Abstract
The persistence of large blocks of homologous synteny and a high frequency of breakpoint reuse are distinctive features of mammalian chromosomes that are not well understood in evolutionary terms. To gain a better understanding of the evolutionary forces that affect genome architecture, synteny relationships among 10 amniotes (human, chimp, macaque, rat, mouse, pig, cattle, dog, opossum, and chicken) were compared at <1 human-Mbp resolution. Homologous synteny blocks (HSBs; N = 2233) and chromosome evolutionary breakpoint regions (EBRs; N = 1064) were identified from pairwise comparisons of all genomes. Analysis of the size distribution of HSBs shared in all 10 species' chromosomes (msHSBs) identified three (>20 Mbp) that are larger than expected by chance. Gene network analysis of msHSBs >3 human-Mbp and EBRs <1 Mbp demonstrated that msHSBs are significantly enriched for genes involved in development of the central nervous and other organ systems, whereas EBRs are enriched for genes associated with adaptive functions. In addition, we found EBRs are significantly enriched for structural variations (segmental duplications, copy number variants, and indels), retrotransposed and zinc finger genes, and single nucleotide polymorphisms. These results demonstrate that chromosome breakage in evolution is nonrandom and that HSBs and EBRs are evolving in distinctly different ways. We suggest that natural selection acts on the genome to maintain combinations of genes and their regulatory elements that are essential to fundamental processes of amniote development and biological organization. Furthermore, EBRs may be used extensively to generate new genetic variation and novel combinations of genes and regulatory elements that contribute to adaptive phenotypes.
The modern evolutionary synthesis attempts to explain Darwinian concepts of “descent with modification” and “natural selection” by applying quantitative methods to describe the behavior of chromosomes, genes, and their variants in populations of organisms. However, such methods, which focus primarily on variations in nucleic acids and proteins, have failed to adequately explain phenotypes found in nature. By largely overlooking the importance of chromosomes, a dynamic and pervasive feature of biology and the ultimate purveyor of genetic information, evolutionary science may have missed a key component of the mechanism for generating phenotypic variation used by natural selection. As such, an unresolved issue in evolutionary biology is whether chromosome rearrangements associated with speciation have adaptive value or are evolutionarily neutral (Ohno 1973; Ayala and Coluzzi 2005). The “chromosomal speciation” model posits that chromosome rearrangements contribute to reproductive isolation between geographically separated populations and promulgate speciation (Ayala and Coluzzi 2005). For example, a reciprocal translocation in yeast that is associated with resistance to sulfite concentrations was shown to be adaptive (Pérez-Ortín et al. 2002), whereas in insects (for review, see Ayala and Coluzzi 2005), chromosome inversions lead to reproductive isolation and thus contribute to speciation (Noor et al. 2001). However, reports supporting the chromosomal speciation model in higher taxa have been controversial (Lu et al. 2003; Navarro and Barton 2003). It is now possible to address this problem in vertebrate genomes from a different perspective because of the recent advances in comparative genomics, data visualization, and DNA sequence availability (Murphy et al. 2005; Ma et al. 2006).
An important theoretical insight into how chromosomes evolve was made by Nadeau and Taylor (1984), who proposed that chromosome breakage in evolution is random. This model of genome evolution was supported by the size distribution of synteny blocks found shared in the human and mouse genomes (Nadeau and Taylor 1984). However, like meiotic recombination, the random breakage model turned out to be a generalization that did not hold up when comparative genome organization was examined in finer detail by using direct DNA sequence comparisons (Pevzner and Tesler 2003) and high resolution chromosome (Larkin et al. 2003) or whole genome (Murphy et al. 2005) maps. These studies revealed that many sites where interchromosomal and intrachromosomal breakages occur in evolution are “reused,” which led to a new “fragile site” breakage model of chromosome evolution. For identification of breakpoint reuse, Larkin et al. (2003) and later Murphy and coworkers (2005) used empirical evidence, i.e., direct identification and counting of overlapping breakpoint regions in multigenome synteny-based comparisons, whereas Pevzner and Tesler (2003) used an algorithmic approach that identified an excess of small synteny blocks that could be explained by breakpoint reuse. Although there has been debate in the literature concerning the algorithmic approach, and whether reuse is nonrandom (Sankoff and Trinh 2005; Peng et al. 2006), the verification of breakpoint reuse by direct observation leaves no doubt as to its validity. Whether breakpoint reuse is nonrandom or due to chance is more controversial because resolution of the comparisons and how data are analyzed will affect the results. For example, resolution of breakpoints at the nucleotide level will produce a very different reuse frequency than resolution at the megabase level as defined by either synteny or sequence-only approaches. Furthermore, relatively high resolution maps are necessary to avoid the problem of “breakpoint chaining” that can produce more overlaps and thus apparent reuse in multigenome comparisons (Murphy et al. 2005). However, with either low or high resolution comparisons, there is no question that the organization of chromosomes in extant species is due at least in part to the independent occurrence of breakpoints at the same chromosomal sites in different vertebrate lineages.
This leads to an obvious question: Are there defining DNA sequence or chromosome features that might account for breakpoint use and reuse in chromosome evolution? It was shown that evolutionary breakpoint regions (EBRs) in chromosomes are gene-rich (Everts-van der Wind et al. 2004, 2005; Ma et al. 2006), are associated with the repositioning of centromeres and telomeres, and contain a higher than expected frequency of segmental duplications, among other features (Murphy et al. 2005; Bulazel et al. 2007). Evolutionary breakpoint regions are also frequently associated with chromosome fragile sites (Ruiz-Herrera et al. 2006) and chromosome rearrangements frequently found in certain cancers (Murphy et al. 2005; Darai-Ramqvist et al. 2008). The high frequency of segmental duplications and/or repetitive elements in EBRs (Bailey et al. 2004; Murphy et al. 2005; Schibler et al. 2006) specific to different lineages of mammals led to the hypothesis that EBRs are evolutionarily unstable regions that promote chromosome rearrangements by nonallelic homologous recombination (Murphy et al. 2005). These studies provided the first evidence for the distinguishing features of EBRs while suggesting a mechanism for use and reuse of specific sites in chromosome evolution. However, a comprehensive analysis of sequence features and functions of genes in EBRs compared with homologous synteny blocks (HSBs), i.e., regions of shared synteny between two or more genomes, is lacking. A better understanding of these features can help to explain not only processes related to chromosome evolution, e.g., whether breakpoint reuse is random or nonrandom, but also factors that are necessary or predisposing to many human and animal diseases.
The relationship of EBRs to various sequence features associated with evolutionary processes, as well as the evidence cited above for the chromosome speciation model, has stimulated a growing interest in chromosomal evolution and its relationship to phenotypic adaptation and diseases. In the present study, genomic resources, data visualization, and annotation tools were used to identify, taxonomically classify, and compare the functional gene content of HSBs and EBRs in genomes of 10 amniote species separated by more than 300 Myr of evolution. These comparisons permitted a first examination of the relationship between chromosome organization, genome rearrangements, and natural selection.
Results
Identification of HSBs and EBRs
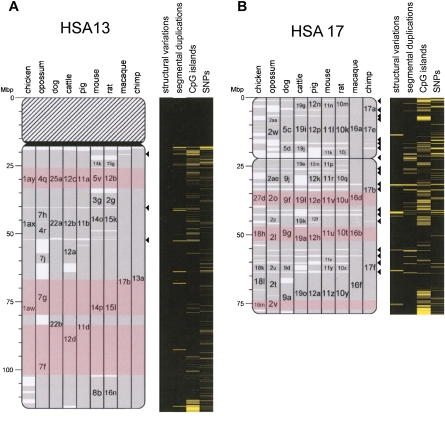
Using the human genome as the reference, pairwise HSBs were defined for representative species of four orders of eutherian mammals (Rodentia, Carnivora, Cetartiodactyla, and Primates), one methatherian (marsupial), and one member of the class Aves. We followed the rules proposed by Murphy et al. (2005) to define HSBs using orthologous genes and BAC-end sequences. Human genome coordinates of the first and last marker in each HSB were used to define the HSB boundaries. We identified 1769 HSBs exceeding the resolution of >500 human-kbp set in our analysis, which have a median size of 4.6 Mbp in all mammals. Excluding the metatherian opossum, there are 1376 eutherian HSBs with a median size of 5.8 Mbp. Addition of the chicken genome to the comparison (all amniotes) resulted in the definition of 2233 HSBs having a median size of 3.8 Mbp. On the basis of HSB definitions and their boundaries, we then identified the positions of 1064 EBRs within all species' chromosomes (Table 1). These EBRs have a median size across all genomes of 295.9 kbp. Eight hundred seventy-seven EBRs are <1 human-Mbp and cover ∼10% of genome size. The maps generated correspond to >90% average comparative genome coverage for all pairwise comparisons (see Supplemental Table 1). As examples, positions of pairwise HSBs overlaid onto HSA13 and HSA17 are presented in Figure 1. These two chromosomes represent opposite extremes in the number of chromosomal rearrangements in primate lineages.
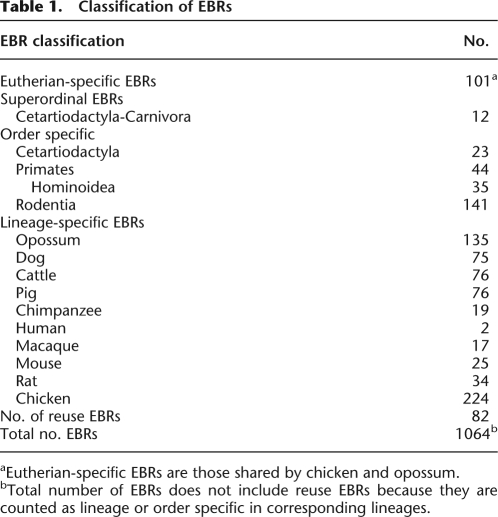
Table 1.
Classification of EBRs
aEutherian-specific EBRs are those shared by chicken and opossum.
bTotal number of EBRs does not include reuse EBRs because they are counted as lineage or order specific in corresponding lineages.
Figure 1.
Multispecies comparative chromosome architecture of HSA13 and HSA17. Multispecies alignment of HSBs on HSA13 (A) and HSA17 (B) as visualized with the Evolution Highway comparative chromosome browser. HSA13 and HSA17 were selected for display because they represent extremes in terms of chromosome rearrangements in primates and because of their differences in sequence feature distribution. The remaining multispecies maps showing full representation of all HSBs and sequence feature heat maps can be visualized using Evolution Highway (http://evolutionhighway.ncsa.uiuc.edu). Gray blocks indicate HSBs, with the species chromosome number indicated inside the bars. The identification of smaller HSBs is hidden in order to improve visualization and data interpretation. The lowercase letters indicate the sequential order of the HSB in that species' chromosome (in alphabetical order, with second alphabet used for chicken HSBs). A new alphabet is used for each chromosome. The borders of the red-shaded blocks indicate the sequence boundaries of the largest msHSBs on each chromosome. The EBRs are represented by white areas between HSBs. Sequence feature heat maps are to the right of each chromosome idiogram. Primate-specific EBRs are indicated by arrowheads. The underlying data for the sequence features were taken from the UCSC Genome Browser. Statistical comparisons for the 10 sequence features analyzed are given in Table 2. The selected sequence features are significantly more dense in either EBRs or HSBs than the average in the remainder of the genome for each comparison (Bonferroni adjusted P < 0.05). Copy number variants and indels (structural variants track from the UCSC Genome Browser) and segmental duplication can be observed to align with primate specific EBRs. The segmental duplications track is shown to illustrate consistency with previous results (Murphy et al. 2005). The heat maps show visually that HSA17 is more gene-dense and CpG islands are clustered in and around EBRs and telomeres.
The largest fraction of EBRs (66.5%) is lineage specific, with the greatest numbers appearing in the deepest branching (chicken and opossum) and most highly rearranged (dog, cattle, mouse, rat) genomes (Table 1). The smallest number of lineage-specific EBRs is in primate species, with human having only two. Muroid rodents have a large number of order-specific EBRs, whereas the cetartiodacyl chromosomes have the fewest. We found 101 EBRs that define the eutherian split from Marsupialia and Aves, whereas 12 unique super-ordinal EBRs were identified in ferungulates (cattle, pig, and dog). If an EBR was found to overlap with EBRs in species from a different clade but was not present in all species within the same clade it is termed as a “reuse” EBR (Murphy et al. 2005). Among 1064 EBRs defined in amniotes, the frequency of EBR reuse is 7.7% (Table 1).
Conservation of multispecies HSBs in amniote genomes
The number of HSBs >1 kbp shared among all species were identified for amniotes (Fig. 1; Supplemental Table 2). These multispecies HSBs (msHSBs) were defined by first determining pairwise HSBs for all species chromosomes in reference to the human genome, followed by identification of chromosome regions not interrupted by EBRs in the chromosomes of any species studied. The distribution of msHSB lengths >1 kbp (N = 823) approximates an exponential distribution (see Supplemental Fig. 1). We found eight msHSBs present in all 10 amniote species that exceed the expected maximum size of 16.3 Mbp (see Supplemental Table 2); three msHSBS, one each on HSA1, HSA2, and HSA4, are too large to be present by chance alone (P < 0.05). The 22.6-Mbp msHSB in HSA1 contains a large heterochromatin block (19.1 Mbp) and was eliminated from further consideration. The others are in HSA2 (22.9 Mbp) and HSA4 (23.6 Mbp). These results demonstrate that chromosome breakage is nonrandom in amniote genome evolution, producing some HSBs that are larger than expected in addition to the small HSBs that result from breakpoint reuse (Pevzner and Tesler 2003).
Gene networks in msHSBs and EBRs
The distributions of gene networks within msHSBs and EBRs were analyzed to determine if genes for specific functional pathways are found preferentially in evolutionarily stable and unstable regions. For this analysis, the distributions of gene functions within or ±100 kbp from 877 amniote EBRs (<1 human-Mbp) and 194 amniote msHSBs >3 human-Mbp were determined. The 3-Mbp threshold for msHSBs was chosen so as to avoid regions that would be affected by genomic features specific for EBRs (see results below). The 194 amniote msHSBs cover 1.2 Gbp (42.9%) of the human genome and contain 4134 human RefSeq genes (23% of total) of which 1315 are annotated in the MetaCore process networks database. The distribution of gene networks was compared for msHSBs and for EBRs using all human RefSeq genes with annotations in MetaCore (N = 5988) as a whole genome reference list. Compared with the reference list, msHSBs are significantly enriched for genes controlling key molecular and cellular processes related to development (N = 387; FDR < 5%; Fig. 2). As a control, we compared the remaining RefSeq genes in chromosomal regions not in msHSBs >3 human-Mbp and not in EBRs (N = 3166) to the whole genome reference list and found no enrichment for network processes at 5% FDR (see Supplemental Fig. 2).
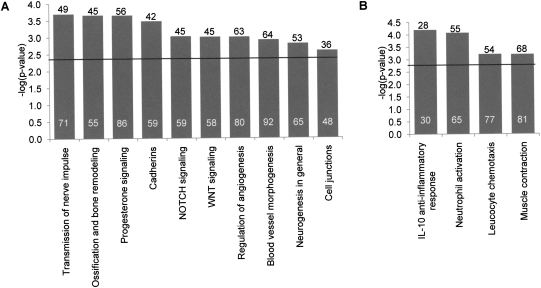
Figure 2.
Enrichment for gene networks in msHSBs and EBRs. The distribution of GeneGO MetaCore process network terms (Ekins et al. 2007) was determined for msHSBs >3 human-Mbp (A) and EBRs <1 human-Mbp (B). Numbers above the bars indicate the number of different msHSBs (A) and EBRs (B) containing genes associated with each process network. The number of genes in each process network in msHSBs and EBRs is given within bars. Only the gene networks that are significantly different in msHSBs or EBRs when compared to the rest of the human genome (FDR < 5%) are shown. The significance threshold for FDR = 5% is indicated by a horizontal bar.
When the functions of genes located in all msHSBs >3 Mbp were analyzed, gene networks related to the development of neurons, the central nervous system, bone, and blood vessels were found in much greater-than-expected frequencies in msHSBs than in the whole genome reference list. For example, the network process “neurogenesis in general” is represented by 71 distinct genes in 58 msHSBs, and the related process “transmission of nerve impulse” was represented by 76 genes in 52 msHSBs (FDR <5%). This finding was supported by an independent analysis of Gene Ontology (GO) processes that identified nervous system development as a highly significant term that is represented by 390 distinct genes in 150 msHSBs (FDR < 5%, raw P < 10−14; Supplemental Table 3). In addition, genes involved in the ancient and conserved Notch- and Wnt-signaling pathways, both key to developmental processes, cadherins, ancient molecules that are fundamental to tissue organization (and associated with Wnt-signaling), and gene networks involved in the formation and maintenance of cell junctions are significantly enriched in msHSBs. Lastly, genes within the network process “progesterone signaling,” which is essential for oocyte maturation, are significantly enriched in msHSBs.
Next we investigated genes in EBRs for enrichment of specific functional networks. Within or near (defined as ±100 kbp) the 877 chromosome EBRs comprising 256 Mbp (9.0%) of the human genome there are 4669 human RefSeq genes. Among these RefSeq genes, 1507 are annotated in the GeneGO MetaCore database. Genes belonging to four MetaCore process networks were enriched in the EBRs compared with their distribution in the whole genome (FDR <5%), three associated with inflammatory response and one with muscle contraction. Inflammatory responses and muscle contractility are both traits that involve an organism's response to external stimuli (Gillis et al. 2007; Li and Flavell 2008). These results were supported by an independent analysis of GO terms that identified significant differences for the terms “response to stimulus” and “immune system response” (see Supplemental Table 4).
Genomic features within primate EBRs and msHSBs
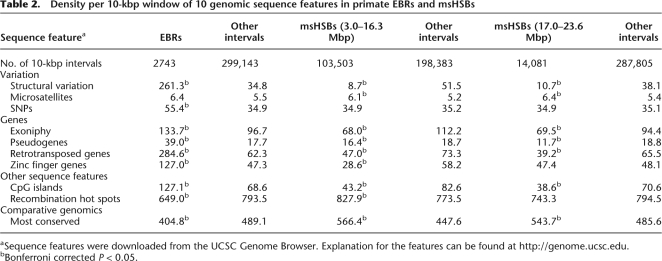
A detailed analysis of the genome landscape within and near the 95 primate ordinal and primate lineage-specific EBRs defined at <1 human-Mbp resolution (see Supplemental Table 5) was performed by statistical evaluation of 10 human sequence features downloaded from the UCSC Genome Browser (hg 17; NCBI build 35). Density values in 10-kbp intervals for each sequence feature were then compared for primate EBRs against all other parts of the genome. An identical analysis of sequence features was done for msHSBs 3.0–16.3 human Mbp and the remaining seven largest msHSBs (17.0–23.6 human Mbp) for comparison. Striking differences (Bonferroni P < 0.05) were found between EBRs and msHSBs for several evolutionarily important sequence features (given below in quotation marks), whereas the two groups of msHSBs were similar (Table 2). Compared with msHSBs 3.0–16.3 human-Mbp in size, the densities of “structural variants,” “retrotransposed genes,” and “zinc finger genes” are 30.0-, 6.0-, and 4.4-fold greater in EBRs. In addition, we found greater exoniphy, a measure of gene density, and lower density of most conserved sequences in EBRs than in msHSBs. Single nucleotide polymorphisms (SNPs) are found more densely located in EBRs. The density of meiotic recombination hot spots are 1.3-fold and 1.2-fold lower in EBRs than in msHSBs 3.0–16.3 human-Mbp in size and in the seven largest msHSBs, respectively.
Table 2.
Density per 10-kbp window of 10 genomic sequence features in primate EBRs and msHSBs
aSequence features were downloaded from the UCSC Genome Browser. Explanation for the features can be found at http://genome.ucsc.edu.
bBonferroni corrected P < 0.05.
Discussion
The 1376 eutherian HSBs found in this study are larger than the 1159 in a previous study that also used synteny analysis (Murphy et al. 2005), but similar to the 1338 identified using DNA sequence alignments of the human, mouse, rat, and dog genomes (Ma et al. 2006). These results generally reflect differences in the number of species examined and the resolution of the comparisons, which range from megabases for the synteny-based comparisons to kilobases for sequence-based comparisons. The most critical factor in comparing different studies is the resolution of EBR distances. The synteny-based interpretations are more conservative than sequence-based comparisons because the latter tend to overestimate chromosome rearrangements due to errors in local assemblies and unoriented contigs (Bhutkar et al. 2006). Nevertheless, the results of synteny-based comparisons and sequence-based comparisons are highly correlated when the resolution of EBR sizes is similar (Ma et al. 2006; Mikkelsen et al. 2007).
Precise determination of the boundaries of HSBs is essential for accurate estimation of the rates of evolution in different lineages (Murphy et al. 2005), reconstruction of ancestral genomes (Murphy et al. 2005; Ma et al. 2006), and identifying sequence features associated with EBRs (Murphy et al. 2005; Schibler et al. 2006). As expected, we found the largest fraction of EBRs to be lineage specific and to be in the most basal amniote genomes (Table 1). The fewest lineage-specific EBRs were found in primates, with human having only two, consistent with earlier observations (Gibbs et al. 2007). Thus, after divergence of human and chimpanzee, the human lineage has had a surprisingly stable genome compared with chimpanzee. With the use of high-quality physical maps of the cattle (Snelling et al. 2007) and pig (Humphray et al. 2007) chromosomes, we were able to identify twice as many Cetartiodactyla-specific EBRs than previously (Murphy et al. 2005). This was largely due to the increased number of pig-specific EBRs identified using the integrated physical map versus the RH map alone. The number of mouse-specific EBRs was found to be less than the number of rat-specific EBRs (25 and 34, respectively), similar to results reported by Murphy et al. (2005) but different in magnitude than results reported by Ma et al. (2006), who found 83 mouse-specific EBRs and 623 rat-specific EBRs using direct sequence-based comparison at 50-kbp resolution. Many of the rat-specific rearrangements found by Ma et al. (2006) and in the present study are small inversions <1 Mbp in size that could result from genome assembly errors. Such errors can greatly affect EBR counts. Lastly, we found 393 human-opossum HSBs, a number remarkably similar to the 367 large-scale synteny blocks obtained from whole-genome sequence alignment (Mikkelsen et al. 2007). The concordant results produced using the different methods for HSB definition give a high level of confidence to earlier conclusions as well as those obtained in the present study.
If an EBR was found to overlap with EBRs in species from a different clade but was not present in all species within the same clade, it is termed as a “reuse EBR” (Murphy et al. 2005). Among 1064 EBRs defined in amniotes, the frequency of EBR reuse was found in the present study to be 7.7% (Table 1), which is lower than a previous map-based estimate of 20% (Murphy et al. 2005) but similar to the 8% estimate based on sequence alignment (Ma et al. 2006). The lower estimate is due to the higher resolution of the map comparisons, which results in fewer overlaps of closely spaced but distinct EBRs. These reuse EBRs may represent unstable sites in chromosomes that predispose to recurrent chromosome rearrangements (Pevzner and Tesler 2003; Larkin et al. 2003; Murphy et al. 2005).
An important question is whether large HSBs remain intact for long evolutionary time periods by chance or due to selection. We investigated this by analyzing the size distribution of msHSBs present in all amniote species. Amniote msHSBs represent the regions of chromosomes where synteny and order of genes have been maintained for over 1 billion yr of collective, independent evolution and date back 310 Myr to the divergence of synapsida and diapsida. We found three msHSBs that are too large to have been maintained by chance alone (P < 0.05). The 22.9-Mbp segment on HSA2 contains the developmentally important HOXD gene cluster (Dollé et al. 1989). The HOXD genes are coordinately regulated, and it has been shown that disruption of gene contiguity in this cluster can cause major phenotypic abnormalities in mice (Spitz et al. 2005; Tarchini et al. 2005). In addition to HOXD, several other developmentally important genes are located within this 22.9-Mbp msHSB, including five genes encoding voltage-gated sodium channel proteins associated with brain and nervous system function and two members of the DLX family of homeobox genes involved in craniofacial, limb, and bone development. Other examples of functionally related genes and coregulated gene clusters can be found on each of the large conserved msHSBs (see Supplemental Table 6), including the SLITRK paralogous cluster on HSA13 encoding neurotrophin-like receptors associated with Tourette syndrome (Abelson et al. 2005). Two zinc finger protein-encoding paralogs are also located in the large HSA13 msHSB, one of which (ZIC2) is highly expressed in cerebellum and is associated with holoprosencephaly. The clustering of developmentally important genes in the largest of the evolutionary conserved HSBs indicates that breakages in some chromosomal regions may lower fitness, and that these regions may span tens of millions of base pairs of DNA.
The analysis of gene process networks in all msHSBs >3 Mbp strongly supports the results and conclusions drawn above from the seven largest msHSBs. On the basis of the highly significant enrichment for gene networks associated with developmental processes found in 194 amniote msHSBs, some that are as old as 310 Myr, it appears that chromosome organization may not evolve in ways that will cause major disruption to molecular pathways essential to the development of the vertebrate body plan and tissue morphogenesis. In contrast to the results for msHSBs, the analysis of gene process networks enriched in EBRs indicates that EBRs are enriched for genes involved in inflammatory response and muscular contraction. Both traits are involved in an organism's response to external stimuli. Thus, as opposed to msHSBs, which we show are subjected to selection, our results suggest that at least some evolutionary chromosome rearrangements may have adaptive value by creating novel configurations of structural and regulatory loci involved in responses to environmental challenges.
Our feature analysis of the genomic landscape in and around evolutionary chromosome breakpoints has yielded important insights into the possible mechanism of breakpoint use, reuse, and genome evolution. Evolutionary breakpoint regions have been shown previously to be gene-dense (Everts-van der Wind et al. 2005; Murphy et al. 2005; Ma et al. 2006), which was further supported by our finding of greater exoniphy and higher density of CpG islands. In primates, EBRs contain significantly more segmental duplications (Fig. 1; Murphy et al. 2005; Kehrer-Sawatzki and Cooper 2008) that appear to promote rather than be a consequence of chromosome rearrangements within the primate lineage (Murphy et al. 2005). Newly discovered sequence features of EBRs described in the present work include higher densities of structural variants (copy number polymorphisms and indels) and SNPs and lower densities of highly conserved sequences and meiotic recombination hot spots. These features suggest that selectable variation in EBRs is created by multiple types of mutations. The lower density of SNPs in msHSBs fits nicely with our finding that msHSBs are enriched for developmentally important genes whose regulation/function likely cannot be disrupted by rearrangements. Similarly, the higher density of structural variants and retrotranspositions within EBRs provides a plausible mechanism for the diversification of the adaptive genes found to be enriched in EBRs (Dunham et al. 2002). For example, enrichment of zinc finger transcription factor genes in EBRs and their association with lineage-specific gene duplications is a feature previously noted in a comparison of HSA19 with mouse orthologous chromosomes (Dehal et al. 2001). Furthermore, genes associated with immune responses, which we found enriched in EBRs and are known to contribute to adaptive phenotypes, are highly correlated with gene duplications and deletions (She et al. 2008). The dramatic differences in human sequence features found in EBRs and msHSBs demonstrate that they are evolving in distinctly different ways and directly support the finding of enriched gene functions within these regions.
In summary, chromosome breakage in evolution is nonrandom, resulting in segments that are conserved over hundreds of millions of years, whereas other regions of the genome are unstable and are more likely to be involved in rearrangements because of their underlying sequence features. EBRs appear to be hotspots of evolutionary activity where genes are created, amplified, and destroyed by a variety of molecular mechanisms. Such cauldrons of genomic variation, with their increased density of zinc finger transcriptional regulators, segmental duplications, copy number variants, and retrotransposed genes, may act as a major reservoir for producing adaptive phenotypes by evolutionary chromosome rearrangements. In humans, it is well known that somatic and germline genomic rearrangements involving evolutionary chromosome breakpoints may cause major disease phenotypes, including a variety of cancers and developmental disorders (Murphy et al. 2005; Lindsay et al. 2006; Darai-Ramqvist et al. 2008). We therefore propose that macroevolutionary change may be linked to relatively rare evolutionary chromosome rearrangements that have adaptive value and are thus subject to positive selection. These results are consistent with the chromosomal speciation model (Ayala and Coluzzi 2005). Continued advances in gene annotation, detailed maps of newly sequenced genomes, and chromosome engineering will contribute greatly toward an improved understanding of the role of chromosome rearrangements in adaptation and speciation.
Methods
Identification of HSBs
The mammalian genomes analyzed included all genomes in the public domain at the time this study was performed that were sequenced at high enough coverage for comparative chromosome analysis. For definition of HSBs, we created a Perl script to implement the rule set described by Murphy et al. (2005). Briefly, each HSB is defined by a minimum of two adjacent markers on the same chromosome in two compared species without interruption. The program uses orthologous gene pairs derived from assembled whole-genome sequence and radiation hybrid (RH) maps as input. The program's output contains the chromosomal position and coordinates of all HSBs defined in the reference and target genomes. For the present work, HSBs were defined for eight completely sequenced genomes (Homo sapiens, Mus musculus, Pan troglodytes, Macaca mulatta, Rattus norvegicus, Canis familiaris, Monodelphis domestica, and Gallus gallus), and two genomes for which high resolution (∼1 Mbp) RH and BAC fingerprint maps are available (Bos taurus and Sus scrofa). For sequenced genomes, the coordinates of orthologous gene pairs were downloaded from Ensembl (BioMart database v.38). The human genome was used as the reference genome for the comparison. The program was set so that HSBs would have a minimum size of 500 kbp. Inversions were defined using a minimum of three markers each ≥300 kbp apart such that the resolution for detecting inversions was 600 kbp.
In addition to the sequenced genomes, the IL-TX 5000 Rad whole-genome cattle RH and comparative maps (Everts-van der Wind et al. 2005) and pig fingerprint physical map (Humphray et al. 2007) (ftp://ftp.sanger.ac.uk/pub/S_scrofa/marc/) were used as primary data sources. For HSB definition, comparative maps were first generated using BAC-end sequences (BESs) anchored to the cattle (Snelling et al. 2007) and pig (Humphray et al. 2007) BAC fingerprint contigs. The cattle and pig BAC fingerprint contigs were ordered on chromosomes according to their positions in the cattle RH and pig fingerprint maps (Everts-van der Wind et al. 2005; Humphray et al. 2007) following the procedure described by Everts-van der Wind et al. (2005). Similarity of the anchored BESs to the human genome (NCBI build 35) was then determined (BLASTN) using E = e−10 as a significance threshold. Next, syntenic BESs with BLASTN E-values below the threshold were separated from nonsyntenic BESs for each fingerprint contig. Homologous synteny blocks were defined for the resulting sets of ordered anchor points in the integrated cattle and pig RH-fingerprint maps and the human genome using the same distance thresholds as applied to the sequenced genomes (described above).
Visualization of HSBs and evolutionary breakpoint regions (EBRs)
The Evolution Highway comparative chromosome browser (Murphy et al. 2005) (http://evolutionhighway.ncsa.uiuc.edu) was used to visualize comparative genome organization and to identify and visualize the different types of evolutionary breakpoints in chromosomes, e.g., lineage specific, ordinal, superordinal, and reuse. The rules for evolutionary breakpoint classification were as described by Murphy et al. (2005). Upgrades to Evolution Highway were made to accommodate the display of additional information. For the identification and visualization of chromosome EBRs, the user selects a species with sequenced genome for the breakpoint identification (the reference species) and one or more genomes for comparison. For each pairwise genome comparison, all evolutionary breakpoints are identified and displayed relative to chromosomes of the reference species. For display of the EBRs, Evolution Highway takes the coordinates of the adjacent HSB boundaries +1 bp (for upstream HSB) or minus 1 bp (for downstream HSB) in the reference genome coordinates. The sequence coordinates of all HSBs in each reference genome are available on the Evolution Highway website (http://evolutionhighway.ncsa.uiuc.edu).
If two or more species are selected in the browser, Evolution Highway will find and display all overlapping EBRs in the reference genome relative to the chromosomes of the species selected. Each EBR must be less than or equal to the maximum threshold and nonoverlapping with any other EBR of less than or equal to the maximum threshold size. The boundary coordinates in the reference genome of the overlapping breakpoint region defined in all selected species are taken as the coordinates of the resulting EBR. If the species selected belong to the same clade, the EBR will be classified as clade specific, e.g., ordinal, superordinal, ferungulate, eutherian, etc. If the species having overlapping breakpoints belong to distinct clades, the breakpoint identified is classified as a reuse breakpoint (Murphy et al. 2005). A special case is the identification of EBRs specific to the reference genome. Such EBRs were identified as those common to all genomes or all genomes except one species from a different clade (to allow for a small number of possible errors in EBR coordinates) relative to the reference species.
For the count of EBRs in Table 1, 35 of the pig-specific EBRs that were found in SSC5 and SSCX were ignored. These 35 EBRs were excluded because they are the likely result of misorientation of small fingerprint contigs. However, these EBRs were not excluded from msHSB definition because many of them overlapped with EBRs in other species and, therefore, did not significantly affect the size distribution of msHSBs.
Identification and analysis of the size distribution of msHSBs
Evolutionary Highway displays HSBs in a visual format that permits the identification of HSBs that overlap in two or more species relative to the same reference genome. We term HSBs completely overlapping in at least three species (including the reference genome) as msHSBs. The msHSBs were identified for all amniote species, which included all mammal genomes and chicken. For the purposes of this analysis, msHSBs had to be at least 1 kbp in size. The expected minimum and maximum msHSB sizes were calculated assuming an exponential distribution (see Supplemental Fig. 1), and the distribution was compared to the observed values for msHSBs that exceeded the maximum expected size. We could not test the distribution of small HSBs due to limitations in the resolution of our gene-based method for defining HSBs <10 kbp. Higher resolution tends to produce more spurious breakpoint regions because of local assembly errors and incorrect ordering of small sequence contigs.
Functional annotation of genes located within or near EBRs and within msHSBs
Sequence coordinates of all genes in human RefSeq 20 (NCBI) were downloaded from the UCSC Genome Browser. The set of RefSeq genes was filtered to remove duplicates (different gene names with the same genome coordinates). The LocusLink identification number (ID) of each gene was extracted from the NCBI LocusLink database and added to the RefSeq gene entry, using the RefSeq ID in LocusLink as a matching criterion. The genes were then assigned to msHBSs or EBRs on the basis of their coordinates in the reference genome. To be assigned to an EBR, gene coordinates had to fall within an EBR in at least one species in comparison to the human genome to be located within or ±100 kbp from an EBR boundary. The LocusLink IDs of genes found in msHSBs and of those found in EBRs were submitted separately to the GeneGo MetaCore database (http://www.genego.com v. 4.7 build 12996) for functional annotation. The well-annotated GeneGo process networks database and classification system hosted on MetaCore (Ekins et al. 2007) was selected as the basis for functionally annotating genes. The distribution of genes in process network categories (N = 127) was then compared for msHSBs and EBRs against the RefSeq reference list using MetaCore Functional Ontogeny Enrichment Tool (http://www.genego.com) as described in the main text. A false discovery rate of 5% was used as a minimal significance threshold.
Analysis of sequence features EBRs and HSBs
Fifteen sequence feature tracks were downloaded from the UCSC Genome Browser (hg17; NCBI build 35; http://genome.ucsc.edu). These features were processed to verify that sequences were not duplicated and the resulting sequence features were assigned to either primate-specific EBRs (defined by a combination of human-specific, chimp, macaque, Hominidae, and Hominidae + Cercopithecidae EBRs), or to the remainder of human chromosomes. The human genome sequence was divided into 10-kbp windows, and the number of bases specific for each sequence feature was counted in each 10-kbp window. Next, the average number of bases for each feature was calculated separately for the windows located within primate EBRs and the remainder of human chromosomes. The average number of feature-specific bases was calculated for 10-kbp windows located in the EBRs and compared with the average number of feature-specific bases in 10-kbp windows found in non-EBR regions using a t-test with unequal variances as described previously (Skovlund and Fenstad 2001; Smith et al. 2005). The same procedure was used to compare densities of sequence features in msHSBs versus the rest of the human genome. The Bonferroni correction was used to control for multiple comparisons. Ten sequence features downloaded from the UCSC Genome Browser for which an average number of bases was more than five in at least one of the 10-kbp sets compared are shown in Table 2. For visualization of sequence features in Evolution Highway, an algorithm was implemented to display sequence features as data tracks. The algorithm calculates the feature frequency in 500-kbp windows across each chromosome and the average of the feature over the entire chromosome. Then, the feature frequency in each 500-kbp window is compared to the chromosome average for the feature and the value converted to yellow color on the basis of increasing feature density. The greater the color intensity, the more frequent the feature is in that chromosomal region.
Acknowledgments
Funding for this research was provided in part by the United States Department of Agriculture Cooperative State Research Education and Extension Service, Livestock Genome Sequencing Initiative (AG 2005-34480-15939). We thank Dr. William Murphy and Dr. Lisa Stubbs for critical review of the manuscript and two anonymous referees who provided many helpful suggestions.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.086546.108.
References
- Abelson J.F., Kwan K.Y., O'Roak B.J., Baek D.Y., Stillman A.A., Morgan T.M., Mathews C.A., Pauls D.L., Rasin M.R., Gunel M., et al. Sequence variants in SLITRK1 are associated with Tourette's syndrome. Science. 2005;310:317–320. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- Ayala F.J., Coluzzi M. Chromosome speciation: Humans, Drosophila, and mosquitoes. Proc. Natl. Acad. Sci. 2005;102:6535–6542. doi: 10.1073/pnas.0501847102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J.A., Baertsch R., Kent W.J., Haussler D., Eichler E.E. Hot spots of mammalian chromosomal evolution. Genome Biol. 2004;5:R23. doi: 10.1186/gb-2004-5-4-r23. http://genomebiology.com/2004/5/4/R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutkar A., Russo S., Smith T.F., Gelbart W.M. Techniques for multi-genome synteny analysis to overcome assembly limitations. Genome Inform. 2006;17:152–161. [PubMed] [Google Scholar]
- Bulazel K.V., Ferreri G.C., Eldridge M.D., O'Neill R.J. Species-specific shifts in centromere sequence composition are coincident with breakpoint reuse in karyotypically divergent lineages. Genome Biol. 2007;8:R170. doi: 10.1186/gb-2007-8-8-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darai-Ramqvist E., Sandlund A., Muller S., Klein G., Imreh S., Kost-Alimova M. Segmental duplications and evolutionary plasticity at tumor chromosome break-prone regions. Genome Res. 2008;18:370–379. doi: 10.1101/gr.7010208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal P., Predki P., Olsen A.S., Kobayashi A., Folta P., Lucas S., Land M., Terry A., Ecale Zhou C.L., Rash S., et al. Human chromosome 19 and related regions in mouse: Conservative and lineage-specific evolution. Science. 2001;293:104–111. doi: 10.1126/science.1060310. [DOI] [PubMed] [Google Scholar]
- Dollé P., Izpisúa-Belmonte J.C., Falkenstein H., Renucci A., Duboule D. Coordinate expression of the murine Hox-5 complex homoeobox-containing genes during limb pattern formation. Nature. 1989;342:767–772. doi: 10.1038/342767a0. [DOI] [PubMed] [Google Scholar]
- Dunham M.J., Badrane H., Ferea T., Adams J., Brown P.O., Rosenzweig F., Botstein D. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 2002;99:16144–16149. doi: 10.1073/pnas.242624799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekins S., Nikolsky Y., Bugrim A., Kirillov E., Nikolskaya T. Pathway mapping tools for analysis of high content data. Methods Mol. Biol. 2007;356:319–350. doi: 10.1385/1-59745-217-3:319. [DOI] [PubMed] [Google Scholar]
- Everts-van der Wind A., Kata S.R., Band M.R., Rebeiz M., Larkin D.M., Everts R.E., Green C.A., Liu L., Natarajan S., Goldammer T., et al. A 1463 gene cattle–human comparative map with anchor points defined by human genome sequence coordinates. Genome Res. 2004;14:1424–1437. doi: 10.1101/gr.2554404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everts-van der Wind A., Larkin D.M., Green C.A., Elliott J.S., Olmstead C.A., Chiu R., Schein J.E., Marra M.A., Womack J.E., Lewin H.A. A high-resolution whole-genome cattle-human comparative map reveals details of mammalian chromosome evolution. Proc. Natl. Acad. Sci. 2005;102:18526–18531. doi: 10.1073/pnas.0509285102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs R.A., Rogers J., Katze M.G., Bumgarner R., Weinstock G.M., Mardis E.R., Remington K.A., Strausberg R.L., Venter J.C., Wilson R.K., et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Gillis T.E., Marshall C.R., Tibbits G.F. Functional and evolutionary relationships of troponin C. Physiol. Genomics. 2007;32:16–27. doi: 10.1152/physiolgenomics.00197.2007. [DOI] [PubMed] [Google Scholar]
- Humphray S.J., Scott C.E., Clark R., Marron B., Bender C., Camm N., Davis J., Jenks A., Noon A., Patel M., et al. A high utility integrated map of the pig genome. Genome Biol. 2007;8:R139. doi: 10.1186/gb-2007-8-7-r139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H., Cooper D.N. Molecular mechanisms of chromosomal rearrangement during primate evolution. Chromosome Res. 2008;16:41–56. doi: 10.1007/s10577-007-1207-1. [DOI] [PubMed] [Google Scholar]
- Larkin D.M., Everts-van der Wind A., Rebeiz M., Schweitzer P.A., Bachman S., Green C., Wright C.L., Campos E.J., Benson L.D., Edwards J., et al. A cattle-human comparative map built with cattle BAC-ends and human genome sequence. Genome Res. 2003;13:1966–1972. doi: 10.1101/gr.1560203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.O., Flavell R.A. Contextual regulation of inflammation: A duet by transforming growth factor-beta and interleukin-10. Immunity. 2008;28:468–476. doi: 10.1016/j.immuni.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Lindsay S.J., Khajavi M., Lupski J.R., Hurles M.E. A chromosomal rearrangement hot spot can be identified from population genetic variation and is coincident with a hotspot for allelic recombination. Am. J. Hum. Genet. 2006;79:890–902. doi: 10.1086/508709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Li W.H., Wu C.I. Comment on “Chromosomal speciation and molecular divergence-accelerated evolution in rearranged chromosomes.”. Science. 2003;302:988. doi: 10.1126/science.302.5647.988a. [DOI] [PubMed] [Google Scholar]
- Ma J., Zhang L., Suh B.B., Raney B.J., Burhans R.C., Kent W.J., Blanchette M., Haussler D., Miller W. Reconstructing contiguous regions of an ancestral genome. Genome Res. 2006;16:1557–1565. doi: 10.1101/gr.5383506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen T.S., Wakefield M.J., Aken B., Amemiya C.T., Chang J.L., Duke S., Garber M., Gentles A.J., Goodstadt L., Heger A., et al. Genome of the marsupial Monodelphis domestica reveals innovation in noncoding sequences. Nature. 2007;447:167–177. doi: 10.1038/nature05805. [DOI] [PubMed] [Google Scholar]
- Murphy W.J., Larkin D.M., Everts-van der Wind A., Bourque G., Tesler G., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., et al. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science. 2005;309:613–617. doi: 10.1126/science.1111387. [DOI] [PubMed] [Google Scholar]
- Nadeau J.H., Taylor B.A. Lengths of chromosomal segments conserved since divergence of man and mouse. Proc. Natl. Acad. Sci. 1984;81:814–818. doi: 10.1073/pnas.81.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro A., Barton N.H. Chromosomal speciation and molecular divergence-accelerated evolution in rearranged chromosomes. Science. 2003;300:321–324. doi: 10.1126/science.1080600. [DOI] [PubMed] [Google Scholar]
- Noor M.A., Grams K.L., Bertucci L.A., Reiland J. Chromosomal inversions and the reproductive isolation of species. Proc. Natl. Acad. Sci. 2001;98:12084–12088. doi: 10.1073/pnas.221274498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Ancient linkage groups and frozen accidents. Nature. 1973;244:259–262. doi: 10.1038/244259a0. [DOI] [PubMed] [Google Scholar]
- Peng Q., Pevzner P.A., Tesler G. The fragile breakage versus random breakage models of chromosome evolution. PLoS Comput. Biol. 2006;2:e14. doi: 10.1371/journal.pcbi.0020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Ortín J.E., Querol A., Puig S., Barrio E. Molecular characterization of a chromosomal rearrangement involved in the adaptive evolution of yeast strains. Genome Res. 2002;12:1533–1539. doi: 10.1101/gr.436602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevzner P., Tesler G. Human and mouse genomic sequences reveal extensive breakpoint reuse in mammalian evolution. Proc. Natl. Acad. Sci. 2003;100:7672–7677. doi: 10.1073/pnas.1330369100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Herrera A., Castresana J., Robinson T.J. Is mammalian chromosomal evolution driven by regions of genome fragility? Genome Biol. 2006;7:R115. doi: 10.1186/gb-2006-7-12-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankoff D., Trinh P. Chromosomal breakpoint reuse in genome sequence rearrangement. J. Comput. Biol. 2005;12:812–821. doi: 10.1089/cmb.2005.12.812. [DOI] [PubMed] [Google Scholar]
- Schibler L., Roig A., Mahe M.F., Laurent P., Hayes H., Rodolphe F., Cribiu E.P. High-resolution comparative mapping among man, cattle and mouse suggests a role for repeat sequences in mammalian genome evolution. BMC Genomics. 2006;7:194. doi: 10.1186/1471-2164-7-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She X., Cheng Z., Zöllner S., Church D.M., Eichler E.E. Mouse segmental duplication and copy number variation. Nat. Genet. 2008;40:909–914. doi: 10.1038/ng.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skovlund E., Fenstad G.U. Should we always choose a nonparametric test when comparing two apparently nonnormal distributions? J. Clin. Epidemiol. 2001;54:86–92. doi: 10.1016/s0895-4356(00)00264-x. [DOI] [PubMed] [Google Scholar]
- Smith A.V., Daryl J., Heather T.H., Munro M., Abecasis G.R. Sequence features in regions of weak and strong linkage disequilibrium. Genome Res. 2005;15:1519–1534. doi: 10.1101/gr.4421405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snelling W.M., Chiu R., Schein J.E., Hobbs M., Abbey C.A., Adelson D.L., Aerts J., Bennett G.L., Bosdet I.E., Boussaha M., et al. A physical map of the bovine genome. Genome Biol. 2007;8:R165. doi: 10.1186/gb-2007-8-8-r165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F., Herkenne C., Morris M.A., Duboule D. Inversion-induced disruption of the Hoxd cluster leads to the partition of regulatory landscapes. Nat. Genet. 2005;37:889–893. doi: 10.1038/ng1597. [DOI] [PubMed] [Google Scholar]
- Tarchini B., Huynh T.H., Cox G.A., Duboule D. HoxD cluster scanning deletions identify multiple defects leading to paralysis in the mouse mutant Ironside. Genes & Dev. 2005;19:2862–2876. doi: 10.1101/gad.351105. [DOI] [PMC free article] [PubMed] [Google Scholar]