Abstract
Natural killer (NK) cells contribute to the essential functions of innate immunity and reproduction. Various genes encode NK cell receptors that recognize the major histocompatibility complex (MHC) Class I molecules expressed by other cells. For primate NK cells, the killer-cell immunoglobulin-like receptors (KIR) are a variable and rapidly evolving family of MHC Class I receptors. Studied here is KIR3DL1/S1, which encodes receptors for highly polymorphic human HLA-A and -B and comprises three ancient allelic lineages that have been preserved by balancing selection throughout human evolution. While the 3DS1 lineage of activating receptors has been conserved, the two 3DL1 lineages of inhibitory receptors were diversified through inter-lineage recombination with each other and with 3DS1. Prominent targets for recombination were D0-domain polymorphisms, which modulate enhancer function, and dimorphism at position 283 in the D2 domain, which influences inhibitory function. In African populations, unequal crossing over between the 3DL1 and 3DL2 genes produced a deleted KIR haplotype in which the telomeric “half” was reduced to a single fusion gene with functional properties distinct from its 3DL1 and 3DL2 parents. Conversely, in Eurasian populations, duplication of the KIR3DL1/S1 locus by unequal crossing over has enabled individuals to carry and express alleles of all three KIR3DL1/S1 lineages. These results demonstrate how meiotic recombination combines with an ancient, preserved diversity to create new KIR phenotypes upon which natural selection acts. A consequence of such recombination is to blur the distinction between alleles and loci in the rapidly evolving human KIR gene family.
Among the most polymorphic and structurally diverse human loci are genes related to immune function (Redon et al. 2006; Frazer et al. 2007; Korbel et al. 2007). A principle example is the KIR locus, which displays both polymorphic and structural diversity throughout all human populations (Parham 2005; Bashirova et al. 2006). The protein products, the killer cell immunoglobulin-like receptors (KIR), recognize determinants of conserved and polymorphic major histocompatibility complex (MHC) Class I molecules (Boyington et al. 2001). Interaction of KIR on immune-system cells with MHC Class I on other cell types allows the health of tissues to be monitored and responded to when compromised by infection or malignant transformation. In the human MHC, the HLA complex, each of the highly polymorphic Class I genes—HLA-A, HLA-B, and HLA-C—has some alleles that encode KIR ligands. KIR are principally found on the surface of natural killer (NK) cells, lymphocytes that function in the early, or innate, immune response to virus infection (Lanier 2008), but they also contribute to an early stage of reproduction, when they remodel the maternal blood vessels that will supply the placenta and nourish the fetus (Moffett and Loke 2006). KIR are also expressed on some T-lymphocytes, cells that are central to the adaptive immune response to infection (Snyder et al. 2004; Moretta et al. 2006).
In humans and other primates, the KIR are encoded by a diverse and rapidly evolving gene family that exhibits considerable species specificity due to continual gene turnover (Parham 2005; Bashirova et al. 2006). In contrast, in mice, the widely used animal model in immunology, the KIR genes are few in number (two) and do not encode NK cell receptors for MHC Class I, those functions having been assumed by the independently evolved KLRA1 (also known as Ly49) receptors (Kelley et al. 2005). This lability and plasticity in genes encoding NK cell receptors likely reflects the strengths of the different and sometimes conflicting selections imposed by the needs of immune defense and placental reproduction, but also by the functional and genetic complexity of matching polymorphic ligands and receptors encoded by unlinked genes (Parham 2005; Moffett and Loke 2006; Lanier 2008).
The KIR locus is part of the leukocyte receptor complex (LRC) on human chromosome 19, which comprises several families of cell-surface receptors expressed by cells of the immune system (Wilson et al. 2000). The KIR genes are flanked on the centromeric side by the leukocyte immunoglobulin-like receptor (LILR) gene family and on the telomeric side by FCAR, the gene encoding the receptor for immunoglobulin A that is expressed on phagocytic cells. Human KIR haplotypes vary in gene content, having between seven and 15 genes (Uhrberg et al. 1997). Each KIR haplotype is divided into two parts by three conserved framework regions. The centromeric part contains KIR2D genes encoding HLA-C receptors, and the telomeric part contains KIR3D genes encoding HLA-A and -B receptors (Bashirova et al. 2006). The latter two genes, comprising KIR3DL1/S1 and KIR3DL2, are the subject of this study. KIR3DL1/S1 recognizes sequence motifs at residues 77–83 of HLA-A and HLA-B allotypes that form the Bw4 antigen, or epitope, defined by HLA serology (Cella et al. 1994; Gumperz et al. 1995; Thananchai et al. 2007), and KIR3DL2 recognizes HLA-A3 and –A11 (Dohring et al. 1996; Pende et al. 1996).
Like HLA-A and -B, KIR3DL1/S1 is a highly polymorphic protein with more than 60 allotypes defined (Robinson et al. 2006). The natural variation affects receptor function by altering the frequency of cellular expression, abundance at the cell surface (Pando et al. 2003; Thomas et al. 2008), avidity and specificity for ligand (O'Connor et al. 2007; Thananchai et al. 2007), and the nature—either inhibitory or activating—of the intracellular signals generated upon ligand engagement (Carr et al. 2007). Furthermore, a person's combination of HLA-A, HLA-B, and KIR3DL1/S1 allotypes influences the development and function of the NK cell repertoire (Foley et al. 2008; Yawata et al. 2008), and in the population, such combinations are associated with disease susceptibility and progression, notably for HIV infection (Martin et al. 2007). The foundation for 3DL1/S1 variety is three ancient lineages of alleles—3DS1 lineage encoding activating receptors and 3DL1-005 and 015 lineages encoding inhibitory receptors—maintained by balancing selection for >3 million years and present in all modern human populations (Norman et al. 2007). Of the three lineages, 3DS1 is essentially homogeneous, whereas both 3DL1 lineages have been extensively diversified by point mutation and recombination. Because recombination with other KIR genes and between KIR3DL1/S1 lineages has the potential to erode the lineage distinctions, we examined the impact that meiotic recombination has had on the KIR3DL1/S1 locus and on human NK cell functional diversity.
Results
Generation of KIR3DL1/S1 diversity by intergenic recombination
In humans, the hominoid KIR lineage II is represented by two genes: 3DL1/S1 encoding NK cell receptors for the Bw4 epitopes of HLA-A and HLA-B; and 3DL2 encoding NK-cell receptors specific for HLA-A*03 and HLA-A*11 (Rajalingam et al. 2004). Not fitting with this picture is the 3DL1/2v cDNA, which encodes extracellular domains like 3DL1 and intracellular domains like 3DL2 (Shilling et al. 2002). To distinguish if the 3DL1/2v cDNA arises from transcription of a single gene or the splicing together of transcripts from both 3DL1 and 3DL2, we analyzed genomic DNA from three healthy donors having different 3DL1/2v variants and one 3DL1/2v donor who lacked 3DL1/S1 because of deletion of this locus from the other KIR haplotype (Norman et al. 2004). The results unequivocally demonstrated that 3DL1/2v represents a unique hybrid gene for which exons 1–5 and associated introns are like 3DL1, and exons 6–9 and associated introns are like 3DL2 (Fig. 1A, upper haplotype). In all four donors, the 3DL1/2v gene was shown to be flanked by 2DL4 on the upstream (centromeric) side and by FCAR on the downstream (telomeric) side. This gene organization is unusual, differing from the more common situation (Wilson et al. 2000) where 3DL1 is downstream from 2DL4, 3DL2 is upstream of FCAR, and 2DS4 lies between 3DL1 and 3DL2 (Fig. 1A, lower haplotype). These results raised the possibility that 3DL1/2v arose through a non-homologous recombination between 3DL1 and 3DL2 that deleted the 3′ part of 3DL1, the entire 2DS4 gene, and the 5′ part of 3DL2. If true, then individuals carrying 3DL1/2v should never be heterozygous for exons 1–5 of 3DL2 or exons 6–9 of 3DL1/S1. Population analysis was performed to test this hypothesis.
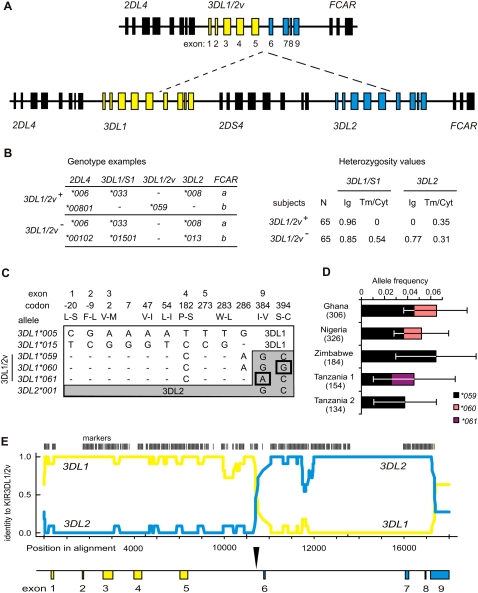
Figure 1.
The 3DL1/2v fusion gene is allelic to 3DL1/S1 and 3DL2. (A, top) Schematic of the four 3DL1/2v haplotypes that were sequenced here (Supplemental Fig. S2). (Bottom) A KIR A haplotype; (dashed lines) the genomic segment absent from 3DL1/2v haplotypes; (yellow) 3DL1; (blue) 3DL2. Exons 1–5 encode the leader peptide and Ig domains (Ig), exon 6 the stalk, and exons 7–9 the transmembrane and cytoplasmic domains (Tm/Cyt). (B, left) Representative genotypes from the group of 65 subjects who carry 3DL1/2v (3DL1/2v+) and the group of 65 who do not (3DL1/2v−). (Right) Comparisons of heterozygosity observed in the 3DL1/2v+ and 3DL1/2v− groups from segments of 3DL1 and 3DL2. All of the 3DL1/2v+ subjects are hemizygous for exons 6–9 of 3DL1/S1 and exons 1–5 of 3DL2, which corresponds to the portion absent from the KIR A haplotype. This shows the 3DL1/2v+ subjects have 3DL1 and 3DL2 on one haplotype and 3DL1/2v on the other. (C) Shown are the nucleotide differences in exons 1–5 that distinguish 3DL1/2v (3DL1*059, 3DL1*060, and 3DL1*061) from (white) 3DL1 and in exons 7–9 that distinguish 3DL1/2v from (gray) 3DL2. The many nucleotide differences (N = 102) that distinguish 3DL1 and 3DL2 are not shown. Shown are 3DL1*00501 and 3DL1*01502, which represent the 005 and 015 lineages of inhibitory receptors; exons 1–5 of 3DL1/2v are related to 3DL1*00501. In exons 7–9, 3DL1*059 is identical to 3DL2*001; 3DL1*060 and 3DL1*061 being distinguished by the SNPs boxed. Codons are numbered according to the mature protein, and amino acid changes are indicated by single letter code. (D) Shown are the frequencies of three 3DL1/2v alleles (3DL1*059–61) in the five sub-Saharan African populations where they were detected. The number of haplotypes examined from each population is shown in parentheses; error bars show the 95% confidence interval of the allele frequency measurements. (E) Shown is a pairwise identity plot from alignment of genomic sequences. (Yellow line) Signifies identity of 3DL1/2v with 3DL1; (blue line) the identity of 3DL1/2v with 3DL2; (vertical bars) the SNP markers used in this analysis. (Shown below by the vertical arrow) The crossover occurred in intron 5 during the interval from 386 to 356 bp upstream from exon 6. A continuous sequence trace that spans the crossover is shown in Supplemental Figure S2.
KIR3DL1/2v is restricted to sub-Saharan Africans and related populations such as African-Americans and Afro-Caribbeans, in which it is present at frequencies of up to 6.5% (Norman et al. 2007). Panels of 65 3DL1/2v+ and 65 3DL1/2v− Africans were genotyped for heterozygosity in the sequences encoding the extracellular and intracellular domains of 3DL1 and 3DL2, and also for 2DL4 as a control. Whereas the 3DL1/2v− Africans exhibited significant heterozygosity in both the 3′ and 5′ parts of the 3DL1 and 3DL2 genes, the 3DL1/2v+ Africans lacked heterozygosity in the 3′-exons of 3DL1/S1 and the 5′-exons of 3DL2 (Fig. 1B). These results, showing that 3DL1/2v is not on the same haplotypes as 3DL1 or 3DL2, are consistent with 3DL1/2v having been formed by a recombination-mediated deletion that fused exons 1–5 of 3DL1 with exons 6–9 of 3DL2. As a consequence, 3DL1/2v has an allelic relationship with both 3DL1 and 3DL2 (Supplemental Fig. S1). Because 3DL1/2v is overall more structurally similar to 3DL1 than 3DL2, the three 3DL1/2v variants were assigned names in the 3DL1/S1 series: 3DL1*059, 3DL1*060, and 3DL1*061 (Robinson et al. 2006). The encoded allotypes, which differ by one or two amino acid substitutions, have extracellular domains that are part of the 3DL1-005 lineage, as opposed to the 3DL1-015 lineage (Fig. 1C). That 3DL1*059 is geographically most widespread, being present in all six African populations studied, compared to only one and two populations for 3DL1*060 and 3DL1*061, respectively (Fig. 1D), suggests that 3DL1*059 was the progenitor 3DL1/2v from which 3DL1*060 and 3DL1*061 were independently derived by single point substitutions. Further supporting this evolutionary model, the 3DL1*059 sequence corresponds to a fusion of sequences from common 3DL1 (e.g., *00501) and 3DL2 (e.g., *001) alleles, whereas 3DL1*060 and 3DL1*061 are each distinguished by a unique and different substitution. From the genomic sequences, the recombination that fused parts of the 3DL1 and 3DL2 genes to form 3DL1/2v can be located to a 30-bp sequence within intron 5 that is located 356 bp upstream of exon 6, and 2780 bp downstream from exon 5 encoding the D2 domain (Fig. 1E; Supplemental Fig. S2).
The genomic location of 3DL1/2v between 2DL4 and FCAR is the same as that observed for Pt-KIR3DL1/2, the single chimpanzee lineage II KIR gene (Rajalingam et al. 2004; Sambrook et al. 2005), which has structural and functional properties in common with both human 3DL1 and 3DL2 (Khakoo et al. 2000). Although these similarities raised the possibility that 3DL1/2v and Pt-KIR3DL1/2 share a common origin that predated the human–chimpanzee separation, several lines of evidence point to 3DL1/2v having evolved independently by recombination during human evolution, subsequent to the split from chimpanzees. First, the inclusion of exons 1–5 of 3DL1/2v in the 005 lineage of 3DL1/S1 alleles (Fig. 1C), and not in either the 015 or 3DS1 lineages, indicates that 3DL1/2 was formed after lineage divergence, which occurred subsequent to the human–chimpanzee separation (Norman et al. 2007). Thus in phylogenetic analysis, human 3DL1/2v was never an out-group to 3DL1 when compared with Pt-KIR3DL1/2 and other hominoid KIR lineage II genes (Supplemental Fig. S3A). Second, the 3DL1/2v Ig-domain is estimated to have arisen ∼0.74 million years ago (Mya) (95% CI: 0.27–1.55 Mya) (Supplemental Fig. S3B), after the three allelic lineages of 3DL1/S1 split. As the Ig-domain divergence preceded the genomic deletion event, 3DL1/2v arose at a time potentially close to the emergence of modern humans (∼200,000 yr ago) (Relethford 2008). Lastly, our finding that all three variants of 3DL1/2v (3DL1*059, 3DL1*060, 3DL1*061) were flanked by identical sequences and 2DL4 (*00801) and FCAR (*001) alleles, despite genome-wide lack of LD in sub-Saharan Africans (Campbell and Tishkoff 2008), points to a recent origin for 3DL1/2v.
In conclusion, the results are all consistent with 3DL1/2v having been formed during a meiotic recombination in a human ancestor that deleted DNA from the telomeric region of a KIR haplotype to fuse the 5′ part of a 3DL1 allele with the 3′ part of a 3DL2 allele. Although the size of the deletion is uncertain because of gene-content variability, the most likely event would have involved the A haplotype configuration shown in Figure 1A with deletion of ∼30 kb containing all of 2DS4 and parts of 3DL1 and 3DS1. That the telomeric part of most chimpanzee KIR haplotypes has similarly, but independently, evolved to resemble that of the KIR3DL1/2v-containing haplotypes, points to this simplified structure having conferred advantage in at least two different circumstances.
Generation of KIR3DL1/S1 diversity by interlineage recombination
Domain-by-domain phylogenetic analysis of 61 3DL1/S1 alleles shows that a minimum of 12 (∼20%) alleles arose through intragenic meiotic recombination (Fig. 2). Most of these events (nine) involved recombination between two of the three different 3DL1/S1 lineages: 3DL1-005, 3DL1-015, and 3DS1. The two regions of the molecule most affected by recombination are the D0 domain (five events) and position 283 (three events) near the C-terminal end of the D2 domain (Fig. 2A). Both these regions bear a strong signature of positive selection in human populations (Norman et al. 2007) and have distinct functions that affect the strength and specificity of the KIR3DL1/S1 interaction with MHC Class I. The D1 and D2 domains form the ligand-binding site, and the D0 domain is an enhancer that modulates the strength of the ligand–receptor interaction (Khakoo et al. 2002).
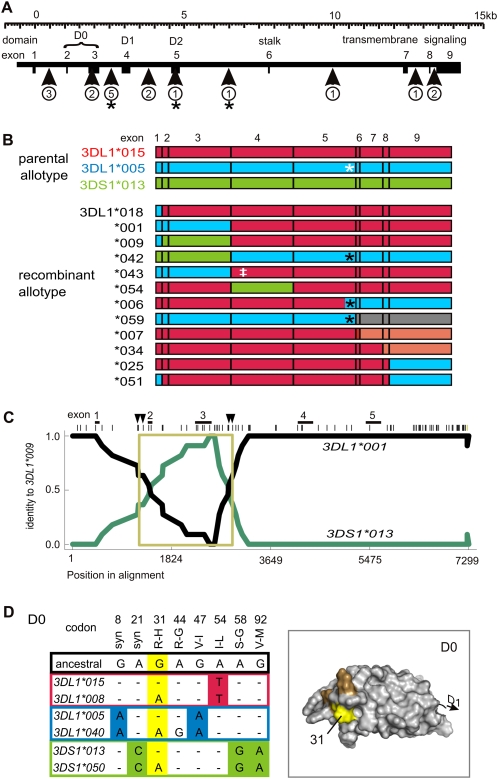
Figure 2.
Motif and domain shuffling between three lineages diversifies 3DL1/S1. (A) Shown to scale is the genomic organization of the 3DL1/S1 locus. (Boxes) Exons; exons 2–5 encode the three Ig domains, D0, D1, and D2. (Vertical arrows) The genomic regions and the number of recombination events detected from comparison of 3DL1/S1 alleles. (*) Recombination event that placed residue leucine 283 onto a different background allotype. (B) Schematic of 12 recombinant alleles of 3DL1/S1 that were identified using domain-by-domain phylogenetic analysis. The recombinant allotypes are represented by segments colored according to allelic lineage: (red) 015, (blue) 005, (green) 3DS1. The allotypes shown at the top of the panel are encoded by the most common modern alleles: 3DL1*01502, 3DL1*00501, and 3DS1*01301. For example, 3DL1*001 is identical to 3DL1*00501 in exons 1–3 and 3DL1*01501 in exons 4–9. (Gray) Recombination from another locus (3DL2); (pink) within-lineage recombinants. (White asterisk) Tryptophan–leucine substitution at residue 283; (black asterisk) recombination has replaced tryptophan at residue 283 with leucine; (‡) threonine 118 that distinguishes 3DL1*043 from 3DL1*001 and is shared with 3DL1*038. (C) The pairwise identity plot shows that 3DL1*009 formed by gene conversion. (Black line) Identity of 3DL1*009 with 3DL1*001; (green line) identity of 3DL1*009 with 3DS1*01301. The recombination included exons 2 and 3, which encode D0. (Across the top, vertical bars) SNP markers; (vertical arrows) the minimum and maximum limits of the gene conversion. Genomic sequences used for the RDP analysis include representatives of the three allelic lineages of 3DL1/S1 (listed in Methods). The 3DL1*009 cDNA sequence that was independently obtained (Middleton et al. 2007) corresponds precisely to the reading frame of the locus characterized here. (D) Shown for each of the 3DL1/S1 lineages is a pair of alleles that are dimorphic at (yellow) codon 31. Nucleotide differences in D0 are shown, and those that distinguish the three lineages are colored as in panel A. (Top) Amino acid substitutions with the ancestral residue first and the ancestral nucleotide immediately below. (Right panel) A homology model of D0; residue 31 (yellow) occurs in a patch of positively selected residues (orange) that were identified in Norman et al. (2007).
Three 3DL1/S1 allotypes likely arose by meiotic gene conversion. Genomic sequence comparison indicates that 3DL1*009 was formed by conversion between 3DL1*001 and 3DS1*01301 that involved a sequence of 1.4–1.5 kb containing exons 2 and 3 (Fig. 2C; Supplemental Fig. S4). Consequently, 3DL1*009 combines the D0 domain from 3DS1*013 with the other domains from 3DL1*001. In analogous fashion, 3DL1*054 combines the D1 domain from 3DS1*013 with the other domains from 3DL1*002 (Thomas et al. 2008). The third candidate for gene conversion involves the arginine/histidine dimorphism at position 31 in the D0 domain. Uniquely, this dimorphism has been introduced into all three 3DL1/S1 lineages, through conversions involving a maximum of 70–110 bp of DNA (Fig. 2D). Residue 31 is part of a positively selected cluster of surface residues implicated in the enhancing function of the D0 domain (Khakoo et al. 2002). In contrast to these three products of interlineage gene conversion, 3DL1*042 combines the leader peptide and D0 domain from 3DS1*013 with the other domains from 3DL1*005 and was likely the product of a simple crossover between the two alleles (Fig. 2B). Similarly, 3DL1*001 and 3DL1*043 combine a D0 domain derived from the 3DL1-005 lineage with the other domains from the 015 lineage. Although the three lineages and their distinctive differences have been maintained by balancing selection over several million years, we also see that these features have regularly been recombined to produce 3DL1/S1 allotypes with distinctive functions that presumably confer some selective advantage in the short term.
KIR3DL1/2v is an inhibitory receptor with distinctive specificity for Bw4 epitopes of HLA-A and -B
A characteristic of 3DL1/S1 gene expression is that each allele is expressed only by a fraction of NK cells, and the fraction varies with the allele (Gardiner et al. 2001; Chan et al. 2003; Trundley et al. 2007; Thomas et al. 2008). We therefore assessed cell surface expression of 3DL1/2v. The KIR3DL1-specific antibody, DX9, bound to a subset (17%) of peripheral blood NK cells from a donor heterozygous for 3DL1*059 and 3DL1*004. Because 3DL1*004 is not expressed at the cell surface (Pando et al. 2003), this DX9 binding had to be due to the expression of 3DL1*059 (Fig. 3A). Comparison of peripheral blood NK cells from different donors, and expressing different 3DL1 allotypes, showed that the level of DX9 binding to 3DL1*059 was intermediate between that observed for 3DL1*001 and 3DL1*005 (Fig. 3A). This binding hierarchy correlates with the two dimorphisms that distinguish the extracellular domains of the three allotypes. High-binding 3DL1*001 has proline 182 and tryptophan 283, whereas low-binding 3DL1*005 has serine 182 and leucine 283; 3DL1*059 is a hybrid of the two that combines proline 182 with leucine 283.
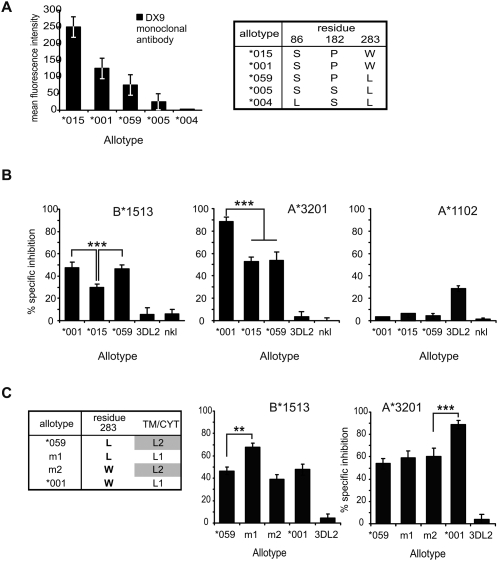
Figure 3.
3DL1/2v mediates HLA-allotype specific inhibition of NK cells. (A) Binding of the 3DL1-specific monoclonal antibody DX9 to NK cells shows that 3DL1/2v is expressed at the cell surface at intermediate level. Shown are FACS analyses of peripheral-blood NK cells from donors who express single 3DL1 allotypes. (Right) The table shows amino acid differences that correspond to the 3DL1 expression level. 3DL1*004 is not expressed because of leucine-for-serine substitution at residue 86 (Pando et al. 2003). 3DL1*015 has further differences in D0 that are shown in Figure 1. Error bars indicate the standard deviation of DX9-PE fluorescence intensity, from all of the lymphocytes that stained positive with DX9. (B,C) Results from Cr51-release cytotoxicity assays. The basis for this assay is killing of HLA Class I–deficient cells (221) by an NK cell line (NKL). Inhibition of cytotoxicity occurs if NKL is transduced with inhibitory KIR and 721.221 is transfected with cognate HLA Class I ligand. Blocking the interaction with specific antibody restores target killing to that obtained using non-transduced NKL. The specificity of inhibition was determined in replicate assays using DX9, which specifically blocks 3DL1, or DX31, which specifically blocks 3DL2. The percent specific inhibition that is shown was calculated from killing in the presence of control/specific-blocking antibody. Results are mean (±SE) of five experiments at an effector:target ratio of 20:1. (***) P < 0.001; (**) P < 0.01 from Student's t-test for comparison of means. All of the effectors killed HLA-negative targets. Each of these assays shows different combinations of KIR and ligand. (B) The degree of inhibition mediated by natural KIR allotypes; these are 3DL1*001, 3DL1*015, 3DL1/2v (3DL1*059), and 3DL2*001 or no KIR (nkl). Target cells were 721.221 cells transfected with (left) HLA-B*1513, (center) A*3201, and (right) A*1102. B*1513 and –A*3201 are ligands for 3DL1, and A*1102 is a ligand for 3DL2. (C, left) The table shows the composition of the mutant KIR allotypes. Tail-swap mutant m1 (*001-L283) has the 3DL1*059 Ig-domain with a 3DL1*001 tail. m2 (*059-W283) has the 3DL1*001 Ig-domain with a 3DL2*001 tail. Results of the cytotoxicity analysis are shown for (center) HLA-A*3201 and (right) HLA-B*1513.
We next assessed the capacity of 3DL1/2v to function as an inhibitory receptor. To do this, we used an in vitro assay in which the NKL cell line kills the HLA Class I–deficient 221 cell line. This killing can be inhibited if the NKL cells are transduced with an inhibitory KIR and the 221 target cells are transfected with a complementary HLA Class I ligand (Gumperz et al. 1995). Transduction of the NKL cell line with 3DL1*059 cDNA gave an NK cell for which 3DL1*059 (3DL1/2v) is the only KIR expressed at the cell surface. The specificity and function of 3DL1*059 were examined in cytotoxicity assays and compared to the 3DL1 and 3DL2 “parental” receptors. NKL cells expressing 3DL1*059, 3DL1*001, or 3DL1*01502 killed HLA Class I–deficient 221 cells as effectively as untransfected NKL cells. When transfected 221 cells expressing Bw4+ HLA-B*1513 were used as the target, reduced lysis was observed for NKL cells expressing 3DL1*059, 3DL1*001, or 3DL1*015, compared to lysis by NKL cells that express no KIR or KIR3DL2 that has no affinity for HLA-B*1513 (Fig. 3B, left). These inhibitory effects were abrogated by the inclusion of either anti-3DL1 (DX9) (Fig. 3B) or anti-HLA Class I (DX17) (data not shown) monoclonal antibodies, showing that the inhibition observed for the 3DL1-transduced NKL cells was dependent on the combination of 3DL1 on the NK cell and HLA-B*1513 on the target cell.
The inhibition mediated by the interaction of B*1513 with 3DL1*059 was comparable to that observed for 3DL1*001 and greater than that seen for 3DL1*015. Thus, the Bw4 epitope of HLA-B is demonstrated to serve as a ligand for 3DL1*059. Similar experiments showed that interaction of 3DL1*059 with the Bw4+ HLA-A*3201 allotype also inhibited the lysis of transfected 221 cells, to an extent comparable to 3DL1*015, but less than 3DL1*001 (Fig. 3B, center). In contrast, the Bw4− HLA-A*1102 allotype did not engage 3DL1, but interacted with 3DL2*001 to inhibit the lysis of 221 cells (Fig. 3B, right), in a manner consistent with its HLA-A3, HLA-A11 specificity (Dohring et al. 1996; Pende et al. 1996; Hansasuta et al. 2004).
In summary, these results demonstrate that 3DL1*059 is a functional, inhibitory cell-surface receptor with a specificity for Bw4+ HLA Class I that is characteristic of KIR3DL1. The unusual 3DL2-like signaling domain of 3DL1*059 (Fig. 4) generates inhibitory signals that are within the range of those obtained from conventional 3DL1 signaling domains, but, as expected, does not skew the specificity of 3DL1*059 toward the HLA-A*1102 recognized by KIR3DL2. Distinguishing 3DL1*059 from the other 3DL1 allotypes examined here is the similar strength of the inhibition resulting from interactions with Bw4 epitopes on HLA-A and HLA-B “backgrounds” (Fig. 3B).
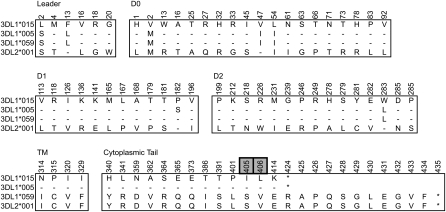
Figure 4.
KIR3DL1 and KIR3DL2 differ at 68 amino acid residues spread throughout the polypeptide. Shown are the amino acid differences that distinguish 3DL1, 3DL2, and 3DL1/2v (3DL1*059). (Gray boxes) Differences that occur in the second ITIM motif of the cytoplasmic tail. Residue numbers are those of the mature protein. The 3DL1*059 sequence that was independently obtained from cDNA (Shilling et al. 2002) is identical to the amino acid sequence encoded by the 3DL1/2v gene characterized here (Fig. 1).
KIR3DL1/2v generates weaker inhibition than KIR3DL1
To examine further the effect of the 3DL2-like transmembrane region and cytoplasmic tail of 3DL1*059, we made a mutant (m1) in which they were replaced by the homologous domains of 3DL1*001. Reciprocally, a second mutant (m2) paired the extracellular domains of 3DL1*001 with the transmembrane region and cytoplasmic tail of 3DL1*059 (Fig. 3C, left). NKL transductants expressing these two mutants and their natural allotypes were compared in cytotoxicity assays against 221 target cells and 221 transfectants expressing A*3201 or B*1513. This analysis showed that the combination of 3DL1*059 and B*1513 gave a weaker inhibition than the combination of m1 and B*1513 (Fig. 3C, center). A similar trend was seen when the ligand was A*3201, but the difference was much smaller (Fig. 3C, right). For the reciprocal pair, 3DL1*001 gave a stronger inhibition than m2, a difference that was greater with A*3201 as the ligand than with B*1513. These results show that in all combinations, the trend is to weaker inhibition when the transmembrane region and cytoplasmic domain are derived from 3DL2. Thus another potential selective advantage of 3DL1/2v is its capacity to generate a weaker inhibitory signal than its progenitor 3DL1 allotype.
Because 3DL1*059 and m2 only differ at position 283, as do 3DL1*001 and m1, this mutational analysis also showed how the dimorphism at position 283 affects 3DL1 function. Substitution of tryptophan for leucine at position 283 increased the inhibitory signal on binding to A*3201 but decreased the response to B*1513 (Fig. 3C). For 3DL1*059, the trend was similar—tryptophan 283 favored interaction with A*3201, leucine 283 its interaction with B*1513—but the difference was smaller. Thus a potential advantage of the recombinations that change residue 283 is that they can increase the compatibility of 3DL1/S1 with the particular Bw4-bearing HLA-A and HLA-B allotypes present in a population.
Gene duplication by unequal crossing over allows individuals to carry all three 3DL1/S1 lineages
The three lineages of 3DL1/S1 alleles (3DL1-005, 3DL1-015, and 3DS1) have been maintained by balancing selection for more than 3 million years (Norman et al. 2007). However, most individuals carry only one or two of the three lineages. Family studies identified a KIR haplotype in Europeans that contains both 3DS1 and 3DL1 and likely was formed by unequal crossing over, which duplicated both the KIR2DL4 and 3DLl/S1 genes (Martin et al. 2003; Williams et al. 2003; Gomez-Lozano et al. 2005). The potential benefit to such duplication haplotypes is that they permit some individuals to carry all three 3DL1/S1 allelic lineages. To assess the prevalence of the duplication haplotype, we used a newly developed pyrosequencing method that detects 3DL1/S1 copy-number variation (Fig. 5) to study a panel of 2800 ethnically diverse blood donors. Eighty individuals (2.9%) were demonstrated to have a KIR haplotype with two copies of 3DL1/S1 (Fig. 6A). By genotyping these individuals for 2DL4, 3DL1/S1, and 3DL2 alleles, we defined seven different duplication KIR haplotypes (Fig. 6B). Each duplication haplotype was differentially distributed; as a group, they are most common in South and East Asians, with frequencies up to 10%, less common in Caucasians, rare in Africans, and absent from Amerindians. Among the 80 individuals carrying a duplication haplotype, 23 of them (28.8%) carried one allele from each of the three KIR3DL1/S1 lineages.
Figure 5.
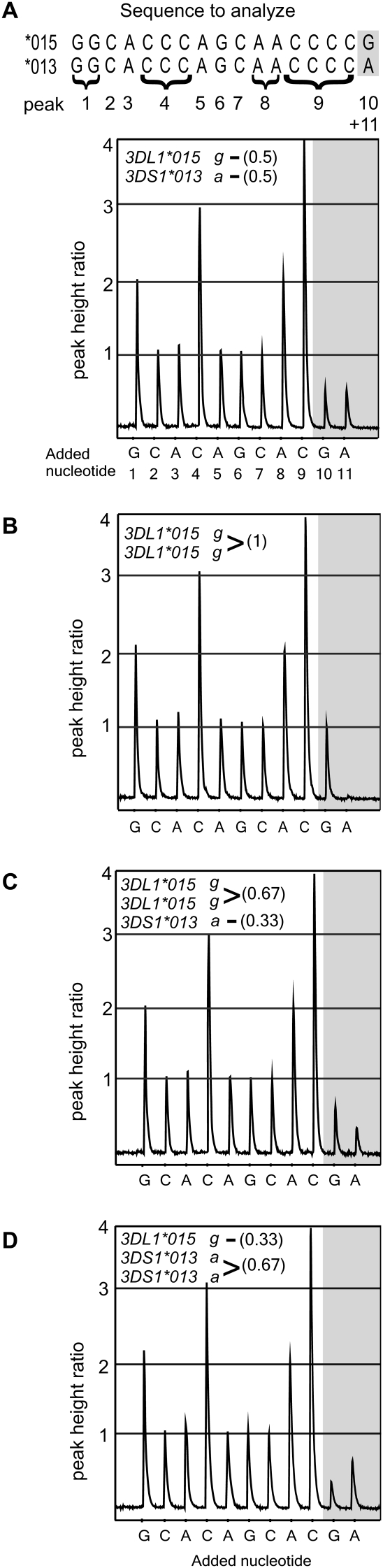
Simultaneous detection of KIR3DL1/S1 polymorphism and copy-number using pyrosequencing. (A, top) The diploid sequence of a 17-bp fragment of exon 3 from an individual heterozygous for 3DL1*01502 and 3DS1*01301. (Gray) SNP g336a. Underneath is a pyrogram obtained from the same individual. To generate the pyrogram, nucleotides were added to a single-strand template in the sequence shown, 1–11, and correspond to peaks of the same number. The peak height is proportional to the quantity of nucleotides that were incorporated, as shown by peak 1 (the sequence is gg), which is twice the height of peak 6 (g). Each peak is a sum of the haplotypes present, so that the monomorphic positions (relative peak height = 1) are used for calibration. The combined peak height at the heterozygous position shown (peaks 10 + 11) is equal to the peak from a single monomorphic position. Shown are four different pyrograms; (top left of each diagram) the derived genotype; (brackets) the peak-height ratio compared with the single monomorphic peak. (A) At SNP g336a, there is one peak for g and one for a (peaks 10 and 11), and each peak is half the height of a single peak (peak 6). This individual has one copy of 3DL1*01502 and one copy of 3DS1*01301 (0.5g: 0.5a). (B) At SNP g336a, there is one peak for g (peak 10) that is the same height as a single monomorphic position (peak 6) and no peak on addition of nucleotide a-11. This individual is homozygous, having two copies of 3DL1*01502 (1g: 0a). (C) There are two peaks as for A, but g is twice the height of a (0.67g: 0.33a), and their sum is the same as peak 6. This individual has two copies of 3DL1*01502 and one 3DS1*01301. (D) This individual has one copy of 3DL1*01502 and two of 3DS1*01301 (0.33g: 0.67a).
Figure 6.
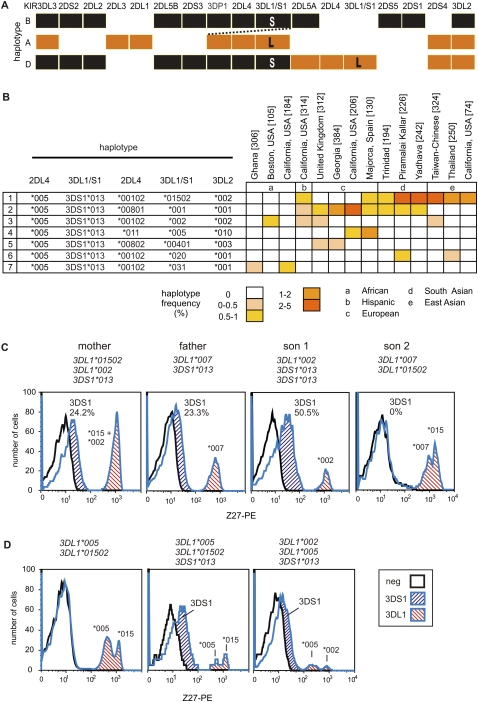
Duplicating KIR loci diversifies the NK cell repertoire in quantity and quality. (A) Schematic of the donor haplotypes and duplication haplotype. (Across the top) The KIR loci; (shaded boxes) indicate presence of the locus. The donor KIR haplotypes B (black) and A (orange); the composite duplication haplotype D is colored accordingly. (Dotted line) Non-allelic homologous recombination that was mediated by sequence similarity in the 5′-regions of 3DP1 and 2DL5A (Martin et al. 2003; Gomez-Lozano et al. 2005). (B) The seven different 2DL4, 3DL1, and 3DL2 haplotypes deduced from analysis of 80 individuals who have the duplication, and their haplotype frequencies in the populations where they were detected. The number of haplotypes analyzed from each population is indicated in parentheses. Georgia is the country. The Piramalai Kaller and the Yadheva are two distinct populations from Tamil Nadu in Southern India. (C) Shown are FACS analyses of freshly isolated 3DL1/S1-expressing NK cells from a European family with a duplicate haplotype. (At the top of each plot) The pyrosequencing genotype. Son 1 inherited 3DS1*013 and 3DL1*002 (haplotype 3, panel B) from the mother and 3DS1*013 from the father. Monoclonal antibody Z27 discriminates 3DL1/S1 allotypes, having high (3DL1*002 and 3DL1*015), low (3DL1*005 and 3DL1*007), and very low (3DS1*013) staining patterns. Showing that 3DL1/S1 expression depends on the number of copies of 3DS1 present, Son 1 has twice the number of 3DS1+ NK cells as the mother or father. Son 2 has inherited the opposite pair of haplotypes to Son 1, and expresses two distinct populations of high (*015) and low (*007) 3DL1 staining and no 3DS1. Further differences among allotypes are quantitative, with *015 being expressed by more cells than *002 (Chan et al. 2003), as observed here by comparing 3DL1 expression of the mother and Son 1. (D) Three unrelated individuals chosen for their 3DL1/S1 genotype. (Left) An individual heterozygous for 3DL1*015 and 3DL1*005 is shown as a negative control for the very low-staining 3DS1*013 peak. (Center and right) Two individuals who both express all three lineages of 3DL1/S1 receptors; these FACS analyses are from NK cells that had been stored frozen. A comparison of fresh and frozen NK cell staining is shown in Supplemental Figure S6.
All seven duplicated haplotypes have the same general structure, containing two copies of 2DL4 and 3DL1/S1 (Supplemental Fig. S5). Common to the seven haplotypes are the 2DL4*005 and 3DS1*01301 alleles; these are associated with three other 2DL4 alleles, four 3DL2 alleles, and seven different 3DL1 alleles embracing both 3DL1 lineages and their full ranges of functional diversity. Each duplication haplotype contains a pair of 3DL1 and 3DL2 alleles that are common and in linkage disequilibrium in the populations where the duplication haplotypes are present (Norman et al. 2004; Middleton et al. 2007). This property, combined with the constancy of 2DL4*005 and 3DS1*01301, supports an evolutionary model in which all the duplication haplotypes originated with a single event of unequal crossing over (Fig. 6A), which was subsequently followed by six independent events of homologous recombination that diversified the 3DL1 and 3DL2 genes of the duplication haplotype. The restricted geographical distribution of the duplication haplotypes, largely outside of Africa, and the linkage disequilibrium of their 3DL1 and 3DL2 genes point to the emergence of the duplication haplotypes during the last 60,000 yr, and since humans left Africa to populate Europe and Asia (Campbell and Tishkoff 2008).
We identified a family in which a son inherited a duplicated haplotype containing 3DS1*01301 and 3DL1*002 from his mother. He inherited a second copy of 3DS1*01301 from his father (Fig. 6C). Using the monoclonal antibodies DX9, specific for KIR3DL1, and Z27 that binds much more strongly to 3DL1 than to 3DS1 (O'Connor et al. 2007; Trundley et al. 2007), we determined 3DL1/S1 expression by the peripheral blood NK cells of members of this family. The frequency of NK cells expressing 3DS1 in the son (50.5%) was more than the sum (47.5%) of those cells in the mother (24.2%) and father (23.3%) (Fig. 6C). This indicates that both 3DS1*013 alleles inherited by the son are expressed by NK cells. The son also expressed 3DL1*002 on 6.8% of his NK cells, consistent with previous observation that this allele is usually expressed at low frequency by NK cells (Chan et al. 2003). Thus, each of the son's three KIR3DL1/S1 alleles is expressed by NK cells. A second son, who inherited two different KIR haplotypes from the parents, had no 3DS1-expressing NK cells and exhibited a bimodal distribution of cells with high-expressing 3DL1*015 inherited from the mother and low-expressing 3DL1*007 inherited from the father (Fig. 6C). Although the mother's NK cells showed a high expression of 3DL1, we could not ascertain whether both 3DL1*002 and 3DL1*1502 were expressed, because they cannot be distinguished using available monoclonal antibodies. Two unrelated donors carrying alleles of each 3DL1/S1 lineage—3DS1*01301 (3DS1 lineage), 3DL1*00501 (005 lineage) and 3DL1*002 or 3DL*01502 (015 lineage)—were therefore identified. Because 3DL1*005 and 3DL1*015 are readily distinguished by their low and high binding to Z27, respectively, we could demonstrate that all three alleles were expressed by subsets of NK cells in these individuals (Fig. 6D). In conclusion, individuals carrying three 3DL1/S1 alleles representing the three allelic lineages express all three allotypes as NK cell receptors.
Discussion
NK cells provide an early defense against infection, particularly viral infections (Lanier 2008), and at an early stage in reproduction, they remodel maternal blood vessels that provide the fetus with nutrients via the placenta (Moffett and Loke 2006). In performing these functions, NK cell receptors have become highly diverse, rapidly evolving and largely species-specific (Kelley et al. 2005). Unlike the B-cell and T-cell receptors that diversify function through somatic recombination and mutation, NK cell receptors can only diversify through meiotic mutation and recombination. And in general, immune system genes have become more diversified by meiotic recombination than other functional types of human gene (Redon et al. 2006; Frazer et al. 2007) and display greater variation as assessed by copy number and segmental insertion/deletions (Dumas et al. 2007; Korbel et al. 2007).
The state of the killer cell immunoglobulin-like receptor (KIR) locus in the leukocyte receptor complex (LRC) varies dramatically between species. In some, like the seals, it is a single conserved and nonpolymorphic gene, and in others, such as the dog, it appears to have been deleted (Hammond et al. 2009). In contrast, in the higher primates, the KIR locus has evolved to become a diverse multigene family encoding MHC Class I receptors (Khakoo et al. 2000; Rajalingam et al. 2004; Sambrook et al. 2005; Guethlein et al. 2007). On the basis of current information, KIR diversity appears greatest in the human species, where the combined effects of gene-content diversity and allelic polymorphism are such that unrelated individuals have distinct KIR genotypes (Shilling et al. 2002). The extent of KIR genotype variability approaches that of HLA class I, with the functionally important combination of KIR and HLA genotypes being even more diverse (Parham 2005). Unequal crossing over is the likely mechanism that expanded the primate KIR locus from its origin as a single-copy gene, and it is clearly implicated in the diversification of gene content in the modern human KIR haplotypes (Khakoo et al. 2000; Wilson et al. 2000; Martin et al. 2003). Further diversity is generated by homologous recombination, in particular, by the reassortment of allelic and gene-content motifs in the centromeric and telomeric parts of the locus (Wilson et al. 2000; Norman et al. 2004; Parham 2005). Here we have explored the role that meiotic recombination has played in the diversification of an individual human KIR gene, KIR3DL1/S1. This gene encodes highly polymorphic NK cell receptors that recognize HLA-A and HLA-B, the most polymorphic MHC genes.
Balancing selection and coevolution with HLA are consistently recognized features of the KIR (Hiby et al. 2004; Norman et al. 2004; Gendzekhadze et al. 2006; Single et al. 2007; Yawata et al. 2008). One of the strongest indicators for balancing selection is the long-term preservation of alleles without fixation or loss (Kimura and Ota 1969); another is the retention of alleles through periods of restricted population size. The 3DL1/S1 locus comprises three ancient lineages of alleles that have been maintained by balancing selection over more than 3 million years and are present in all modern human populations (Norman et al. 2007), despite severe bottlenecks for some (Hey 2005). Contrasting with the conservation of the 3DS1 lineage, the 3DL1-005 and 3DL1-015 lineages of inhibitory receptors are now highly diverse. Of the approximately 60 alleles defined, 12 are recombinants, most of which have been formed by recombination or conversion between alleles from different lineages. Two main targets for recombination have been polymorphic motifs in the D0 domain, the enhancer that modulates the strength of the binding site formed by the D1 and D2 domains (Khakoo et al. 2002) and a dimorphism at position 283 in D2 that we show alters the strength of the inhibitory signal transduced upon binding of 3DL1 to Bw4.
Only the three lineage-defining 3DL1/S1 alleles—3DS1*01301, 3DL1*00501, and 3DL1*01502—are common to all human populations, the vast majority of the others being specific to particular populations, geographical regions, and ethnic groups (Norman et al. 2007). Since humans originated in Africa and eventually went on to populate other continents (Goldstein and Chikhi 2002), unequal crossing over has produced novel recombinant alleles and haplotypes that mark particular populations. For example, the 3DL1*009 recombinant allotype, in which the D0 domain of 3DL1*015 is replaced with that of 3DS1*013, is restricted to European and some South Asian populations (Middleton et al. 2007; Norman et al. 2007). That we consistently observe population stratification of 3DL1/S1 alleles points to recent or ongoing selection for variation. Up to 10% of sub-Saharan African populations have a KIR haplotype that was formed within 0.74 million years by an unequal crossing over that caused the deletion of >30 kb from the telomeric part of the KIR locus and created a novel hybrid gene, by fusing exons 1–5 from 3DL1*005 with exons 7–9 of the 3DL2*001 gene. The fusion protein encoded by this 3DL1/2v allele (3DL1*059) combines the extracellular, Ig-like domains of 3DL1*005 with the transmembrane and cytoplasmic domains of 3DL2*001. As expected, 3DL1*059 has specificity for Bw4, but it has a more even response to HLA-A and HLA-B allotypes having the Bw4 epitope, and transduces a weaker inhibitory signal, than other 3DL1 allotypes. Since the formation of 3DL1*059 by recombination, subsequent point substitutions in the cytoplasmic tail produced 3DL1*060 and 3DL1*061 allotypes. Valine 384 that distinguishes 3DL1*061 is close to an ITIM motif (Feng et al. 2005), and the substitution of cysteine for serine at residue 394 in 3DL1*060 that eliminates a protein kinase C phosphorylation site is predicted to increase inhibitory signals (Alvarez-Arias and Campbell 2007). Thus, the variants of 3DL1/2v have the potential for functional difference. That all hominoid KIR haplotypes have a lineage II gene with an exon 9 orthologous to that of 3DL2, despite much recombination, points to the importance of the signaling domain it encodes (Rajalingam et al. 2001; Sambrook et al. 2005; Guethlein et al. 2007).
Gene fusion is a recognized means for generating genetic novelty (Bailey and Eichler 2006), but to our knowledge, 3DL1/2v is the first demonstration of the phenomenon occurring in humans. Among the 600 gene fusions described for eukaryotes (Kummerfeld and Teichmann 2005), all those identified as human-specific are associated with disease or abnormality. One possible exception is the family of X chromosome gene fusions of OPN1MW/LW that cause anomalous color detection in males (Nathans et al. 1986), but may benefit female carriers (Deeb 2006). As shown for other SNPs at these loci, heterozygous females have enhanced color discrimination (Jameson et al. 2001).
Whereas the unequal crossing over that produced 3DL1/2v reduced the size of a KIR haplotype, in Eurasia a different event of unequal crossing over served to duplicate the 3DL1/S1 locus such that the haplotype had one copy of 3DS1 and one copy of 3DL1. Subsequent events of reciprocal crossing over have diversified this duplicated haplotype so that at least seven different 3DL1 alleles, including most of the more common alleles of both the 005 and 015 lineages, are associated with 3DS1*013. Although we do not know which 3DL1 allele was present on the original duplicated haplotype, that containing 3DL1*01502 appears the most prevalent.
When first characterized from cDNA, 3DL1 and 3DS1 were assigned to different loci based on the divergent sequence and function of their transmembrane and cytoplasmic domains (Robinson et al. 2006). Subsequent family and population studies showed that 3DL1 and 3DS1 usually segregate as alleles (Middleton et al. 2007), and sequence comparison indicated that 3DS1 was formed from 3DL1 by recombination with a gene encoding an activating KIR (Rajalingam et al. 2004). Consequently, 3DS1 and 3DL1 are generally considered to be alleles of a single locus: 3DL1/S1. The duplicated haplotypes are exceptional because they have two genes of which one is always 3DS1 and the other is always 3DL1. So far no duplicated haplotype containing two copies of 3DL1 and only one with two copies of 3DS1 (Gomez-Lozano et al. 2005) have been identified, indicating that such recombination is rare or that such events are selected against. Thus within the context of the duplicated haplotypes 3DS1 and 3DL1 are two different genes. This illustrates a more general complexity of the KIR system, namely, imprecision or blurring in the distinction between loci and alleles. At the KIR, MHC, and other immunological loci, an understanding of the synergy between allelic polymorphism and copy-number variability that generates diversity has required intensive, focused analysis (Bergstrom et al. 1999; Chung et al. 2002; Hollox et al. 2008). To determine the extent to which the findings apply to other gene families will require analyses of similar commitment and intensity (Dumas et al. 2007; Perry et al. 2008).
Throughout human history, balancing selection has maintained the 3DS1, 3DL1-005, and 3DL1-015 lineages of alleles. Today, the only three KIR3DL1/S1 alleles present in all the populations studied are 3DS1*01301, 3DL1*00501, and 3DL1*01502. This observation indicates that the three lineages have distinct and complementary functions that have facilitated the survival and propagation of human populations, a corollary being that populations that lost a 3DL1/S1 lineage failed to compete and survive over the long term. Despite the benefit of three allelic lineages for populations, most modern humans carry alleles from only one or two 3DL1/S1 lineages and suffer no obvious disadvantage in either survival or reproduction. In this context, the potential advantage of the duplicated KIR haplotype is that it is a mechanism by which individuals can carry all three 3DL1/S1 lineages and, as we have shown, express them as NK cell receptors. In sub-Saharan African populations, the balance between the three 3DL1/S1 lineages appears to have given way to selection for the 3DL1-015 lineage and against the 3DS1 and 3DL1-005 lineages (Norman et al. 2007). A further characteristic of sub-Saharan Africans is that a significant proportion of the 3DL1-005 lineage allotypes is 3DL1*059, 3DL1*060, and 3DL1*061, products of the gene fusion between 3DL1*005 and 3DL2. Selection for these alleles, might also reflect selection against conventional 3DL1*005 and for the modified functions imposed on its extracellular domains by the distinctive transmembrane and cytoplasmic domains of 3DL2.
KIR3DL1 and 3DL2 that recognize determinants of HLA-A and -B are encoded by lineage II KIR genes, which are represented in hominoid species by two or more genes. Exceptional is the chimpanzee, which has one lineage II KIR gene (Pt-KIR3DL1/2) that encodes a protein with structural and functional similarities to both 3DL1 and 3DL2. In chimpanzee KIR haplotypes, Pt-KIR3DL1/2 is the only gene on the telomeric side of Pt-KIR2DL4, an arrangement like that of the human haplotypes containing 3DL1/2v (3DL1*059, 3DL1*060, and 3DL1*061) in which 3DL1 and 3DL2 have been reduced to a single fusion gene. We have shown that the similarities of 3DL1/2v and Pt-KIR3DL1/2 are not due to common ancestry but to convergence, further emphasizing the dynamic nature of the KIR gene family and continuing cycles of gene expansion and contraction through meiotic recombination.
Methods
Subjects
The subjects studied here are as described by Norman et al. (2007). Blood samples were collected and the research conducted with approval from the appropriate Institutional Review Boards. The two Tanzanian populations are from (1) a village that lives on the coastal plain near the Indian Ocean and (2) a village at an altitude of 1700 m in the West Usambara Mountains.
KIR nomenclature
KIR genes and alleles were named by the KIR Nomenclature Committee (Marsh et al. 2003), formed from the WHO Nomenclature Committee for factors of the HLA system and the HUGO Genome Nomenclature Committee. A curated database is available at http://www.ebi.ac.uk/ipd/kir/ (Robinson et al. 2006). <D> denotes the number of Ig-like Domains; <L> a Long, inhibitory, cytoplasmic tail; <S> a Short, activating, tail; and <P> a Pseudogene. For each locus, the alleles are named in the order of their discovery, with the first three digits distinguishing alleles encoding different proteins. The second two digits are used to indicate synonymous variation. As an example, KIR3DL1*01501 and KIR3DL1*01502 are synonymous variants of the 3DL1*015 allele, and both encode the 3DL1*015 allotype—an inhibitory receptor having three Ig-like domains. When the meaning is unambiguous, KIR or even KIR3DL1 can be dropped from allele names.
PCR/DNA sequencing
PCR was performed using a Perkin-Elmer 9600 thermal cycler with a 3-min denaturing step at 94°C, 10 cycles of (10 sec at 94°C; 60 sec at 65°C) and 20 cycles of (10 sec at 94°C, 50 sec at 61°C, 30 sec at 72°C). When the expected amplicons were >3.5 kb, long range-PCR (LR-PCR) was performed using TripleMaster/HiFi reagents (Eppendorf) and the oligonucleotide primers described in Supplemental Figure S2. LR-PCR conditions were 3 min at 94°C followed by 28 cycles of (20 sec at 94°C, 10 sec at 61°C, 4 min at 68°C) completed with 15-min extension at 68°C. Products were cloned using the pCR 2.1-TOPO vector (Invitrogen) and sequenced using internal primers. Standard DNA sequencing was performed in forward and reverse directions using BigDye Terminator v3.1 and analyzed using an ABI-377 sequencer (ABI). Three clones of the desired allele were sequenced from each individual.
Characterizing the KIR3DL1/2v gene sequence
The KIR3DL1/2v gene sequence was characterized from one donor using multiple PCR primer pairs (Supplemental Fig. S7) and from four donors using six overlapping PCR fragments that spanned the haplotype from 2DL4 to FCAR (Supplemental Fig. S2). The four donors were selected to represent all three different 3DL1/2v sequences and also to have a distinct second KIR haplotype. Unambiguous sequence data were generated from one of the donors, who is hemizygous for 3DL1/2v so has no second 3DL1/S1 allele (Supplemental Fig. S2). For each heterozygous individual, sufficient SNPs were present to distinguish two haplotypes upon cloning, except for two of the reactions that were allele-specific (bands A and C) (Supplemental Fig. S2). Each amplicon was cloned using pCR 2.1 and sequenced in triplicate for each allele present. All of the PCR primers were designed to be KIR gene- or allele-specific (Supplemental Fig. S2; Supplemental Table S1). Accession numbers for the newly generated 3DL1*059–61 genomic DNA sequences are EU267269–71 and FJ459734.
The 3DL1/S1*059 donor was an African-American from California (genotype: 3DL1*031, 3DL1*059, 3DL2*001). The 3DL1/S1*060 donor was an African from Nigeria (genotype: 3DL1*035, 3DL1*60, 3DL2*001). The 3DL1/S1*061 donor was an African-American from California (genotype: 3DL1*040, 3DL1*061, 3DL2*010). The hemizygous donor was an African from Nigeria who has a KIR haplotype that lacks 3DP1, 2DL4, and 3DL1/S1 but retains 3DL2 (Norman et al. 2004). This individual has one 3DL1/S1 allele and one 3DL2 allele (genotype: 3DL1*059, 3DL2*006).
DNA sequences and recombination analysis
Recombinant sequences were identified from a domain-by-domain phylogenetic analysis. Gene conversion was identified using RDP (Recombination Detection Programs) version 2 (Martin et al. 2005) with a sliding window of 20 bp, with Bonferroni correction and a threshold of P = 0.001. Coding sequences of 3DL1/S1 used for alignments were obtained from the IPD database (http://www.ebi.ac.uk/ipd/kir/index.html) (Robinson et al. 2006) and those described in Belle et al. (2008) and Thomas et al. (2008). cDNA from 3DS1*050 (EF582383) was fully characterized here using methods previously described (Shilling et al. 2002). Genomic DNA sequences that spanned exons 1–5 (encoding the extracellular domains) and exons 7–9 (encoding the cytoplasmic domain) were characterized by LR-PCR from the following alleles: 3DL1*001, 3DL1*00501, 3DL1*009, 3DL1*01501, 3DL1*022, 3DL1*029, 3DL1*030, 3DL1*031, 3DL1*035, 3DL1*036, and 3DL1*040 (GenBank: FJ158650–60). 3DS1*01301 and 3DL2*007 sequences were extracted from AL133414 (3DS1a) (Wilson et al. 2000) and AY320039 (3DS1b). A second 3DL1*001 genomic sequence, and 2DL4*00801 and 3DL2*001 were obtained from AC011501 (Wilson et al. 2000). For the analysis indicated in Supplemental Figure 3B, a further 2 kb of sequence that surrounds the crossover point in intron 5 was obtained from 3DL1*00501, 3DL1*01501, and 3DL1*040; for 3DL1*002 the equivalent sequence was obtained from CU151839 and 3DL2*010 from CU464063 (Horton et al. 2006). pt2DL4 and pt3DL1/2 genomic DNA sequence was obtained from BX842589 (Sambrook et al. 2005) and Popy2DL4 and Popy3DLa from EF014479 (Guethlein et al. 2007).
DNA sequence analysis
Nucleotide sequences were aligned using ClustalX (Thompson et al. 1997) and BioEdit (www.mbio.ncsu.edu/BioEdit/bioedit.html). Neighbor-joining phylogenetic analyses were conducted using the Tamura-Nei method of MEGA3.1 and 500 bootstrap replicates (Kumar et al. 2004). PAML was used to reconstruct the ancestral sequences at specified nodes, using the marginal reconstruction method and the M8 model (Yang 2007).
Divergence time estimations
Divergence times were estimated using the MCMCTREE program from PAML 4 (Yang 2007), which uses Markov-chain Monte Carlo analysis for estimating mean posterior divergence times and their 95% credibility interval (CI). The Markov chain was sampled 5000 times every 10 cycles, and the burn-in stage was set to 5000 cycles. MODELTEST 3.7 (Posada and Crandall 1998) was used to gauge the nucleotide-substitution model (HKY95) and the α-distribution of this substitution (0.83 and 1.03 for the sequences 5′ and 3′ from the 3DL1/2v crossover, respectively). Calibration points were set according to fossil data and the divergence times: CI = 23–33 Mya for macaque/hominoid, CI = 10–18 Mya (mean 12) orangutan/hominid, and CI = 6–8 Mya (mean 7) for chimpanzee/hominid (Benton and Donoghue 2007).
Genotyping by pyrosequencing
Using primers described in Supplemental Table 1, individual exons from 2DL4 and 3DL2 were amplified and subjected to pyrosequencing. Exon-specific PCR was performed using a Perkin-Elmer 9600 thermal cycler with a 3-min denaturing step at 94°C, 10 cycles of (10 sec at 94°C, 60 sec at 65°C), and 20 cycles of (10 sec at 94°C, 50 sec at 61°C, 30 sec at 72°C). Identical conditions were used for a second specific reaction but with 35 cycles in the final step. Samples were purified using strepdavidin-coated Sephadex beads, and pyrosequencing reactions were performed using the Pyro-Gold reagents and a PSQ96A machine fitted with Capillary Dispensing Tips (Biotage Ltd.). 3DL1/S1 genotyping was performed as described (Norman et al. 2007). The genotyping procedures were performed in parallel with previously described family and control samples (Gardiner et al. 2001; Shilling et al. 2002; Artavanis-Tsakonas et al. 2003; Norman et al. 2004, 2007). To further validate the techniques, a random selection of 25% donors was also re-sequenced from a freshly generated amplicon. According to the most recent sequence alignments (http://www.ebi.ac.uk/ipd/kir/index.html), the combination of techniques discriminates 11 2DL4 alleles, 52 3DL1/S1 alleles, 22 3DL2 alleles, three 3DL1/2v alleles, and copy-number variation for each locus.
Detection of KIR3DL1/2v by genotyping 3DL1/S1 and 3DL2
Because no SNP uniquely defines the 3DL1/2v sequences, the pyrosequencing protocol was designed to detect a unique combination of SNPs in exons 1–5 that distinguishes 3DL1/2v from 3DL1/S1 (Fig. 1). Three different 3DL1/2v alleles (3DL1*059, 3DL1*060, 3DL1*061) were distinguished by one further SNP in exon 5 and two SNPs in exon 9. At the cDNA level, 3DL1*059 (Shilling et al. 2002) and 3DL1*060 (Artavanis-Tsakonas et al. 2003) have been described previously, and 3DL1*061 was discovered during the present study. Two other 3DL1/2v sequences reported previously (AY366254-5) were not observed in the present study. Heterozygosity was calculated using Arlequin (Schneider et al. 2000).
Detecting copy-number variation using pyrosequencing
Pyrosequencing uses enzyme-coupled reactions to generate light from the pyrophosphate molecules released upon DNA elongation (Ronaghi et al. 1996). Nucleotides are added sequentially so that the reaction proceeds only when the correct base is added. Light emission is plotted on a Pyrogram that has peak height proportional to the number of pyrophosphate molecules released. The technique is semiquantitative because the combined peak heights from a polymorphic position equal the height of the single peak at a monomorphic position (Fig. 5). With this method, we were able to determine the presence of three 3DL1/S1 alleles in individuals having a duplication of the 3DL1/S1 locus on one haplotype even when two of the alleles were identical in sequence, because peak-height ratios were 2:1 in favor of the allele present in two copies. All genotyping methods were designed so that each genotype gave a distinct pattern of several SNPs dispersed through the locus and did not rely on single marker SNPs. We were thus able to accurately assign three-copy genotypes by virtue of consistency in peak-height ratios throughout the locus.
To validate the copy-number detection method, two families shown previously by segregation analysis to have duplicated 3DL1/S1 (Martin et al. 2003; Norman et al. 2004) were used as standards. Furthermore, for eight of the 80 individuals in whom the duplication was detected, PCR products that span exons 3–5 of 2DL4 were generated, cloned, and sequenced (12 clones) to verify the presence of three different 2DL4 alleles and thus of the duplication. Some individuals may possess two copies of a 3DL1/S1-duplicated haplotype, and if the 3DL1/S1 pairs on the two haplotypes were identical, we would not have distinguished them from a normal heterozygote. However, the observed maximum frequency of 10% for duplicated-3DL1/S1 haplotypes implies that, assuming Hardy-Weinberg equilibrium, only one or two individuals from the entire cohort had two identical duplicated-3DL1/S1 haplotypes.
Haplotype composition and frequency were estimated in the following way: Allele phase was assigned per population by linkage disequilibrium using Arlequin (Schneider et al. 2000). Genotypes found in only one or two individuals were first compared with other populations of similar origin, and then with the entire sample set, to derive the most parsimonious interpretation. This approach was designed to estimate the minimum number of different haplotypes that could be distinguished based on the composition of 2DL4, 3DL1/S1, and 3DL2 alleles. After such iterative deductions, the genotypes of only two individuals remained ambiguous. Fifty-one of the 80 individuals having a duplication haplotype were genotyped using locus-specific primers to determine their KIR locus genotype (Uhrberg et al. 1997).
Flow cytometry
Peripheral blood mononuclear cells (PBMC) were isolated on ficoll gradients. Monoclonal antibodies used for flow cytometry were CD3-PE-Cy5, CD56-FITC, 3DL1(DX9)-PE, and IgG1 isotype controls (all BD Biosciences) and 3DL1(Z27)-PE (Beckman Coulter). CD3−CD56+ NK cells were monitored for expression of 3DL1/S1 using DX9, which reacts with 3DL1, and Z27, which reacts with both 3DL1 and 3DS1 (Supplemental Fig. S6; Gardiner et al. 2001; Carr et al. 2007; Trundley et al. 2007; Thomas et al. 2008). Thus, NK cells from individuals carrying 3DS1 and 3DL1 give two peaks of Z27 staining, but only one peak of DX9 staining. 3DL1*004 is not expressed at the cell surface and binds neither antibody (Pando et al. 2003). Flow-cytometry data were analyzed using FlowJo software.
Generation of NK effector and target cell lines
Effector cells were NKL cells transduced to express wild-type and mutant 3DL1 allotypes. Full-length KIR cDNA sequences were amplified from PMBC and cloned into a pCR 2.1 vector (Invitrogen). Site-directed mutagenesis was performed using the QuikChange mutagenesis kit (Stratagene) according to the manufacturer's instructions. Full-length coding regions were then cloned into the pBMN retroviral vector and transferred by retrovirus to NKL cells. Recombinant retrovirus was generated by transfection into Phi-NX cells, supernatants were used to infect NKL, and the cells were sorted for KIR expression level using a FACSVantage cell sorter (BD Biosciences), as previously described (Moesta et al. 2008).
Target cells were the Class I–deficient B-cell line 221 and transfectants of 221 expressing HLA allotypes: A*1102, A*3201, or B*1513. A*3201 cDNA was obtained from the WT47 cell line. A*1102 cDNA was generated by site-directed mutagenesis of A*1101 cDNA, which had been obtained from the KT17 cell line. HLA-E can present peptides that are derived from the leader peptides of HLA Class I molecules and protect 221 cells from lysis, owing to interaction of HLA-E/peptide with the CD94/NKG2A inhibitory receptor that is expressed by NK cells (Braud et al. 1998). Recombinant PCR was used to replace the HLA-A leader peptide fragment with VMAPVTLLLLL; the R5V mutation (underlined) abrogates interaction of the HLA-E/peptide with CD94/NKG2A (Michaelsson et al. 2002). HLA Class I cDNA was transfected into 221 cells using pBJneo as previously described (Gumperz et al. 1995).
NK cytotoxicity assays
Standard chromium-release assays were used to assess the degree of inhibition conferred by specific 3DL1 allotypes. NKL cells expressing 3DL2*001 and 721.221 cells expressing HLA-A*1102 were used as controls to investigate the specificity of 3DL1/2v-mediated inhibition. Two blocking agents were used in parallel experiments as controls: DX9 (anti-3DL1) was used to block the 3DL1/HLA interaction, and DX31 (anti-3DL2) was used to block the 3DL2/HLA-A11 interaction. Effector cells were mixed with 51Cr loaded target cells for 4 h at 37°C at ratios ranging from 40:1 to 10:1. Following incubation, supernatants were harvested and 51Cr was quantified using a Wallac β-scintillation counter. Percent specific lysis was calculated using the formula [(Specific Release − Spontaneous Release)/(Total Release − Spontaneous Release) × 100]. Experiments were conducted in triplicate for each condition, and each experiment was repeated independently five times. Comparison of means at the 20:1 ratio was performed using the Student's t-test (8 df).
Acknowledgments
We thank Lewis Lanier for supplying the DX17 and DX31 antibodies.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GenBank (http://www.ncbi.nlm.nih.gov/Genbank/) under accession nos. EF582383, EU267269–71, FJ158650–60, and FJ459734.]
Article is online at http://www.genome.org/cgi/doi/10.1101/gr.085738.108.
References
- Alvarez-Arias D.A., Campbell K.S. Protein kinase C regulates expression and function of inhibitory killer cell Ig-like receptors in NK cells. J. Immunol. 2007;179:5281–5290. doi: 10.4049/jimmunol.179.8.5281. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas K., Eleme K., McQueen K.L., Cheng N.W., Parham P., Davis D.M., Riley E.M. Activation of a subset of human NK cells upon contact with Plasmodium falciparum-infected erythrocytes. J. Immunol. 2003;171:5396–5405. doi: 10.4049/jimmunol.171.10.5396. [DOI] [PubMed] [Google Scholar]
- Bailey J.A., Eichler E.E. Primate segmental duplications: Crucibles of evolution, diversity and disease. Nat. Rev. Genet. 2006;7:552–564. doi: 10.1038/nrg1895. [DOI] [PubMed] [Google Scholar]
- Bashirova A.A., Martin M.P., McVicar D.W., Carrington M. The killer immunoglobulin-like receptor gene cluster: Tuning the genome for defense. Annu. Rev. Genomics Hum. Genet. 2006;7:277–300. doi: 10.1146/annurev.genom.7.080505.115726. [DOI] [PubMed] [Google Scholar]
- Belle I., Hou L., Chen M., Steiner N.K., Ng J., Hurley C.K. Investigation of killer cell immunoglobulin-like receptor gene diversity in KIR3DL1 and KIR3DS1 in a transplant population. Tissue Antigens. 2008;71:434–439. doi: 10.1111/j.1399-0039.2008.01017.x. [DOI] [PubMed] [Google Scholar]
- Benton M.J., Donoghue P.C. Paleontological evidence to date the tree of life. Mol. Biol. Evol. 2007;24:26–53. doi: 10.1093/molbev/msl150. [DOI] [PubMed] [Google Scholar]
- Bergstrom T.F., Erlandsson R., Engkvist H., Josefsson A., Erlich H.A., Gyllensten U. Phylogenetic history of hominoid DRB loci and alleles inferred from intron sequences. Immunol. Rev. 1999;167:351–365. doi: 10.1111/j.1600-065x.1999.tb01404.x. [DOI] [PubMed] [Google Scholar]
- Boyington J.C., Brooks A.G., Sun P.D. Structure of killer cell immunoglobulin-like receptors and their recognition of the class I MHC molecules. Immunol. Rev. 2001;181:66–78. doi: 10.1034/j.1600-065x.2001.1810105.x. [DOI] [PubMed] [Google Scholar]
- Braud V.M., Allan D.S., O'Callaghan C.A., Soderstrom K., D'Andrea A., Ogg G.S., Lazetic S., Young N.T., Bell J.I., Phillips J.H., et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391:795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- Campbell M.C., Tishkoff S.A. African genetic diversity: Implications for human demographic history, modern human origins, and complex disease mapping. Annu. Rev. Genomics Hum. Genet. 2008;9:403–433. doi: 10.1146/annurev.genom.9.081307.164258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr W.H., Rosen D.B., Arase H., Nixon D.F., Michaelsson J., Lanier L.L. Cutting Edge: KIR3DS1, a gene implicated in resistance to progression to AIDS, encodes a DAP12-associated receptor expressed on NK cells that triggers NK cell activation. J. Immunol. 2007;178:647–651. doi: 10.4049/jimmunol.178.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cella M., Longo A., Ferrara G.B., Strominger J.L., Colonna M. NK3-specific natural killer cells are selectively inhibited by Bw4- positive HLA alleles with isoleucine 80. J. Exp. Med. 1994;180:1235–1242. doi: 10.1084/jem.180.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan H.W., Kurago Z.B., Stewart C.A., Wilson M.J., Martin M.P., Mace B.E., Carrington M., Trowsdale J., Lutz C.T. DNA methylation maintains allele-specific KIR gene expression in human natural killer cells. J. Exp. Med. 2003;197:245–255. doi: 10.1084/jem.20021127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung E.K., Yang Y., Rennebohm R.M., Lokki M.L., Higgins G.C., Jones K.N., Zhou B., Blanchong C.A., Yu C.Y. Genetic sophistication of human complement components C4A and C4B and RP-C4-CYP21-TNX (RCCX) modules in the major histocompatibility complex. Am. J. Hum. Genet. 2002;71:823–837. doi: 10.1086/342777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeb S.S. Genetics of variation in human color vision and the retinal cone mosaic. Curr. Opin. Genet. Dev. 2006;16:301–307. doi: 10.1016/j.gde.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Dohring C., Scheidegger D., Samaridis J., Cella M., Colonna M. A human killer inhibitory receptor specific for HLA-A1,2. J. Immunol. 1996;156:3098–3101. [PubMed] [Google Scholar]
- Dumas L., Kim Y.H., Karimpour-Fard A., Cox M., Hopkins J., Pollack J.R., Sikela J.M. Gene copy number variation spanning 60 million years of human and primate evolution. Genome Res. 2007;17:1266–1277. doi: 10.1101/gr.6557307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Garrity D., Call M.E., Moffett H., Wucherpfennig K.W. Convergence on a distinctive assembly mechanism by unrelated families of activating immune receptors. Immunity. 2005;22:427–438. doi: 10.1016/j.immuni.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley B.A., De Santis D., Van Beelen E., Lathbury L.J., Christiansen F.T., Witt C.S. The reactivity of Bw4+ HLA-B and HLA-A alleles with KIR3DL1: Implications for patient and donor suitability for haploidentical stem cell transplantations. Blood. 2008;112:435–443. doi: 10.1182/blood-2008-01-132902. [DOI] [PubMed] [Google Scholar]
- Frazer K.A., Ballinger D.G., Cox D.R., Hinds D.A., Stuve L.L., Gibbs R.A., Belmont J.W., Boudreau A., Hardenbol P., Leal S.M., et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner C.M., Guethlein L.A., Shilling H.G., Pando M., Carr W.H., Rajalingam R., Vilches C., Parham P. Different NK cell surface phenotypes defined by the DX9 antibody are due to KIR3DL1 gene polymorphism. J. Immunol. 2001;166:2992–3001. doi: 10.4049/jimmunol.166.5.2992. [DOI] [PubMed] [Google Scholar]
- Gendzekhadze K., Norman P.J., Abi-Rached L., Layrisse Z., Parham P. High KIR diversity in Amerindians is maintained using few gene-content haplotypes. Immunogenetics. 2006;58:474–480. doi: 10.1007/s00251-006-0108-3. [DOI] [PubMed] [Google Scholar]
- Goldstein D.B., Chikhi L. Human migrations and population structure: What we know and why it matters. Annu. Rev. Genomics Hum. Genet. 2002;3:129–152. doi: 10.1146/annurev.genom.3.022502.103200. [DOI] [PubMed] [Google Scholar]
- Gomez-Lozano N., Estefania E., Williams F., Halfpenny I., Middleton D., Solis R., Vilches C. The silent KIR3DP1 gene (CD158c) is transcribed and might encode a secreted receptor in a minority of humans, in whom the KIR3DP1, KIR2DL4 and KIR3DL1/KIR3DS1 genes are duplicated. Eur. J. Immunol. 2005;35:16–24. doi: 10.1002/eji.200425493. [DOI] [PubMed] [Google Scholar]
- Guethlein L.A., Older Aguilar A.M., Abi-Rached L., Parham P. Evolution of killer cell Ig-like receptor (KIR) genes: Definition of an orangutan KIR haplotype reveals expansion of lineage III KIR associated with the emergence of MHC-C. J. Immunol. 2007;179:491–504. doi: 10.4049/jimmunol.179.1.491. [DOI] [PubMed] [Google Scholar]
- Gumperz J.E., Litwin V., Phillips J.H., Lanier L.L., Parham P. The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. J. Exp. Med. 1995;181:1133–1144. doi: 10.1084/jem.181.3.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond J.A., Guethlein L.A., Abi-Rached L., Moesta A.K., Parham P. Evolution and survival of marine carnivores did not require a diversity of KIR or Ly49 NK cell receptors. J. Immunol. 2009;182:3618–3627. doi: 10.4049/jimmunol.0803026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansasuta P., Dong T., Thananchai H., Weekes M., Willberg C., Aldemir H., Rowland-Jones S., Braud V.M. Recognition of HLA-A3 and HLA-A11 by KIR3DL2 is peptide-specific. Eur. J. Immunol. 2004;34:1673–1679. doi: 10.1002/eji.200425089. [DOI] [PubMed] [Google Scholar]
- Hey J. On the number of New World founders: A population genetic portrait of the peopling of the Americas. PLoS Biol. 2005;3:e193. doi: 10.1371/journal.pbio.0030193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiby S.E., Walker J.J., O'Shaughnessy K.M., Redman C.W., Carrington M., Trowsdale J., Moffett A. Combinations of maternal KIR and fetal HLA-C genes influence the risk of preeclampsia and reproductive success. J. Exp. Med. 2004;200:957–965. doi: 10.1084/jem.20041214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollox E.J., Barber J.C., Brookes A.J., Armour J.A. Defensins and the dynamic genome: what we can learn from structural variation at human chromosome band 8p23.1. Genome Res. 2008;18:1686–1697. doi: 10.1101/gr.080945.108. [DOI] [PubMed] [Google Scholar]
- Horton R., Coggill P., Miretti M.M., Sambrook J.G., Traherne J.A., Ward R., Sims S., Palmer S., Sehra H., Harrow J., et al. The LRC haplotype project: A resource for killer immunoglobulin-like receptor-linked association studies. Tissue Antigens. 2006;68:450–452. doi: 10.1111/j.1399-0039.2006.00697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson K.A., Highnote S.M., Wasserman L.M. Richer color experience in observers with multiple photopigment opsin genes. Psychon. Bull. Rev. 2001;8:244–261. doi: 10.3758/bf03196159. [DOI] [PubMed] [Google Scholar]
- Kelley J., Walter L., Trowsdale J. Comparative genomics of natural killer cell receptor gene clusters. PLoS Genet. 2005;1:129–139. doi: 10.1371/journal.pgen.0010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakoo S.I., Rajalingam R., Shum B.P., Weidenbach K., Flodin L., Muir D.G., Canavez F., Cooper S.L., Valiante N.M., Lanier L.L., et al. Rapid evolution of NK cell receptor systems demonstrated by comparison of chimpanzees and humans. Immunity. 2000;12:687–698. doi: 10.1016/s1074-7613(00)80219-8. [DOI] [PubMed] [Google Scholar]
- Khakoo S.I., Geller R., Shin S., Jenkins J.A., Parham P. The D0 domain of KIR3D acts as a major histocompatibility complex class I binding enhancer. J. Exp. Med. 2002;196:911–921. doi: 10.1084/jem.20020304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., Ota T. The average number of generations until extinction of an individual mutant gene in a finite population. Genetics. 1969;63:701–709. doi: 10.1093/genetics/63.3.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbel J.O., Urban A.E., Affourtit J.P., Godwin B., Grubert F., Simons J.F., Kim P.M., Palejev D., Carriero N.J., Du L., et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Kummerfeld S.K., Teichmann S.A. Relative rates of gene fusion and fission in multi-domain proteins. Trends Genet. 2005;21:25–30. doi: 10.1016/j.tig.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Lanier L.L. Evolutionary struggles between NK cells and viruses. Nat. Rev. Immunol. 2008;8:259–268. doi: 10.1038/nri2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh S.G., Parham P., Dupont B., Geraghty D.E., Trowsdale J., Middleton D., Vilches C., Carrington M., Witt C., Guethlein L.A., et al. Killer-cell immunoglobulin-like receptor (KIR) nomenclature report, 2002. Immunogenetics. 2003;55:220–226. doi: 10.1007/s00251-003-0571-z. [DOI] [PubMed] [Google Scholar]
- Martin M.P., Bashirova A., Traherne J., Trowsdale J., Carrington M. Cutting edge: Expansion of the KIR locus by unequal crossing over. J. Immunol. 2003;171:2192–2195. doi: 10.4049/jimmunol.171.5.2192. [DOI] [PubMed] [Google Scholar]
- Martin D.P., Williamson C., Posada D. RDP2: Recombination detection and analysis from sequence alignments. Bioinformatics. 2005;21:260–262. doi: 10.1093/bioinformatics/bth490. [DOI] [PubMed] [Google Scholar]
- Martin M.P., Qi Y., Gao X., Yamada E., Martin J.N., Pereyra F., Colombo S., Brown E.E., Shupert W.L., Phair J., et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat. Genet. 2007;39:733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelsson J., Teixeira de Matos C., Achour A., Lanier L.L., Karre K., Soderstrom K. A signal peptide derived from hsp60 binds HLA-E and interferes with CD94/NKG2A recognition. J. Exp. Med. 2002;196:1403–1414. doi: 10.1084/jem.20020797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton D., Meenagh A., Gourraud P.A. KIR haplotype content at the allele level in 77 Northern Irish families. Immunogenetics. 2007;59:145–158. doi: 10.1007/s00251-006-0181-7. [DOI] [PubMed] [Google Scholar]
- Moesta A.K., Norman P.J., Yawata M., Yawata N., Gleimer M., Parham P. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. J. Immunol. 2008;180:3969–3979. doi: 10.4049/jimmunol.180.6.3969. [DOI] [PubMed] [Google Scholar]
- Moffett A., Loke C. Immunology of placentation in eutherian mammals. Nat. Rev. Immunol. 2006;6:584–594. doi: 10.1038/nri1897. [DOI] [PubMed] [Google Scholar]
- Moretta L., Bottino C., Pende D., Castriconi R., Mingari M.C., Moretta A. Surface NK receptors and their ligands on tumor cells. Semin. Immunol. 2006;18:151–158. doi: 10.1016/j.smim.2006.03.002. [DOI] [PubMed] [Google Scholar]
- Nathans J., Piantanida T.P., Eddy R.L., Shows T.B., Hogness D.S. Molecular genetics of inherited variation in human color vision. Science. 1986;232:203–210. doi: 10.1126/science.3485310. [DOI] [PubMed] [Google Scholar]
- Norman P.J., Cook M.A., Carey B.S., Carrington C.V., Verity D.H., Hameed K., Ramdath D.D., Chandanayingyong D., Leppert M., Stephens H.A., et al. SNP haplotypes and allele frequencies show evidence for disruptive and balancing selection in the human leukocyte receptor complex. Immunogenetics. 2004;56:225–237. doi: 10.1007/s00251-004-0674-1. [DOI] [PubMed] [Google Scholar]
- Norman P.J., Abi-Rached L., Gendzekhadze K., Korbel D., Gleimer M., Rowley D., Bruno D., Carrington C.V., Chandanayingyong D., Chang Y.H., et al. Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nat. Genet. 2007;39:1092–1099. doi: 10.1038/ng2111. [DOI] [PubMed] [Google Scholar]
- O'Connor G.M., Guinan K.J., Cunningham R.T., Middleton D., Parham P., Gardiner C.M. Functional polymorphism of the KIR3DL1/S1 receptor on human NK cells. J. Immunol. 2007;178:235–241. doi: 10.4049/jimmunol.178.1.235. [DOI] [PubMed] [Google Scholar]
- Pando M.J., Gardiner C.M., Gleimer M., McQueen K.L., Parham P. The protein made from a common allele of KIR3DL1 (3DL1*004) is poorly expressed at cell surfaces due to substitution at positions 86 in Ig domain 0 and 182 in Ig domain 1. J. Immunol. 2003;171:6640–6649. doi: 10.4049/jimmunol.171.12.6640. [DOI] [PubMed] [Google Scholar]
- Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat. Rev. Immunol. 2005;5:201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- Pende D., Biassoni R., Cantoni C., Verdiani S., Falco M., di Donato C., Accame L., Bottino C., Moretta A., Moretta L. The natural killer cell receptor specific for HLA-A allotypes: A novel member of the p58/p70 family of inhibitory receptors that is characterized by three immunoglobulin-like domains and is expressed as a 140-kD disulphide-linked dimer. J. Exp. Med. 1996;184:505–518. doi: 10.1084/jem.184.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry G.H., Ben-Dor A., Tsalenko A., Sampas N., Rodriguez-Revenga L., Tran C.W., Scheffer A., Steinfeld I., Tsang P., Yamada N.A., et al. The fine-scale and complex architecture of human copy-number variation. Am. J. Hum. Genet. 2008;82:685–695. doi: 10.1016/j.ajhg.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D., Crandall K.A. MODELTEST: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Rajalingam R., Hong M., Adams E.J., Shum B.P., Guethlein L.A., Parham P. Short KIR haplotypes in pygmy chimpanzee (Bonobo) resemble the conserved framework of diverse human KIR haplotypes. J. Exp. Med. 2001;193:135–146. doi: 10.1084/jem.193.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajalingam R., Parham P., Abi-Rached L. Domain shuffling has been the main mechanism forming new hominoid killer cell Ig-like receptors. J. Immunol. 2004;172:356–369. doi: 10.4049/jimmunol.172.1.356. [DOI] [PubMed] [Google Scholar]
- Redon R., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relethford J.H. Genetic evidence and the modern human origins debate. Heredity. 2008;100:555–563. doi: 10.1038/hdy.2008.14. [DOI] [PubMed] [Google Scholar]
- Robinson J., Waller M.J., Fail S.C., Marsh S.G. The IMGT/HLA and IPD databases. Hum. Mutat. 2006;27:1192–1199. doi: 10.1002/humu.20406. [DOI] [PubMed] [Google Scholar]
- Ronaghi M., Karamohamed S., Pettersson B., Uhlen M., Nyren P. Real-time DNA sequencing using detection of pyrophosphate release. Anal. Biochem. 1996;242:84–89. doi: 10.1006/abio.1996.0432. [DOI] [PubMed] [Google Scholar]
- Sambrook J.G., Bashirova A., Palmer S., Sims S., Trowsdale J., Abi-Rached L., Parham P., Carrington M., Beck S. Single haplotype analysis demonstrates rapid evolution of the killer immunoglobulin-like receptor (KIR) loci in primates. Genome Res. 2005;15:25–35. doi: 10.1101/gr.2381205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider S., Roessli A., Excoffier L. Arlequin ver. 2.0: A software for population genetics data analysis. Genetics and Biometry Laboratory, University of Geneva; Geneva, Switzerland: 2000. [Google Scholar]
- Shilling H.G., Guethlein L.A., Cheng N.W., Gardiner C.M., Rodriguez R., Tyan D., Parham P. Allelic polymorphism synergizes with variable gene content to individualize human KIR genotype. J. Immunol. 2002;168:2307–2315. doi: 10.4049/jimmunol.168.5.2307. [DOI] [PubMed] [Google Scholar]
- Single R.M., Martin M.P., Gao X., Meyer D., Yeager M., Kidd J.R., Kidd K.K., Carrington M. Global diversity and evidence for coevolution of KIR and HLA. Nat. Genet. 2007;39:1114–1119. doi: 10.1038/ng2077. [DOI] [PubMed] [Google Scholar]
- Snyder M.R., Nakajima T., Leibson P.J., Weyand C.M., Goronzy J.J. Stimulatory killer Ig-like receptors modulate T cell activation through DAP12-dependent and DAP12-independent mechanisms. J. Immunol. 2004;173:3725–3731. doi: 10.4049/jimmunol.173.6.3725. [DOI] [PubMed] [Google Scholar]
- Thananchai H., Gillespie G., Martin M.P., Bashirova A., Yawata N., Yawata M., Easterbrook P., McVicar D.W., Maenaka K., Parham P., et al. Cutting Edge: Allele-specific and peptide-dependent interactions between KIR3DL1 and HLA-A and HLA-B. J. Immunol. 2007;178:33–37. doi: 10.4049/jimmunol.178.1.33. [DOI] [PubMed] [Google Scholar]
- Thomas R., Yamada E., Alter G., Martin M.P., Bashirova A.A., Norman P.J., Altfeld M., Parham P., Anderson S.K., McVicar D.W., et al. Novel KIR3DL1 alleles and their expression levels on NK cells: convergent evolution of KIR3DL1 phenotype variation? J. Immunol. 2008;180:6743–6750. doi: 10.4049/jimmunol.180.10.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trundley A., Frebel H., Jones D., Chang C., Trowsdale J. Allelic expression patterns of KIR3DS1 and 3DL1 using the Z27 and DX9 antibodies. Eur. J. Immunol. 2007;37:780–787. doi: 10.1002/eji.200636773. [DOI] [PubMed] [Google Scholar]
- Uhrberg M., Valiante N.M., Shum B.P., Shilling H.G., Lienert-Weidenbach K., Corliss B., Tyan D., Lanier L.L., Parham P. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7:753–763. doi: 10.1016/s1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- Williams F., Maxwell L.D., Halfpenny I.A., Meenagh A., Sleator C., Curran M.D., Middleton D. Multiple copies of KIR 3DL/S1 and KIR 2DL4 genes identified in a number of individuals. Hum. Immunol. 2003;64:729–732. doi: 10.1016/s0198-8859(03)00089-2. [DOI] [PubMed] [Google Scholar]
- Wilson M.J., Torkar M., Haude A., Milne S., Jones T., Sheer D., Beck S., Trowsdale J. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc. Natl. Acad. Sci. 2000;97:4778–4783. doi: 10.1073/pnas.080588597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yawata M., Yawata N., Draghi M., Partheniou F., Little A.M., Parham P. MHC class I-specific inhibitory receptors and their ligands structure diverse human NK cell repertoires towards a balance of missing-self response. Blood. 2008;112:2369–2380. doi: 10.1182/blood-2008-03-143727. [DOI] [PMC free article] [PubMed] [Google Scholar]