Abstract
Population structure can provide novel insight into the human past, and recognizing and correcting for such stratification is a practical concern in gene mapping by many association methodologies. We investigate these patterns, primarily through principal component (PC) analysis of whole genome SNP polymorphism, in 2099 individuals from populations of Northern European origin (Ireland, United Kingdom, Netherlands, Denmark, Sweden, Finland, Australia, and HapMap European-American). The major trends (PC1 and PC2) demonstrate an ability to detect geographic substructure, even over a small area like the British Isles, and this information can then be applied to finely dissect the ancestry of the European-Australian and European-American samples. They simultaneously point to the importance of considering population stratification in what might be considered a small homogeneous region. There is evidence from FST-based analysis of genic and nongenic SNPs that differential positive selection has operated across these populations despite their short divergence time and relatively similar geographic and environmental range. The pressure appears to have been focused on genes involved in immunity, perhaps reflecting response to infectious disease epidemic. Such an event may explain a striking selective sweep centered on the rs2508049-G allele, close to the HLA-G gene on chromosome 6. Evidence of the sweep extends over a 8-Mb/3.5-cM region. Overall, the results illustrate the power of dense genotype and sample data to explore regional population variation, the events that have crafted it, and their implications in both explaining disease prevalence and mapping these genes by association.
Patterns of genetic variation within and between human populations have long provided novel insights into the origin and history of different groups. The advent of whole genome association (WGA) mapping has also highlighted the practical importance of identifying and understanding these patterns. A mismatch in the ancestry of individuals in a simple case/control association paradigm can lead to false positives and/or reduced power to detect associations.
Studies of population-level whole genome (WG) polymorphism were initially restricted to the International HapMap populations (Yoruban, Japanese, Chinese, and European-Americans) but provided valuable information on intercontinental variation across the human genome, including structural variation, recombination, and selection (International HapMap Consortium 2005, 2007). The whole genome approach has now begun to be applied to more nuanced intracontinental variation within Europe. First generation studies using European-Americans or small numbers of in situ Europeans (but with relatively few markers) quickly identified a clear North–South split in the continent's population and hinted at further structure (Seldin et al. 2006; Bauchet et al. 2007; Price et al. 2008; Seldin and Price 2008; Tian et al. 2008). Analysis of WG variation in larger numbers of individuals sampled in situ from multiple European populations has recently extended these findings. Using principal component analysis (PCA) of up to ≈300,000 SNPs, they have shown a remarkable correlation of an individual's position in genetic space to their geographic origin (Heath et al. 2008; Lao et al. 2008; Novembre et al. 2008).
We continue this progression by exploring subcontinental WG (300K) variation in 2099 individuals from six Northern European populations (Ireland, United Kingdom, Netherlands, Sweden, Denmark, and Finland), as well as two descendent New World populations (European-Australians and the European-American HapMap sample). The data demonstrate and confirm an ability to dissect regional to subregional geographic structure and also point to the discernable impact of differential natural selection on the recently diverged Northern European populations. Both of these observations have important present-day consequences in explaining disease incidence and in mapping complex traits through association methods.
Results
Principal component analysis of Northern Europeans
We initially investigated the genetic structure of eight European population samples (Netherlands, Sweden, Denmark, Ireland, United Kingdom, Finland, HapMap European-American [CEU], and Australia) using principal component analysis (PCA) of 296,553 autosomal SNPs. We did not prune the genotypes with respect to linkage disequilibrium (LD) prior to PCA since this allows us to investigate both population substructure and large genomic structural/haplotypic variation. The novel genotype data from this study are deposited at the European Genotype Archive (see Methods).
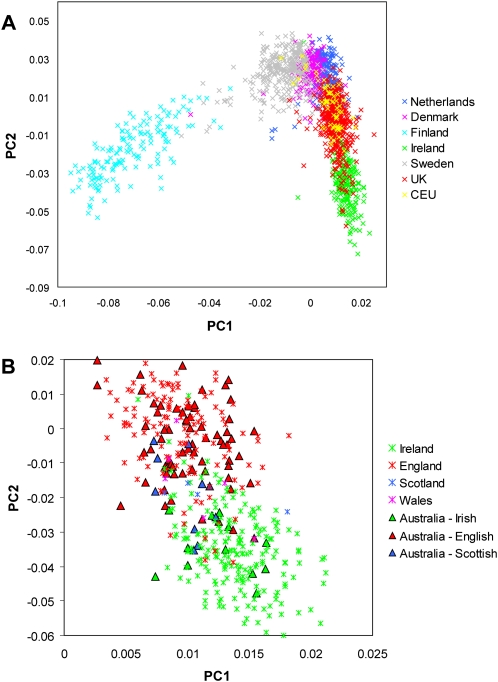
The top 100 PCs were generated using the EIGENSOFT package (Patterson et al. 2006; Price et al. 2006); however, we focused on the top five since the eigenvalues remain relatively constant in subsequent PCs (Supplemental Fig. S1). PC1 and PC2 are plotted together in Figure 1A. Individuals from the different populations are largely separable, although there is clearly some overlap, suggesting that variation is generally continuous rather than discrete. Mantel testing (Mantel 1967) confirms that differences in individual PC1 or PC2 scores are strongly correlated with the geographic distance between sampled individuals (r = 0.76, P < 0.0001 and r = 0.36, P < 0.0001, respectively). PC1 most obviously separates the Finnish individuals from the other Northern Europeans and, more subtly, these from each other. Remarkably, given the smaller geographic distances, this pattern is apparent even within Finland once regional origins are considered (Supplemental Fig. S2). PC2 tends to separate the insular Irish and U.K. populations from each other and from their closest continental neighbors. We repeated PCA excluding the Finns, given their relatively outlying positions, and were still able to discern good separation between the remaining populations with the originally observed PC2 becoming the most prominent pattern in the data (PC1 in Supplemental Fig. S3).
Figure 1.
PCA of Northern European population structure. (A) PC1 versus PC2 from 2051 individuals genotyped for 296,553 autosomal SNPs. PCA was conducted including the Australian sample (n = 451), but these are not shown here (see Supplemental Fig. S4). (B) PC1 versus PC2 focused on the United Kingdom, Irish, and Australian populations. For ease of illustration, PC1 has been constrained to between 0.02 and −0.06 and PC2 to between 0 and 0.025. Only those U.K. samples with birthplace information are displayed (n = 143), and these are distinguished as England, Scotland, and Wales. Australian samples whose four grandparental ancestries are from one country are also shown.
As previous studies have noted (Bauchet et al. 2007; Lao et al. 2008), the position of CEU HapMap individuals in the genetic space is consistent with their putative Northwest European ancestry. This illustrates the potential for ancestry reconstruction given sufficient numbers of markers and appropriate comparative populations, an application we used next to investigate the origin of a large Australian population sample.
Population structure of Australia and the British Isles
Approximately 85% of current Australians are descendents of European settlers who began arriving in 1788. The vast majority of these originated in Britain or Ireland but were joined by smaller numbers of Germans, Greeks, Italians, and Eastern Europeans (among others) in the 19th and 20th Centuries.
Fourteen individuals from our Australian sample (n = 465) were identified as outliers in eigenvector analysis, and subsequent checks revealed that these had self-reported non-European or Southern European ancestry. Virtually all of the remaining Australians are distributed in the PC1–PC2 regions occupied by the United Kingdom (noting its overlap with the Dutch sample) and Irish populations (Fig. 1B; Supplemental Fig. S4). PC2 is most informative at distinguishing the United Kingdom and Irish populations. The mean Australian PC2 value (−0.01) is closer to the United Kingdom (−0.003) than Ireland (−0.036), with the difference between Australia and the United Kingdom ∼22% of the total distance between the United Kingdom and Irish means along this axis. Interestingly, this figure coincides with the fraction of all British Isles self-reported ancestry mentions that are Irish (Source: 2006 Census, Australia Bureau of Statistics; http://www.abs.gov.au/).
Several previous studies, using single marker systems and most recently whole genome SNP data (Wellcome Trust Case Control Consortium 2007), have noted a gradient of genetic variation running approximately Southeast to Northwest across Britain or the British Isles. Although we did not have detailed regional ancestry information for the United Kingdom or Irish samples, we used place of birth, which was available for a subset of the United Kingdom sample (n = 143) to divide these into English (n = 133), Scottish (n = 4), and Welsh (n = 6) cohorts. Although numbers are small, the mean PC2 values for the Scottish and Welsh cohorts are intermediate between those of England and Ireland (Fig. 1B).
We also availed of detailed self-reported ancestry for the Australian sample to distinguish three further cohorts whose four-grandparental ancestries are English (n = 61), Irish (n = 12), or Scottish (n = 10). There is a significant correspondence of this self-reported ancestry with PC2 scores (one-way ANOVA, P < 0.0001). Once again the average intermediate position of the Australian-Scots (mean PC2 score of −0.019 vs. −0.005 in the English and −0.035 in Irish-Australian cohorts) is evidence that further subregional patterns can be discerned across the British Isles (Fig. 1B).
Ancestry informative markers and structure
We next explored whether smaller numbers of SNPs, which are most differentiated between our populations, are effective at capturing the variation revealed by the full 296K markers. Panels of Ancestry Informative Markers (AIMs) could have important practical applications in detecting and correcting for variation in individual ancestry that can confound some association gene mapping methods by increasing false positive results and/or reducing power. This is particularly the case in candidate gene-driven studies or follow-up replication attempts where a full suite of genome polymorphism is not available.
The Northern European sample population was divided in half to give a discovery (n = 770) and a test panel (n = 1281, which included Australia and CEU). FST values between populations in the discovery panel were used to select 500, 1500, 2500, and 5000 top-ranked SNP markers as AIM sets with the additional condition that no marker was within 250 kb of another, a measure taken to minimize LD between SNPs. The performance of these AIM panels was then assessed in the test data set. All panels returned PC1 scores for individuals that were significantly correlated to the values revealed by the full data set, both when the Finns were included and excluded (r ≥ 0.91 and r ≥ 0.67, respectively, with minimum r values observed using the 500 SNP sets). All sets performed substantially better than random marker sets of the same size especially as AIM panel size decreased (Supplemental Fig. 5A). In a similar analysis of PC2, the 5000 AIM set preserved a moderate correlation (r = 0.649, P < 0.05) to the full SNP test set, but further reductions in AIM number led to a steep decline in correlation, and they are no longer significant for sets of 1500 or 500 markers either FST-based or randomly selected (Supplemental Fig. 5A).
We also explored the performance of the AIM panels using an alternative Bayesian modeling approach implemented in the structure package (Pritchard et al. 2000; Falush et al. 2003). We examined each AIM panel over K = 2 to K = 5 in the test sample set, where K is the predefined number of populations into which the data are to be split (Supplemental Figs. 5B and 6). At K = 2, all AIM panels were able to distinguish the Finnish samples from other populations. At K = 3, there was little obvious coherent division of the remaining European samples for AIM sets of 500 to 2500 SNPs. However, at 5000 markers, some distinction between the British Isles (especially the Irish) and non-Finnish continental samples became apparent (Supplemental Fig. 5B). This is reminiscent of PC2, and the results again appear to demonstrate the majority British Isles ancestry of the Australian sample. We also repeated the AIM selection and structure analysis for 5000 SNPs excluding the Finnish sample and discerned some tendency for the separation of the British Isles (especially Irish) from the continental Europeans over various K values (Supplemental Fig. 5C). Overall, the results illustrate the potential to develop a single AIM set that will be informative on even subtle Northern European variation, an ability that will increase as bigger sets of initially genotyped markers (in more populations) become available. (A list of the top 10,000 SNPs by FST value is given in Supplemental Table 1.
Extent of population stratification
Mismatched ancestry between cases and control groupings in a standard whole genome association study is a potential source of Type 1 and 2 errors. However, balanced against this concern are the potentials to increase power by combining (often rare) cases across populations and achieve significant cost savings by reusing standard sets of controls. The genomic control inflation factor (λgc) is one way to quantify the impact of population stratification between cases and controls (Devlin and Roeder 1999). However, this is highly dependent on sample sizes as well as the level of ancestry mismatch between groups as measured by FST. As theory suggests (and as we empirically observe—see Supplemental Fig. 7), the relationship between λgc, FST, and total sample size (n individuals) is approximately E(λgc) ∼ 1 + (n * FST) when FST is small (such as the levels between European populations) and the numbers of “cases” and “controls” are the same.
As an illustration of the potential impact of Northern European stratification, we calculated expected pairwise λgc values between populations (equivalent to a situation in which cases and controls come entirely from different populations) given the FST values in Table 1 and assuming a total sample size of 1000 individuals. These show that a random United Kingdom population sample is well matched to the Australians (λgc = 1.025) after obvious outliers are removed from both, either by self-reported ancestry or by EIGENSTRAT. However, other combinations show substantial inflation and corresponding potential for false positives (Table 1).
Table 1.
Population pairwise FST (above the diagonal) and expected λgc values (below the diagonal)
Expected λgc values based on a case-control experiment of 500 cases and 500 controls, using E(λgc) = 1 + 1000 * FST.
Differential natural selection among Northern Europeans
While the differentiation/structure observed between our populations is expected to be mainly a consequence of neutral evolution (drift) and demographic processes like migration, it is also possible that natural selection has played a role in shaping the diversity of some SNPs. One simple expected signature of differential positive selection—the action of a selective force in some populations but not others—is the overrepresentation of SNPs with high FST values in genic regions, since these are a (albeit imperfect) proxy for functional polymorphism. A recent study demonstrated this effect for an FST comparison between the HapMap populations (Barreiro et al. 2008).
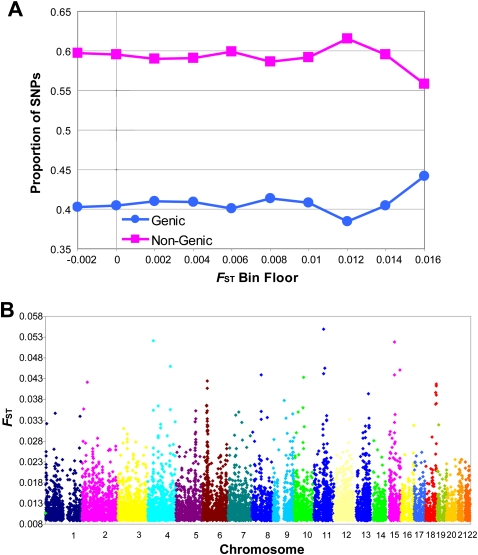
We divided FST values calculated at each autosomal SNP in our six in situ European populations (Denmark, Ireland, Netherlands, Finland, United Kingdom, and Sweden) into 10 bins of increasing (0.002 increment) FST floor and noted a significant excess of genic SNPs in the top FST (>0.016) category (χ2 = 7.4; d.f. = 1; P = 0.0071; Fig. 2A). The χ2 test may be nonconservative in these circumstances given the potential nonindependence of markers due to LD. However, this is mitigated to some extent by the “tagging” criteria inherent in the selection of SNPs for the genotyping platform. Furthermore, stringent pruning for LD would bias the genic/nongenic test against detecting selection since the “hitchhiking” effect is a key footprint of its action. The effect was still detectable but weaker after the Finnish population was removed (χ2 = 4.9; d.f. = 1; P = 0.028). However, given the relative genetic distinction of the Finnish, the absolute FST value for each SNP is generally lower after their exclusion, and only 50 SNPs exceed FST of 0.016 (compared to 1358 including Finland). Rescaling to a cut-off of FST > 0.01 still preserves the enrichment trend (χ2 = 4.0; d.f. = 1; P = 0.046).
Figure 2.
Northern European FST values by genomic location. (A) Proportion of SNPs that are inside (genic) or outside (nongenic) genes by FST bin category. X-axis FST values refer to the lower boundary of the bin. (B) Genome-wide distribution of the top 10,000 FST (>0.00888) values by chromosome and genomic position.
Interpretation of these findings requires a consideration of any ascertainment bias underlying the inclusion of genic versus nongenic SNPs on the genotyping platform and whether any systematic bias in the relationship of minor allele frequency (MAF) to FST exists. The overall genic/nongenic distribution of SNPs is very weakly but significantly different (Kolmogorov–Smirnov test: D = 0.0117, P = 7.5 × 10−9; Supplemental Fig. S8A) owing to a slight excess of genic SNPs in the lower MAF categories. We therefore randomly sampled from the nongenic SNPs to ensure that the number in each MAF bin was exactly equal (even though this involves discarding ≈20% of the SNPs and potentially reducing the power to detect any effect). Based on 1000 such replicate analyses, we continue to observe a similar trend of genic enrichment in relation to high FST (P = 0.017). Furthermore, the average FST estimate was similar across most FST categories, suggesting that difference in MAF could not explain the finding (Supplemental Fig. S8B). Indeed, there is some evidence of systematically lower FST values in lower MAF categories, which would bias against detecting the observed excess (since genic SNPs are somewhat overrepresented in these lower categories).
The genomic locations of the top 10,000 SNPs by FST value are shown in Figure 2B (with a more detailed view available in Supplemental Table 1). There is clustering of high FST values at certain locations, and, while it is impossible to definitively ascribe any particular region of high FST values to positive selection, these may be considered candidate regions for its action. We identified the most prominent of these (which had a peak FST within the top 100 ranked values or FST > ≈0.028; Table 2). Consistent with similar analysis in the United Kingdom population (Wellcome Trust Case Control Consortium 2007), the human-leukocyte-antigen (HLA) region of chromosome 6 is the most obvious candidate. However, the enrichment of genic SNPs in the top FST category remains significant (χ2 = 6.2; d.f. = 1; P = 0.011) even when a large region around the top HLA SNP is removed from the analysis. There is also a prominent peak on chromosome 5 (around rs9784675/position 132.1 Mb). Several genes in this area are, like many of those in the HLA region, involved in immune response including several interleukins and their receptors. A further peak on chromosome 4 is close to several Toll-like receptors and was also identified as highly differentiated between United Kingdom regions (Wellcome Trust Case Control Consortium 2007). Guided by these anecdotal observations, we tested whether SNPs in the highest FST category (>0.016) were overrepresented among genes of a particular functional class by using the PANTHER gene ontology database (Thomas et al. 2003). The “Immunity and Defense” (BP00148) category is the only one of the 23 ontological terms (with sufficient numbers to be tested) to show a significant enrichment of high FST SNPs after correction for multiple testing (P = 0.0012, adjusted significance level = 0.0022). As a contrast, we examined the ontology of the 1358 SNPs with the lowest FST values (FST ≈ 0) and found that no category was close to either nominal or multiple test adjusted significance.
Table 2.
Genomic regions showing high FST clustering
aRegions were identified from a visual inspection of a plot of the top 10,000 FST values against genomic position. Reported SNP and FST values are the highest observed in that cluster.
bNCBI 36 build of the human genome.
cGenes within 250 kb either side of the peak SNP are listed. Where the SNP falls within a gene, the gene symbol is indicated in bold.
dA further 59 genes within this region are in proximity (±250 kb) to the peak SNP: AIF1, APOM, ATP6V1G2, BAT1, BAT2, BAT3, BAT4, BAT5, C2, C4A, C4B, C6orf21, C6orf25, C6orf26, C6orf27, C6orf47, C6orf48, CCHCR1, CDSN, CFB, CLIC1, CSNK2B, CYP21A2, DDAH2, DOM3Z, EHMT2, HCG27, HCP5, HLA-B, HLA-C, HSPA1A, HSPA1B, HSPA1L, LSM2, LST1, LTA, LTB, LY6G5B, LY6G5C, LY6G6C, LY6G6D, MCCD1, MICA, MICB, MSH5, NCR3, NEU1, NFKBIL1, POU5F1, PSORS1C2, RDBP, SKIV2L, SLC44A4, STK19, TCF19, TNF, TNXB, VARS, ZBTB12.
While this suggests that the selective signal is focused on immune function, it does not preclude the action of differential selection at some other individual loci. For example, the distribution of top FST points to one cluster of high values centered on the HERC2 gene on chromosome 15. HERC2 is close to OCA2, which substantially controls eye color (Sturm et al. 2008) and has been identified in several studies as a selection target in Europeans (Voight et al. 2006). Several additional peak SNPs do not fall within 250 kb of known (or validated) genes. These may simply be poorly localized by the peak SNP, false positive signatures of selection, or a sign that remote regulatory elements/and or other noncoding DNAs are also selective targets. The lactase gene (LCT) on chromosome 2, which is a well-established target of selection, is not prominent in this survey, an observation probably explained by a similar strength of selection across Northern Europe.
PC3 and PC4 reflect large-scale genome structural variation
PC5 displays a significant correlation with geographic distance, although it is substantially weaker (Mantel test: r = 0.067; P < 0.0001) than those observed with PC1 and PC2. Accordingly, the geographic patterning is less obvious and defies simple description (Supplemental Figs. S3 and S9). It appears to somewhat separate Ireland and Sweden from the remaining populations. The explanation for this is unclear although it does not seem to be a genotyping artifact since individual PC5 scores are not correlated with missing data, nor is the observation consistent with any potential batch effect. Further PCs (6–10) were found not to correlate with geography.
Interestingly, PC3 and PC4 were also not stratified by population label, but, rather, PC4 shows a neat three-way split of individuals, a pattern reminiscent of that seen in the third PC of a similarly genotyped European-American data set (Fig. 3A; Tian et al. 2008). This reflected the division of individuals based on the three possible genotypes of a large (≈4 Mb), polymorphic inversion on chromosome 8p23.1 (between 8 and 12 Mb). We confirmed that the same feature was responsible for our PC4 by dividing individuals into two extreme cohorts based on their PC4 score (<−0.02, n = 385 and >0.01, n = 647) and calculating the allele frequency difference (δ) between groups for each SNP. The top 554 SNPs localize to chromosome 8 between 8.135 and 11.90 Mb.
Figure 3.
PC3 and PC4 in Northern European populations. (A) PC3 versus PC4 derived from 2051 individuals genotyped for 296,553 autosomal SNPs. (B) An 8-Mb section of chromosome 8 (6 Mb to 14 Mb) showing the distribution of SNP δ values derived from extreme PC4 cohorts. (C) A 10-Mb section of chromosome 6 (24 Mb to 34 Mb) showing the distribution of SNP δ values derived from extreme PC3 cohorts. Whereas δ values remain high over the entire chromosome 8 inversion region (8 Mb to 12 Mb), those on chromosome 6 show a gradual decay upstream and downstream from the peak δ value observed at rs2508049 (position 29.99 Mb).
PC3 also appears to split individuals, regardless of population origin, into at least two groups. Reasoning that it might also reflect large structural variation, we divided samples into two extreme PC3 score cohorts (high: >0.02, n = 376 and low: <0, n = 1256) and again calculated δ for each autosomal SNP. Of the top 1000 δ values, 975 fall within an 8-Mb region of chromosome 6 (25.1 Mb–33.2 Mb) encompassing the HLA region. However, the distribution of values is distinct from that observed on chromosome 8 (Fig. 3B,C). The highest δ values in the chromosome 8 region are distributed across the 4-Mb block consistent with a discrete inversion structure. Those in the chromosome 6 region, however, display a marked decrease both 5′ and 3′ of the maximum δ value observed at the rs2508049 SNP (position 29931862), consistent with a large haplotype decaying with increased recombination distance from this core.
There is a strong association between rs2508049 genotype and PC3 scores (ANOVA, P = 3.2 × 10−293; Supplemental Fig. S10) with copy number of the G allele (present at a frequency of 18.3% in our entire population sample) correlated with higher PC3 scores. Deleting SNPs from a 20-Mb region of chromosome 6 (20 Mb to 40 Mb) abolishes PC3 as originally observed.
Selective sweep around rs2508049 in Northern Europeans
We investigated the structure of this region further by phasing a 20-Mb segment of chromosome 6 centered on rs2508049 (6p22.3–21.2 from 20 Mb to 40 Mb, encompassing 3080 SNPs), followed by extended haplotype homozygosity (EHH) analysis. EHH measures the probability that two haplotypes with the same core allele (in our case, either G or A at rs2508049) will be identical to a defined distance or particular SNP (Sabeti et al. 2002). Figure 4A shows EHH scores for each core allele across a 5-cM (9-Mb) region of chromosome 6 for all the Northern European populations as well as the African (YRI) and Asian (ASN) HapMap populations. The rs2508049-G core shows substantially higher levels of EHH than its sister rs2508049-A allele in all European samples over a region spanning ∼3.5 cM/8 Mb. While European populations and the African and Asian samples share a similar rs2508049-A EHH pattern, the low frequency of the G allele in the Asian population (1%, or two chromosomes out of 178) and its absence in the West African Yoruba sample preclude any meaningful analysis of the rs2508049-G EHH in these populations.
Figure 4.
EHH patterns around rs2508049 on chromosome 6. (A) EHH for the rs2508049 G and A alleles over a 5-cM (or ≈9 Mb, from 25.06 Mb to 33.93 Mb) region around the SNP in all Northern European populations. The EHH pattern for rs2508049-A in the HapMap Yoruban and Asian (Chinese and Japanese) is also shown, although the absence or very low frequency of the G allele precludes similar analysis of it in these populations. (B) EHH for the rs2508049-A and G alleles using the pre-phased CEU HapMap data, over a 4-cM region (or ≈8 Mb, from 25.06 Mb to 33.07 Mb). A 95 percentile boundary curve from an empirical EHH distribution of 15,325 chromosome 6 SNPs (30650 observations), calculated at 0.1-cM intervals over a 2-cM distance from the cores, is also indicated. Population codes are as follows: (ASN) East Asian HapMap; (AUS) Australia; (CEU) European-American HapMap; (DEN) Denmark; (FIN) Finland; (IRL) Ireland; (NET) Netherlands; (SWE) Sweden; (UK) United Kingdom; (YRI) Yoruba HapMap.
To determine whether the structure around rs2508049 is unusual in the Northern European genome, we used an empirical “significance” approach using the pre-phased chromosome 6 CEU HapMap data (see Methods). A total of 15,325 SNP alleles were identified with a similar frequency (±2.5%) to the rs2052089-G allele (18.3%). The empirical distributions of the 30,650 EHH observations (an upstream and downstream value for each SNP), over a range of distances from the cores, were used to construct a 95% boundary curve. The EHH pattern around rs2508049 in these data is similar to that observed in the other European, including the CEU, populations that were phased separately (Fig. 4B). Furthermore, the EHH values of the rs2508049-G allele run substantially ahead of the empirically derived 95 percentile over a 2-cM region 3′ of the core SNP. The 5′ pattern is more complex with rs2508049-G EHH values running consistently ahead up to 0.167 cM from the core and then again from 0.34 cM to 1.1 cM. In the interval it is close to the 95th percentile, criss-crossing slightly above and under at various distances. However, the overall pattern is supportive of an usually long rs2508049-G haplotype over at least a 3.1-cM span (≈7.5 Mb, from position 25.5 Mb to 33 Mb).
In a previous analysis of about 3.4 million autosomal SNPs in the HapMap populations, rs2508049 showed a locus-specific branch length (an FST-based measure of population-specific divergence) within the top 5% of values observed in Europeans (McEvoy et al. 2006). The virtual absence of the rs2508049-G allele outside of Europe together with its extreme haplotype structure within the continent is consistent with a strong positive selective sweep in which an advantageous variant was driven to a higher frequency, dragging linked variation along with it.
Discussion
We examined whole genome polymorphism for several population samples either from, or largely descendent from, Northern Europe. In common with recent similar studies, we demonstrated that it is possible to discern clear differences among individuals from very closely related populations and even to observe stratification within the same population. The nature and relative importance/order of PCs will, of course, be affected by the populations and individuals included. Within our data, the distinctive position of Finland in PC1 may be the consequence of migrational contact with Northern Asians or admixture from the Saami population, who are known European genetic outliers. The differentiation apparent within Finland along a Northeast to Southwest axis is consistent with such a genetic diffusion.
PC2 largely separates Ireland from Britain and these from mainland Europe, and it is possible to discern further British Isles structure within this trend despite our limited information on the local ancestry of these individuals. This pattern is similar to the culmination of the Southeast to Northwest continental wide trends observed with Y chromosomes (Hill at al. 2000; Rosser et al. 2000; Capelli et al. 2003) and classical gene frequencies (Cavalli-Sforza et al. 1994). Although its origin is debated, it may be consistent with suggestions that Western European fringe populations, like Ireland, retain a closer genetic continuity to an earlier, perhaps pre-Neolithic European population (McEvoy et al. 2004). However, caution is warranted in interpreting these patterns since PCA trends need not be the result of a specific grand migration but can also occur under simpler demographic scenarios such as isolation by distance models (Novembre and Stephens 2008). Whatever their ultimate causes in the past, these patterns have important present-day practical ramifications in gene mapping by association. Our example of pairwise genome inflation factors between these populations is a reminder to consider structure even when combining individuals from a small and prima facie homogeneous area. As sample sizes are increased to detect associated variants of smaller effects, so the risk of confounding by even subtle stratification increases (Marchini et al. 2004).
In addition to the expected predominant role of neutral evolution (random genetic drift proportional to divergence time), and possibly migration, it appears that differential selection has had a discernable action between these Northern European populations. A previous study pioneered a similar genic/nongenic FST SNP test to demonstrate the role of selection between the HapMap populations (Barreiro et al. 2008). However, these populations diverged tens of thousands of years ago and have since occupied distinct geographic areas. The close relatedness of Northern Europeans and similar ecological range might suggest less opportunity for different exogenous conditions to exert a selective influence. In addition, the present study used up to 10-fold fewer SNPs than were available for the HapMap comparison, potentially weakening the power to detect such signals.
Accordingly, the general signal of positive selection in our intracontinental analysis is very modest compared to the intercontinental HapMap-based study. The top FST bin floor is much less here (>0.016 vs. >0.85), for instance, and the genic fraction never exceeds the nongenic fraction in Europeans as seen with the HapMap populations. The observation that the genic enrichment among SNPs with high FST is largely focused on genes from a single ontology category (“immunity and defense”) is support that the signal, while subtle, is real and robust. Furthermore, this category simultaneously provides a biologically plausible explanation for the detectable selection signal. Variability in the geographic extent of infectious disease outbreaks as well as potentially strong selection coefficients associated with such epidemics are a simple means by which selection could have a detectable effect within such a limited temporal and geographic range.
While the genic FST test is collective evidence for positive selection, our study identified one specific example of a probable selective event/sweep around the rs2508049-G allele in the HLA region of chromosome 6. The exceptionally long span of haplotype homozygosity suggests relatively recent and/or intense selection across all Northern Europeans (although we cannot distinguish whether this was before or after population divergence). Unusual patterns of genetic variation within this region are well established, and it has likely been affected by a complex palimpsest of processes that seem to include positive and balancing selection (de Bakker at el. 2006; Voight et al. 2006; Sabeti et al. 2007).
It is somewhat difficult to compare homozygosity results across studies since the number of markers genotyped can influence the result. However, it appears that the rs2508049-G haplotype is one of the largest spans at appreciable frequency yet observed. To illustrate, we examined EHH, using similar marker density and the CEU data set, around the rs4988235 SNP (also known as −13910 G/T) associated with the lactase (LCT) persistence trait in Europeans. The region is thought to have been intensely selected during the Neolithic period (<10,000 yr ago) to allow the population to avail of milk and other dairy foods into adulthood (Bersaglieri et al. 2004). The like-for-like span (defined as the interval between the 5′ and 3′ points from the core where the EHH falls below 0.25) for the persistence allele, rs4988235-T (1.3 Mb/0.4 cM), is substantially less than that observed with rs2508049-G (2 Mb/1.1 cM). However, rs4988235-T is nearly fourfold more frequent in the CEU population sample than rs2508049-G. These observations (longer span and lower allele frequency) may suggest that any selective pressure underlying the rs2508049 region was both more recent and/or of shorter duration than that at LCT.
The unusual characteristics of the rs2508049-G region are consistent with an episode of positive selection, and the pattern of haplotype decay is consistent with a target in the vicinity of rs2508049. The SNP is located 25 kb downstream from the HLA-G gene, one of the HLA class I heavy chain paralogs that play a role in antigen presentation and recognition and that has also been associated with asthma (Nicolae et al. 2005), HIV susceptibility (Lajoie et al. 2006), and various pregnancy outcome phenotypes (Favier et al. 2007). Two other genes of the same family (HLA-A, HLA-F) are also located close by. However, the region (±250 kb) also encompasses loci with various other roles including a gamma-aminobutyric acid neurotransmitter receptor (GABBR1). Therefore, while one selective explanation is a response to infectious disease, it is clearly not the only possible scenario. The region has also been repeatedly implicated in many disorders, many of which like asthma, multiple sclerosis, and schizophrenia, occur at a higher frequency in Europeans. While these disorders are likely to be complex, it is interesting to speculate that some of the risk may be an evolutionary hangover of selective hitchhiking.
Conclusions
Our analysis of several Northern European populations demonstrates once again the remarkable ability of dense data, in terms of both genome and population coverage, to dissect a range of events from selection to migration over a recent timeframe and across very limited geographic areas. The results also highlight the present-day legacy of the recent population past both in potential disease risk and in attempts to map complex trait loci through whole genome association.
Methods
Samples and genotyping
This study primarily used members of national twin cohorts that are part of the GenomEUtwin project (Peltonen 2003) (http://www.GenomEUtwin.org). Genotyped individuals are one member of a monozygotic twin pair from Australia, United Kingdom, Denmark, Sweden, Netherlands, or Finland (n = 1862). We also included an Irish population sample used as controls in a previous case/control study of Amyotrophic lateral sclerosis (Cronin et al. 2008), as well as the European-American HapMap population (CEU) (n = 60). Subsets of the HapMap data for the East Asian (ASN) (Japanese + Chinese) and West African Yoruban (YRI) populations were also used for some analysis. All samples were collected with informed consent and appropriate ethical approval. The novel genotype data are deposited in the European Genotype Archive (http://www.ebi.ac.uk/ega/page.php), under the accession number EGAS00000000033, where it may be retrieved for legitimate research purposes.
Twin samples were genotyped using the Infinium II assay on the HumanHap300-Duo Genotyping Beadchips (Illumina, Inc.). Around two-thirds of the subjects were genotyped at the Finnish Genome Center (Helsinki) and the remainder at the SNP Technology Platform, Uppsala University (Uppsala, Sweden). In total, 318,237 SNPs were genotyped. There was a 99.99% reproducibility rate between SNPs in 14 duplicate samples typed at both sites. Within each center, 26 samples were genotyped in duplicate with a 99.99% consistency rate. SNPs and individuals with >10% missing data were excluded for the purposes of the analysis described herein. The Irish sample had been typed using the Illumina Infinium II 550 K SNP platform. When integrating the GenomEUtwin, Irish, and HapMap samples, we included only those SNPs genotyped in all three groups where unambiguous allele matching could be made. We checked allele flipping errors in this process by screening for gross outliers in pairwise SNP FST values between the GenomEUtwin and the Irish or HapMap populations. A total of 305,320 SNPs passed this process, but only those with an autosomal location were included in most analysis (296,553 SNPs).
FST and δ
Genetic distances as FST values were calculated for each SNP according to Weir and Cockerham (see Weir 1996) and averaged to obtain a single estimate for each pairwise population combination or a global value over all populations. FST values normally range between 0 and 1, but small negative values are possible. As previously noted (Barreiro et al. 2008), this reflects the predominance of sampling error over very weak population subdivision and has no biological interpretation. δ is defined as the absolute difference in allele frequency between any two predefined groups. FST and δ were calculated using purpose written Perl scripts.
Principal component analysis (PCA)
We used the EIGENSOFT package (Patterson et al. 2006; Price et al. 2006), and its default parameters, to calculate up to 100 eigenvectors or principal components (PC). We first ran an exploratory analysis to identify and remove individuals who were greater than 6 standard deviations from the mean along any of the top 10 PCs (n = 48). Using detailed self-reported ancestries where available (from the United Kingdom and Australian populations), it could be shown that these individuals typically had at least partial non-European or Southern European ancestry. The total (n = 2051) cleaned sample sizes are: Australia (n = 451), United Kingdom (n = 433), Denmark (n = 161), Sweden (n = 302), Netherlands (n = 284), Finland (n = 149), Ireland (n = 211), and CEU-HapMap (n = 60). As a quality control measure, we tested for any significant correlation between individual scores across the top 10 PCs and individual rates of missing genotype data but observed no significant relationship.
We investigated the correspondence of PC values and geography using Mantel tests of correlation between two matrices (Mantel 1967). A matrix of geographic distances between individuals (in kilometers) was generated from generic coordinates for each European population (excluding Australia and CEU) as follows (Latitude/Longitude): Ireland: 54/−7; United Kingdom: 53/−1; Netherlands: 52/5; Denmark: 56/10; Sweden: 60/−15; Finland: 63/−27. Negative longitude values indicate Western Hemisphere locations. For each PC, a matrix of absolute interindividual differences in PC score was created. Mantel correlation significance was quantified by random permutation of matrix elements over 10,000 replicate analyses.
Ancestry informative markers
To develop and test sets of ancestry informative markers (AIMs), we divided, at random, the complete sample data set into discovery and test cohorts. The discovery data set (n = 770) consisted of half of each of the United Kingdom, Denmark, Sweden, Finland, Ireland, and Netherlands samples, thus preserving the relative population sample sizes of the full data set. A global FST value was calculated for each SNP using the discovery set and values ranked. Sets of the top 500, 1500, 2500, and 5000 SNPs were selected as AIM panels with the additional proviso that no marker was within 250 kb of another, a measure taken to minimize LD between SNPs. These sets were used in PCA and structure analysis of the test data set. We included all the Australian and CEU-HapMap samples in the test data set (n = 1281) since, as admixed groups, they might skew FST calculations, and, secondly, we are interested in investigating their ancestry in the Northern European genetic context.
structure
In addition to PCA, we applied an alternative Bayesian clustering approach, as implemented in the program structure (v2.1), to investigate population stratification (Pritchard et al. 2000; Falush et al. 2003). The method does not rely on a priori population labels but instead infers a specified K number of population clusters from the genotype data essentially using departures from Hardy–Weinberg equilibrium. We applied the conditions used in a similar previous analysis of European-Americans (Tian et al. 2008). Briefly, this involved using the admixture model (which allows fractional assignment of an individual's genome to different clusters) without using any population label information. To facilitate computational tractability, the entire data set (296,553 SNPs) was not used, but, rather, we examined the performance of each of the previously described AIM sets (500, 1500, 2500, and 5000 SNPs) in the test sample set (composed of half of the Europeans and all Australians and HapMap European-American individuals). A single run, consisting of a burn-in of 5000 replicates followed by 10,000 iterations, was carried out for each AIM set over K = 2 to K = 5.
Extended haplotype homozygosity
Extended haplotype homozygosity (EHH) is the probability that two randomly chosen chromosomes with the same allele at a particular core (be it a single SNP or a small haplotype) will be identical (at all genotyped SNPs) to a certain distance, either upstream or downstream, from that core (Sabeti et al. 2002). As such, it can be used to identify a selective sweep signature. In order to compute EHH scores, we first phased a large region (up to 20 Mb) around a core SNP of interest, for each population separately, using the fastPHASE 1.2 algorithm and its default settings (Scheet and Stephens 2006). EHH scores were then calculated using a purpose written Perl script.
EHH is often expressed as a ratio of the value at an allele of interest to the average at all other alleles, as a method of controlling for variation in recombination rate when comparing EHH scores across loci (relative EHH or rEHH). Where the core is a single SNP, this is simply the ratio of allele A to allele B. However, once the EHH score for the comparative allele reaches zero, the ratio becomes undefined, and this problem increases with distance from the core. To avoid this, we work with raw EHH scores, matched for recombination distance when comparing across loci using genetic map information from the HapMap (release #22) CEU and YRI populations.
EHH significance
There are two general approaches to gauge the significance of an observed EHH score based on either simulated or observed data. Simulation requires specification of demographic parameters (bottlenecks, expansions, etc.) that are generally unknown. Empirical data, in contrast, are the realization of these events, and loci that stand apart from the distribution of observed EHH values are possible targets of selection. While not a formal test of significance, we used an empirical approach to assess the “unusualness” of the rs2508049 EHH result on chromosome 6. The pre-phased genotype information for chromosome 6 in the CEU population was retrieved (Release #22), and those alleles with a similar frequency to the initial core allele of interest were identified. Treating each of these as a core in turn, EHH was calculated at 0.1-cM intervals up to 2 cM upstream and downstream of this position. The genotype coverage was pruned (reduced) by a factor of 12 during EHH calculations to achieve similar SNP density to that in the 305K data set. The empirical distribution of the values (including two—upstream and downstream—observations per SNP) at each distance was used to construct a 95th percentile boundary curve.
Gene annotation and selection analysis
To investigate a general signature of selection across the Northern European genome, we examined the distribution of SNP FST values conditioned on their location inside or outside genes. We used SNP annotation information previously compiled for Affymetrix, Illumina, and Perlegen genotyping platforms (https://slep.unc.edu/evidence/files/README_annotations.pdf). These SNP annotations were created using the TAMAL database (Hemminger et al. 2006) based chiefly on UCSC Genome Browser files, HapMap, and dbSNP. A genic SNP was defined as one falling anywhere in the transcribed portion of a gene.
We further investigated the distribution of SNP FST values with respect to the function of the genes they fall in by using the PANTHER gene ontology database, which assigns many genes to one or more “biological processes” (Thomas et al. 2003). The full list of 25,431 human genes (and their genomic locations) classed under 31 different ontological terms was retrieved from http://www.pantherdb.org/. Analysis was restricted to those genes (21,407) with a given autosomal map location. We used the proportion of the 296,553 autosomal SNPs in or in close proximity (±10 kb) to genes in each ontology class as the basis for the expected value in a χ2 test against the numbers observed in the top FST (>0.016) category.
Acknowledgments
The GenomEUtwin project is supported by the European Commission under the program “Quality of Life and Management of the Living Resources” of 5th Framework Programme (QLG2-CT-2002-01254). This work was funded by the Center of Excellence in Complex Disease Genetics of the Academy of Finland (J.K., L.P., M.P., and S.R.), Nordic Center of Excellence in Disease Genetics, and The Finnish Cultural Foundation. The SNP Technology Platform in Uppsala is funded by Uppsala University and Uppsala University Hospital and the Swedish Wallenberg Foundation. We thank the staff of the SNP Platform for their contributions to genotyping. We also acknowledge financial support from the Wellcome Trust, the United Kingdom Department of Health via the National Institute for Health Research (NIHR) comprehensive Biomedical Research Centre award to Guy's and St. Thomas' NHS Foundation Trust in partnership with King's College London, Biotechnology and the Biological Sciences Research Council (BBSRC). B.P.M., A.R.M., G.W.M., and P.M.V. are supported by the National Health and Medical Research Council of Australia.
Footnotes
[Supplemental material is available online at www.genome.org. The genotype data from this study have been submitted to the European Genotype Archive (http://www.ebi.ac.uk/ega/page.php), under accession no. EGAS00000000033.]
Article is online at http://www.genome.org/cgi/doi/10.1101/gr.083394.108.
References
- Barreiro L.B., Laval G., Quach H., Patin E., Quintana-Murci L. Natural selection has driven population differentiation in modern humans. Nat. Genet. 2008;40:340–345. doi: 10.1038/ng.78. [DOI] [PubMed] [Google Scholar]
- Bauchet M., McEvoy B., Pearson L.N., Quillen E.E., Sarkisian T., Hovhannesyan K., Deka R., Bradley D.G., Shriver M.D. Measuring European population stratification with microarray genotype data. Am. J. Hum. Genet. 2007;80:948–956. doi: 10.1086/513477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersaglieri T., Sabeti P.C., Patterson N., Vanderploeg T., Schaffner S.F., Drake J.A., Rhodes M., Reich D.E., Hirschhorn J.N. Genetic signatures of strong recent positive selection at the lactase gene. Am. J. Hum. Genet. 2004;74:1111–1120. doi: 10.1086/421051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelli C., Redhead N., Abernethy J.K., Gratrix F., Wilson J.F., Moen T., Hervig T., Richards M., Stumpf M.P., Underhill P.A., et al. A Y chromosome census of the British Isles. Curr. Biol. 2003;27:979–984. doi: 10.1016/s0960-9822(03)00373-7. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza L.L., Menozzi P., Piazza A. The history and geography of human genes. Princeton University Press; Princeton, NJ: 1994. [Google Scholar]
- Cronin S., Berger S., Ding J., Schymick J.C., Washecka N., Hernandez D.G., Greenway M.J., Bradley D.G., Traynor B.J., Hardiman O. A genome-wide association study of sporadic ALS in a homogenous Irish population. Hum. Mol. Genet. 2008;17:768–774. doi: 10.1093/hmg/ddm361. [DOI] [PubMed] [Google Scholar]
- de Bakker P.I., McVean G., Sabeti P.C., Miretti M.M., Green T., Marchini J., Ke X., Monsuur A.J., Whittaker P., Delgado M., et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat. Genet. 2006;38:1166–1172. doi: 10.1038/ng1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B., Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- Falush D., Stephens M., Pritchard J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favier B., LeMaoult J., Rouas-Freiss N., Moreau P., Menier C., Carosella E.D. Research on HLA-G: An update. Tissue Antigens. 2007;69:207–211. doi: 10.1111/j.1399-0039.2006.00757.x. [DOI] [PubMed] [Google Scholar]
- Heath S.C., Gut I.G., Brennan P., McKay J.D., Bencko V., Fabianova E., Foretova L., Georges M., Janout V., Kabesch M., et al. Investigation of the fine structure of European populations with applications to disease association studies. Eur. J. Hum. Genet. 2008;16:1413–1429. doi: 10.1038/ejhg.2008.210. [DOI] [PubMed] [Google Scholar]
- Hemminger B.M., Saelim B., Sullivan P.F. TAMAL: An integrated approach to choosing SNPs for genetic studies of human complex traits. Bioinformatics. 2006;22:626–627. doi: 10.1093/bioinformatics/btk025. [DOI] [PubMed] [Google Scholar]
- Hill E.W., Jobling M.A., Bradley D.G. Y chromosome variation and Irish origins. Nature. 2000;404:351–352. doi: 10.1038/35006158. [DOI] [PubMed] [Google Scholar]
- International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajoie J., Hargrove J., Zijenah L.S., Humphrey J.H., Ward B.J., Roger M. Genetic variants in nonclassical major histocompatibility complex class I human leukocyte antigen (HLA)-E and HLA-G molecules are associated with susceptibility to heterosexual acquisition of HIV-1. J. Infect. Dis. 2006;193:298–301. doi: 10.1086/498877. [DOI] [PubMed] [Google Scholar]
- Lao O., Lu T.T., Nothnagel M., Junge O., Freitag-Wolf S., Caliebe A., Balascakova M., Bertranpetit J., Bindoff L.A., Comas D., et al. Correlation between genetic and geographic structure in Europe. Curr. Biol. 2008;18:1241–1248. doi: 10.1016/j.cub.2008.07.049. [DOI] [PubMed] [Google Scholar]
- Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- Marchini J., Cardon L.R., Phillips M.S., Donnelly P. The effects of human population structure on large genetic association studies. Nat. Genet. 2004;36:512–517. doi: 10.1038/ng1337. [DOI] [PubMed] [Google Scholar]
- McEvoy B., Richards M., Forster P., Bradley D.G. The Longue Durée of genetic ancestry: Multiple genetic marker systems and Celtic origins on the Atlantic facade of Europe. Am. J. Hum. Genet. 2004;75:693–702. doi: 10.1086/424697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEvoy B., Beleza S., Shriver M.D. The genetic architecture of normal variation in human pigmentation: an evolutionary perspective and model. Hum. Mol. Genet. 2006;15:R176–R181. doi: 10.1093/hmg/ddl217. [DOI] [PubMed] [Google Scholar]
- Nicolae D., Cox N.J., Lester L.A., Schneider D., Tan Z., Billstrand C., Kuldanek S., Donfack J., Kogut P., Patel N.M., et al. Fine mapping and positional candidate studies identify HLA-G as an asthma susceptibility gene on chromosome 6p21. Am. J. Hum. Genet. 2005;76:349–357. doi: 10.1086/427763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novembre J., Stephens M. Interpreting principal component analyses of spatial population genetic variation. Nat. Genet. 2008;40:646–649. doi: 10.1038/ng.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novembre J., Johnson T., Bryc K., Kutalik Z., Boyko A.R., Auton A., Indap A., King K.S., Bergmann S., Nelson M.R., et al. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N., Price A.L., Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltonen L. GenomEUtwin: A strategy to identify genetic influences on health and disease. Twin Res. 2003;6:354–360. doi: 10.1375/136905203770326358. [DOI] [PubMed] [Google Scholar]
- Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Price A.L., Butler J., Patterson N., Capelli C., Pascali V.L., Scarnicci F., Ruiz-Linares A., Groop L., Saetta A.A., Korkolopoulou P., et al. Discerning the ancestry of European Americans in genetic association studies. PLoS Genet. 2008;4:e236. doi: 10.1371/journal.pgen.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J.K., Stephens M., Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosser Z.H., Zerjal T., Hurles M.E., Adojaan M., Alavantic D., Amorim A., Amos W., Armenteros M., Arroyo E., Barbujani G., et al. Y chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language. Am. J. Hum. Genet. 2000;67:1526–1543. doi: 10.1086/316890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti P.C., Reich D.E., Higgins J.M., Levine H.Z., Richter D.J., Schaffner S.F., Gabriel S.B., Platko J.V., Patterson N.J., McDonald G.J. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;24:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- Sabeti P.C., Varilly P., Fry B., Lohmueller J., Hostetter E., Cotsapas C., Xie X., Byrne E.H., McCarroll S.A., Gaudet R., et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheet P., Stephens M.A. Fast and flexible statistical model for large-scale population genotype data: Applications to inferring missing genotypes and haplotypic phase. Am. J. Hum. Genet. 2006;78:629–644. doi: 10.1086/502802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seldin M.F., Price A.L. Application of ancestry informative markers to association studies in European Americans. PLoS Genet. 2008;4:e5. doi: 10.1371/journal.pgen.0040005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seldin M.F., Shigeta R., Villoslada P., Selmi C., Tuomilehto J., Silva G., Belmont J.W., Klareskog L., Gregersen P.K. European population substructure: Clustering of northern and southern populations. PLoS Genet. 2006;15:e143. doi: 10.1371/journal.pgen.0020143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm R.A., Duffy D.L., Zhao Z.Z., Leite F.P., Stark M.S., Hayward N.K., Martin N.G., Montgomery G.W. A single SNP in an evolutionary conserved region within intron 86 of the HERC2 gene determines human blue-brown eye color. Am. J. Hum. Genet. 2008;82:424–431. doi: 10.1016/j.ajhg.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P.D., Campbell M.J., Kejariwal A., Mi H., Karlak B., Daverman R., Diemer K., Muruganujan A., Narechania A. PANTHER: A library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C., Plenge R.M., Ransom M., Lee A., Villoslada P., Selmi C., Klareskog L., Pulver A.E., Qi L., Gregersen P.K., et al. Analysis and application of European genetic substructure using 300 K SNP information. PLoS Genet. 2008;4:e4. doi: 10.1371/journal.pgen.0040004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight B.F., Kudaravalli S., Wen X., Pritchard J.K. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir B.S. Genetic data analysis II. Sinauer; Sunderland, MA: 1996. pp. 176–179. [Google Scholar]
- Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]