Abstract
Currently, little is known about histone modifications and molecular mechanisms of negatively regulated transcription. In pituitary cells, thyroid hormone (T3) decreased transcription, and surprisingly increased histone acetylation, of TSHα promoter. This increase was mediated directly by thyroid hormone receptor. Histone acetylation of H3K9 and H3K18 sites, two modifications usually associated with transcriptional activation, occur in negative regulation of TSHα promoter. T3 also caused release of a corepressor complex composed of histone deacetylase 3 (HDAC3), transducin β-like protein 1, and nuclear receptor coprepressor (NCoR)/ silencing mediator for retinoic and thyroid hormone receptor from TSHα promoter in chromatin immunoprecipitation assays. NCoR and HDAC3 overexpression selectively increased ligand-independent basal transcription. Two histone acetyltransferase inhibitors increased overall transcription but did not abrogate negative regulation or NCoR/HDAC3 complex release by T3. Chromatin immunoprecipitation analyses of an endogenous positively regulated target gene showed increased histone acetylation and corepressor complex release with T3 treatment. Finally, microarray analyses suggested there is a subset of negatively regulated genes with increased histone acetylation. These findings demonstrate the critical role of NCoR/HDAC3 complex in negative regulation of TSHα gene expression and show that similar complexes and overlapping epigenetic modifications can participate in both negative and positive transcriptional regulation.
The negative regulation of TSHα gene transcription by thyroid hormone surprisingly occurs via histone H3 acetylation and loss of a corepressor/HDAC3 complex at its promoter.
Thyroid hormone receptors (TRs) belong to a superfamily of nuclear hormone receptors that act as ligand-regulatable transcription factors (1,2). There are two major TR isoforms, TRα and TRβ, encoded on separate genes. TRs bind to thyroid hormone response elements in the promoters of target genes to regulate their transcription. In positively regulated target genes, unliganded TRs bind to corepressors such as nuclear receptor corepressor (NCoR) or silencing mediator for retinoic and thyroid hormone receptors (SMRT) that form corepressor complexes containing transducin β-like protein 1 (TBL1) and histone deacetylase 3 (HDAC3), and mediate basal transcriptional repression by unliganded thyroid hormone receptor in positively regulated target genes (3,4,5). In the presence of T3, corepressor complexes are released from liganded TRs that, in turn, associate with coactivator complexes that contain steroid receptor coactivator (SRCs), cAMP response element-binding protein (CREB)-binding protein (CBP), and P/CAF. These complexes cause increased histone acetylation near the TRE of the promoter (1,2,6). ATP-dependent chromatin remodeling complexes similar to the SWI/SNF family in yeast that contains the adenosine triphosphatase subunit, Brahma-related gene 1, also are recruited to the promoter (7,8) and critical for transcriptional activation. Another major complex, Mediator complex, also can interact with the promoter and serves to recruit RNA polymerase II (RNA pol II) (9,10,11). Recently, chromatin immunoprecipitation (ChIP) studies have suggested that liganded TRs and nuclear hormone receptors recruit coactivators in a cyclical pattern on the promoters of some target genes (12,13,14,15,16).
In contrast to positively regulated target genes, negatively regulated genes can be stimulated in the absence of T3 and decreased by its presence. The mechanism(s) for negative transcriptional regulation by T3 is not well understood; however, corepressors and coactivators may be involved. NCoR and SMRT can increase basal transcription of some target genes in the absence of T3 (17,18,19,20). Coactivators also can play an apparently paradoxical role in T3-dependent negative regulation of some target genes (21). On the other hand, it appears that HDACs are recruited by TRs during ligand-dependent negative regulation in other cases (22). Thus, cofactor-associated changes in histone acetylation, and alterations in chromatin structure, may be involved in T3-mediated negative regulation. Of note, not all negatively regulated target genes are activated in the absence of ligand, suggesting that cofactors may be differentially recruited to promoters of negatively regulated target genes (23).
TSH is a heterodimer composed of two subunits: TSHα and TSHβ. TSHα, known as glycoprotein hormone α common subunit, also is a subunit for several other glycoprotein hormones such as LH, FSH, and human choriogonadotropic hormone, whereas TSHβ is unique to TSH. T3 negatively regulates TSH by decreasing both TSHα and TSHβ gene as well as TRH gene transcription (17,19,20). These genes have been studied as models of negatively regulated gene transcription by T3. From a physiological perspective, their negative regulation is critical for feedback control of the hypothalamic/pituitary/thyroid axis.
To better understand the mechanism for negative regulation of gene transcription by T3, we established a pituitary cell line in which stably expressed luciferase was under the control of the TSHα promoter and negatively regulated by T3. This line was generated because there currently are no cell culture models for TSHα-negative regulation, and endogenous TSHα is not expressed in GH3 cells. Surprisingly, the negative regulation of TSHα is associated with increased histone acetylation of the TSHα promoter. The mechanism for this negative regulation is dependent upon corepressor and HDAC3 release from the promoter. Our findings show that histone acetylation occurs in positively, as well as negatively, regulated target genes via overlapping molecular mechanisms and suggest that histone acetylation of the promoter per se in not sufficient to predict the direction of transcription in a given target gene.
Results
Establishment of a stable rat pituitary GH3 cell line
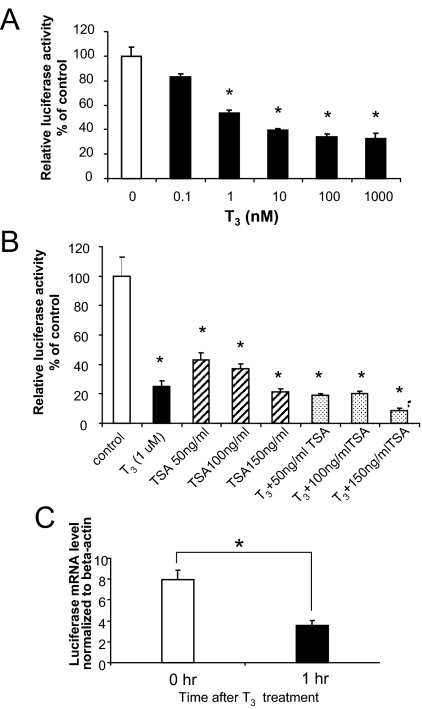
We generated a stable rat pituitary GH3 cell line, α-23, containing the human TSHα promoter (−840 to +1) in tandem with a luciferase reporter cDNA to study negative regulation by T3 in a native chromatin context. This construct was shown previously to be sufficient for negatively regulated transcription by T3 in transient transfection studies (24). As expected, α-23 cells exhibited a dose-dependent decrease in luciferase activity by T3 (Fig. 1A). A decrease was observed at 1 nm T3 and a maximal decrease (∼90%) was seen at 0.1 μm T3 (Fig. 1A). These results demonstrate that the stably integrated TSHα promoter is negatively regulated by T3 in the GH3 cells, and mimics the ligand-dependent negative-feedback regulation of the TSHα gene by the pituitary in vivo. We found minimal expression of endogenous TSHα gene in the absence or presence of T3 by microarray analyses and quantitative RT-PCR so it apparently is silenced in parent GH3 cells.
Figure 1.
T3 regulation of transcription and histone acetylation by TSHα promoter in α-23 cells. A, Negative regulation of TSHα promoter activity by T3. α-23 cells were plated in 24-well plates and cultured in T3-free 10% FBS-DMEM for 3 d. Cells were then incubated with indicated concentrations of T3 in 10% FBS-DMEM overnight. The cell lysates were prepared and luciferase activities were measured as described in Materials and Methods. Shown are the mean of triplicate samples ± sd normalized to control as 100%. Similar findings were found in two other experiments. *, P < 0.05 difference from control using ANOVA. B, Effects of the HDAC inhibitor, TSA, on the TSHα promoter activity. α-23 cells were cultured in T3-free 10% FBS-DMEM medium for 3 d before adding TSA (at indicated concentrations), 0.1 μm T3, or TSA/T3. Cells were cultured overnight before cell lysates were prepared, and luciferase activities were measured as described in Fig. 1A. Shown are the mean of triplicate samples ± sd. Similar findings were observed in three other experiments. Bars 2–8 had P < 0.05 difference from control using ANOVA. White bar, Control; black bar, T3; striped bar, TSA; and stippled bar, T3 + TSA. C, Luciferase mRNA expression from TSHα promoter after 1 h T3 treatment. Cells were harvested at 0 and 1 h after treatment with 0.1 μm T3. RNA was isolated and both luciferase and β-actin mRNA expression were determined by quantitative RT-PCR as described in Materials and Methods. Shown are mean values from triplicate samples normalized to β-actin. Paired Student’s t test performed; *, P < 0.01. Results were confirmed in three consecutive experiments.
Inhibition of TSHα promoter activity by trichostatin A (TSA)
We next examined the effects of the histone deacetylase inhibitor, TSA, and T3 on luciferase activity in α-23 cells. Surprisingly, TSA inhibited luciferase activity in α-23 cells in a dose-dependent fashion, suggesting that negative transcriptional regulation by the TSHα promoter is mediated by histone acetylation (Fig. 1B). Indeed, TSA stimulated histone acetylation of TSHα promoter in ChIP assay (data not shown). Moreover, because coadministration of T3 with TSA led to only a slightly further decrease in transcription, T3-mediated negative regulation of TSHα may occur by the same mechanism as TSA, i.e. histone acetylation. At 1 h, T3 also negatively regulated luciferase mRNA from TSHα promoter (Fig. 1C).
Histone H3 acetylation of the TSHα promoter after T3 treatment
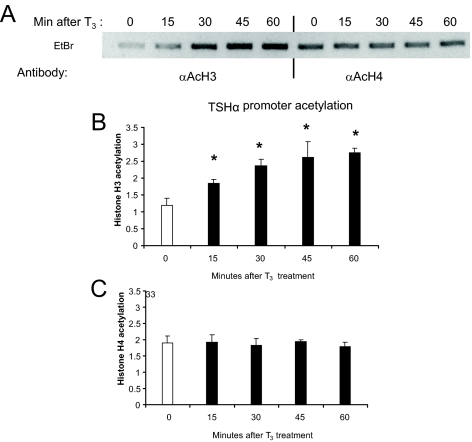
To examine whether T3 induced histone acetylation of TSHα promoter, we performed ChIP assay. T3 induced a rapid time-dependent increase in histone H3 acetylation of the TSHα promoter that could be detected after 15 min but did not significantly change histone H4 acetylation (Fig. 2, A–C). Thus, T3 increased histone acetylation of a negatively regulated target gene.
Figure 2.
T3 acetylation of histones H3 and H4 in the TSHα promoter region. α-23 cells were grown in T3-free 10% FBS-DMEM for 3 d cells and then were treated with 0.1 μm T3 for the time periods indicated. Cells were collected and ChIP assay was performed as described in Materials and Methods using antibodies against acetylated histones H3 or H4. A, PCR products from ChIP assay using anti-acetyl H3 and anti-acetyl H4 antibodies. Shown are bands from gel electrophoresis. Similar findings were observed in two other experiments. In these time course studies, ChIP with control H3 antibody showed no changes at indicated time points after T3 addition. Quantitative RT-PCR analyses of PCR products from three ChIP experiments using anti-acetyl H3 (B) and anti-acetyl H4 (C) antibodies. The statistical analysis was performed utilizing ANOVA. Values are expressed as the mean ± sd. *, P < 0.05 difference from control.
Histone acetylation of TSHα promoter mediated directly by TR
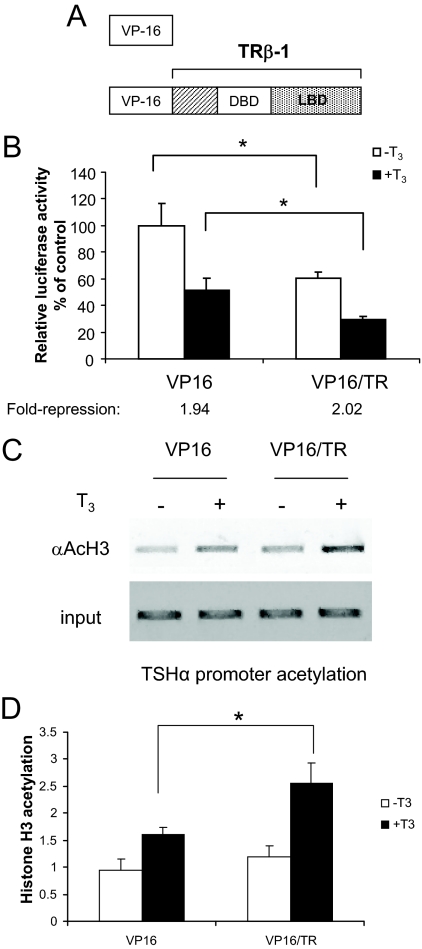
These unexpected findings prompted us to investigate further the mechanism of histone acetylation. To increase histone acetylation on TSHα promoter, we transiently transfected α-23 cells with a vector encoding VP16/TRβ, a dominant positive fusion protein containing the activation domain of Herpes simplex 1 virus (VP16) fused to the amino terminus of full-length human TRβ (Fig. 3Α). Previous studies have shown that this fusion protein had constitutive transcriptional activity in positively regulated target genes associated with increased histone acetylation (25). In the case of the TSHα, VP16/TRβ further decreased transcriptional activity both in the absence and presence of T3 while still exhibiting negative regulation by T3 (Fig. 3B). Significantly, ChIP analysis showed that VP16/TRβ mediated increases in histone H3 acetylation that were associated with decreased TSHα gene transcription both in the absence and presence of hormone (Fig. 3B), although there was a net increase in histone acetylation in the latter (Fig. 3, C and D). These findings further suggest that decreased transcriptional activity by the TSHα promoter is associated with increased histone acetylation. Moreover, they also strongly suggest that TR complex directly mediates the histone acetylation changes on the promoter by T3 in a cis manner as recruitment of another enhancer, namely the dominant positive VP16/TR, led to a T3-dependent increase in histone H3 acetylation that was associated with a concomitant decrease in transcriptional activity.
Figure 3.
VP16/TRβ effects on transcription and histone acetylation. A, VP16 and VP16/TRβ constructs. VP16/TRβ contains the activation domain of VP16 (78-amino acid carboxy terminus) fused to full-length hTRβ. B, Effects of VP16/TRβ on TSHα promoter activity. Triplicate wells of α-23 cells were transiently transfected with 100 ng VP16/TRβ and the molar equivalent of empty vector VP16 as control. Cells were incubated with 0.1 μm T3, harvested, and analyzed in three independent experiments as described for Fig. 1. C, ChIP analysis of the α-23 cells transfected with VP16 and VP16/TRβ. Experiment was performed as described in Fig. 1C and Materials and Methods. Similar results were seen in two other experiments. D, Quantitative RT-PCR analyses of PCR products from using anti-acetyl H3 antibody (n = 3). α-23 cells were treated for 1 h with T3 before harvest for ChIP assay. Statistical analyses were performed as in Fig. 1. *, P < 0.05. DBD, DNA-binding domain; LBD, ligand-binding domain.
Dissociation of cofactors from TSHα promoter after T3 treatment
We next investigated the cofactors that participated in the T3-dependent histone acetylation of TSHα promoter by ChIP analysis (Fig. 4A). TRβ bound to TSHα promoter in both the absence and presence of T3, although its binding increased with T3 treatment. NCoR, SMRT, TBL1, and HDAC3 also bound to TSHα promoter in the absence of hormone; however, in contrast to TRβ, their binding decreased with T3 treatment. These proteins previously were shown to form a corepressor complex in positively regulated target genes (3,4,5). Moreover, HDAC3 appears to be a specific part of the corepressor complex that dissociated upon T3 treatment because no changes in HDAC1 or HDAC2 binding to the promoter were observed. Consistent with Fig. 1 data showing that luciferase expression was negatively regulated by T3 in α-23 cells, RNA pol II binding to TSHα promoter decreased with T3 treatment.
Figure 4.
Corepressor and HDAC3 binding and release from the TSHα promoter and associated effects on transcriptional activity and histone acetylation. A, Corepressor complex and coactivator binding to TSHα promoter in the absence and presence of T3. α-23 cells were treated for 1 h ± 0.1μm T3 and harvested. ChIP assays were performed similar to Fig. 1. Antibodies used in ChIP assay are indicated in panel A and described in Materials and Methods. Similar results were obtained in two other experiments. B, Effects of NCoR and HDACs on TSHα promoter activity. α-23cells were transiently transfected with 100 ng/well of pCMX/NCoR, PCMX/HDAC vectors, or the molar equivalent of empty vector as a negative control. Cells were incubated overnight with 0.1 μm T3, harvested, and analyzed as in Fig. 1. Shown are the mean of triplicate samples ± sd normalized to control as 100%. Similar findings were found in two other experiments. *, P < 0.05 difference from control using ANOVA. C, Quantitative RT-PCR analyses of PCR products from using anti-acetyl H3 antibody (n = 3). α-23 cells were treated for 1 h with T3 before harvest for ChIP assay. Statistical analyses were performed as in Fig. 2. #, P < 0.05; *, P < 0.01 from untreated and treated controls, respectively, using ANOVA.
There was no additional recruitment of SRC-1 coactivator with T3 treatment (Fig. 4A). Similar findings also were observed for SRC-2, SRC-3, CREB-binding protein (CBP)/p300 and P/CAF (data not shown). Taken together, our ChIP findings are not only consistent with the observed increase in histone H3 acetylation after T3 treatment but also suggest that the release of corepressors, NCoR and SMRT, as well as HDAC3 after T3 treatment, rather than the recruitment of known p160 coactivators and CBP with histone acetyltransferase activity, contribute to the observed increase in histone acetylation. Additionally, receptor-interacting protein 140, a coregulator that blocks steroid receptor- and TR-mediated transcriptional activity as well as recruits HDACs and histone methyltransferases (26,27,28), did not change its binding to the TSHα promoter in the absence or presence of T3.
Effects of NCoR and HDACs on TSHα promoter basal activity
To determine whether corepressors and HDAC3 had specific functional roles on the negative-regulation of TSHα gene, we examined the effects of NCoR and HDAC overexpression on TSHα promoter activity in transient transfection assays. Both NCoR and HDAC3 overexpression induced a significant increase in basal transcription in the absence of T3 and fold repression (ratio of basal transcription level/ligand-dependent transcription level). The HDAC3 effect was specific because HDAC1 and HDAC2 overexpression had no significant effects on either basal transcription level or fold repression (Fig. 4B). The increased transcriptional activity by NCoR and HDAC3 overexpression was associated with decreased histone H3 acetylation (Fig. 4C).
Effects of P/CAF and CBP/300 HAT inhibitors on TSHα promoter transcriptional activity
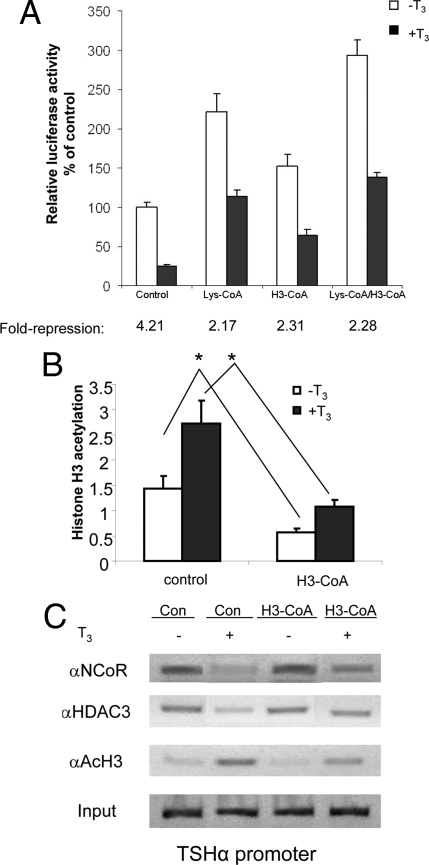
To further examine the role of corepressors and HDAC3 in mediating negative regulation, we examined the effects of two specific peptide histone acetyl transferase (HAT) inhibitors on TSHα promoter activity: Lys-coenzyme A (CoA) for CBP/p300 and H3-CoA for P/CAF (6) (Fig. 5A). These two inhibitors increased basal transcription levels in the absence and presence of T3, presumably by decreasing histone acetylation. However, T3-mediated negative-regulation still occurred when cells were treated with these HAT inhibitors, albeit at a higher set point than cells treated with control peptide. Interestingly, whereas H3-CoA led to higher transcription levels, it also was associated with decreased histone H3 acetylation both in the absence and presence of T3. ChIP analysis showed T3-dependent release of NCoR and HDAC3 from the TSHα promoter even with concomitant H3-CoA treatment (Fig. 5C, lanes 3 and 4). Taken together, these data provide further evidence for the importance of the corepressor complex loss in mediating negative regulation of the TSHα gene even though basal levels may be determined by overall histone H3 acetylation.
Figure 5.
TSHα transcripitonal activity and corepressor complex binding after blocking with P/CAF and CBP/p300 HAT inhibitors. A, Effects of two HAT inhibitors on TSHα promoter activity. α-23 cells were seeded in 24-well plates and cultured in T3-free 10% FBS-DMEM medium for 3 d. Ten microliters of digitonin solution (50 mg/ml) were added to the wells 10 min before the addition of the HAT inhibitors, Lys-CoA and H3-CoA. T3 (0.1 μm) was added to the wells, and cells were cultured overnight. The cell lysates were prepared, and luciferase activities were measured as in Fig. 1. Shown are the mean of triplicate samples ± sd normalized to control as 100%. Similar findings were observed in two other experiments. *, P < 0.05 difference between indicated samples using ANOVA. B, Quantitative RT-PCR analyses of PCR products from using anti-acetyl H3 antibody (n = 3). α-23 cells were treated with H3-CoA overnight as above and then treated for 1 h with T3 before harvest for ChIP assay. Statistical analyses were performed as in Fig. 1. *, P < 0.05. C, Effects of a HAT inhibitor on NCoR and HDAC3 binding to TSHα promoter. α-23 cells were grown in T3-free 10% FBS-DMEM for 3 d and then treated with P/CAF inhibitor, H3-CoA, and H3-AC (Con) overnight as in Fig. 4B. Cells then were treated for 1 h with 0.1 μm T3 before harvest, and ChIP assay was performed as described in Materials and Methods using NCoR and HDAC3 antibodies. Similar findings were observed in two other experiments.
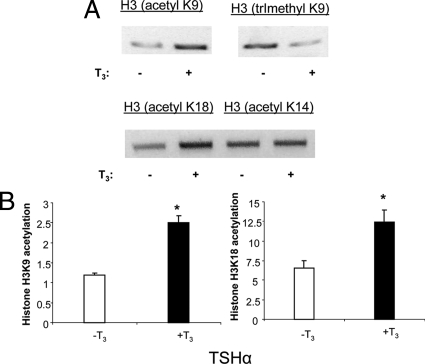
H3K9 and H3K18 are the sites for increased histone acetylation of TSHα promoter by T3
We performed ChIP assay using site-specific antibodies against histone-acetylated lysines. H3K9 and H3K18 acetylation increased, whereas H3K14 was unaffected (Fig. 6, A and B). Thus, it appears that H3K9 and H3K18 are major contributors to histone H3 acetylation. Of note, H3K9 trimethylation decreased concomitantly with H3K9 acetylation.
Figure 6.
ChIP assays of specific histone H3 acetylation sites on the TSHα promoter. α-23 cells were treated for 1 h with T3 before harvest for ChIP assay. A, PCR products from ChIP assay using indicated anti-acetyl H3 antibodies. Shown are bands from gel electrophoresis. Similar findings were observed in two other experiments. B, Quantitative RT-PCR analyses of PCR products from using anti-acetyl H3K9 and H3K18 antibodies (n = 3). Statistical analyses were performed as in Fig. 1. *, P < 0.05.
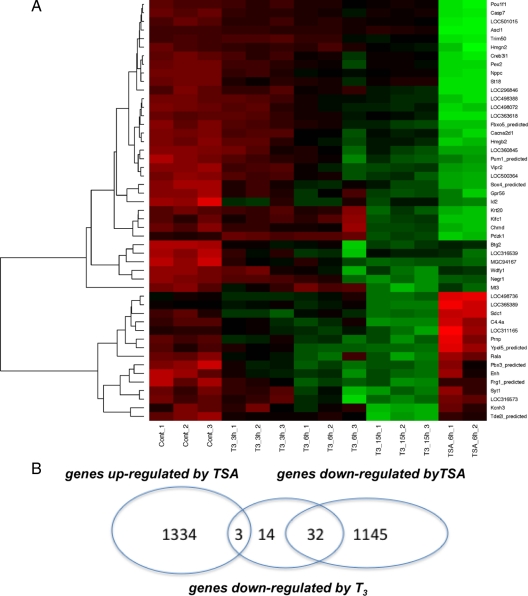
Microarray analyses of genes that are coregulated by TSA and T3
To determine whether increased histone acetylation could potentially be associated with negative regulation of other target genes by T3 other than TSHα, we performed microarray analysis of α-23 cells treated with T3 for 0, 3, 6, and 15 h and TSA for 6 h. Triplicate samples were obtained for the T3-treated samples at each time point. RNA expression was measured using the Illumina rat ref12 genome chip (Fig. 7A). A total of 10,874 genes was expressed in α-23 cells. Among them, 1334 (12.2%) were up-regulated greater than 1.5-fold by TSA, and 1145 (10.5%) were negatively regulated greater than 60% by TSA. Forty nine genes were negatively regulated by T3 at 15 h. Figure 7 shows gene expression of negatively regulated target genes (red to green over time) with a major cluster at the top (green with TSA) and a smaller one (red with TSA) on the bottom of the heat map, representing negative and positive regulation with TSA, respectively. Among the 49 negatively regulated target genes, 32 also were negatively regulated by TSA compared with only three that were up-regulated by TSA (Fig. 7B) Although it is not known definitively whether all these negatively regulated genes have increased histone acetylation of their promoters after T3 addition, these findings are consistent, nonetheless, with the notion that there may be a subclass of negatively regulated target genes for which this occurs. Detailed studies of the histone acetylation of promoters of these genes would be needed to confirm this. Interestingly, among 72 positively-regulated target genes, 30 were down-regulated, five were up-regulated, and 37 were not regulated by TSA (supplemental Fig. 1 published as supplemental data on The Endocrine Society’s Journals Online web site at http://mend. endojournals.org). The latter findings suggest that there may be some genes positively regulated by T3 associated with decreased histone acetylation.
Figure 7.
Microarray analyses of α-23 cells treated with T3 or TSA. A, Heat map of α-23 cells treated with 10−6 m T3 for 0, 3, 6, and 15 h or 100 ng/ml TSA for 6 h. T3 samples were performed in triplicate and TSA samples were performed in duplicate. RNA was isolated and expression measured on Illumina rat Beadchip microarrays. Red denotes increased expression and green indicates decreased expression. B, Venn diagram showing overlap of negatively regulated target genes by T3 with genes up-regulated or down-regulated by TSA.
Corepressor complex release from positively regulated target gene promoters
Previously, we found that T3 increased acetylation of histone H3 and H4 on the promoters of endogenous target genes, including phosphoenolpyruvate carboxykinase (PEPCK) in GH3 rat pituitary cells (16). These effects were associated with cyclical recruitment of p160 coactivators. Currently, little is known about corepressor recruitment to endogenous target genes that are positively regulated by T3. We thus examined NCoR, SMRT, and HDAC3 binding to the PEPCK promoter in the absence and presence of T3 by ChIP assay (Fig. 8A). Similar to negatively regulated TSHα, T3 induced NCoR, SMRT, and HDAC3 release from the PEPCK promoter (Fig. 8A). Both histone H3 and H4 acetylation of the PEPCK promoter also increased; however, in contrast to TSHα promoter, RNA pol II binding to the PEPCK promoter increased. A control study showed that PEPCK mRNA increased after 1 h of T3 treatment (Fig. 8B). Similar findings also were observed in two other positively regulated target genes, cholesterol 7α hydroxylase (cyp7) and sarcoplasmic endoplasmic reticulum calcium-adenosine triphosphatase in which we previously characterized their coactivator recruitment pattern to their promoters (data not shown) (16). These findings suggest that T3-mediated dissociation of corepressor complex from TSHα promoter is similar to its dissociation from positively regulated target genes even though it leads to an opposite effect on transcriptional activity.
Figure 8.
Corepressor complex binding to PEPCK promoter. α-23 cells were treated for 1 h ± 0.1 μm T3 and harvested. A, ChIP assays were performed as in Fig. 1 with primers specific for the PEPCK promoter region. Antibodies used in the ChIP assay are indicated in the figure. Similar results were obtained in two other experiments. B, PEPCK mRNA expression after T3 treatment. Cells were harvested at 0 h and 1 h after treatment with 0.1 μm T3. RNA was isolated and PEPCK and β actin mRNA expression was determined by quantitative RT-PCR as described in Materials and Methods. Shown are values from triplicate samples normalized to β-actin. Paired Student’s t test was performed; *, P < 0.01. Results were confirmed in three consecutive experiments.
Discussion
Previous studies have shown that transcriptional activation is associated with increased histone acetylation (1,2). Our current study demonstrates that T3-mediated negative regulation of the TSHα gene also is associated with increased histone H3 acetylation in a rat pituitary cell line. This surprising finding suggests that increased histone acetylation does not inevitably lead to transcriptional activation but also can be associated with transcriptional repression in some instances. Of note, it also has been reported that TSA can block transcriptional activation of the mouse mammary tumor virus promoter by progesterone, suggesting that histone acetylation per se is not always correlated with increased transcription (29).
Our studies also showed that the corepressor complex that is commonly found on positively regulated target genes plays an important role in mediating negative regulation of TSHα gene and contributes to the increased histone acetylation (Fig. 7). NCoR, SMRT, HDAC3, and TBL1 bind to the TSHα promoter in the absence of T3. The corecruitment of TBL1 suggests the formation of a corepressor complex on the TSHα promoter because TBL1 has been shown to interact preferentially with hypoacetylated histones to help stabilize the NCoR-HDAC3 complex on chromatin (30). In this case, “corepressor complex” is actually a misnomer because it is associated with ligand-independent transcription and will be referred to as NCoR-HDAC3 complex. Similar to corepressor complexes that cause repression of many positively regulated target genes in the absence of T3 (5,31), NCoR-HDAC3 complex on the TSHα promoter specifically contains HDAC3 rather than other HDACs. Moreover, specific overexpression of NCoR and HDAC3 led to increased basal transcriptional activity. CBP and P/CAF HAT inhibitors also increased ligand-independent activation and ligand-dependent transcription of TSHα; however, they did not abrogate negative regulation by T3, and HAT inhibitor did not affect T3-mediated NCoR and HDAC3 release from the TSHα promoter. Currently, it is not known whether there are other activator proteins that may be recruited by NCoR-HDAC3 complex to TSHα promoter. We also observed decreased RNA pol II recruitment by T3 was associated with decreased transcription. It is not known whether T3 modulates other RNA pol II functions such as initiation, elongation, or termination to contribute to the decreased transcription.
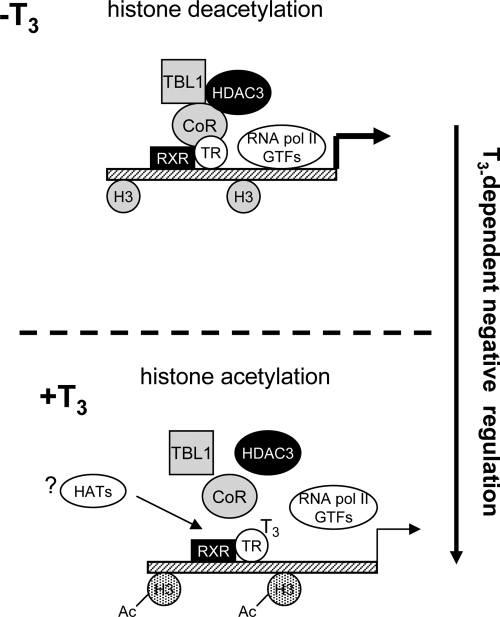
Taken together, our findings show that T3-induced loss of the NCoR-HDAC3 complex from TSHα promoter may be the key switch for its negative transcriptional regulation (see Fig. 9). In this connection, NCoR appears to be involved in ligand-independent transcription of TRH and CD44, target genes that are negatively regulated by T3 (32,33). Moreover, we previously observed that a dominant negative NCoR targeted to liver in transgenic mice decreased ligand-independent activation of the negatively regulated target gene, sialyltransferase in vivo (34). The histone modifications of these negatively regulated target genes are not known; however, our microarray studies suggest that there may be a subset of target genes that undergo both negative regulation and histone acetylation after T3 treatment (Fig. 7). In the case of TSHα promoter, it appears histone acetylation occurs primarily on H3K9 and H3K18. These sites have generally been thought to be associated with transcriptional activation (35); however, our findings suggest that there may be examples where this is not the case. In particular, histone methylations and phosphorylations also may contribute to the overall transcriptional activity, and further characterization is being performed by our laboratory. Of note, we have observed that H3K4 is demethylated by T3 (Wang, D., and P.M. Yen, unpublished results), and methylation at this site is often associated with transcriptional activation.
Figure 9.
Model for T3-mediated negative regulation of TSHα promoter activity. RXR, Retinoic X receptor; GTFs, general transcription factors.
We also observed that NCoR, SMRT, and HDAC3 binding to the promoters of several positively regulated target genes (cyp7, PEPCK, and sarcoplasmic endoplasmic reticulum calcium-adenosine triphosphatase) is decreased after T3 treatment in GH3 cells (Fig. 8 and our unpublished results). These findngs are consistent with other studies of positively regulated target genes which also show that NCoR, SMRT, and HDAC3 can form a corepressor complex with TR in the absence of hormone (3,4,5). In the presence of T3, this corepressor complex dissociates from the promoter whereas coactivators with HAT activity are recruited, most likely in a cyclical pattern (16). Thus, T3-induced corepressor loss leading to histone acetylation seems to be a common feature of positively regulated target genes and the negatively regulated TSHα gene.
Previous studies in SRC-1 knockout mice and TRβ activation function 2 domain mutant knockin mice suggest that coactivators may be important for negative regulation of TSH (36,37). Our data did not show significant recruitment of SRCs. CBP/p300, or P/CAF to the TSHα promoter. Still it is possible that P/CAF and CBP/p300 may make modest contributions to negative regulation (despite their lack of binding on ChIP assay) because the fold repression by T3 slightly decreased after treatment with inhibitors (Fig. 5). However, absolute transcription levels increased in both the absence and presence of T3, and negative regulation was maintained (Fig. 5). Further evidence supporting a contributory role for HATs is our observation that VP16/TRβ can increase histone acetylation and decrease transcription in the absence of ligand (Fig. 2) and also is known to recruit p300/CBP (38). Although the loss of NCoR/HDAC3 binding appears to be sufficient to increase histone acetylation and mediate negative regulation of TSHα, it is possible that an unknown HAT could still be recruited to TSHα promoter in the presence of T3 (Fig. 9). However, our studies with the HDAC inhibitor, TSA, suggest that decreased histone deacetylation is a critical feature of negative regulation of TSHα promoter (Fig. 1).
Our findings that histone acetylation is involved in the negative regulation of TSHα gene contrast with earlier studies of TSHα promoter activity in transiently transfected human embryonal kidney cells (21), which showed that T3 caused deacetylation of histone H4. T3 also has been shown to recruit HDACs to the TRH and TSHβ promoters (22,39); therefore histone deacetylation may also be an important mechanism for the negative regulation of other target genes by T3. Several groups have suggested that CREB or other positive transcription factors such as activator protein 1, Pit-1, and GATA may be involved in the negative transcriptional regulation by T3 (21,40,41,42). Furthermore, human TSHα promoter activity is positively regulated by T3 and TSA in Xenopus oocytes (43). Thus, it is possible that hormone-dependent changes in histone modifications observed on the TSHα promoter may depend on stable vs. transient transfection, costimulation by enhancers, as well as cell type- and gene-specific contexts.
In summary, our findings show that histone acetylation associated with loss of corepressor complex with HDAC3 from the promoter can occur not only in positive regulation of transcription by T3 but also in negative regulation. Thus, histone acetylation of the promoter by itself in not sufficient to predict the direction of transcription in a given target gene. It also is likely that similar events may occur in other target genes that are negatively regulated by nuclear hormone receptors and transcription factors. Our findings show that employment of similar epigenetic modifications and transcription complexes can lead to opposite transcriptional consequences in positively and negatively regulated target genes and, thus, shed light on the complexity, flexibility, and diversity of changes in the promoter that determine the ultimate direction of gene expression.
Materials and Methods
Reagents
Trichostatin A (TSA) was purchased from Upstate Biotechnology, Inc. (Lake Placid, NY). H3-AC-20 control, H3-CoA-20, Lys-CoA peptides have been previously described (6). cDNAs for human NCoR, HDAC1, HDAC2, and HDAC3 were cloned into pcDNA 3.1 plasmids (Invitrogen, Carlsbad, CA).
Antibodies
Antibody against TRβ-1 was obtained from Affinity BioReagents, Inc. (Golden, CO) and antibodies against NCoR(C-20), SMRT(C-19), HDAC3(H-99), SRC-1 (C-20), receptor-interacting protein 140, Brahma-related gene 1 (H-88), and RNA Pol II (N-20) were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Antibodies against acetyl H3K9, H3K14, H3K18, and trimethyl H3K9 were obtained from Abcam (Cambridge, MA).
Establishment of α-23 cell line
A luciferase reporter construct under the control of the TSH α-subunit promoter (−840 to +1), pGL2/ TSHα-840, was kindly given by Dr. Larry Jameson (Northwestern University School of Medicine,) (24). pGL2/ TSHα-840 was mixed with pEGFPc1 (CLONTECH Laboratories, Inc., Palo Alto, CA) (4:1 ratio) and used to transfect rat pituitary GH3 cells, and selectively cultured in the presence of G418 (1 mg/ml). Resistant clones were expanded in G418 medium and analyzed for negative-regulation luciferase activity by 0.1 μm T3 with the clone (α-23) exhibiting the highest amount of inhibition chosen for further study.
Cell culture, transient transfection, and reporter analyses
Monolayer cultures of clone α-23 cells were grown in DMEM (Life Technologies, Inc., Gaithersburg, MD) supplemented with 10% heat-inactivated fetal calf serum (Biofluids, Rockville, MD) and maintained in 5% CO2 atmosphere at 37 C. Cells were transfected with 300 ng DNA/well in 12-well plates with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Cell culture media were changed 6 h after transfection to antibiotic-free DMEM plus 10% charcoal-dextran-treated fetal bovine serum. T3 was added to the cells 48 h later, and cells were harvested, lysed and assayed for reporter gene activity the next day using dual luciferase assay reagents according to the manufacturer’s instructions (Promega Corp., Madison, WI).
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was performed as previously described (16). Briefly, α-23 cells were grown to 90% confluence in phenol red-free DMEM supplemented with 10% charcoal DEXTRAN-stripped fetal bovine serum (FBS) for at least 3 d. After addition of 10−6 m T3 at various time intervals, ChIP assays were performed according to manufacturer’s protocol (Upstate Biotechnology) with some minor modifications. After treatment with T3, chromatin was cross-linked in 1% formaldehyde in PBS, and nuclei were extracted. Chromatin was sonicated to yield 500- to 1000-bp DNA fragments, and the supernatant containing precleared chromatin was then incubated at 0 C overnight with different antibodies or rabbit IgG control. After reverse cross-linking by heating the samples at 65 C overnight, and treating with Proteinase K, DNA was purified by QIAGEN PCR Purification Kit (QIAGEN, Chatsworth, CA) per manufacturer’s instructions. PCR was performed to visualize the enriched DNA fragments using primers that amplify the promoter region of TSHα in pGL2/TSHα Plasmid and endogenous rat PEPCK promoters were previously described (16,21). Conventional PCR signals stained with ethidium bromide in 2% agarose gels.
Real-time PCR
For quantitative real-time PCR of ChIP products, we employed the SYBR green system (Applied Biosystems) using an Applied Biosystems 7300 sequence detector. We used the same sets of primers as conventional PCR and confirmed that the set of the primers for the real-time PCR yielded a single peak in the 40-cycle procedure. Relative values (mean ± sd) normalized to input levels were compared with those obtained with IgG.
Microarrays
Triplicate samples of RNA from separate α-23 cell wells were isolated, labeled, hybridized, and scanned from microarrays according to the manufaturer’s protocol (Illumina Inc., San Diego, CA). The Illumina Rat Ref-12 expression BeadChip was used, which contains more than 22,000 probes selected primarily from the NCBI RefSeq database. Expressed genes were analyzed and normalized with the lumi and limma packages in R/Bioconductor software for data preprocessing and analysis. The vst method was used for data transformation and the rsv for normalization.
Nonexpressed genes were excluded if detected P values were greater than 0.01. Differentially expressed genes were selected if adjusted P values were less than 0.01 by BH method (44) and fold changes greater than 1.5-fold. Statistics were done with the limFit and eBayes methods in the limma package.
Supplementary Material
Footnotes
This research was supported in part by grants from the Intramural Research Program of National Institute of Child Health and Human Development, National Institutes of Health (to Y.S.) and National Cancer Institute (to P.M.) and by R01 NIDDK 1R01DK069899 (to P.M.Y.).
Disclosure Summary: The authors have nothing to disclose.
First Published Online February 5, 2009
Abbreviations: CBP, CREB-binding protein; ChIP, chromatin immunoprecipitation; CoA, coenzyme A; CREB, cAMP response element binding protein; FBS, fetal bovine serum; HAT, histone acetyl transferase; HDAC3, histone deacetylase 3; NCoR, nuclear receptor corepressor; PEPCK, phosphoenolpyruvate carboxykinase; RNA Pol II, RNA polymerase II; SMRT, silencing mediator for retinoic and thyroid hormone receptor; SRC, steroid receptor coactivator; TBL1, transducin β-like protein 1; TR, thyroid hormone receptor; TSA, trichostatin A.
References
- Zhang J, Lazar MA 2000 The mechanism of action of thyroid hormones. Annu Rev Physiol 62:439–466 [DOI] [PubMed] [Google Scholar]
- Yen PM 2001 Physiological and molecular basis of thyroid hormone action. Physiol Rev 81:1097–1142 [DOI] [PubMed] [Google Scholar]
- Tomita A, Buchholz DR, Shi YB 2004 Recruitment of N-CoR/SMRT-TBLR1 corepressor complex by unliganded thyroid hormone receptor for gene repression during frog development. Mol Cell Biol 24:3337–3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HG, Chan DW, Huang ZQ, Li J, Fondell JD, Qin J, Wong J 2003 Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. EMBO J 22:1336–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka T, Lazar MA 2003 The N-CoR/histone deacetylase 3 complex is required for repression by thyroid hormone receptor. Mol Cell Biol 23:5122–5131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KC, Li J, Cole PA, Wong J, Kraus WL 2003 Transcriptional activation by thyroid hormone receptor-β involves chromatin remodeling, histone acetylation, and synergistic stimulation by p300 and steroid receptor coactivators. Mol Endocrinol 17:908–922 [DOI] [PubMed] [Google Scholar]
- Lemon B, Inouye C, King DS, Tjian R 2001 Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature 414:924–928 [DOI] [PubMed] [Google Scholar]
- Yan Z, Cui K, Murray DM, Ling C, Xue Y, Gerstein A, Parsons R, Zhao K, Wang W 2005 PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes. Genes Dev 19:1662–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fondell JD, Guermah M, Malik S, Roeder RG 1999 Thyroid hormone receptor-associated proteins and general positive cofactors mediate thyroid hormone receptor function in the absence of the TATA box-binding protein-associated factors of TFIID. Proc Natl Acad Sci USA 96:1959–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachez C, Freedman LP 2001 Mediator complexes and transcription. Curr Opin Cell Biol 13:274–280 [DOI] [PubMed] [Google Scholar]
- Park SW, Li G, Lin YP, Barrero MJ, Ge K, Roeder RG, Wei LN 2005 Thyroid hormone-induced juxtaposition of regulatory elements/factors and chromatin remodeling of Crabp1 dependent on MED1/TRAP220. Mol Cell 19:643–653 [DOI] [PubMed] [Google Scholar]
- Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M 2000 Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103:843–852 [DOI] [PubMed] [Google Scholar]
- Burakov D, Crofts LA, Chang CP, Freedman LP 2002 Reciprocal recruitment of DRIP/mediator and p160 coactivator complexes in vivo by estrogen receptor. J Biol Chem 277:14359–14362 [DOI] [PubMed] [Google Scholar]
- Reid G, Hübner MR, Métivier R, Brand H, Denger S, Manu D, Beaudouin J, Ellenberg J, Gannon F 2003 Cyclic, proteasome-mediated turnover of unliganded and liganded ERα on responsive promoters is an integral feature of estrogen signaling. Mol Cell 11:695–707 [DOI] [PubMed] [Google Scholar]
- Sharma D, Fondell JD 2002 Ordered recruitment of histone acetyltransferases and the TRAP/Mediator complex to thyroid hormone-responsive promoters in vivo. Proc Natl Acad Sci USA 99:7934–7939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Xia X, Fondell JD, Yen PM 2006 Thyroid hormone-regulated target genes have distinct patterns of coactivator recruitment and histone acetylation. Mol Endocrinol 20:483–490 [DOI] [PubMed] [Google Scholar]
- Hollenberg AN, Monden T, Wondisford FE 1995 Ligand-independent and -dependent functions of thyroid hormone receptor isoforms depend upon their distinct amino terminal. J Biol Chem 270:14272–14280 [DOI] [PubMed] [Google Scholar]
- Chatterjee VK, Lee JK, Rentoumis A, Jameson JL 1989 Negative regulation of the thyroid-stimulating hormone α gene by thyroid hormone: receptor interaction adjacent to the TATA box. Proc Natl Acad Sci USA 86:9114–9118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenner DL, Mroczynski MA, Weintraub BD, Radovick S, Wondisford FE 1991 A detailed functional and structural analysis of a major thyroid hormone inhibitory element in the human thyrotropin β-subunit gene. J Biol Chem 266:21666–21673 [PubMed] [Google Scholar]
- Tagami T, Madison LD, Nagaya T, Jameson JL 1997 Nuclear receptor corepressors activate rather than suppress basal transcription of genes that are negatively regulated by thyroid hormone. Mol Cell Biol 17:2642–2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami T, Park Y, Jameson JL 1999 Mechanisms that mediate negative regulation of the thyroid-stimulating hormone α gene by the thyroid hormone receptor. J Biol Chem 274:22345–22353 [DOI] [PubMed] [Google Scholar]
- Sasaki S, Lesoon-Wood LA, Dey A, Kuwata T, Weintraub BD, Humphrey G, Yang WM, Seto E, Yen PM, Howard BH, Ozato K 1999 Ligand-induced recruitment of a histone deacetylase in the negative-feedback regulation of the thyrotropin β gene. EMBO J 18:5389–5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen PM, Feng X, Flamant F, Chen Y, Walker RL, Weiss RE, Chassande O, Samarut J, Refetoff S, Meltzer PS 2003 Effects of ligand and thyroid hormone receptor isoforms on hepatic gene expression profiles of thyroid hormone receptor knockout mice. EMBO Rep 4:581–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison LD, Ahlquist JA, Rogers SD, Jameson JL 1993 Negative regulation of the glycoprotein hormone α gene promoter by thyroid hormone: mutagenesis of a proximal receptor binding site preserves transcriptional repression. Mol Cell Endocrinol 94:129–136 [DOI] [PubMed] [Google Scholar]
- Buchholz DR, Tomita A, Fu L, Paul BD, Shi YB 2004 Transgenic analysis reveals that thyroid hormone receptor is sufficient to mediate the thyroid hormone signal in frog metamorphosis. Mol Cell Biol 24:9026–9037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treuter E, Albrektsen T, Johansson L, Leers J, Gustafsson JA 1998 A regulatory role for RIP140 in nuclear receptor activation. Mol Endocrinol 12:864–881 [DOI] [PubMed] [Google Scholar]
- Augereau P, Badia E, Balaguer P, Carascossa S, Castet A, Jalaguier S, Cavaillès V 2006 Negative regulation of hormone signaling by RIP140. J Steroid Biochem Mol Biol 102:51–59 [DOI] [PubMed] [Google Scholar]
- Kiskinis E, Hallberg M, Christian M, Olofsson M, Dilworth SM, White R, Parker MG 2007 RIP140 directs histone and DNA methylation to silence Ucp1 expression in white adipocytes. EMBO J 26:4831–4840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MA, Ricci AR, Deroo BJ, Archer TK 2002 The histone deacetylase inhibitor trichostatin A blocks progesterone receptor-mediated transactivation of the mouse mammary tumor virus promoter in vivo. J Biol Chem 277:15171–15181 [DOI] [PubMed] [Google Scholar]
- Yoon HG, Choi Y, Cole PA, Wong J 2005 Reading and function of a histone code involved in targeting corepressor complexes for repression. Mol Cell Biol 25:324–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang J, Nawaz Z, Liu JM, Qin J, Wong J 2000 Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J 19:4342–4350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenberg AN, Monden T, Madura JP, Lee K, Wondisford FE 1996 Function of nuclear co-repressor protein on thyroid hormone response elemets is regulated by the receptor A/B domain. J Biol Chem 271:28516–28520 [DOI] [PubMed] [Google Scholar]
- Kim SW, Ho SC, Hong SJ, Kim KM, So EC, Christoffolete M, Harney JW 2005 A novel mechanism of thyroid hormone-dependent negative regulation by thyroid hormone receptor, nuclear receptor corepressor (NCoR), and GAGA-binding factor on the rat cD44 promoter. J Biol Chem 280:14545–14555 [DOI] [PubMed] [Google Scholar]
- Feng X, Jiang Y, Meltzer P, Yen PM 2001 Transgenic targeting of a dominant negative corepressor to liver blocks basal repression by the thyroid hormone receptor and increases cell proliferation. J Biol Chem 276:15066–15072 [DOI] [PubMed] [Google Scholar]
- Berger SL 2007 The complex language of chromatin regulation during transcription. Nature 447:407–412 [DOI] [PubMed] [Google Scholar]
- Weiss RE, Xu J, Ning G, Pohlenz J, O'Malley BW, Refetoff S 1999 Mice deficient in the steroid receptor co-activator 1 (SRC-1) are resistant to thyroid hormone. EMBO J 18:1900–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiga-Carvalho TM, Shibusawa N, Nikrodhanond A, Oliveira KJ, Machado DS, Liao XH, Cohen RN, Refetoff S, Wondisford FE 2005 Negative regulation by thyroid hormone receptor requires an intact coactivator-binding surface. J Clin Invest 115:2517–2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu TK, Palhan VB, Wang Z, An W, Cole PA, Roeder RG 2000 Activator-dependent transcription from chromatin in vitro involving targeted histone acetylation by p300. Mol Cell 6:551–561 [DOI] [PubMed] [Google Scholar]
- Ishii S, Yamada M, Satoh T, Monden T, Hashimoto K, Shibusawa N, Onigata K, Morikawa A, Mori M 2004 Aberrant dynamics of histone deacetylation at the thyrotropin-releasing hormone gene in resistance to thyroid hormone. Mol Endocrinol 18:1708–1720 [DOI] [PubMed] [Google Scholar]
- Wondisford FE, Steinfelder HJ, Nations M, Radovick S 1993 AP-1 antagonizes thyroid hormone receptor action on thyrotropin β-subunit gene. J Biol Chem 268:2749–2754 [PubMed] [Google Scholar]
- Méndez-Pertuz M, Sánchez-Pacheco A, Aranda A 2003 The thyroid hormone receptor antagonizes CREB-mediated transcription. EMBO J 22:3102–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K, Matsushita A, Sasaki S, Misawa H, Nishiyama K, Kashiwabara Y, Nakamura H 2004 Thyroid-hormone-dependent negative regulation of thyrotropin β gene by thyroid hormone receptors: study with a new experimental system using CV1 cells. Biochem J 378:549–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collingwood TN, Urnov FD, Chatterjee VK, Wolffe AP 2001 Chromatin remodeling by the thyroid hormone receptor in regulation of the thyroid-stimulating hormone α-subunit promoter. J Biol Chem 276:34227–34234 [DOI] [PubMed] [Google Scholar]
- Hochberg Y, Benjamini Y 1990 More powerful procedures for multiple significance testing. Stat Med 9:811–818 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.