Abstract
Molecular cytogenetics provides a visual, pictorial record of the tree of life, and in this respect the fusion origin of human chromosome 2 is a well-known paradigmatic example. Here we report on a variant chromosome 6 in which the centromere jumped to 6p22.1. ChIP-chip experiments with antibodies against the centromeric proteins CENP-A and CENP-C exactly defined the neocentromere as lying at chr6:26,407–26,491 kb. We investigated in detail the evolutionary history of chromosome 6 in primates and found that the primate ancestor had a homologous chromosome with the same marker order, but with the centromere located at 6p22.1. Sometime between 17 and 23 million years ago (Mya), in the common ancestor of humans and apes, the centromere of chromosome 6 moved from 6p22.1 to its current location. The neocentromere we discovered, consequently, has jumped back to the ancestral position, where a latent centromere-forming potentiality persisted for at least 17 Myr. Because all living organisms form a tree of life, as first conceived by Darwin, evolutionary perspectives can provide compelling underlying explicative grounds for contemporary genomic phenomena.
One of the major tenets of Darwin's theory of evolution is that all forms of life are connected by descent from common ancestors. Extant species represent the endpoint of branches on the tree of life. Paleontology, comparative anatomy, embryology, and more recently comparative genomics, drew trees in which, as in a million-pieces puzzle, each species was placed into a particular position. Bioinformatic sequence comparisons, which can evaluate billions of characters, are robust in scientific terms, but not very accessible to the general public. The molecular cytogenetic approach to the tree of life is more adapt for public viewing because it provides images “that speak.” The most renowned example in this respect is the fusion of two hominid ancestral chromosomes that generated human chromosome 2 and reduced the total number of human chromosomes from 48 found in great apes to 46 (Yunis and Prakash 1982).
Between 17 and 23 million yeas ago (Mya) the centromere of chromosome 6 repositioned to its current location in a common ancestor of the Hominoids (lesser apes [gibbon and siamang], great apes [orangutan, gorilla, and chimpanzee], and humans). Its original position corresponded to human 6p22.1, which (we show here) is the ancestral centromere location for primates. In this report we demonstrate that a human variant chromosome 6, segregating in a three-generational family of normal individuals, has a centromere that repositioned back to the ancestral primate location. Knowledge of the evolutionary past provides compelling underlying explicative grounds for contemporary genomic phenomena.
Results
Family studies
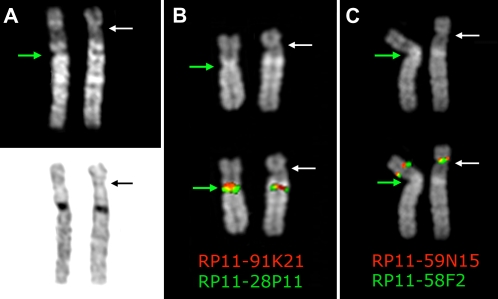
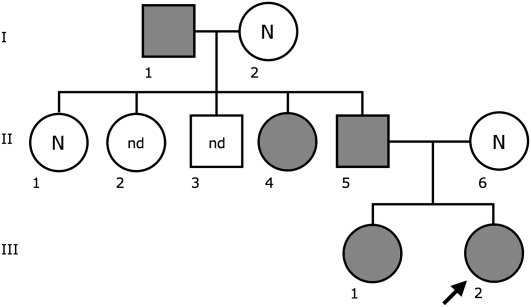
In a prenatal diagnosis due to maternal age (performed in a different laboratory) the fetus was diagnosed with an abnormal chromosome 6 originally interpreted as a pericentric inversion of the short arm (6p21.3–6q12). The father was also diagnosed with the same “inversion.” The couple came to our observation when they requested a prenatal cytogenetic analysis for a subsequent pregnancy. After C-banding showed that the centromeric heterochromatic block retained the original position, we used fluorescence in situ hybridization (FISH) analysis using appropriate bacterial artificial chromosome (BAC) clones to determine whether the centromere shift was the result of an inversion or a centromere repositioning. The BAC–FISH analysis showed that there was no marker order change along the chromosome and ruled out an inversion. Further, reiterative FISH experiments defined the position of the neocentromere as lying between BACs RP11-59N15 (chr6:26,015,628–26,168,047) and RP11-150E2 (chr6:26,546,515–26,703,802) (BAC mapping is according to UCSC hg18, March 2006 release). Examples of relevant FISH results are summarized in Figure 1. The anomaly was therefore interpreted as a centromere repositioning event. The analysis was then extended to the family. The repositioned centromere was found in five individuals in three generations (Fig. 2). The father of the proposita (II-5) was studied in detail. Peripheral blood culture analysis revealed a monosomy of the abnormal chromosome 6 (neocen6) in 18% of metaphases and a neocen6 trisomy in 7% (100 metaphases analyzed). Multiple copies of the neocen6, up to nine, were occasionally observed.
Figure 1.
Chromosome 6 homologs from metaphases belonging to the father of the proposita, (A) after Q-banding top and C-banding bottom; (B) hybridized with BAC clones RP11-28P11 and RP11-91K21, which flank the normal centromere; (C) hybridized with BAC clones RP11-58F2 and RP11-59N15, which flank the repositioned neocentromere. Note that marker order is conserved in the variant chromosome, thus excluding an inversion. The top part of B and C shows the DAPI banding of the homologs to better show the morphology of the chromosomes, the primary constriction in particular, indicated by the arrows. The map position of the BAC clones used in B and C is reported in Table 1. The white arrow indicates the repositioned centromere, while the green arrow indicates the normal centromere.
Figure 2.
Pedigree of the family. Individuals in gray have the repositioned centromere. (N) normal karyotype; (nd) no data.
ChIP-chip analysis
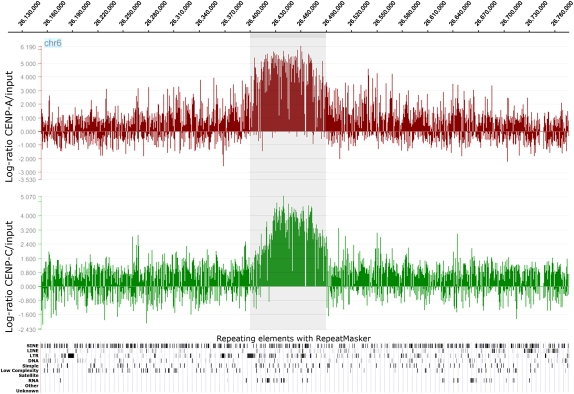
In order to define the position of the neocentromere at the sequence level, we performed ChIP-chip experiments using two rabbit polyclonal antibodies directed against CENP-A or CENP-C human centromeric proteins. These DNA-binding proteins are required for kinetochore function and are exclusively targeted to functional centromeres (for review, see Carroll and Straight 2006). Thus, the immunoprecipitation of the DNA bound to these proteins allows the isolation of centromeric sequences, including those of the neocentromere. The immunoprecipitated and purified DNA was amplified using the Whole Genome Amplification kit (Sigma-Aldrich) and hybridized to a NimbleGen custom tiling array, which has an average resolution of about 100 bp. The enrichment of ChIP DNA, before and after amplification, was validated by real-time PCR (Supplemental Fig. 1). The analysis showed a clear-cut and unique peak at 6p22.1 (chr6:26,407–26,491 kb for CENP-A, and at chr6:26,415–26,491 kb for CENP-C), using very stringent conditions (98th percentile threshold and P < 0.0001; Fig. 3).
Figure 3.
Partial view of the ChIP-chip analysis data on chromosome 6, using anti-CENP-A and anti-CENP-C antibodies. Results are presented as the log2 ratio between the hybridization signal obtained with immunoprecipitated DNA using anti-CENP-A or CENP-C antibodies and that from the input DNA sample. The x-axis shows the genomic position of each oligo on chromosome 6. The data are visualized by the SignalMap software (NimbleGen Systems, Inc.). Details of the microarray structure are reported at the NimbleGen site (http://www.nimblegen.com). The CENP-A and CENP-C domains (the shaded area) clearly map at chr6:26,407–26,491 kb and chr6:26,415–26,491 kb, respectively. Below is shown the RepeatMasker analysis of the interspersed repetitive DNA elements as deducted by the UCSC Genome Browser. The “RNA” lane includes the tRNA elements.
Evolution of chromosome 6
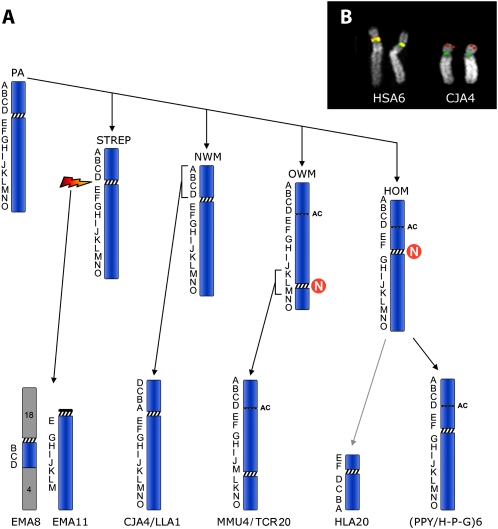
The evolution of chromosome 6 in primates, previously outlined by Eder et al. (2003), was refined using the BACs reported in Table 1. The results (Fig. 4A) strongly suggest that both Hominoidea and Old World Monkeys (OWM) centromeres are evolutionarily new. The position of the centromere was conserved in marmoset (CJA) and in woolly monkey (LLA) (New World Monkeys, NWM), but the short arm underwent, in these species, a paracentric inversion that apparently encompassed the entire short arm, with one breakpoint inside the centromere and the second break at the telomere. A FISH example is reported in Figure 1B.
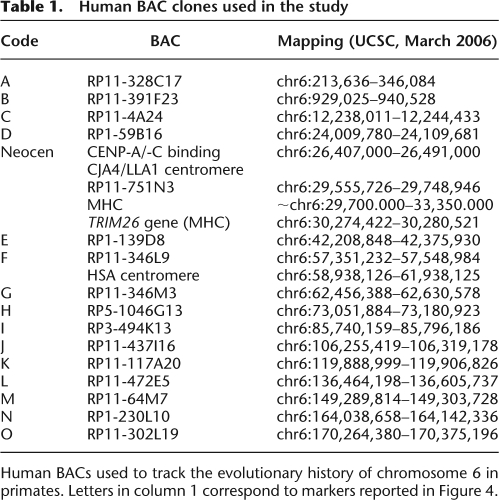
Table 1.
Human BAC clones used in the study
Human BACs used to track the evolutionary history of chromosome 6 in primates. Letters in column 1 correspond to markers reported in Figure 4.
Figure 4.
(A) The figure graphically summarizes the evolutionary flow of chromosomal changes of human chromosome 6 in primates. ENCs (Evolutionary New Centromere) are indicated by the N in a red circle. The letters on the left of each chromosome indicate the BACs used in the study that are reported in Table 1. The acronyms indicate the primate species as reported in the Methods. The rearrangements that intervened between the Hominoidea ancestral form to HLA20 are not illustrated (for details, see Misceo et al. 2008). The Lar gibbon chromosome bearing the remaining portion of chromosome 6 (HLA3) is not reported because it is not relevant for the position of the centromere. (B) Examples of FISH experiments, in human and in marmoset, of the two BAC clones RP11-297M4 (red) and RP11-1021F13 (green), showing that the clone RP11-1021F13 maps on the tip of the CJA4 short arm because of an inversion (for details, see text). (PA) Primate Ancestor; (STREP) Strepsirrhini; (HLA) Hylobates lar; (HOM) Hominoidea; (H-P-G) Homo-Pan-Gorilla group; (AC) Ancestral Centromere.
To characterize in detail the region encompassing the ancestral centromere and the human neocentromere (as defined by the ChIP-chip analysis), we selected a panel of 25 almost-overlapping human BAC clones, starting at chr6:26,015,628 (telomeric to the neocentromere) and ending at chr6:29,748,946 (on the 6q side of the ancestral centromere). In humans the panel covered a region of about 3.73 Mb. Each clone was hybridized in situ to marmoset metaphases. The results are summarized in Table 2. BAC clone RP11-751N3 (chr6:29,259,359–29,405,414) was the clone closest to the CJA4 centromere on the side facing the long arm. In humans it maps telomerically to the Major Histocompatibility Complex (MHC), which maps at ∼chr6:29,700.000–33,350.000. A high proportion of these BACs gave faint signal or failed to yield any FISH signal. Most of the failing clones were very close to the neocentromere domain.
Table 2.
Human BACs spanning the ancestral centromere and the neocentromere
FISH results, on marmoset metaphases, of a panel of 25 almost-overlapping human BAC clones. (nor) Normal FISH signal; (ns) no signal; (MHC) Major histocompatibility complex. Many probes map to CJA4ptel (telomeric region of the CJA4 short arm) because of the inversion of this arm in marmoset (for details, see text and Fig. 4B).
Segmental Duplications (SD) are usually present at inactivated centromeres, as remains of SD clusters that typically flank active centromeres (Ventura et al. 2003; Hillier et al. 2005), while satellite DNA arrays are quickly and completely lost, with the only known exception represented by the relatively recently inactivated centromere at 2q13 (Baldini et al. 1993; Hillier et al. 2005). An SD cluster of about 303 kb is present at chr6:26,775,197–27,078,328, very close to the position of the neocentromere (∼285 kb apart). Altogether, our fine mapping data and SD analysis suggest that the neocentromere locus was seeded in a region corresponding to the pericentromeric domain of the chromosome 6 ancestral centromere.
6p22.1 Sequence features
A very peculiar feature of the region chr6:26,394–29,064 kb, which includes the CENP-A/C domain (chr6:26,407–26,491 kb), is a massive clustering of tRNA (included in the “RNA” lane of the “Repeating Elements by RepeatMasker” track in UCSC browser) (also, see the bottom of Fig. 3). The CENP-A/C domain, in addition, showed an AT content of 57.24% (average genome: 57.2%). The spanning of the different types of repeat elements in the CENP-A/C domain, in the flanking regions, on the entire chromosome 6, and in the human genome, is reported in Table 3. Within the CENP-A/C domains there is the BTN3A2 gene (chr6:26,473,377–26,486,527). This gene encodes a member of the immunoglobulin superfamily, containing two Ig domains with similarity to Ig variable and Ig constant domains. The BTN3A2 expression, evaluated by reverse real-time PCR in the lymphoblastoid cell line derived from the father of the proposita, was found to be very similar to two other lymphoblastoid cell lines taken as a reference (see Supplemental Fig. 2). This result agrees with the previous studies on two neocentromere cases, which have shown that neocentromere formation does not affect the expression of genes that are located inside or near the CENP-A/CENP-C domain (Saffery et al. 2003; Lam et al. 2006).
Table 3.
Repeat element distribution in the CENP-A/C domain and in the two flanking regions
Alu and LINE spanning in the CENP-A/C domain, in the 300 kb flanking, on both sides, this domain, with reference to the entire chromosome 6 and to the entire genome.
Discussion
Here we report on a human chromosome 6 neocentromere segregating in a three-generational normal family. It was discovered serendipitously, during a prenatal diagnosis. The variant chromosome was present also in the grandfather of the proposita. We do not know whether the centromere repositioning occurred de novo in the grandfather or whether it dates back to even more distant generations, because we were unable to examine more distant relatives.
Reports on centromere repositioning in humans are rare because of the lack of phenotypic consequences. If the chromosome Y is not considered, this is only the third finding of centromere repositioning in humans (Amor et al. 2004; Ventura et al. 2004). Supernumerary chromosomal fragments with neocentromeres, on the contrary, are not infrequently found, because these produce abnormal phenotypes, which do not escape the clinical filter. Yet, the relatively high number of evolutionary centromere repositioning events implies a corresponding high basal level of this phenomenon. Comparison shows that about half the centromeres between macaques and humans have evolutionarily new centromeres (Ventura et al. 2007). In this context it is also worth noting that all evolutionary novel centromeres, in macaque in particular, acquired large blocks of satellite DNA that probably stabilized centromeric function. In the present case, indeed, as well as in many clinical neocentromere cases, the centromeric functionality is not optimized, as demonstrated by the frequently reported somatic mosaicism for the chromosome bearing the neocentromere (see Marshall et al. 2008).
Evolutionary history of chromosome 6
The marker order of the short arm of chromosome 6 in OWM and Hominoidea (Fig. 4) was identical even if the centromere positions were different. The marker order of NWM differed for a single inversion encompassed by markers A–D (Fig. 4). Eder et al. (2003) noted that the NWM order was similar to that of the cat and suggested that this marker order was ancestral. Studies on MHC provided the most pertinent information for reconstructing the ancestral primate organization of HSA6p. They suggested an alternative hypothesis. In the cat the MHC was disrupted by an inversion whose centromeric breakpoint was close to the TRIM26 gene (chr6:30,274,422–30,280,521) (Beck et al. 2005). These data clash with data from the vast majority of species in which the MHC synteny was conserved in a single uninterrupted block (Beck et al. 2005). The interrupted MHC in the cat appears, therefore, as a derived character. More precise data on the inversion that generated chromosome CJA4 and LLA1 in NWM, the mapping of the BAC RP11-751N3 in particular, indicated that, unlike the cat, the MHC in CJA was not disrupted by the inversion. The two inversions were therefore independent and do not support the previous hypothesis that the inversion was already present in the primate ancestor.
Another line of evidence in favor of the second hypothesis was provided by molecular cytogenetic studies in Strepsirrhini (Muller et al. 1997; Cardone et al. 2002). In black lemurs (Eulemur macaco, EMA, Strepsirrhini) human chromosome 6 synteny was disrupted into two segments. One segment is composed by EMA chromosome 11 which is homologous to the long arm of marmoset chromosome 4 (CJA4), as if a break occurred at the centromere (Fig. 4). The remaining portion of CJA4 is associated with human 18 and human 4 (18-cent-6/4) to form EMA8 (data from Eder et al. [2003]). The original interpretation was that an ancestral chromosome corresponding to CJA4 was fissioned at the centromere as the initial rearrangement leading to EMA8 and EMA11. The centromere of EMA8 is on the same side of the chromosome 6 segment that corresponds to CJA4p, lending credence to the hypothesis that the centromere of EMA8 derives from the homolog to human chromosome 6. This scenario would provide support for an ancestral inverted marker order compared with humans. However, an alternative interpretation seems more plausible if we include molecular cytogenetic data from Eulemur fulvus and Lemur catta (Muller et al. 1997; Cardone et al. 2002) (see, in particular, Fig. 4 of Cardone et al. 2002). E. fulvus has a chromosome (EFU13) composed only of the 6/4 association. That this chromosome was ancestral to EMA8 is supported by the fact that the same association without the 18 association is also found in Lemur catta chromosome 4. Apparently, in EMA, an apomorphic fusion of this chromosome with the homolog to human 18 led to EMA8. It is significant that in E. fulvus the centromere for the 6/4 association is provided by chromosome 4. The most likely interpretation is that 6/4 plus 18 fusion in EMA was a centromere to telomere fusion. The 18 centromere remained active and the chromosome 4 centromere was inactivated. Therefore, EMA8 provides no support for an inverted marker order in the primate ancestor.
The most parsimonious interpretation is that the marker order in the ancestral primate chromosome 6 was identical to humans. Further, the centromere in EMA11 almost certainly derives from chromosome 6 and provides good support that the ancestral primate centromere position, given that an identical position is found in both NWM and Strepsirrhines, was at 6p22.1.
The peculiar feature of our finding, therefore, is that the repositioning took place in a domain where an ancestral centromere inactivated, as if a latent potentiality was dormant for at least 17 Myr. The ancestral centromere, indeed, was inactivated, very likely after OWM and before gibbon divergence, in the range of from ∼17 to 23 Mya (Raaum et al. 2005). SD remains are clearly present at this domain (chr6:26,840,000–27,078,000), while almost absent around the normal human centromere, indicating its recent emergence (She et al. 2004).
The fine mapping by ChIP-chip analysis of a number of human neocentromeres has allowed a precise sequence comparison among different seeding domains (for review, see Capozzi et al. 2008; Marshall et al. 2008). The analysis, however, did not disclose any shared critical sequence features that could predict this potentiality, with the only exception of a satellite DNA in human that corresponds to an evolutionary new centromere on OWM chromosome 18 (Carbone et al. 2008). In our case, we have noticed a massive clustering of tRNAs in the region (see the “Repeating Elements by RepeatMasker” track in UCSC browser) (Fig. 3, bottom). These findings are typical of some pericentromeric and telomeric regions. In fission yeast, a tRNA has been reported as a functional barrier separating the heterochromatin central core from outer heterochromatin (Scott et al. 2006). The CENP-A/C domain showed an AT content of 57.24% (average genome: 57.2%). Repeat element distribution within the CENP-A/C domain, in the 300 kb flanking the domain on both sides, as well as in the entire chromosome 6 and in the human genome, are reported in Table 3. AluY and AluSx are separately reported because they appear to be the more active Alu elements (Bennett et al. 2008). In evaluating the differences between the region under study against the entire chromosome 6 and the entire genome, it has to be kept in mind that Alu and LINE elements are unevenly distributed along the human genome. Alu are particularly abundant in GC- and gene-rich regions (Lander et al. 2001) and show a strong correlation with R-banding (Baldini and Ward 1991).
A number of cases with trisomy 6p have been reported in literature (Domínguez et al. 2003), but no clinical neocentromere 6p22.1 have been described. However, most of the extra chromosomes harboring neocentromeres consist of an inverted duplication (invdup), which results in a partially tetrasomic karyotype (Marshall et al. 2008). Tetrasomy of 6p is probably incompatible with a normal embryonic development.
Concluding remarks
Reuse of chromosomal breakpoint domains in evolution is well documented (Pevzner and Tesler 2003). Some clinical neocentromeres arose in the same sequence domain where ancestral centromeres were seeded (Ventura et al. 2003; Cardone et al. 2006; Capozzi et al. 2008) or where an ancestral centromere was inactivated (Ventura et al. 2004). In the present case, mapping data and sequence features suggest that the neocentromere locus was seeded in a region corresponding to the pericentromeric domain of the ancestral centromere. It represents the first instance, in primates, of a centromere repositioning event bringing the centromere back to the ancestral position, providing an example clearly showing that our genome shares its history with other extant species because they are branches of a unique tree of life, as first conceived by Darwin.
Methods
Fluorescence in situ hybridization (FISH)
Metaphase preparations from familial studies were from standard blood cultures, except for the prenatal diagnosis that utilized amniotic fluid cell culture. Metaphases from nonhuman primates were obtained from lymphoblastoid or fibroblast cell lines of the following species: great apes: common chimpanzee (Pan troglodytes, PTR); gorilla (Gorilla gorilla, GGO); Borneo orangutan (Pongo pygmaeus pygmaeus, PPY); Lar gibbon (Hylobates lar, HLA, Hylobatidae); OWM: rhesus monkey (Macaca mulatta, MMU, Cercopithecinae); silvered leaf-monkey (Trachypithecus cristatus, TCR, Colobinae); New World Monkeys (NWM): common marmoset (Callithrix jacchus, CJA, Callitrichinae); woolly monkey (Lagothrix lagothricha, LLA, Callicebinae).
DNA extraction from BACs and FISH experiment protocols were reported previously. Digital images were obtained using a Leica DMRXA2 epifluorescence microscope equipped with a cooled CCD camera (Princeton Instruments). Cy3-dCTP, FluorX-dCTP, DEAC, Cy5-dCTP, and DAPI fluorescence signals, detected with specific filters, were recorded separately as grayscale images. Pseudocoloring and merging of images were performed using Adobe Photoshop software.
ChIP-chip analysis
To identify the sequences bound by CENP-A, native chromatin immune-precipitation (N-ChIP) analysis was performed, as previously described (Umlauf et al. 2004). Briefly, lymphoblastoid cells derived from the father were processed and the native chromatin was prepared by nuclease digestion of cell nuclei, then the immunoprecipitation was performed using polyclonal antibodies against the centromeric protein CENP-A. Cross-linked chromatin immune-precipitation (X-ChIP) analysis, as previously described (Wells and Farnham 2002), was performed to identify the sequences bound by CENP-C. Briefly, cells were cross-linked in situ by adding formaldehyde to a 1% final concentration directly to the culture medium, and chromatin was immunoprecipitated with an anti-CENP-C polyclonal antibody (S. Trazzi, G. Perini, R. Bernardoni, M. Zoli, J.C. Reese, A. Musacchio, and G. Della Valle, in prep.). In both methods, purified DNA fragments were amplified using the Whole Genome Amplification kit (Sigma-Aldrich). The labeled ChIP and total DNAs were cohybridized to a NimbleGen custom tiling array (specific for chromosome 6-masked sequences from build hg18), which has an average resolution of 100 bp. DNA-binding peaks were identified by using the statistical model and methodology described at http://chipanalysis.genomecenter.ucdavis.edu/cgi-bin/tamalpais.cgi (Bieda et al. 2006).
BTN3A2 expression
Total RNA was extracted from the lymphoblastoid cells using TriReagent (Sigma-Aldrich). Total RNA was treated with DNase I (New England BioLabs, Inc.) to remove possible genomic contamination, and DNA-free RNA was retrotranscribed with SuperScript III (Invitrogen). Diluted RT reaction was used for Real Time PCR using the IQ SYBR Green Supermix (Bio-Rad) performed on an IQ5 Real-time PCR machine (Bio-Rad). Primers for BTN3A2 were the following: AAGACAGCCAGCATTTCCAT (BTN3A2_1s), GAGAAGCAGCAGCAAGATAGG (BTN3A2_1as), GCAACAGAGCGGGAAATAAG (BTN3A2_2s), and ACGAAGACTCCTCTCCACGA (BTN3A2_2as). Expression of two housekeeping genes (GUSB and ACTB) were used for normalization.
Acknowledgments
This project was supported by PRIN 2006 funded by MUR (Ministero della Universita' e della Ricerca). We thank S. Trazzi for anti-CENP-A and anti-CENP-C sera (S. Trazzi, G. Perini, R. Bernardoni, M. Zoli, J.C. Reese, A. Musacchio, and G. Della Valle, in prep.). S.P. was supported by a fellowship from European Union–Programma Regionale per la Ricerca Industriale.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article is online at http://www.genome.org/cgi/doi/10.1101/gr.085688.108.
References
- Amor D.J., Bentley K., Ryan J., Perry J., Wong L., Slater H., Choo K.H. Human centromere repositioning “in progress.”. Proc. Natl. Acad. Sci. 2004;101:6542–6547. doi: 10.1073/pnas.0308637101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldini A., Ward D.C. In situ hybridization banding of human chromosomes with Alu-PCR products: A simultaneous karyotype for gene mapping studies. Genomics. 1991;9:770–774. doi: 10.1016/0888-7543(91)90374-n. [DOI] [PubMed] [Google Scholar]
- Baldini A., Ried T., Shridhar V., Ogura K., D'Aiuto L., Rocchi M., Ward D.C. An alphoid DNA sequence conserved in all human and great ape chromosomes: Evidence for ancient centromeric sequences at human chromosomal regions 2q21 and 9q13. Hum. Genet. 1993;90:577–583. doi: 10.1007/BF00202474. [DOI] [PubMed] [Google Scholar]
- Beck T.W., Menninger J., Murphy W.J., Nash W.G., O'Brien S.J., Yuhki N. The feline major histocompatibility complex is rearranged by an inversion with a breakpoint in the distal class I region. Immunogenetics. 2005;56:702–709. doi: 10.1007/s00251-004-0742-6. [DOI] [PubMed] [Google Scholar]
- Bennett E.A., Keller H., Mills R.E., Schmidt S., Moran J.V., Weichenrieder O., Devine S.E. Active Alu retrotransposons in the human genome. Genome Res. 2008;18:1875–1883. doi: 10.1101/gr.081737.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieda M., Xu X., Singer M.A., Green R., Farnham P.J. Unbiased location analysis of E2F1-binding sites suggests a widespread role for E2F1 in the human genome. Genome Res. 2006;16:595–605. doi: 10.1101/gr.4887606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capozzi O., Purgato S., Verdun di Cantogno L., Grosso E., Ciccone R., Zuffardi O., Della Valle G., Rocchi M. Evolutionary and clinical neocentromeres: Two faces of the same coin? Chromosoma. 2008;117:339–344. doi: 10.1007/s00412-008-0150-z. [DOI] [PubMed] [Google Scholar]
- Carbone L., Misceo D., D'Addabbo P., Vessere G., De Jong P., Rocchi M. A satellite-like sequence, representing a “clone gap” in the human genome, was likely involved in the seeding of a novel centromere in macaque. Chromosoma. 2008 doi: 10.1007/s00412-008-0196-y. [DOI] [PubMed] [Google Scholar]
- Cardone M.F., Ventura M., Tempesta S., Rocchi M., Archidiacono N. Analysis of chromosome conservation in Lemur catta studied by chromosome paints and BAC/PAC probes. Chromosoma. 2002;111:348–356. doi: 10.1007/s00412-002-0215-3. [DOI] [PubMed] [Google Scholar]
- Cardone M.F., Alonso A., Pazienza M., Ventura M., Montemurro G., Carbone L., de Jong P.J., Stanyon R., D'Addabbo P., Archidiacono N., et al. Independent centromere formation in a capricious, gene-free domain of chromosome 13q21 in Old World monkeys and pigs. Genome Biol. 2006;7:R91. doi: 10.1186/gb-2006-7-10-r91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll C.W., Straight A.F. Centromere formation: From epigenetics to self-assembly. Trends Cell Biol. 2006;16:70–78. doi: 10.1016/j.tcb.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Domínguez M.G., Wong-Ley L.E., Rivera H., Vásquez A.I., Ramos A.L., Sánchez-Urbina R., Morales J.A., Figuera L.E. Pure partial trisomy 6p due to a familial insertion (16;6)(p12;p21.2p23) Ann. Genet. 2003;46:45–48. doi: 10.1016/s0003-3995(03)00004-2. [DOI] [PubMed] [Google Scholar]
- Eder V., Ventura M., Ianigro M., Teti M., Rocchi M., Archidiacono N. Chromosome 6 phylogeny in primates and centromere repositioning. Mol. Biol. Evol. 2003;20:1506–1512. doi: 10.1093/molbev/msg165. [DOI] [PubMed] [Google Scholar]
- Hillier L.W., Graves T.A., Fulton R.S., Fulton L.A., Pepin K.H., Minx P., Wagner-McPherson C., Layman D., Wylie K., Sekhon M., et al. Generation and annotation of the DNA sequences of human chromosomes 2 and 4. Nature. 2005;434:724–731. doi: 10.1038/nature03466. [DOI] [PubMed] [Google Scholar]
- Lam A.L., Boivin C.D., Bonney C.F., Rudd M.K., Sullivan B.A. Human centromeric chromatin is a dynamic chromosomal domain that can spread over noncentromeric DNA. Proc. Natl. Acad. Sci. 2006;103:4186–4191. doi: 10.1073/pnas.0507947103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Marshall O.J., Chueh A.C., Wong L.H., Choo K.H. Neocentromeres: New insights into centromere structure, disease development, and karyotype evolution. Am. J. Hum. Genet. 2008;82:261–282. doi: 10.1016/j.ajhg.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misceo D., Capozzi O., Roberto R., Dell'oglio M.P., Rocchi M., Stanyon R., Archidiacono N. Tracking the complex flow of chromosome rearrangements from the Hominoidea ancestor to extant Hylobates and Nomascus gibbons by high-resolution, synteny mapping. Genome Res. 2008;18:1530–1537. doi: 10.1101/gr.078295.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S., O'Brien P.C.M., Ferguson-Smith M.A., Wienberg J. Reciprocal chromosome painting between human prosimisans (Eulemur macaco macaco and E. fulvus mayottensis) Cytogenet. Cell Genet. 1997;78:260–271. doi: 10.1159/000134669. [DOI] [PubMed] [Google Scholar]
- Pevzner P., Tesler G. Human and mouse genomic sequences reveal extensive breakpoint reuse in mammalian evolution. Proc. Natl. Acad. Sci. 2003;100:7672–7677. doi: 10.1073/pnas.1330369100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaum R.L., Sterner K.N., Noviello C.M., Stewart C.B., Disotell T.R. Catarrhine primate divergence dates estimated from complete mitochondrial genomes: Concordance with fossil and nuclear DNA evidence. J. Hum. Evol. 2005;48:237–257. doi: 10.1016/j.jhevol.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Saffery R., Sumer H., Hassan S., Wong L.H., Craig J.M., Todokoro K., Anderson M., Stafford A., Choo K.H. Transcription within a functional human centromere. Mol. Cell. 2003;12:509–516. doi: 10.1016/s1097-2765(03)00279-x. [DOI] [PubMed] [Google Scholar]
- Scott K.C., Merrett S.L., Willard H.F. A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. Curr. Biol. 2006;16:119–129. doi: 10.1016/j.cub.2005.11.065. [DOI] [PubMed] [Google Scholar]
- She X., Horvath J.E., Jiang Z., Liu G., Furey T.S., Christ L., Clark R., Graves T., Gulden C.L., Alkan C., et al. The structure and evolution of centromeric transition regions within the human genome. Nature. 2004;430:857–864. doi: 10.1038/nature02806. [DOI] [PubMed] [Google Scholar]
- Umlauf D., Goto Y., Feil R. Site-specific analysis of histone methylation and acetylation. Methods Mol. Biol. 2004;287:99–120. doi: 10.1385/1-59259-828-5:099. [DOI] [PubMed] [Google Scholar]
- Ventura M., Mudge J.M., Palumbo V., Burn S., Blennow E., Pierluigi M., Giorda R., Zuffardi O., Archidiacono N., Jackson M.S., et al. Neocentromeres in 15q24-26 map to duplicons which flanked an ancestral centromere in 15q25. Genome Res. 2003;13:2059–2068. doi: 10.1101/gr.1155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M., Weigl S., Carbone L., Cardone M.F., Misceo D., Teti M., D'Addabbo P., Wandall A., Björck E., de Jong P., et al. Recurrent sites for new centromere seeding. Genome Res. 2004;14:1696–1703. doi: 10.1101/gr.2608804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M., Antonacci F., Cardone M.F., Stanyon R., D'Addabbo P., Cellamare A., Sprague L.J., Eichler E.E., Archidiacono N., Rocchi M. Evolutionary formation of new centromeres in macaque. Science. 2007;316:243–246. doi: 10.1126/science.1140615. [DOI] [PubMed] [Google Scholar]
- Wells J., Farnham P.J. Characterizing transcription factor binding sites using formaldehyde crosslinking and immunoprecipitation. Methods. 2002;26:48–56. doi: 10.1016/S1046-2023(02)00007-5. [DOI] [PubMed] [Google Scholar]
- Yunis J.J., Prakash O. The origin of man: A chromosomal pictorial legacy. Science. 1982;215:1525–1530. doi: 10.1126/science.7063861. [DOI] [PubMed] [Google Scholar]