4). In addition, ERα regulates many genes by interacting with transcription factors that bind sequences that are not directly recognized by ERα (5,6,7). Finally, recent attention has been devoted to a class of rapid actions of estrogen that ultimately impact upon gene transcription but are not mediated by nuclear ERα action (nongenomic or rapid responses to estrogen) (Ref. 8 and reviewed by Refs. 9 and 10). The diverse regulators of ERα function have, in part, been defined by identification of an array of protein-binding partners of ERα.
Microtubules play a vital role in cell functions, including cellular trafficking and motility. In addition, microtubules regulate signaling cascades by serving as protein scaffolds or by facilitating function-altering translocation of signaling components. Specifically, microtubules participate in nuclear functions through scaffolding and titrating nuclear factors such as p53 (11), SMAD-related proteins (12), Myc-interacting zinc finger protein-1 (MIZ-1) (13), and catenins (14). Glucocorticoid receptor (15), nuclear factor-κB (NFκB) (16,17), and nuclear factor of activated T cells (NFAT) (18) nuclear translocation depends on an intact microtubule transport system; the dynein-microtubular transport system has been implicated in the retrograde transport of proteins from the axon to the cell body in peripheral nerves (19). In addition, transcription factors such as RUNX may be regulated by microtubule dynamics; the treatment of cancer cells with taxol, which stabilizes microtubules, results in nuclear export of RUNX (20).
Several groups have demonstrated an interaction between the microtubule system and estrogen. Estrogen decreases microtubule polymerization in vitro and in cultured hippocampal neurons (21), MCF-7, and MDA-MB-231 cells (22); in addition, estrogen has been shown, in vitro, to bind to tubulin from cell extracts (23). Blaustein (24) described the accumulation of ERα in neuronal cell bodies in response to the microtubule depolymerizer colchicine and argued that microtubules participate in the transport of ERα to axons and dendrites, sites of potential nongenomic ERα action (25,26,27). The activation function 1 domain of ERα binds to tubulin when expressed at the cell surface, implying a possible link between microtubule function and nongenomic actions of estrogen (28). A direct transcriptional role of tubulin is suggested by experiments showing that nuclear β-tubulin binds and activates nuclear Notch activity and associates with chromosomal Notch target genes (29).
Dynein-dynactin complexes mediate many of the motor functions of microtubules. Dynein light chain (DLC1), a motor protein that is also found in the nucleus, has been shown to bind ERα, to potentiate transcription, and to immunoprecipitate with estrogen-responsive promoters (30). This study supports an indirect effect of DLC1 on transcriptional potency of ERα, because knockdown of DLC1 protein resulted in inhibition of ERα nuclear translocation and subsequent reduction of ERα transcriptional potency.
Recently, hematopoietic PBX-interacting protein (HPIP), a microtubule-associated protein, was shown to bind to ERα and to facilitate estrogen-dependent AKT and MAPK activation (31). Interestingly, these studies also indicated that microtubule-depolymerizing agents increase nuclear ERα function. These data suggest a model of microtubule regulation of ERα action in which ERα is scaffolded in the cytoplasm to subserve nongenomic functions; it was suggested that one possible mechanism by which microtubule-depolymerizing agents increase ERα action could be through facilitating increases in nuclear ERα. Thus, microtubular regulation of ERα may depend on cytoplasmic scaffolding that enhances nongenomic signaling and, in some cell types, represses nuclear function.
Recently, Hu et al. (32) demonstrated that estrogen- responsive genes rapidly mobilize within the nucleus after exposure to estrogen. Gene knockdown experiments implicate nuclear coactivators and, surprisingly, several components of the cytoskeletal network, including actin/myosin and DLC1, play an essential role in this process. Their studies confirm that cytoskeletal motor proteins may indeed modulate steroid hormone responses through novel mechanisms that dynamically alter physical location of chromosomal targets. Moreover, this study underscores the concept that cytoskeletal proteins, previously believed to be strictly cytoplasmic factors, are found in the nucleus and can be ascribed nuclear functions.
In this study, we describe experiments that suggest that the dynactin subunit p150/glued influences the action of ERα without affecting nuclear/cytoplasmic partitioning of ERα. We provide evidence that the microtubule motor protein p150/glued is present in the nucleus of cells, increases ER-dependent transcription, and is recruited to estrogen-responsive genes.
Results
ERα binds the C-terminal domain of p150/glued
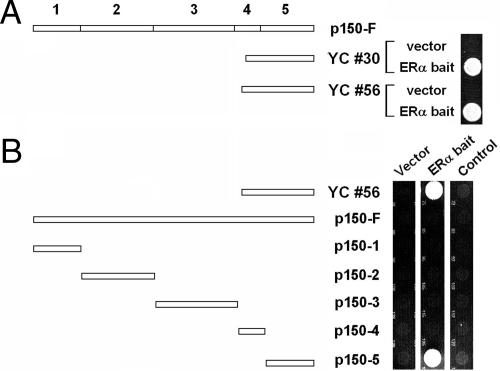
We performed a yeast two-hybrid screen using full-length human ERα as bait and identified two independent human cDNA clones expressing the largest subunit of dynactin, p150/glued. p150/glued is composed of five major domains (33). The clones identified correspond to part of the fourth and the entire fifth domain (at the C-terminal end of the protein; Fig. 1A). To map the interaction between ERα and p150/glued to the fifth domain, we subcloned the fifth domain of rat p150/glued as a GAL4 activation fusion, and determined that this was able to specifically interact with ERα; other fragments of p150/glued failed to interact with ERα in yeast (Fig. 1B). Thus, it appeared that domain 5 of p150/glued is sufficient for physical interactions with ERα. Full-length p150 fused to the GAL4 activation domain failed to rescue ERα-expressing yeast. This may have been due to poor expression of the full-length p150 fusion, because independent transfection experiments using the full-length clone fused to the GAL4 activation domain failed to produce significant amounts of full-length protein in transfected cells (data not shown).
Figure 1.
ERα interacts with the C terminus of p150/glued in yeast. A, A yeast two-hybrid screen resulted in two independent clones encoding the C-terminal 326 and 333 amino acids of p150/glued. These clones, YC #30 and #56, reproducibly rescued yeast carrying GAL4-ERα but not empty control plasmids (vector expressing only GAL4 DNA binding domain). These clones correspond to part of domain 4 and all of domain 5 of p150/glued (see schematic of p150/glued domain structure at top of figure). B, Domain mapping. PCR-generated fragments of p150/glued (from rat; fused to the GAL4 activation domain) were tested for their ability to rescue yeast-expressing ERα. A rat clone encoding the C-terminal domain of p150/glued, but not other domains of the protein, was able to rescue GAL4-ERα-expressing yeast (center panel). Rescue was dependent on ERα; there was no growth with vector (left panel) or when an irrelevant cDNA was used as bait (MAFB transcription factor; right panel). These experiments were done without the addition of estrogen to the media.
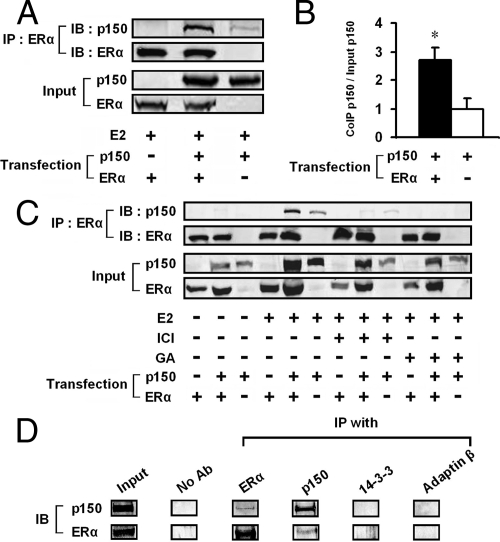
We next examined whether ERα could interact with p150/glued in transiently transfected mammalian cells. Immunoblot analysis of immunoprecipitates using an ERα antibody demonstrated p150/glued association with ERα (Fig. 2A). There was some p150/glued in lysates immunoprecipitated without ERα; we therefore quantitated multiple replicates of this experiment to demonstrate that there was immunoprecipitation between ERα and p150/glued. This analysis demonstrated a significant 2.7-fold increase over baseline in the amount of p150/glued pelleted with ERα (Fig. 2B) (P < 0.05). To demonstrate the specificity of this response, we performed coimmunoprecipitation experiments in the presence of pharmacological modifiers of ERα function. Coimmunoprecipitation of p150/glued was not seen under estrogen-free conditions, and the effect of estrogen was blocked by ICI 182,780, a specific inhibitor of ERs (Fig. 2C). In addition, geldanomycin, which blocks ERα maturation by interactions with heat shock proteins, completely prevented the coimmunoprecipitation (Fig. 2C). We are uncertain why estrogen is required for the interaction in mammalian cells but not in yeast; we cannot rule out the possibility that estrogen-like compounds are found in bacteriological media used or that the protein fragments in yeast are folded differently from native p150/glued in mammalian cells.
Figure 2.
p150/glued binds to ERα in mammalian cells. A, HEK 293 cells were transiently transfected with plasmids encoding ERα and/or p150/glued. ERα antibodies were used to immunopurify ERα-containing complexes in cells treated with estrogen. Immunoprecipitated proteins were blotted and probed for p150/glued to detect molecular association (top panel). Lysate volumes were adjusted to normalize for equal protein content in each lane. We used a rabbit antiserum [no. EM49 (52)] which recognized transfected rat p150/glued, but not endogenous human protein. A small background band that migrated with p150/glued was consistently seen without transfection of ERα. However, there was a very consistent increase in the precipitated p150/glued found when ERα was included in the precipitates. B, Quantitation of seven coimmunoprecipitations indicated a 2.7-fold increase in precipitated p150/glued in the presence of transfected p150/glued and ERα (with estrogen; compared with control lysates without ERα). *, P < 0.05 for immunoprecipitation with transfected ERα compared with without transfected ERα. C, Complex formation was dependent on estrogen (E2; 10 nm) and inhibited by ICI 182,780 (ICI; 100 nm) and geldanomycin (GA; 1 μm). HEK 293 cells were transfected with cDNAs shown; lysates were analyzed as in panel A. D, p150 and ERα interact in MCF-7 cells. Coimmunoprecipitation was performed in cells treated with DTBP (5 mm in PBS). Buffer or antibodies against ERα, p150, 14-3-3, or adaptin-β were used to pull down protein complexes. Only p150 and ERα antibodies pulled down complexes containing both p150 and ERα. IB, Immunoblotting; IP, immunoprecipitation.
To investigate whether physiologically expressed p150 interacts with ERα, we performed coimmunoprecipitations in MCF-7 cells (Fig. 2D). Proteins from cells treated with the cell-permeable cross-linker dimethyl 3,3′-dithiobispropionimidate- 2Hcl (DTBP) were immunopurified with antibodies against a variety of targets. Both p150 and ERα were detected in immunoprecipitates generated from antibodies to p150 and ERα, but control antibodies failed to pull down complexes containing these proteins, suggesting that under physiological conditions, p150 and ERα are found in the same molecular complex. We were unable to identify complexes in the absence of cross-linker, suggesting that the interactions are transient in nature.
Overexpressed p150/glued enhances the action of ERα
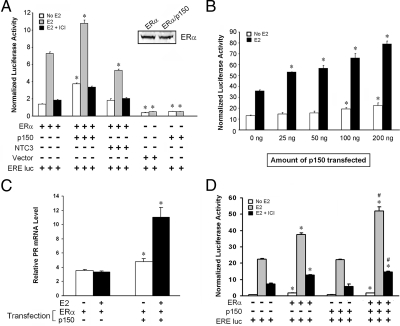
To determine whether p150/glued modulates ERα transcriptional potency, we cotranfected p150/glued and ERα in human embryonic kidney (HEK) 293 cells together with an ERE-luciferase reporter construct (34). The transcriptional potency of ERα was stimulated by estrogen, and this was significantly enhanced by overexpression of p150/glued (Fig. 3A). In control experiments, p150/glued, transfected by itself without ERα, demonstrated no enhancement of ERE-luciferase activity; in addition, transfection of p150/glued did not change the protein levels of ERα (Fig. 3A; inset Western blot). The degree of stimulation was directly related to the amount p150/glued plasmid transfected (Fig. 3B). Cotransfection of p150 together with ERα resulted in a marked increase in estrogen responsiveness of progesterone receptor (PR) mRNA, a known target of ERα (Fig. 3C). We did not detect activation of other endogenous estrogen-responsive genes (cathepsin D and pS2) in HEK 293 cells transfected with p150, ERα, or both plasmids.
Figure 3.
Transactivation of ERα is enhanced by p150/glued. A, ERα expressed transiently in HEK 293 cells activated ERE-luciferase in a ligand/antagonist-dependent fashion. When p150/glued was overexpressed, the activity of ERα was substantially increased. There was an increase in both baseline and ligand-dependent activation of the reporter. Western blots for ERα (see inset) demonstrate that ERα protein levels were not increased with p150 transfection. Asterisk denotes significant increases (or decreases) in activity in groups with p150 (or Notch3) transfection compared with ERα only control (P < 0.05). B, There was a dose-dependent increase in ERα activity with increasing amounts of p150/glued DNA transfection. Asterisk denotes significant increases in activity in groups with p150 transfection compared control (0 ng DNA; P < 0.05). C, Overexpression of p150/glued resulted in activation of endogenous estrogen target gene, PR. HEK 293 cells transfected with constructs shown and treated with estrogen or vehicle were analyzed for PR mRNA by quantitative RT-PCR. PR transcription was stimulated by cotransfection of p150/glued and ERα. Asterisk denotes significant increases (P < 0.05) in mRNA in groups with p150 transfection compared with ERα only control. D, p150/glued also increased ERα activity assessed using an ERE-luciferase reporter in MCF-7 cells transfected with p150/glued and/or ERα; the enhancement was not seen in cells transfected with p150/glued alone. *, Significant (P < 0.05) increases in activity stimulated by transfection of ERα compared with endogenous ERα activity; #, significant (P < 0.05) increases in activity after transfection of p150/glued, compared with endogenous p150/glued. NTC3, Notch 3.
Enhancement of ERα activity was also observed in ERα-transfected MCF-7 cells (Fig. 3D) and hippocampal neurons (data not shown). However, p150 transfection (without transfected ERα) did not amplify ERE-luciferase responses in MCF-7 cells (Fig. 3D) and hippocampal neurons that express endogenous ERα (data not shown). The reason for this discrepancy is not clear but may depend on stoichiometric ratios of p150 and ERα. Endogenous ERα may already be saturated with endogenous p150. An effect of transfected p150 may only be seen when levels of ERα are increased by cotransfection. If this is the case, p150 knockdown would be predicted to reduce the efficacy of endogenous ERα. Indeed, gene knockdown experiments (see Fig. 8) described later in this study support this possibility.
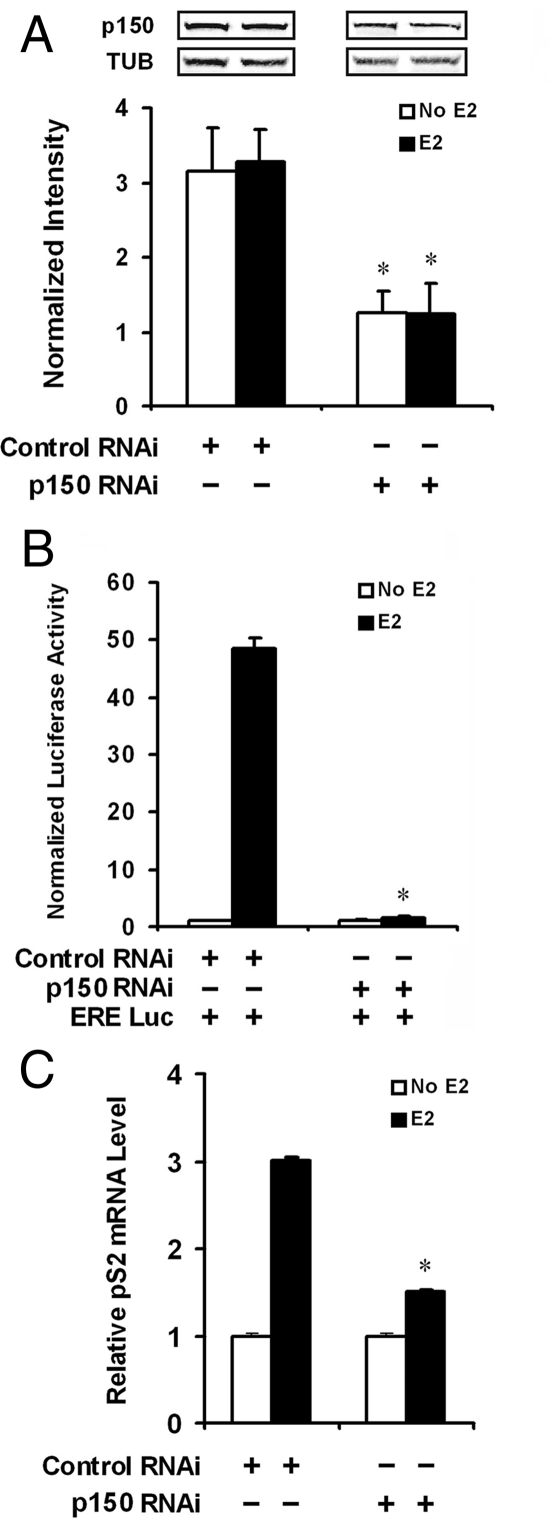
Figure 8.
p150/glued is required for full activation of estrogen-dependent gene expression in MCF-7 cells. We designed RNAi oligonucleotides against human p150/glued (see Materials and Methods), which were transfected into MCF-7 cells. A, Western analysis demonstrated that compared with control RNAi, there was approximately 50% inhibition of p150/tubulin (TUB) protein levels when pair 56 p150-specific RNAi was used (normalized to loading controls). Bands are from the same Western blot of one representative experiment. B, We compared estrogen-stimulated ERE-luciferase expression in MCF-7 cells cotransfected with RNAi (pair 56) 48 h after transfection. As before, Renilla luciferase cotransfection was performed in all groups to normalize transfection efficiencies. This experiment demonstrated a significant decrease in the potency of estrogen-stimulated transcription when p150/glued protein levels were inhibited. Statistical analyses were performed to compare groups with and without p150/glued RNAi; asterisk denotes a significant reduction in estrogen-stimulated luciferase activity with p150 RNAi compared with control RNAi (P < 0.05). C, Inhibition of p150 expression by RNAi impairs endogenous pS2 gene expression. MCF-7 cells were transfected with p150 or control RNAi and treated with estrogen to stimulate endogenous ERα-dependent transcription. Relative pS2 mRNA, measured by quantitative PCR, was significantly suppressed after p150 RNAi treatment. *, Significant reduction in estrogen-stimulated mRNA levels with p150 RNAi compared with control RNAi (P < 0.05).
Microtubule inhibition does not affect ERα localization
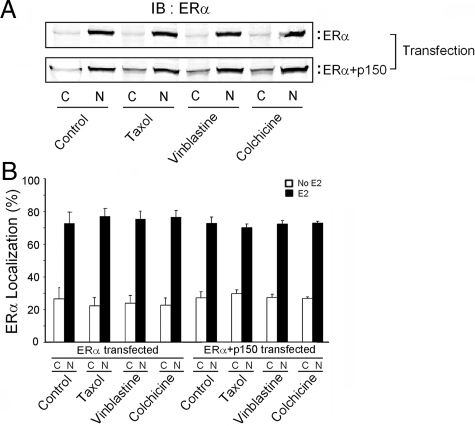
One possible explanation for the action of p150 is that ERα is anchored to the cytoplasm by p150/glued and microtubules, and that overexpression of p150/glued leads to a shift of ERα into the nucleus. To address this possibility, we performed subcellular analysis of ERα protein in vinblastine-, colchcine-, and taxol-treated cells (Fig. 4, A and B). If microtubular proteins anchor ERα to the cytoplasm, we would expect an increase in the nuclear levels of ERα after microtubule disruption, but we did not find evidence of increased nuclear ERα under any of these conditions. We also considered the possibility that ERα could be enhanced by phosphorylation of the receptor after alteration of microtubule dynamics. However, MAPK inhibitors, which block posttranslational activation of ERα in growth factor-stimulated and stress conditions, failed to block the stimulatory effect of p150/glued overexpression (data not shown).
Figure 4.
Microtubule destabilization does not enhance nuclear expression of ERα. ERα levels in the nucleus and cytoplasm were determined by immunoblotting. Transfected HEK 293 cells were treated with estrogen overnight and, when indicated, also treated with microtubule-altering drugs for 6 h. Cells were lysed and nuclei separated from cytoplasm by low-speed centrifugation. Protein fractions were analyzed by immunoblotting (A). The ratio of nuclear to cytoplasmic ERα was the same under all conditions tested (B). Quantitative data shown are from three independent experiments. IB, Immunoblotting.
p150/glued is found in the nucleus
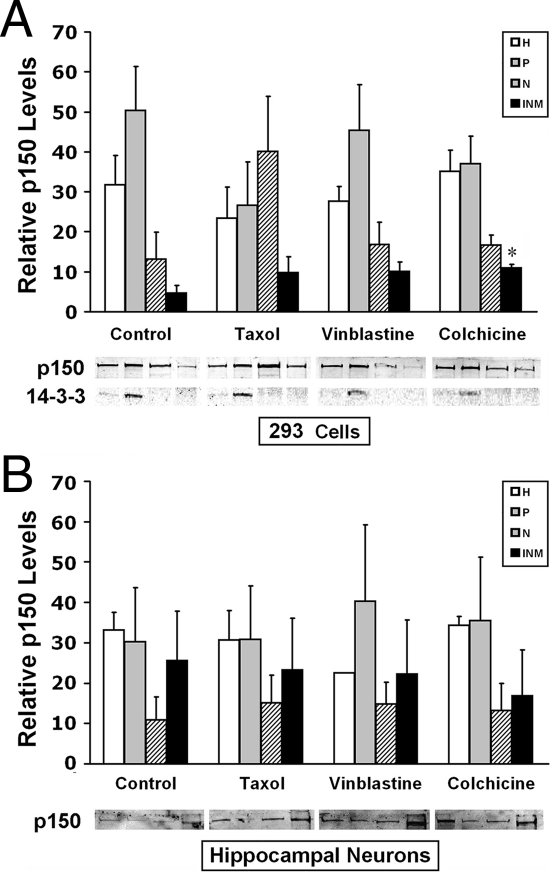
We next addressed the alternative possibility that p150/glued is a nuclear cofactor for ERα. Although p150/glued has clear effects in the cytoplasm, its presence in the nucleus has not been described. We therefore performed subcellular fractionation to quantitate p150/glued subcellular protein content (Fig. 5). Cells were fractionated into cytoplasm, stripped nuclear protein extracts, and insoluble nuclear proteins using high-speed centrifugation and salt extraction (34); these fractions were analyzed for p150/glued content by immunoblotting. In HEK 293A cells, p150/glued was found abundantly in cytoplasmic fractions (P); in addition, the protein was found at modest levels in both soluble (N) and nonextractable [insoluble nuclear matrix (INM)] fractions (Fig. 5A). Analysis of soluble, cytoplasmic 14-3-3 proteins confirmed that the nuclear fractions were not contaminated with cytosol. Microtubules attached to nuclei were not a source of contaminating p150/glued because experiments were performed on ice using detergents used to strip nuclei of endoplasmic reticulum and the outer nuclear membrane, conditions that favor depolymerization of microtubules. However, to rule out the possibility that small amounts of polymerized microtubules were contaminating cellular fractions, we also performed experiments on cells treated with vinblastine and colchicine to completely depolymerize cellular microtubules. As shown in Fig. 5A, p150/glued was present in the nuclear extracts and insoluble nuclear fraction, even in the presence of these drugs.
Figure 5.
p150/glued localizes to the nucleus. To assess the subcellular localization of p150/glued, HEK 293 cells (A) and mouse hippocampal neurons (B) were fractionated, and proteins were analyzed by immunoblotting. Fractions were loaded as follows: whole-cell homogenate (H), postnuclear fraction (P), soluble nuclear extract (N), and nuclear matrix fractions (INM). Localization was determined in cells exposed for 6 h to tubulin depolymerizers, colchicine (C; 10 μm), vinblastine (V; 300 nm), or the microtubule stabilizer taxol (T; 500 nm). Colchicine caused a modest increase in nuclear matrix p150/glued only in HEK 293 cells; other treatments did not cause significant changes in p150/glued distribution. 14-3-3 protein was examined to confirm that nuclear fractions were free of cytoplasmic components. Graphs show quantitation of bands in three experiments. Equal volumes of proteins were loaded for each subcellular fraction; thus, quantitation is displayed in relative units of band intensity (y-axis); only comparisons between treatments can be made; there is, in fact, much less nuclear p150/glued compared with cytoplasmic protein; however, nuclear bands appear strong because of increased nuclear protein concentration after centrifugation of organelles. Representative bands from a single experiment are shown below graphs. Statistical analyses were performed to compare groups relative to cells without drug treatment; *, P < 0.05.
Because p150/glued and other dynactin proteins participate in nuclear envelope breakdown during mitosis (35), it is possible that nuclear fractions contained dynactin proteins transiently associated with the nuclear envelope during cell division. To eliminate this possibility, we performed fractionation experiments in postmitotic hippocampal neurons as well (Fig. 5B). Our results suggested that p150/glued is present in significant quantities in the nucleus of both HEK 293A and hippocampal cultures. In addition, studies using microtubule-targeting drugs demonstrate that p150/glued content is relatively stable. Only colchicine, which destabilizes microtubules, resulted in a significant increase in nuclear p150/glued in HEK 293 cells, whereas microtubule-stabilizing taxol did not change subcellular distributions of p150/glued. In neurons, microtubule depolymerizers and stabilizers did not result in subcellular shifts in p150/glued.
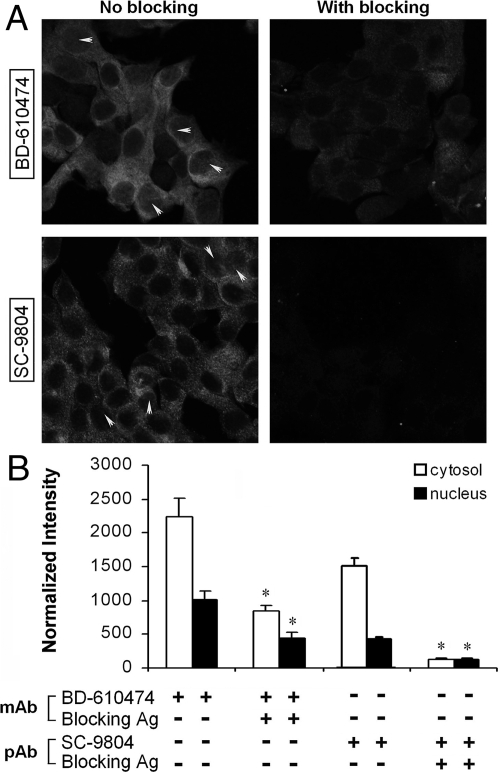
We performed immunocytochemical localization of p150/glued in MCF-7 cells using two different antibodies (Fig. 6 and supplemental Figs. 1 and 2 published as supplemental data on The Endocrine Society’s Journals Online web site at http://mend.endojournals.org). These two antibodies bound avidly to cytosol and modestly to nuclear components. The nuclear staining was present in most cells and had a diffuse ground glass appearance that in some cells appeared clumped (yellow arrows). Fusion protein and peptide blockers of these antibodies significantly reduced both the cytosolic and nuclear binding of these antibodies, confirming that p150 immunoreactivity was specific. Similar results were obtained in HEK 293 cells (see supplemental Figs. 3 and 4). These studies complement biochemical experiments that demonstrate a modest expression of p150 in the nucleus.
Figure 6.
Localization of p150 in MCF-7 cells. Cells were analyzed using two independent antibodies. BD-610474 is a well-characterized mouse monoclonal antibody. SC-9084 is goat polyclonal antibody. Secondary antibodies did not result in detectable staining (data not shown). As an additional negative control, blocking antigen was used included as a control for specificity. A, MCF-7, visualized by confocal microscopy, demonstrated strong staining in the cytoplasm, as has been noted before. In addition, significant, although weaker, staining was observed in the nuclei of many cells; the strongest cells occasionally contained inhomogeneous concentrations of protein denoted by yellow arrows. Quantitative analysis of cytoplasmic and nuclear staining intensities is shown in panel B. Both the nuclear and cytoplasmic staining were attenuated by blocking antigen. Images (with and without peptide) were obtained with identical camera settings for each antibody. See supplemental Figs. 1 and 2 for Z-stack images, which demonstrate nuclear staining through multiple optical planes. Statistical differences were noted between staining levels with and without antigen blocking (*, P < 0.01). mAb, Monoclonal antibody; pAb, polyclonal antibody.
p150/glued recruitment to estrogen-responsive promoters
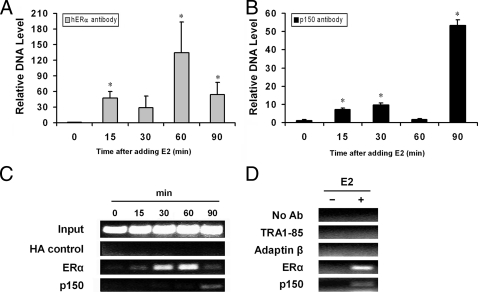
To prove whether p150/glued in the nucleoplasm plays a role in transcription of estrogen-regulated genes, we performed chromatin immunoprecipitation (ChIP) experiments. MCF-7 cells pulsed with estrogen were cross-linked with formaldehyde, and extracts were immunopurified with antibodies to determine whether p150/glued protein is associated with estrogen-responsive genes in MCF-7 cells (Fig. 7). As expected, using an ERα antibody for ChIP, we demonstrated that ERα is physically associated with the well-characterized estrogen-responsive promoter pS2 after acute estrogen exposure (Fig. 7, A and C) (3). p150/glued is also physically associated with estrogen-responsive genes with acute estrogen treatment, showing a delayed temporal pattern after estrogen addition (Fig. 7, B and C). Similar results were obtained with an independent p150 antibody (data not shown), whereas a negative control monoclonal hemagglutinin (HA) antibody did not precipitate detectable amounts of promoter DNA (Fig. 7C). To further confirm the specificity of the ChIP assay, we repeated the procedure using antibodies against a series of cellular targets that were not expected to interact with p150 or ERα. None of these antibodies associated with the pS2 promoter, regardless of whether estrogen was included (Fig. 7D). Taken together with studies demonstrating protein interactions and transcriptional activation studies, our results suggest that nuclear p150/glued participates in estrogen-dependent transcriptional activation in cells by physical interaction with chromatin.
Figure 7.
p150/glued is recruited to the estrogen-responsive pS2 promoter in MCF-7 cells. Protein association with pS2 promoter was assessed using ChIP. MCF-7 cells were plated in estrogen-free DMEM 5% fetal calf serum for 3–5 d and then pulsed with estrogen (10 nm) for periods shown. At the end of each incubation, cells were fixed with formalin and chromatin fragmented by sonication. A, ERα antibodies were used to demonstrate the temporal profile of ERα association with the ERE sites of the pS2 promoter, which was quantified using quantitative PCR (qPCR). B, p150/glued monoclonal antibodies were used to immunoprecipitate protein/DNA adducts, which were subjected to qPCR analysis using primers against the proximal pS2 promoter. The immunoprecipitated pS2 promoter DNA was normalized to the value at time zero. C and D, Endpoint PCR analysis of ChIP assay demonstrates that multiple negative control antibodies failed to precipitate pS2 target gene. Negative control antibodies, which failed to associate with the target gene (with or without estrogen), included HA, TRA1-85, and adaptin-β; in addition, reactions performed without antibody failed to precipitate detectable quantities of pS2 chromatin. This analysis is qualitative and does not precisely mirror qPCR results, which are considered more quantitative. qPCR data shown are from two independent experiments. Endpoint PCR analysis represents four independent experiments in panel C and two experiments in panel D. *, Statistical significance (P < 0.05) relative to unstimulated cells. Ab, Antibody.
p150/glued knockdown reduces potency of estrogen-stimulated transcription
To further demonstrate the role of p150/glued in the regulation of ERα, we tested whether reduction of p150/glued levels in cells changes the potency of estrogen transcriptional regulation. Overexpression studies in Fig. 3 suggested that p150 levels modulate ERα potency. Thus, we predicted that decreasing endogenous p150 levels could lower endogenous ERα potency in MCF-7 cells. RNA interference (RNAi) oligonucleotides specifically targeting human p150/glued were transfected into MCF-7 cells and demonstrated specific ability to reduce the protein levels of p150/glued. The most potent RNAi oligonucleotides reduced p150/glued protein levels by roughly 50% compared with a control sequence (Fig. 8A). When RNAi oligonucleotides were cotransfected with an estrogen reporter plasmid, we measured a marked decrease in estrogen-stimulated transcription compared with the group of cells treated with the control RNA (Fig. 8B). Two less potent sets of RNAi oligonucleotides also modestly reduced ERα activity (data not shown). Our results demonstrate that endogenous p150/glued participates in the stimulation of estrogen-dependent transcription in MCF-7 cells expressing physiologically relevant levels of ERα. In MCF-7 cells treated with p150 RNAi, we also measured significant reduction in pS2 mRNA, demonstrating that physiologically expressed p150 plays a significant role in enhancing estrogen-dependent gene expression (Fig. 8C).
Discussion
The importance of microtubules and dynactin in a number of transcriptional processes is currently emerging. Here, we demonstrate that p150/glued, the largest dynactin subunit, participates in estrogen-dependent gene regulation through action within the nucleus. We find that: 1) ERα binds to p150/glued through physical interactions with the C-terminal domain of p150/glued; 2) p150/glued is found in biologically relevant quantities in the nucleus; and 3) p150/glued is recruited to an estrogen-dependent promoter and is required for maximal estrogen-stimulated transcription.
Mechanisms of p150 enhancement of ERα function
Potential mechanisms for the stimulatory action of p150 in ERα action include: 1) a cytoplasmic mechanism in which p150 enhances ERα nuclear transport; 2) a mechanism involving ERα movement within the nucleus; and 3) a transport-independent nuclear mechanism. We favor either the second or third mechanisms.
We first considered whether p150/glued mediates cytoplasmic transport of ERα, which may influence transcriptional function. Such a mechanism appears to mediate DLC1 enhancement of ERα-dependent transcription, because DLC1 increases the nuclear localization of ERα (30). Moreover, the microtubule-associated protein HPIP inhibits transcription by tethering ERα to the cytoplasm (31). Cytoplasmic motor function participates in the delivery of nuclear factor-κB (NFκB) (16,17), p53 (11), SMAD-related protein (12), and glucocorticoid receptors (15) to the nucleus. Other recent studies have elucidated nuclear actin as an essential regulator of transcriptional cofactor megakaryoblastic leukemia 1 through direct binding between actin and megakaryoblastic leukemia 1 in the nucleus, which blocks nuclear export (36); thus, unexpected localization of cytoskeletal components of the cell to the nucleus can potentially increase nuclear levels of transcription factors. However, analogous mechanisms are unlikely to account for p150/glued-mediated activation of ERα for several reasons. First, p150/glued overexpression is not likely to stimulate dynactin transport function, and may, in fact, inhibit the cytoplasmic motor function because overexpression of motor protein subunits disrupts stoichiometry of protein assemblies. Second, our experiments (data not shown) confirm the observation (31) that microtubule depolymerization agents actually enhance ERα function in HEK 293 and MCF-7 cells in a taxol-sensitive fashion, suggesting that intact microtubular transport may, in fact, repress ERα function. Most importantly, we did not see changes in total or nuclear levels of ERα when we overexpressed p150/glued. We therefore believe that our observations with p150/glued are independent of cytoplasmic transport processes.
Alternatively, p150/glued may act on intranuclear protein dynamics to facilitate the movement of protein complexes within the nucleus. In fact, several reports have characterized intranuclear movement of ERα function in response to ligands and antagonists (37,38). It is not known whether the movement of these factors is essential for transcription or whether the movement is mediated by dynactin. In support of this possibility, it is known that other proteins that participate in motor functions, such as tubulin (29,39,40), dynein light chain (30), and p62 (41) have been found in the nucleus. Recent studies have demonstrated that ERα chromosomal targets are dynamically mobilized within the nucleus after exposure to ligand (32). Molecular inhibition studies indicate that actin, dynein light chain, and myosin may play a role in this process, although other components of this intriguing phenomenon have not been elucidated. Our results here demonstrate that yet another component of the molecular trafficking machinery of the cell could be localized to the nucleus. Future work to address the presence of other dynein/dynactin components and a functional role of dynactin in intranuclear movement are necessary to fully address this mechanism.
A third potential mechanism is that p150/glued modulates ERα function via an intranuclear mechanism that is independent of the known transport function of p150/glued. β-Tubulin is an example of another predominantly cytoplasmic protein that is now thought to actively regulate transcription through association with a nuclear protein (Notch) and association with chromosomal targets (42). Our experiments suggest that p150/glued could potentiate ERα-mediated transcription by binding and chromosomal association. A mechanism for 150/glued coactivation is not clearly defined because it has no homology to known transcriptional activator domains; in addition, p150/glued domains fused to the GAL4 DNA-binding domain fail to induce Gal4-reporter expression, suggesting that p150/glued lacks intrinsic coactivator activity (data not shown). We suggest that if p150/glued regulates ERα function directly, it does so either through binding-induced allosteric changes in ERα or through an adaptor function that bridges interactions between ERα and transactivating domains of other activators. Intriguingly, p150/glued colocalizes with HDAC6 in the cytoplasm and regulates microtubule deacetylation and chemotaxis (43). It is conceivable that p150/glued may regulate transcription through interactions with related nuclear deacetylases, which are known to participate in estrogen-dependent transcription.
Context-dependent activity of p150
ERα expressed in HEK 293 cells activates an ERE-luciferase promoter and the PR gene but not cathepsin D or pS2; likewise, in MCF-7 cells, p150 physically associates with the pS2 gene, but we were not able to detect PR gene colocalization. The potential explanations include, but are not limited to, the possibilities of cell type-specific ERα transcriptional mechanisms (multiple examples (e.g. Refs. 44,45,46,47) and/or gene-specific ERα responsiveness to dynactin complexes. Large-scale genomic analyses have demonstrated that coactivators and associated regulators of transcription may bind with considerable diversity across the large family of estrogen-dependent genes (i.e. there is never 100% concordance between estrogen responsiveness, and coactivator localization (e.g. Refs. 48,49,50). The possibility that p150/glued acts in a cell-specific manner or localizes only to specific coactivator complexes, restricting it to a subset of estrogen-responsive genes, deserves to be tested in future studies.
Potential cytoplasmic functions of p150/ERα interaction
Although our studies focused on the nuclear effects of p150/ERα, we do not exclude a role of this interaction on cytoplasmic functions. An intriguing possibility that is brought up by these studies is that ERα may modulate microtubular dynamics or other p150 functions via protein-protein interactions. The ERα interaction domain of p150/glued has an important physiological role and has been implicated in disease processes. The fifth domain of p150/glued binds directly to membranous cargo (51). Kumar further suggested that conformational changes within the fifth domain of p150/glued regulate the loading and unloading of membranous cargo. Potentially, estrogen-regulated interactions between p150/glued and ERα may limit the accessibility or change the conformation of this domain and thereby alter rates of transport within the cell. A similar mechanism may play a role in macrophages (17), in which ERs inhibit the nuclear localization of NFκB.
Another protein that has been shown to bind to the fifth domain of p150/glued is the Huntington-associated protein (HAP), which also appears to participate in intracellular transport (52). Our finding that ERα binds to the same region of this protein suggests that ERα could potentially influence HAP-mediated regulation of microtubule transport. This could play a critical role in homeostasis of ERα-expressing neurons, because HAP is known to be essential for viability (53).
Pharmacological inhibition of the microtubule system is used in the treatment of cancer and in inflammatory conditions. ERα plays a clear role in breast cancer cell proliferation; thus, as Manavathi (31) points out, clinically used microtubule inhibitors (some of which were tested in this report) may have intrinsically limited efficacy, because they may inhibit mitosis and cell migration via motor inhibition but may also inadvertently enhance mitosis through activation of classical ERα activity. Our results and the study by Rayala et al. (30) demonstrate that, in contrast to tubulin targeting, inhibition of dynein components could attenuate both motor function and classical ERα nuclear function. As such, direct inactivation of dynein motor components such as p150/glued or DLC1 may represent an improved approach to inhibiting ERα-dependent tumor cell growth.
Materials and Methods
Cells and reagents
HEK 293A cells were grown in DMEM supplemented with 10% fetal calf serum, antibiotics, and glutamine. MCF-7 cells were grown in RPMI supplemented with 10% fetal calf serum, antibiotics, and glutamine; before use in experiments, cells were shifted to phenol red free media and charcoal-stripped serum to remove endogenous estrogens. Hippocampal neurons from embryos were prepared from time pregnant C57/B6 mice as described by Xu et al. (27); neurons were maintained in Neurobasal media supplemented with glucose and B27 supplement. Primary cultures were more than 99% neuronal by neurofilament staining and lack of glial fibrillary acidic protein staining. Chemicals were obtained from Sigma (St. Louis, MO) and Tocris Cookson (Ellisville, MO). Antibodies used include the ERα polyclonal H184 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), 14-3-3 polyclonal (Santa Cruz), a rabbit polyclonal against rat p150/glued [a generous gift of Xiaojiang Li, used for immunoblotting in some experiments (52)], a mouse monoclonal against p150/glued [catalog no. 610474; BD Biosciences(Palo Alto, CA), used for immunoblotting, ChIP, and cell staining], and a goat polyclonal (Santa Cruz SC-9804; for immunostaining). Control antibodies for chromatin immunoprecipitation included the anti-HA monoclonal 12CA5 (Santa Cruz). Tubulin and TRA1-85 monoclonal antibodies were obtained from the Developmental Studies Hybridoma Bank at the University of Iowa.
Yeast two-hybrid studies
We followed the Matchmaker protocol (CLONTECH Laboratories, Inc., Palo Alto, CA) to screen for ERα interacting factors. Full-length human ERα was cloned into the bait vector pGBK-T7 and transformed into AH109a. Transformants were selected on synthetic defined (-Trp) plates. Human ERα-expressing yeast were mated en masse to commercially prepared libraries of prey (in strain Y197α encoding human embryonic cDNA clones fused to the GAL4 activation domain in pACT2-AD). Positive clones were selected on Sabouraud dextrose (-Ade-His-Trp-Leu) and further characterized by LacZ staining. Clones were rescued by transformation into bacteria, sequenced, and reintroduced into human ERα-expressing yeast to confirm interactions. We mapped the binding sequences in p150 by subcloning fragments of the rat clone (52) using PCR into pACT2-AD. These were then transformed into human ERα-expressing yeast and replica plated onto selective media to score for growth indicative of protein-protein interactions.
ERα transactivation assays
Methods have been described previously (34). Cells were cotransfected with plasmid mixtures (total DNA, 1 μg for 24-well plates) that include ERE-luciferase (100 ng) and phRG-TK (100 ng; Promega Corp., Madison, WI), which produces Renilla luciferase under constitutive control. p150/glued in pRK5 was a gift from Dr. Xiaojiang Li (52), whereas ERα in pCMV5 was modified from a clone provided by Dr. Benita Katzenellenbogen (54). All transfections included either p150/glued plasmid (100 ng) or an equal mass of pRK5; in addition, either ERα (100 ng) or an equal mass of pCMV5 was used. The remainder of the DNA in all transfections was composed of pCMV-Sport6 (empty vector). Cell lysates were analyzed 1 d after transfection for both firefly and Renilla luciferase activities (Promega). The relative value of firefly to Renilla luciferase is reported as ERE-driven transcriptional activity.
Western blotting was performed by standard methods. Membranes were blocked with milk; IR-dye secondary antibodies were detected with LiCor Odyssey flatbed detector, which is quantitative over at least five logs.
Coimmunoprecipitations were performed as described (55) using the ERα polycolonal antibodies or p150 monoclonal antibody (Becton Dickinson, Franklin Lakes, NJ). For experiments involving transfection, a total of 4 μg of DNA was mixed with Lipofectamine 2000 (Invitrogen) and applied to six-well plates. For each well, we used 400 ng of each expressed plasmid (or matched empty vector). The remainder of the DNA was pCMV-Sport6 and empty expression vector. Briefly, cell lysates were mixed with 2 μg of antibody and then precipitated with 40 μl packed protein G agarose (Upstate Biotechnology, Inc., Lake Placid, NY). Captured proteins were analyzed by Western blotting. For MCF-7 immunoprecipitations, cells were first cross-linked with 5 mm DTBP (Pierce Chemical Co., Rockford, IL) in PBS for 1 h. Negative control antibodies used to assess the specificity of the precipitation included monoclonal antibodies to adaptin-β (Becton Dickinson) and rabbit antisera against 14-3-3 (Santa Cruz).
Cell fractionation experiments have been described in detail (34,56). Briefly, cells were lysed by homogenization in TKM buffer (25 mM Tris-HCl, pH 7.4; 5 mM KCl, and 1 mM MgCl2). An aliquot was saved as homogenate (H). Nuclei were sedimented and then stripped by extraction with TKM supplemented with 1% Nonidet P-40. The supernatant of this extraction is the postnuclear fraction (P) and represents endoplasmic reticulum, Golgi, some plasma membrane protein, and the outer nuclear membrane. Stripped nuclei, devoid of outer nuclear membrane protein, were further purified through a sucrose step gradient. The purified nuclear pellet was extracted with a high-salt solution (420 mm NaCl) and sedimented. The supernatant contained extractable nucleoplasm (N), and the pellet contained insoluble nuclear matrix components (INM).
Chromatin immunoprecipitation
MCF-7 cells were plated in phenol red-free DMEM supplemented with 5% dextran-charcol-treated fetal calf serum. To initiate ER activation, cells were treated with 10 nm 17β-estradiol (E2) or vehicle for a specified amount of time; the reaction was terminated by adding 1% formaldehyde for 10 min, which also cross linked protein-DNA complexes. Cross-linking was quenched by adding 125 mm glycine. The cells were washed with PBS and lysed in 50 mm Tris-HCl, pH 8.0; 150 mm NaCl; 1 mm EDTA; 1% Triton X-100; 0.1% Na-deoxycholate and protease inhibitors (Pierce) followed by ten 10-sec pulses of sonication. Extracts were preclearing at 4 C with 0.05% BSA, 5 μg sheared salmon sperm DNA, and 30 μl protein G agarose (50% slurry; Upstate Biotechnology) under gentle agitation for 2 h at 4 C. The supernatant was transferred to a fresh tube, and 2 μg of antibody was added overnight at 4 C. Antibodies used included the HA monoclonal negative control, p150 mouse monoclonal antibody from BD Bioscience, and ERα polyclonal H184 from Santa Cruz Biotechnologies as a positive control. Protein G agarose was then added and incubated for 1.5 h. The pellets were successively washed for 10 min in 1 ml buffer 1 [20 mm Tris-HCl (pH 8.0); 150 mm NaCl; 2 mm EDTA; 1% Triton X-100; 0.1% sodium dodecyl sulfate (SDS)], 1 ml buffer 2 [20 mm Tris-HCl pH 8.0; 500 mm NaCl; 2 mm EDTA; 1% Triton X-100; 0.1% SDS], 1 ml LiCl buffer [20 mm Tris-HCl, pH 8.0; 250 mm LiCl; 1 mm EDTA; 1% Nonidet P-40; 0.1% Na-deoxycholate], and twice in 1 ml TE [10 mm Tris-HCl, pH 8.0; 1 mm EDTA]. Protein-DNA complexes were eluted in 120 μl elution buffer [TE with 1% SDS] for 30 min, and the cross-links were reversed by incubation at 65 C for 16 h. DNA was purified using a PCR purification kit (QIAGEN, Chatsworth, CA) and eluted in 50 μl. Immunoprecipitated protein-DNA complexes were analyzed for estrogen target gene expression by quantitative PCR for the human pS2 gene using primers flanking established ERE sites: 5′-GGCCATCTCTCACTATGAATCACT-3′ and 5′-GGATTTGCTGATAGACAGAGACGA-3′. Relative DNA immunoprecipitated was defined as the inverse log (base 2) of the difference between the cycle threshold (Ct) of input DNA and the Ct of precipitated DNA. In some experiments, end point PCR (40 cycles of amplification) was performed and analyzed by agarose gel electrophoresis. These were qualitative assays only (Fig. 7C) and used to confirm the validity of positive and negative controls.
RNAi
RNAi duplexes against p150/glued and control (labeled RNAi) were obtained from Invitrogen (Carlsbad, CA). Three pairs of RNAi were tested: pair 56: UUUAGCUUUGCUUUGUCUUCUGCCC and GGGCAGAAGACAAAGCAAAGCUAAA; pair 57: AACAGCACCAGAACGCAGUCAUGGU and ACCAUGACUGCGUUCUGGUGCUGUU; and pair 58: ACUGCAUGAAGUUUCCAGAUCCCGG and CCGGGAUCUGGAAACUUCAUGCAGU. RNAi efficacy was established by immunoblotting to confirm protein knockdown. All three pairs of RNAi interfered with p150 levels. However, pair 56 showed the most efficient reduction and was used for all studies shown. Cells were transfected with Lipofectamine 2000. We mixed RNAi duplexes with plasmid DNAs in cotransfections; cells were analyzed 48 h after transfection.
Quantitative RT-PCR
HEK 293 cells grown in six-well plates were transfected with ERα (or blank vector pCMV5; 5 ng) and p150 (or blank vector pRK5; 400 ng) mixed with 3.595 μg of pCMV-Sport6 blank vector for 24 h with E2 (10 nm in ethanol) or vehicle overnight. Cells were collected in lysis buffer, and total RNA was prepared using an RNeasy column. Random primed reverse transcribed cDNA was then quantitated with primers to estrogen target genes human PR: 5′-TCAGTGGGCAGATGCTGTATTT-3′ and 5′-GCCACATGGTAAGGCATAATGA-3′, cathepsin D: 5′-GTACATGATCCCCTGTGAGAAGGT-3′ and 5′-GGGACAGCTTGTAGCCTTTGC-3′, or pS2; 5′-ATACCATCGACGTCCCTCCA 3′ and 5′-AAGCGTGTCTGAGGTGTCCG-3′ in a thermal cycler detecting SYBR green reactive products. 18S RNA levels were quantitated to control for RNA levels. Primers for 18S RNA were: 5′-TTTGTTGGTTTTCGGAACTGAGGC-3′ and 5′-GGCATCGTTTATGGTCGGAACTACG-3′. The inverse log (base 2) of the difference in Ct between the estrogen target PCR product and actin RNA was used as the relative expression value of the target gene.
Immunofluorescence
Monoclonal antibody staining was done on 4% paraformaldehyde-fixed cells using primary antibodies at 5 μg/ml. Blocking studies were done with a bacterial expressed recombinant p150 fragment (amino acids 1-333) at 7.5 μm (a gift of Erika Holzbauer). Goat polyclonal antibody staining of p150/glued was performed at 4 μg/ml. Blocking peptide from Santa Cruz (SC-9804P) was used at 20 μg/ml. Imaging was performed on an Olympus confocal microscope (Olympus Corp., Lake Success, NY) using a TRITC filter. All groups of experiments with the same cell type and antibody were photographed on identical settings. Pixel numbers of multiple randomly chosen areas of cytoplasm or nucleoplasm (five per cell) were quantitated. The average was used as that cell’s intensity level. Ten cell intensity levels were then grouped and quantitated in Fig. 6 and supplemental Fig. 3.
Statistical analysis
Student’s t test was applied, with P < 0.05 considered significant. All error bars are sem of measurement.
Supplementary Material
Acknowledgments
We thank Jimo Borjigin and the Wang laboratory for valuable discussions. Genggeng Yu provided excellent technical support. We thank Xiaojiang Li, Erika Holzbauer, and Mark Day for providing valuable reagents for these studies. Tubulin and TRA1-85 monoclonal antibodies were obtained from the Developmental Studies Hybridoma Bank at the University of Iowa.
Footnotes
This work was supported by National Institutes of Health Grants NS041342 and NS054724 (to M.M.W.) and a Burroughs Wellcome Fund Career Award in Biomedical Sciences (to M.M.W.).
Disclosure Summary: The authors have nothing to disclose.
First Published Online February 19, 2009
Abbreviations: ChIP, Chromatin immunoprecipitation; Ct, cycle threshold; DLC, dynein light chain; DTBP, dimethyl 3,3′-dithiobispropionimidate-2Hcl; E2, 17β-estradiol; ER, estrogen receptor; ERE, estrogen response element; HA, hemagglutinin; HAP, Huntington-associated protein; HEK, human embryonic kidney; INM, insoluble nuclear matrix; NFκB, nuclear factor-κB; PR, progesterone receptor; RNAi, RNA interference; RUNX, Runt-related transcription factor; SDS, sodium dodecyl sulfate.
References
- Hewitt SC, Korach KS 2002 Estrogen receptors: structure, mechanisms and function. Rev Endocr Metab Disord 3:193–200 [DOI] [PubMed] [Google Scholar]
- McDonnell DP, Norris JD 2002 Connections and regulation of the human estrogen receptor. Science 296:1642–1644 [DOI] [PubMed] [Google Scholar]
- Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M 2000 Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103:843–852 [DOI] [PubMed] [Google Scholar]
- Reid G, Hüner MR, Métivier R, Brand H, Denger S, Manu D, Beaudouin J, Ellenberg J, Gannon F 2003 Cyclic, proteasome-mediated turnover of unliganded and liganded ERα on responsive promoters is an integral feature of estrogen signaling. Mol Cell 11:695–707 [DOI] [PubMed] [Google Scholar]
- Jakacka M, Ito M, Weiss J, Chien PY, Gehm BD, Jameson JL 2001 Estrogen receptor binding to DNA is not required for its activity through the nonclassical AP1 pathway. J Biol Chem 276:13615–13621 [DOI] [PubMed] [Google Scholar]
- Paech K, Webb P, Kuiper GG, Nilsson S, Gustafsson J, Kushner PJ, Scanlan TS 1997 Differential ligand activation of estrogen receptors ERα and ERβ at AP1 sites. Science 277:1508–1510 [DOI] [PubMed] [Google Scholar]
- Wang MM, Traystman RJ, Hurn PD, Liu T 2004 Non-classical regulation of estrogen receptor-α by ICI182,780. J Steroid Biochem Mol Biol 92:51–62 [DOI] [PubMed] [Google Scholar]
- Razandi M, Pedram A, Greene GL, Levin ER 1999 Cell membrane and nuclear estrogen receptors (ERs) originate from a single transcript: studies of ERα and ERβ expressed in Chinese hamster ovary cells. Mol Endocrinol 13:307–319 [DOI] [PubMed] [Google Scholar]
- Kelly MJ, Levin ER 2001 Rapid actions of plasma membrane estrogen receptors. Trends Endocrinol Metab 12:152–156 [DOI] [PubMed] [Google Scholar]
- Vasudevan N, Pfaff DW 2007 Membrane-initiated actions of estrogens in neuroendocrinology: emerging principles. Endocr Rev 28:1–19 [DOI] [PubMed] [Google Scholar]
- Giannakakou P, Sackett DL, Ward Y, Webster KR, Blagosklonny MV, Fojo T 2000 p53 is associated with cellular microtubules and is transported to the nucleus by dynein. Nat Cell Biol 2:709–717 [DOI] [PubMed] [Google Scholar]
- Dong C, Li Z, Alvarez Jr R, Feng XH, Goldschmidt-Clermont PJ 2000 Microtubule binding to Smads may regulate TGF β activity. Mol Cell 5:27–34 [DOI] [PubMed] [Google Scholar]
- Ziegelbauer J, Shan B, Yager D, Larabell C, Hoffmann B, Tjian R 2001 Transcription factor MIZ-1 is regulated via microtubule association. Mol Cell 8:339–349 [DOI] [PubMed] [Google Scholar]
- Roczniak-Ferguson A, Reynolds AB 2003 Regulation of p120-catenin nucleocytoplasmic shuttling activity. J Cell Sci 116:4201–4212 [DOI] [PubMed] [Google Scholar]
- Harrell JM, Murphy PJ, Morishima Y, Chen H, Mansfield JF, Galigniana MD, Pratt WB 2004 Evidence for glucocorticoid receptor transport on microtubules by dynein. J Biol Chem 279:54647–54654 [DOI] [PubMed] [Google Scholar]
- Mackenzie GG, Keen CL, Oteiza PI 2006 Microtubules are required for NF-κB nuclear translocation in neuroblastoma IMR-32 cells: modulation by zinc. J Neurochem 99:402–415 [DOI] [PubMed] [Google Scholar]
- Ghisletti S, Meda C, Maggi A, Vegeto E 2005 17β-Estradiol inhibits inflammatory gene expression by controlling NF-κB intracellular localization. Mol Cell Biol 25:2957–2968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie GG, Oteiza PI 2007 Zinc and the cytoskeleton in the neuronal modulation of transcription factor NFAT. J Cell Physiol 210:246–256 [DOI] [PubMed] [Google Scholar]
- Hanz S, Perlson E, Willis D, Zheng JQ, Massarwa R, Huerta JJ, Koltzenburg M, Kohler M, van-Minnen J, Twiss JL, Fainzilber M 2003 Axoplasmic importins enable retrograde injury signaling in lesioned nerve. Neuron 40:1095–1104 [DOI] [PubMed] [Google Scholar]
- Pockwinse SM, Rajgopal A, Young DW, Mujeeb KA, Nickerson J, Javed A, Redick S, Lian JB, van Wijnen AJ, Stein JL, Stein GS, Doxsey SJ 2006 Microtubule-dependent nuclear-cytoplasmic shuttling of Runx2. J Cell Physiol 206:354–362 [DOI] [PubMed] [Google Scholar]
- Kipp JL, Ramirez VD 2003 Estradiol and testosterone have opposite effects on microtubule polymerization. Neuroendocrinology 77:258–272 [DOI] [PubMed] [Google Scholar]
- Aizu-Yokota E, Ichinoseki K, Sato Y 1994 Microtubule disruption induced by estradiol in estrogen receptor-positive and -negative human breast cancer cell lines. Carcinogenesis 15:1875–1879 [DOI] [PubMed] [Google Scholar]
- Ramirez VD, Kipp JL, Joe I 2001 Estradiol, in the CNS, targets several physiologically relevant membrane-associated proteins. Brain Res Brain Res Rev 37:141–152 [DOI] [PubMed] [Google Scholar]
- Blaustein JD, Olster DH 1993 Colchicine-induced accumulation of estrogen receptor and progestin receptor immunoreactivity in atypical areas in guinea-pig brain. J Neuroendocrinol 5:63–70 [DOI] [PubMed] [Google Scholar]
- Singh M, Sétáló Jr G, Guan X, Warren M, Toran-Allerand CD 1999 Estrogen-induced activation of mitogen-activated protein kinase in cerebral cortical explants: convergence of estrogen and neurotrophin signaling pathways. J Neurosci 19:1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen B, Akama K, Alves S, Brake WG, Bulloch K, Lee S, Li C, Yuen G, Milner TA 2001 Tracking the estrogen receptor in neurons: implications for estrogen-induced synapse formation. Proc Natl Acad Sci USA 98:7093–7100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Traystman RJ, Hurn PD, Wang MM 2003 Neurite-localized estrogen receptor-α mediates rapid signaling by estrogen. J Neurosci Res 74:1–11 [DOI] [PubMed] [Google Scholar]
- Azuma K, Horie K, Inoue S, Ouchi Y, Sakai R 2004 Analysis of estrogen receptor α signaling complex at the plasma membrane. FEBS Lett 577:339–344 [DOI] [PubMed] [Google Scholar]
- Yeh IT, Ludueña RF 2004 The βII isotype of tubulin is present in the cell nuclei of a variety of cancers. Cell Motil Cytoskeleton 57:96–106 [DOI] [PubMed] [Google Scholar]
- Rayala SK, den Hollander P, Balasenthil S, Yang Z, Broaddus RR, Kumar R 2005 Functional regulation of oestrogen receptor pathway by the dynein light chain 1. EMBO Rep 6:538–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavathi B, Acconcia F, Rayala SK, Kumar R 2006 An inherent role of microtubule network in the action of nuclear receptor. Proc Natl Acad Sci USA 103:15981–15986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Kwon YS, Nunez E, Cardamone MD, Hutt KR, Ohgi KA, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG, Fu XD 2008 Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proc Natl Acad Sci USA 105:19199–19204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroer TA 2004 Dynactin. Annu Rev Cell Dev Biol 20:759–779 [DOI] [PubMed] [Google Scholar]
- Xu Y, Traystman RJ, Hurn PD, Wang MM 2004 Membrane restraint of estrogen receptor α enhances estrogen-dependent nuclear localization and genomic function. Mol Endocrinol 18:86–96 [DOI] [PubMed] [Google Scholar]
- Salina D, Bodoor K, Eckley DM, Schroer TA, Rattner JB, Burke B 2002 Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell 108:97–107 [DOI] [PubMed] [Google Scholar]
- Vartiainen MK, Guettler S, Larijani B, Treisman R 2007 Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science 316:1749–1752 [DOI] [PubMed] [Google Scholar]
- Htun H, Holth LT, Walker D, Davie JR, Hager GL 1999 Direct visualization of the human estrogen receptor α reveals a role for ligand in the nuclear distribution of the receptor. Mol Biol Cell 10:471–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenoien DL, Mancini MG, Patel K, Allegretto EA, Smith CL, Mancini MA 2000 Subnuclear trafficking of estrogen receptor-α and steroid receptor coactivator-1. Mol Endocrinol 14:518–534 [DOI] [PubMed] [Google Scholar]
- Walss C, Kreisberg JI, Ludueña RF 1999 Presence of the βII isotype of tubulin in the nuclei of cultured mesangial cells from rat kidney. Cell Motil Cytoskeleton 42:274–284 [DOI] [PubMed] [Google Scholar]
- Woulfe J 2000 Class III β-tubulin immunoreactive intranuclear inclusions in human ependymomas and gangliogliomas. Acta Neuropathol (Berl) 100:427–434 [DOI] [PubMed] [Google Scholar]
- Karki S, Tokito MK, Holzbaur EL 2000 A dynactin subunit with a highly conserved cysteine-rich motif interacts directly with Arp1. J Biol Chem 275:4834–4839 [DOI] [PubMed] [Google Scholar]
- Yeh TS, Hsieh RH, Shen SC, Wang SH, Tseng MJ, Shih CM, Lin JJ 2004 Nuclear βII-tubulin associates with the activated notch receptor to modulate notch signaling. Cancer Res 64:8334–8340 [DOI] [PubMed] [Google Scholar]
- Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP 2002 HDAC6 is a microtubule-associated deacetylase. Nature 417:455–458 [DOI] [PubMed] [Google Scholar]
- Peterson TJ, Karmakar S, Pace MC, Gao T, Smith CL 2007 The silencing mediator of retinoic acid and thyroid hormone receptor (SMRT) corepressor is required for full estrogen receptor α transcriptional activity. Mol Cell Biol 27:5933–5948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endoh H, Maruyama K, Masuhiro Y, Kobayashi Y, Goto M, Tai H, Yanagisawa J, Metzger D, Hashimoto S, Kato S 1999 Purification and identification of p68 RNA helicase acting as a transcriptional coactivator specific for the activation function 1 of human estrogen receptor α. Mol Cell Biol 19:5363–5372 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Eeckhoute J, Carroll JS, Geistlinger TR, Torres-Arzayus MI, Brown M 2006 A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes Dev 20:2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer R, Wolf SS, Obendorf M 2007 PRMT2, a member of the protein arginine methyltransferase family, is a coactivator of the androgen receptor. J Steroid Biochem Mol Biol 107:1–14 [DOI] [PubMed] [Google Scholar]
- Kininis M, Chen BS, Diehl AG, Isaacs GD, Zhang T, Siepel AC, Clark AG, Kraus WL 2007 Genomic analyses of transcription factor binding, histone acetylation, and gene expression reveal mechanistically distinct classes of estrogen-regulated promoters. Mol Cell Biol 27:5090–5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, Fox EA, Silver PA, Brown M 2005 Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122:33–43 [DOI] [PubMed] [Google Scholar]
- Cheng AS, Jin VX, Fan M, Smith LT, Liyanarachchi S, Yan PS, Leu YW, Chan MW, Plass C, Nephew KP, Davuluri RV, Huang TH 2006 Combinatorial analysis of transcription factor partners reveals recruitment of c-MYC to estrogen receptor-α responsive promoters. Mol Cell 21:393–404 [DOI] [PubMed] [Google Scholar]
- Kumar S, Zhou Y, Plamann M 2001 Dynactin-membrane interaction is regulated by the C-terminal domains of p150(Glued). EMBO Rep 2:939–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SH, Gutekunst CA, Hersch SM, Li XJ 1998 Interaction of huntingtin-associated protein with dynactin P150Glued. J Neurosci 18:1261–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SH, Yu ZX, Li CL, Nguyen HP, Zhou YX, Deng C, Li XJ 2003 Lack of huntingtin-associated protein-1 causes neuronal death resembling hypothalamic degeneration in Huntington’s disease. J Neurosci 23:6956–6964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrenn CK, Katzenellenbogen BS 1993 Structure-function analysis of the hormone binding domain of the human estrogen receptor by region-specific mutagenesis and phenotypic screening in yeast. J Biol Chem 268:24089–24098 [PubMed] [Google Scholar]
- Zhang Y, Jia L, Lee SJ, Wang MM 2007 Conserved signal peptide of Notch3 inhibits interaction with proteasome. Biochem Biophys Res Commun 355:245–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer GA, Radsak KD 2000 Identification of a novel signal sequence that targets transmembrane proteins to the nuclear envelope inner membrane. J Biol Chem 275:3857–3866 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.