Abstract
We report an analysis of more than 240,000 loci genotyped using the Affymetrix SNP microarray in 554 individuals from 27 worldwide populations in Africa, Asia, and Europe. To provide a more extensive and complete sampling of human genetic variation, we have included caste and tribal samples from two states in South India, Daghestanis from eastern Europe, and the Iban from Malaysia. Consistent with observations made by Charles Darwin, our results highlight shared variation among human populations and demonstrate that much genetic variation is geographically continuous. At the same time, principal components analyses reveal discernible genetic differentiation among almost all identified populations in our sample, and in most cases, individuals can be clearly assigned to defined populations on the basis of SNP genotypes. All individuals are accurately classified into continental groups using a model-based clustering algorithm, but between closely related populations, genetic and self-classifications conflict for some individuals. The 250K data permitted high-level resolution of genetic variation among Indian caste and tribal populations and between highland and lowland Daghestani populations. In particular, upper-caste individuals from Tamil Nadu and Andhra Pradesh form one defined group, lower-caste individuals from these two states form another, and the tribal Irula samples form a third. Our results emphasize the correlation of genetic and geographic distances and highlight other elements, including social factors that have contributed to population structure.
Microarray technology has generated unprecedented quantities of data on human genetic variation. These data are useful for fine-scaled inferences of human evolutionary history (Jakobsson et al. 2008; Li et al. 2008; Novembre et al. 2008; Tian et al. 2008) and, under some circumstances, the estimation of individual ancestry (Seldin et al. 2006; Bauchet et al. 2007; Price et al. 2008; Tian et al. 2008). In this context, the new data have contributed to a better and more nuanced understanding of the relationship between genetics and “race” (Race, Ethnicity, and Genetics Working Group 2005; Witherspoon et al. 2007). In addition, a more thorough knowledge of between-population genetic variation also has been important in improving the design and interpretation of case-control studies of common diseases (Wellcome Trust Case Control Consortium 2007; Nelson et al. 2008; Price et al. 2008).
For a variety of reasons, most studies have focused primarily on European populations (Seldin et al. 2006; Bauchet et al. 2007; Novembre et al. 2008; Price et al. 2008; Tian et al. 2008), and worldwide coverage of human populations remains incomplete. For example, the Human Genome Diversity Project (HGDP) database, one of the most widely used resources, lacks coverage in the Indian subcontinent. Other major regions, such as Eastern Europe and northern Africa, are also underrepresented in databases of human genetic variation.
Among these underrepresented populations, those of the Indian subcontinent, which contains one-sixth of the world's inhabitants, are of particular interest. The origins of and relationships among Indian populations are the subjects of continuing debate (Bamshad et al. 1998, 2001; Basu et al. 2003; Vishwanathan et al. 2004; Watkins et al. 2005; Rosenberg et al. 2006; Chaubey et al. 2007), but most previous genetic studies of these populations have been based on modest data sets. Indian populations are also used increasingly in linkage and case-control studies of genetic disease (Alcais et al. 2007; Chambers et al. 2008; Holliday et al. 2008). A better understanding of the genetic structure in India will facilitate these studies.
Here, along with another 21 populations from around the world, we analyzed six Indian populations, including five caste populations and one tribal population, from two southern Indian states (Andhra Pradesh and Tamil Nadu). The inclusion of caste populations from different states and with different languages allowed us to assess the effects of social status, geography, and language on genetic structure in Indian populations. We have also included Daghestanis from the Caucasus region and Ibans from Sarawak, Malaysia to improve coverage in other underrepresented regions. Our analysis offers new insights on the genetic affinities and evolution of populations residing between commonly studied populations in sub-Saharan Africa, Europe, and East Asia.
Results
Population samples
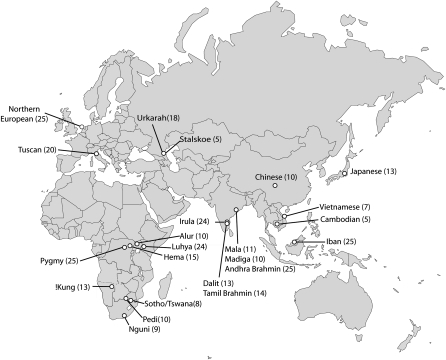
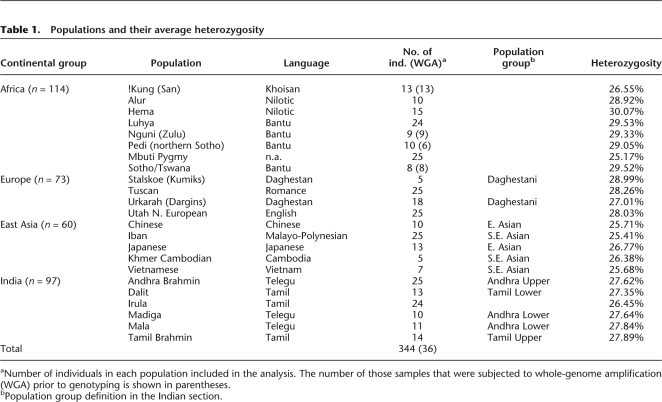
We genotyped 344 individuals from 23 worldwide populations using the Affymetrix 250K NspI and 6.0 SNP mapping array. These samples represent populations from sub-Saharan Africa (8), Europe (4), South Asia (6), and East/Southeast Asia (5) (Fig. 1; Table 1). In addition, the analyses included 210 unrelated individuals from four HapMap populations (YRI, CEU, CHB, and JPT) that were genotyped on the Affymetrix 250K NspI SNP mapping array. The final data set consists of 243,855 autosomal loci genotyped in 554 individuals from 27 populations (see Methods for details of SNP selection criteria).
Figure 1.
Population samples analyzed. Location and number of individuals sampled in each population group.
Table 1.
Populations and their average heterozygosity
aNumber of individuals in each population included in the analysis. The number of those samples that were subjected to whole-genome amplification (WGA) prior to genotyping is shown in parentheses.
bPopulation group definition in the Indian section.
Genetic diversity among populations and continental groups
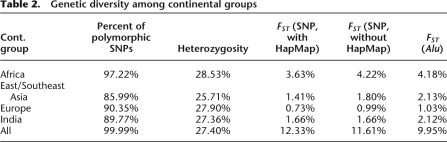
To compare genetic diversity among major continental regions, we grouped the 27 populations into four groups based on their continental or regional origins: Africa, East/Southeast Asia, Europe, and India. Among the four major groups, Africa has the highest heterozygosity (28.5%), while East/Southeast Asia has the lowest (25.7%, Table 2). The heterozygosities in Europe and India are similar (27.9% and 27.4%, respectively). These estimates are in general agreement with other studies but are expected to be influenced to some degree by the ascertainment bias of the SNPs selected for the microarray. This bias could inflate estimates of heterozygosity for populations (e.g., Europeans) in which the SNPs were initially ascertained. Consistent with its higher average heterozygosity, Africa also has the highest proportion (97.2%) of genotyped loci that are polymorphic in the sample (minor allele frequency [MAF] > 0) among the 243,855 SNPs. In contrast, only 86.0% of the SNPs in the East/Southeast Asia group are polymorphic (Table 2).
Table 2.
Genetic diversity among continental groups
Next, we calculated how many polymorphisms are shared among major groups. The vast majority of SNPs are polymorphic (MAF > 0) in multiple groups, with 81.2% of all loci being polymorphic in the four major continental groups, 89.2% polymorphic in at least three groups, and 93.0% polymorphic in at least two groups. Almost all of the SNPs that are polymorphic in only one group are unique to Africa (6.6%), while collectively only 0.39% of the SNPs are unique to any of the other three continental groups (Supplemental Table 2). There were no fixed differences between continental populations at any locus. Thus, these results support the emerging conclusion that most common genetic variation is shared among major human population groups.
To assess the proportion of genetic variation attributable to population subdivision, we estimated FST for the total sample, divided into four major groups (FST = 12.33%). We also estimated FST among populations within each continental group. Africa has the highest value (3.63%), which is more than twice the FST in East/Southeast Asia (1.41%) and India (1.66%) and more than four times that found in Europe (0.73%). Similar continental and overall FST values are obtained when HapMap populations are excluded from the analysis (Table 2). This result is comparable to FST values calculated using Alu insertion polymorphisms in a previous study (Table 2; Watkins et al. 2003). Taken together, these results highlight the larger genetic diversity in Africa compared with other continental groups.
Next, we calculated pairwise FST between populations (Supplemental Table 1). The largest FST values are observed between the African Mbuti Pygmy population and eastern Asian populations (e.g., 0.240 for Pygmy vs. Iban, Pygmy vs. Chinese, and Pygmy vs. HapMap JPT). The smallest FST values are observed between populations that are sampled from the same geographic location (e.g., 0.0004 for Utah Northern European vs. HapMap CEU, 0.0010 for Indian lower caste Mala vs. Madiga, and 0.0014 for Chinese vs. HapMap CHB).
A large proportion of individuals included in this study have been genotyped previously for other types of polymorphisms (Jorde et al. 1997; Watkins et al. 2003, 2005; Wooding et al. 2004). For these individuals, we examined the concordance of between-population genetic distances based on the high-density SNP data and other types of autosomal polymorphisms. Between-population genetic distances estimated from 100 Alu insertions polymorphisms (11 populations, 243 individuals) or 45 Short Tandem Repeats (STRs, 10 populations, 217 individuals) show high, statistically significant correlations with between-population distances estimated from the 243,855 autosomal SNPs in this study (r = 0.98, P < 10−5 and r = 0.90, P < 10−5, respectively, Mantel matrix correlation). The slightly lower correlation for SNPs and STRs compared with that for SNPs and Alus is probably caused by the high STR mutation rate, which can obscure population relationships (Jorde et al. 1997). The correlation between Alu insertion polymorphisms and STRs was also high and significant but lower than the SNP/STR correlation (r = 0.86, P < 10−3).
Principal components analysis
To further investigate genetic structure in our samples, pairwise allele-sharing distances (Mountain and Cavalli-Sforza 1997) were calculated among all pairs of individuals, and principal components analysis (PCA) was applied to the resulting pairwise distance matrix. Figure 2 illustrates the first two principal components (PCs). The first principal component (PC1), which accounts for 75.9% of the total variation, separates Africans from other populations. PC2 (11.9% of the total variation) separates East/Southeast Asians from European populations, with Indians located between the two groups. PC3 separates Indian populations from other populations, and the two African hunter–gatherer groups (Mbuti Pygmy and !Kung) are separated from other African populations on PC4 (Supplemental Fig. 1).
Figure 2.
Principal components analysis of population structure in 554 individuals. First two principal components (PCs) are shown here. Each individual is represented by one dot and the color label corresponding to their self-identified population origin. The percentage of the variation in genetic distances explained by each PC is shown on the axes.
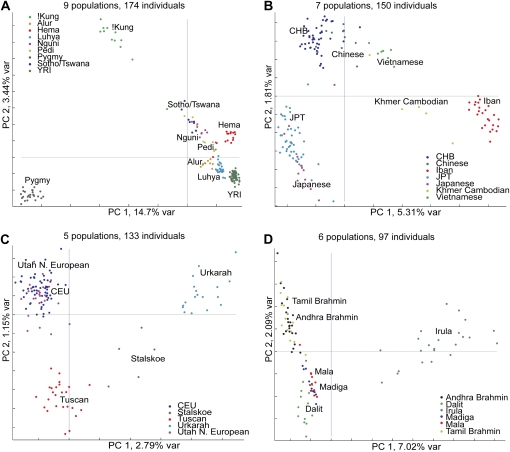
We next examined each of the four regions separately (Fig. 3A–D). When PCA was performed on African populations only (Fig. 3A), PC1 and PC2 separate Mbuti Pygmy and !Kung from other African populations, respectively. Although the remaining African populations are less distinct, a north-to-south gradient can be observed along PC2. PC3 parallels geographic and linguistic difference among African populations. Nilotic-speaking Alur and Hema individuals from the Democratic Republic of the Congo are separated from Bantu-speaking populations (Nguni, Pedi, and Sotho/Tswana) from South Africa (Supplemental Fig. 1B). In the East/Southeast Asian group (Fig. 3B), the Iban from Borneo, Malaysia form a tight cluster and show the largest genetic distance from other East/Southeast Asian populations. As expected, our Chinese and Japanese individuals overlap with the HapMap CHB and JPT individuals, respectively. The Vietnamese and Cambodians fall between Iban and East Asian populations (i.e., Chinese and Japanese). Among European populations (Fig. 3C), PC1 clearly divides the Eastern European Daghestani populations (Urkarah and Stalskoe) from Western European populations. PC2 reflects north-to-south variation within Western Europe (Utah Northern European and HapMap CEU vs. Tuscan) and differences between highland and lowland populations in Daghestan (Urkarah vs. Stalskoe, respectively). For Indian populations (Fig. 3D), PC1 separates the caste groups from the Irula tribal population. PC2 shows subtle but clear separation between the upper-caste (Andhra Brahmins and Tamil Brahmins) and lower-caste (Mala, Madiga, and Dalit) individuals. The tribal Irula individuals are largely distinct from the caste groups and show more between-individual variation compared with the caste populations.
Figure 3.
Principal components analysis of population structure in each continental group. (A) Africa, (B) East/Southeast Asia, (C) Europe, and (D) India. First two PCs are shown. Each individual is represented by one dot and the color label corresponding to their self-reported population origin. The percentage of variation explained by each PC is shown on the axes.
Because extensive genotypic data are available for the HGDP samples (Li et al. 2008), we merged those data with ours, producing a common data set of 47,563 SNPs genotyped in 1494 individuals. Principal components analysis of this data set (Supplemental Fig. 2) demonstrated substantial genetic similarity between our samples and those of the HGDP from the same regions. Within each of our four major regions (Supplemental Fig. 3A–D), interpopulation geographic and genetic distances are generally correlated, with the exception of the Central–South Asia region (Supplemental Fig. 3D). The genetic distances between HGDP Central–South Asia populations (sampled from Pakistan and northwestern China) and South Indian populations appear to be influenced by caste affiliation. The two South Indian upper-caste populations in our samples are located more closely to the HGDP Central Asia populations than are the lower-caste or tribal populations sampled from the same region (Supplemental Fig. 3D). Other notable findings include the genetic similarity between the Iban and Cambodian individuals in our sample and the HGDP sample (Supplemental Fig. 3B) and between the Daghestani individuals and HGDP Adygei individuals from the Caucasus region (Supplemental Fig. 3C).
Individual group membership
We assessed the proportion of each individual's ancestry drawn from a given number of inferred populations (K) using a maximum-likelihood based algorithm implemented in FRAPPE (Tang et al. 2005). Individual ancestries and clusters were inferred without reference to known population designations. When the number of population is set to four (i.e., K = 4), the groups inferred by FRAPPE are identical to the four continental groups (Fig. 4A). The African, East/Southeast Asian, European, and Indian individuals are correctly assigned to their self-identified continental groups without exception.
Figure 4.
Individual grouping inferred by FRAPPE. (A) K = 4; (B) K = 7. Each individual's genome is represented by a vertical bar composed of colored sections, where each section represents the proportion of an individual's ancestry derived from one of K ancestral populations. Individuals are arrayed horizontally and grouped by population (labeled on the bottom of the graph) and continent of origin (labeled on the top of the graph).
Some individuals show evidence of membership in multiple groups. South Indian upper- and lower-caste populations have ∼30% and 10% membership in the inferred European group, respectively. South Indian tribal Irula have a relatively high probability of membership in the inferred Indian cluster. Southeast Asians (Iban, Cambodians, and Vietnamese) have ∼10% membership in the inferred Indian cluster, and the African Hema cluster shares ∼15% membership with the inferred European cluster.
When we increase the assumed number of populations, subcontinental population structure can be detected, and genetically isolated groups are split apart from the continental groups. With K = 5, all Mbuti Pygmy and all but three !Kung individuals form a group distinct from other African individuals, reflecting the relative isolation of these populations (Supplemental Fig. 4A). When K = 6, all but one Irula individuals are separated from other Indian individuals to form an additional group (Supplemental Fig. 4B). When K = 7, Southeast Asian individuals (Iban, Cambodian, and Vietnamese) are separated from East Asians to form a single group, with the exception of one Cambodian and one Vietnamese (Fig. 4B). Among the Southeast Asian populations, the Iban show little influence from East Asia, while all but one of the Cambodian and Vietnamese individuals contain a considerable proportion (>30%) of the East Asian component. The separation of populations in subcontinental groups demonstrates the substantial power provided by the large number of SNPs to cluster individuals into smaller groups. With increasing K, weak within-population structure results in unstable groupings of small numbers of individuals.
Cultural and geographic influence on genetic variation in Indian individuals
The population affiliations and caste ranks of our South Indian samples are: two upper-caste (Andhra Brahmin and Tamil Brahmin), three lower-caste (Mala, Madiga, and Dalit), and one non-caste tribal group (Irula). Caste populations from Andhra Pradesh (Andhra Brahmin, Mala, and Madiga) belong to the Telegu linguistic group, while caste populations from Tamil Nadu (Tamil Brahmin and Dalit) and the tribal Irula population speak languages of the Tamil linguistic group. Because Mala and Madiga show great similarity (Fig. 3D; Supplemental Table 1), we combined the two populations into an Andhra lower-caste group in the following analyses.
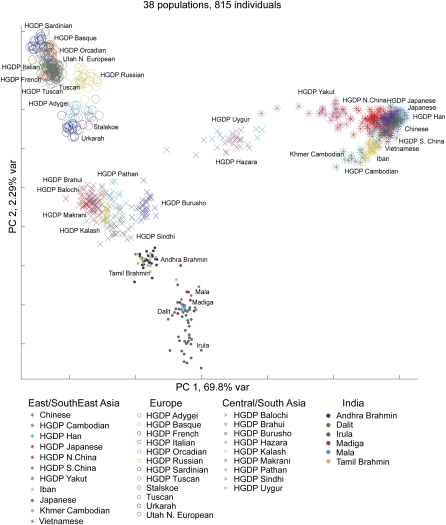
We first sought to examine the relationship between Indian and Eurasian populations. To this end, we calculated pairwise FST distances between populations from Europe, East/Southeast Asia, and India (Table 3). The population relationships based on the FST distances are depicted using a neighbor-joining network (Fig. 5A). Genetic distances between East/Southeast Asian populations and South Indian castes are all larger than distances between European populations and South Indian castes (Table 3). Genetic distances between South Indian castes and European populations are correlated with caste rank. FST distances among Europeans and upper castes (0.033 and 0.032 for Andhra and Tamil upper castes, respectively) are smaller than distances between Europeans and lower castes (0.051 and 0.057 for Andhra and Tamil lower castes, respectively). This genetic cline is more apparent when HGDP samples are added to the data set (Fig. 6). A linear relationship along the second component of the PCA analysis connects populations from northern Europe, southern Europe, the Caucasus region, and Pakistan to the Indian upper-caste, Indian lower-caste, and Indian tribal populations.
Table 3.
FST distances between Eurasians and South Indian populations
aPopulation definition is shown in Table 1.
bStandard error shown in parentheses.
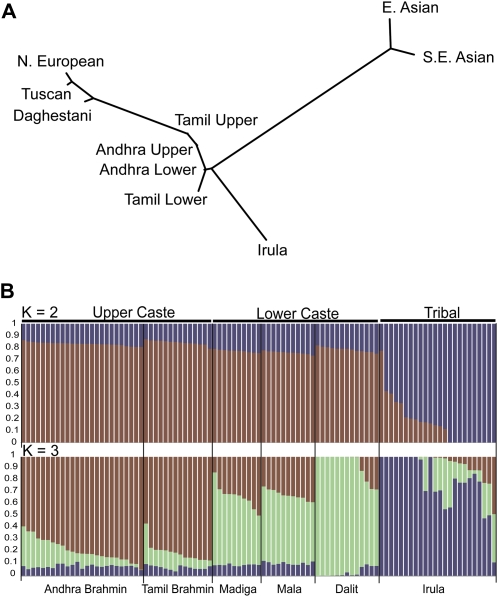
Figure 5.
Genetic variation in South India. (A) Neighbor-joining network of Eurasian and Indian populations based on pairwise FST distances. (B) Individual grouping inferred by FRAPPE in South Indian populations with K = 2 and K = 3. Each individual's genome is represented by a vertical bar composed of colored sections, where each section represents the proportion of an individual's ancestry derived from one of the K ancestral populations. Individuals are arrayed horizontally and grouped by population (labeled on the bottom of the graph) and caste (labeled on the top of the graph).
Figure 6.
Principal components analysis of population structure for 815 Eurasian individuals using 47,563 SNPs. The first two principal components (PCs) are shown. Individuals from East/Southeast Asia (asterisks), Europe (open circles), Central/South Asia (crosses), and India (filled circles) are indicated. The color label of each individual corresponds to his or her self-identified population origin. The percentage of the variation explained by each PC is shown on the axes. For the East/Southeast Asian region, Chinese ethnic groups in the HGDP panel were grouped into Northern China (Oroqen, Daur, Hezhen, Mongola, Xibo, and Tu), Southern China (Dai, Lahu, Miao, Naxi, She, Tujia, and Yi), and Han Chinese (Han and Han-NChina) to improve clarity.
Within the Indian groups, the smallest FST values are observed between the Andhra and Tamil upper-caste samples (0.002). This distance is less than half the distance between upper and lower castes in either state (0.005 for Andhra upper vs. Andhra lower, 0.010 for Tamil upper vs. Tamil lower). The tribal Irula group is substantially differentiated from all caste groups and other Eurasians. FST distances between the Irula and all other caste groups range from 0.026 to 0.032 and are comparable to FST distances between European populations and upper-caste groups (Table 3). This finding is consistent with genetic isolation and drift in the Irula population despite its close geographic proximity to the Tamil castes. Overall, this result is congruent with the principal components analysis (Fig. 3D), where individuals tend to cluster in castes rather than in geographic regions or linguistic groups. Notably, the two upper-caste Brahmin groups are intermingled in the principal components analysis despite their linguistic and geographic separation.
We next determined if the ancestry of each individual could be assigned to a given number of populations using the FRAPPE analysis (Fig. 5B). When the number of presumed ancestral populations is set to two (i.e., K = 2), all caste individuals form one group, while all but one individual from the tribal Irula form another. When K = 3, three clusters corresponding to upper-caste, lower-caste, and tribal populations are identified with the exceptions of one Andhra lower-caste individual and one Irula individual. Using more than three ancestral populations generates less stable patterns within each caste group (i.e., different grouping patterns are generated among multiple runs; data not shown).
Discussion
In 1871, Charles Darwin noted in The Descent of Man, and Selection in Relation to Sex: “It may be doubted whether any character can be named which is distinctive of a race and is constant.” Modern studies of genetic variation, including this one, have supported Darwin's observation, showing that most common variants are shared widely among human populations (Altshuler et al. 2005; Jakobsson et al. 2008). Our data confirm what Darwin believed: We found not a single SNP locus, out of nearly 250,000, at which a fixed difference would distinguish any pair of continental populations. In addition, because population affiliation is not a reliable predictor of an individual's specific genotype or haplotype, a self-identified population is at best loosely correlated with disease phenotypes (Jorde and Wooding 2004; Race, Ethnicity, and Genetics Working Group 2005). Nevertheless, the partial isolation of human populations through time has produced a correlation between geographic ancestry and genetic similarity. This relationship has long been observed at the level of populations (Cavalli-Sforza and Edwards 1963; Jorde 1980; Cavalli-Sforza et al. 1994), and the recent explosion of SNP microarray data has helped to define the ways in which genomic similarities and differences can be measured and analyzed among individuals, rather than populations (Witherspoon et al. 2007; Jakobsson et al. 2008; Li et al. 2008).
In our study, all measures of genetic diversity (heterozygosity, percentage of polymorphic SNPs, and FST) were highest in African populations. Most other genetic surveys show similar results (Yu et al. 2002; Tishkoff and Verrelli 2003; Guthery et al. 2007; Wall et al. 2008). In contrast, some surveys, based on markers that were ascertained for high heterozygosity in specific populations, have shown the highest heterozygosity in non-African populations (Rogers and Jorde 1996). The difference in African and non-African heterozygosity values is smaller in our analysis than are the differences in nucleotide diversity values based on resequencing data in a recent study (Wall et al. 2008), however. These comparisons must be regarded cautiously, because they can be affected by the population sampling scheme. They suggest that heterozygosity estimates based on SNPs in the 250K panel are less affected by ascertainment bias than are those based on other polymorphic systems, but that European heterozygosity, in particular, is still likely to be biased upward and the microarray results here may not have completely assessed the genetic diversity in Africa.
It is encouraging that the genetic distance patterns, as well as overall FST values, obtained from the 250K SNPs were highly similar to the patterns previously seen in these populations for other polymorphisms, such as Alus and STRs, that are generated by very different mutational mechanisms (Jorde et al. 1997, 2000; Bamshad et al. 2003; Watkins et al. 2003; Witherspoon et al. 2006). In addition, the observed genetic distance patterns are similar to those of other studies of worldwide populations that used different population samples and marker panels (Rosenberg et al. 2002, 2005; Jakobsson et al. 2008; Li et al. 2008).
PCA and FRAPPE analyses demonstrate that population structure can be detected at a fine scale with a large number of SNPs. Sub-Saharan African individuals are readily distinguished from non-Africans, and East Asian, European, and Indian individuals are assigned into groups congruent with their places of origin. Using FRAPPE, all 554 individuals in our analysis could be correctly assigned to their continental groups when K was set to 4. It is noteworthy that South Indian upper- and lower-caste populations have ∼30% and 10% membership in the inferred European group, respectively. The lower caste population also has ∼10% East/Southeast Asian group membership. These results reflect the geographic position of India (between Europe and East Asia), the effects of endogamy, and influences from ancient and historical migration events. These patterns are also apparent in the PCA analysis (see Fig. 2). When larger K numbers were used, a few individuals exhibited complex ancestral origin (with >10% genetic membership in at least three groups), and some showed a discrepancy between genetically inferred origin and their self-reported population designation (e.g., three !Kung individuals were assigned to the non-!Kung African group when K = 7). These discrepancies could reflect admixture in these individuals' ancestry. Overall, these results demonstrate the power of the microarray data and are broadly congruent with previous analyses (Rosenberg et al. 2002; Yang et al. 2005; Jakobsson et al. 2008; Li et al. 2008).
Subtle differences that were suggestive previously can be clearly seen with this data set. For example, PCA analysis indicated that the highland Dahgestani Dargins and lowland Kumiks can be separated into distinct groups, substantiating an earlier suggestion that these populations have different histories despite their geographic proximity (Bulayeva et al. 2003; Marchani et al. 2008). PCA and FRAPPE analysis showed that the Daghestani populations are more closely related to Europeans but have some genetic affinity to Asian populations, consistent with evidence that the Caucasus region has served as a migratory gateway between continents (Wells et al. 2001; Bulayeva et al. 2003).
The peopling of Southeast Asia is another topic of ongoing interest in studies of human population history (Bellwood 1997; Barker et al. 2007; Hill et al. 2007; Soares et al. 2008). Our PCA analysis showed that the Iban population, which is located on Borneo in the center of Island Southeast Asia, forms an identifiable group that is most similar to other Southeast Asian populations (Vietnamese and Cambodian; Fig. 3B). The FRAPPE analysis indicated that the Iban population, unlike the Cambodian and Vietnamese samples, shows little genetic contribution from other East Asian populations. Additional studies, with further population sampling, are needed to determine whether this population represents a genetic isolate.
By merging our results with those of the HGDP, we gained insights about population relationships in Central and South Asia. The Central/South Asian populations from the HGDP panel were separated into two groups by PCA (Fig. 6). Seven of these populations fall between European and Indian individuals, while two populations (Uygur and Hazara) show greater similarity to East/Southeast Asia populations. The Uygur were sampled from northwestern China, in contrast to other HGDP South/Central Asia populations that were sampled from Pakistan, while the Hazara are thought to contain a large Mongolian genetic contribution (Kakar 1973; Qamar et al. 2002).
Our analyses also shed additional light on the genetic structure of Indian populations, which has been the subject of much research and debate (Bamshad et al. 1998, 2001; Basu et al. 2003; Vishwanathan et al. 2004; Watkins et al. 2005, 2008; Rosenberg et al. 2006; Chaubey et al. 2007). Our results show relatively larger genetic distances between the caste and tribal populations than among caste populations (Fig. 3D). The tribal Irula population also exhibits more interindividual variation but lower overall heterozygosity than do the caste populations. This difference is likely to reflect the greater degree of genetic drift in small isolated groups within the Irula population (Watkins et al. 2005). Since the Irula individuals in this study were sampled from two locations in southern Andhra Pradesh, it is possible that the genetic structure observed within this population may reflect differences in sampling locality. It is noteworthy that some of the Pakistani groups in the HGDP set show similar levels of genetic diversity to the Irula (Supplemental Fig. 3D).
The upper-caste and lower-caste populations from each Indian state can be distinguished despite being sampled from the same geographic location, speaking the same language, and having a relatively small distance from each other (FST = 0.005 and 0.010 for Andhra Pradesh and Tamil Nadu, upper- vs. lower-caste samples, respectively). This result is consistent with a recent study of Y chromosome, mtDNA, and autosomal STR markers in Tamil and Andhra castes, which also provided evidence for sex differences in gene flow (Watkins et al. 2008). It is noteworthy that while previous studies demonstrated genetic structure for the Andhra Indian caste populations (Bamshad et al. 1998, 2001), this high-density SNP data set clusters individuals into self-identified groups (upper and lower caste) with little overlap between the two groups (Fig. 3D). The resolution necessary to cluster individuals into caste groups based on genetic data alone was not achievable using these populations with a smaller number (45) of STR markers (Watkins et al. 2008).
Consistent with the PCA results, FRAPPE analysis of Indian individuals can correctly place most individuals into upper-caste, lower-caste, or tribal groups when the number of populations is set to three (with the exceptions of two individuals). This result differs somewhat from that of a previous study in which little genetic structure could be detected among 15 Indian populations (Rosenberg et al. 2006). Several factors may be responsible for this difference. First, the number of markers used in the Rosenberg study (1200) was much smaller than the number used in our study. Second, because all individuals sampled in the Rosenberg study were Indians living in the United States, their data set was likely biased toward upper-caste individuals and thus less likely to detect the effects of caste or tribal membership. Finally, the Rosenberg sample contained individuals from all regions of India, while our sample was derived only from South Indian populations.
By investigating unique populations in locations underrepresented in earlier studies, we have discovered previously undetected population structure, especially within the Indian subcontinent. These results emphasize the correlation of genetic and geographic distances and highlight other elements, including social factors that have contributed to population structure. Furthermore, the high-density SNP genotype data generated in this study using a standardized genotyping platform can help to serve as reference for future studies.
Methods
DNA samples and whole-genome amplification
We used DNA samples from 383 individuals in 23 worldwide populations (Fig. 1; Table 1). We sampled eight populations from sub-Saharan Africa, including four Bantu-speaking populations (Luhya, Sotho/Tswana, Pedi, and Nguni), two Nilotic-speaking populations (Alur and Hema), and two hunter–gatherer groups (Mbuti Pygmy and !Kung). We included four European populations: two from Dagehstan (Urkarah and Stalskoe), Utahns of Northern European descent, and Tuscans from Italy. Five populations were sampled from East/Southeast Asia, including Chinese, Japanese, Cambodian, Vietnamese, and the Iban from Sarawak, Malaysia. The South Asian samples were collected from two South Indian states (Andhra Pradesh and Tamil Nadu), including one tribal population (Irula from southern Andhra Pradesh) and five caste groups: Brahmin, Mala, and Madiga from Andhra Pradesh; Tamil Brahmin and Dalit from Tamil Nadu.
Most samples were collected previously in our laboratory (Jorde et al. 1995; Bamshad et al. 1998, 2001; Watkins et al. 1999, 2008; Bulayeva et al. 2003). DNA samples of Luhya from Webuye, Kenya and Tuscans from Italy are part of the International HapMap project and were purchased from the Coriell Cell Repositories (http://ccr.coriell.org/).
Because DNA samples are available only in limited quantity for four African populations, 36 samples from these populations were subjected to whole-genome amplification (WGA) prior to genotyping (Table 1). Four additional samples were also whole-genome amplified as duplicates to assess the quality of the WGA product. WGA was performed on these samples using a REPLI-g mini kit (Qiagen) following the manufacturer's protocol. Ten nanograms of purified genomic DNA was used as template, and the amplification product was normalized to a concentration of 50 ng/μL prior to the microarray experiment.
Genotyping
For all individuals except those from Tamil Nadu, high-throughput microarray genotyping of ∼262,000 SNPs were performed using one array (version NspI) from the Affymetrix GeneChip Human Mapping 500K Array set (Affymetrix). The 27 Tamil samples were genotyped using Affymetrix Genome-Wide Human SNP Array 6.0 (Affymetrix), which contains more than 906,000 SNPs. The recommended protocol as described in the Affymetrix manual was followed. Briefly, DNA libraries were prepared from genomic DNA or WGA product for each platform. Samples were then injected into microarray cartridges and hybridized in a GeneChip Hybridization Oven 640 (Affymetrix), followed by washing and staining in a GeneChip Fluidics Station 450 (Affymetrix). Mapping array images were obtained using the GeneChip Scanner 3000 7G (Affymetrix).
Genotype calling and quality control
For the 250K NspI array, genotypes for each microarray were first called with the Affymetrix Dynamic Model algorithm (Di et al. 2005) to assess the quality of the experiment. Out of 362 total arrays (356 unique individuals plus six duplicates for quality control purposes), 14 had call rates lower than a 90% threshold level and were excluded from further analysis. The remaining 348 arrays (342 unique individuals plus six duplicates) were then called together using the BRLMM algorithm (Affymetrix 2006) with default parameters. Among the six duplicated samples, two are technical duplicates (two experiments performed on the same sample) and four are genomic DNA/WGA DNA duplicates, for which one experiment is performed on genomic DNA and one experiment is performed on the whole-genome amplified DNA sample. All duplicates have very high concordance rate (99.93% for the two technical duplicates and 98.65% for the four DNA/whole-genome amplification DNA duplicates, on average). Detailed comparisons of call rates and concordant rates among duplicates indicate that WGA products perform as well as genomic DNA on microarrays (Xing et al. 2008). Among the 348 samples, six duplicated samples, 13 samples with insufficient population information, and nine samples with <95% BRLMM call rates were excluded from subsequent analyses.
For the 6.0 array, all 27 arrays passed the initial quality control, and genotypes were called using the Birdseed algorithm (version 2) included in the Affymetrix Power Tools (APT) Software Package (http://www.affymetrix.com/). Because the manufacturer recommends a minimum of 44 samples for accurate genotype calling, we included the CEL files of 90 HapMap CEU samples, and genotypes were called for a total of 117 arrays with default parameters.
Next, we merged the two data sets and kept only the SNPs that were present in both platforms, resulting in a total of 254,326 SNPs in 347 samples. We then calculated the pairwise genetic distances between each pair of individuals. Four comparisons showed unusually small genetic distances, indicating close relatedness among these individuals. Three samples were then excluded to remove the possible related individual pairs (one individual was involved in two comparisons). The remaining 344 samples from 23 populations composed our data set.
SNP selection
Several criteria have been applied to select SNPs for the analyses. First, we excluded all SNPs on the X chromosome or SNPs whose chromosomal location is unknown (5589 SNPs). Then, SNPs with >10% missing data were removed (3872 SNPs). We then used hweStrata (Schaid et al. 2006) to test each SNP for deviations from Hardy–Weinberg equilibrium (HWE) while allowing for between-population differences in allele frequencies. The data set was subdivided into smaller groups (respecting continent-level groupings where possible) in order to keep the computational costs manageable. The group-level HWE P-values were combined using Stouffer's Z-average method (Stouffer et al. 1949), and 226 SNPs that deviated from HWE at P < 2 × 10−7 (Bonferroni correction: 0.05/244,865) were excluded from subsequent analyses. When the same procedure applied to the SNPs with >10% missing data, we found they were about 45 times more likely to deviate from HWE at that level than SNPs with higher call rates, indicating a general lower quality of these SNPs.
To combine our data set with HapMap samples, genotypes of the 210 unrelated HapMap samples were obtained from the Affymetrix website (http://www.affymetrix.com/support/technical/sample_data/500k_hapmap_genotype_data.affx), and the same SNP selection criteria were applied to HapMap samples. The filtered HapMap data set was combined with our data set, resulting in a final data set containing 243,855 loci genotyped in 554 individuals from 27 populations. Genotypes of all samples in the final data set are available as a supplemental file on our website (http://jorde-lab.genetics.utah.edu/) under Published Data.
The genotypes of 940 unrelated HGDP samples, belonging the 952 panel of Rosenberg (2006), were obtained from Li et al. (2008) and subjected to the same SNP selection criteria described above. A merged data set was then generated by combining all SNPs present in both data sets, yielding a data set containing 47,563 SNPs genotyped in 1494 individuals.
Data analysis
Pairwise allele-sharing genetic distance calculations and PCA analyses were performed using MATLAB (ver. r2008a). FST estimates between populations were calculated by the method described by Weir and Cockerham (1984). A maximum-likelihood-based algorithm implemented in FRAPPE (Tang et al. 2005) was used to determine the genetic ancestry of each individual in a given number of groups without using his/her population designation. Each individual is assigned to the group in which he/she has the highest proportion of inferred ancestry. Each run was repeated at least three times to assess the stability of the clustering pattern.
To assess the concordance of between-population genetic distances based on the high-density SNP data and other types of polymorphic autosomal markers, pairwise FST distances were calculated for autosomal SNPs or 100 Alu insertion polymorphisms for 11 populations (243 individuals) that include Africans (!Kung, Mbuti Pygmy, South African Bantu-speaking groups, and Nilotic-speaking groups), Europeans (Utah and Daghestani), South Indians (upper-caste, lower-caste, and tribal Irula), and Eastern Asians (Eastern Asians and South Eastern Asians). Pairwise allele-weighed stepwise weighted genetic distances were calculated using STR data and the populations listed above, excluding Daghestanis. The SNP–STR and Alu–STR comparisons were performed using a final data set of 217 samples. A correlation coefficient was calculated for each pair of distance matrices using the Mantel matrix correlation test. Significance values were determined by random permutations of the columns of the matrices.
To investigate the relationship between Eurasian and Indian populations, some populations were grouped to increase the sample size in each population. The composition of each population group is shown in Table 1. Pairwise FST estimates and standard errors between population groups were obtained using the EIGENSTRAT software package (Price et al. 2006).
Acknowledgments
We thank Judith and Kenneth Kidd and Himla Soodyall for contributing DNA samples. We also thank Diane Dunn and Edward Meenen for their technical support during the microarray hybridization and scanning process. We thank Hua Tang for providing the 64-bit version of the FRAPPE program. We thank the anonymous reviewers and Tatum S. Simonson for their valuable comments. This work was supported by grants from the National Science Foundation (BCS-0218370) and the National Institutes of Health (GM-59290 to L.B.J. and DK069513 to S.L.G.).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article is online at http://www.genome.org/cgi/doi/10.1101/gr.085589.108.
References
- Affymetrix. BRLMM: An improved genotype calling method for the GeneChip Human Mapping 500K array set. 2006 http://www.affymetrix.com/support/technical/whitepapers/brlmm_whitepaper.pdf.
- Alcais A., Alter A., Antoni G., Orlova M., Nguyen V.T., Singh M., Vanderborght P.R., Katoch K., Mira M.T., Vu H.T., et al. Stepwise replication identifies a low-producing lymphotoxin-alpha allele as a major risk factor for early-onset leprosy. Nat. Genet. 2007;39:517–522. doi: 10.1038/ng2000. [DOI] [PubMed] [Google Scholar]
- Altshuler D., Brooks L.D., Chakravarti A., Collins F.S., Daly M.J., Donnelly P. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad M.J., Watkins W.S., Dixon M.E., Rao B.B., Naidu J.M., Prasad B.V.R., Reddy P.G., Sung S., Rasanayagam A., Hammer M.F., et al. Female gene flow stratifies Hindu castes. Nature. 1998;395:651–652. doi: 10.1038/27103. [DOI] [PubMed] [Google Scholar]
- Bamshad M., Kivisild T., Watkins W.S., Dixon M.E., Ricker C.E., Rao B.B., Naidu J.M., Prasad B.V., Reddy P.G., Rasanayagam A., et al. Genetic evidence on the origins of Indian caste populations. Genome Res. 2001;11:994–1004. doi: 10.1101/gr.173301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad M.J., Wooding S., Watkins W.S., Ostler C.T., Batzer M.A., Jorde L.B. Human population genetic structure and inference of group membership. Am. J. Hum. Genet. 2003;72:578–589. doi: 10.1086/368061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker G., Barton H., Bird M., Daly P., Datan I., Dykes A., Farr L., Gilbertson D., Harrisson B., Hunt C., et al. The “human revolution” in lowland tropical Southeast Asia: The antiquity and behavior of anatomically modern humans at Niah Cave (Sarawak, Borneo) J. Hum. Evol. 2007;52:243–261. doi: 10.1016/j.jhevol.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Basu A., Mukherjee N., Roy S., Sengupta S., Banerjee S., Chakraborty M., Dey B., Roy M., Roy B., Bhattacharyya N.P., et al. Ethnic India: A genomic view, with special reference to peopling and structure. Genome Res. 2003;13:2277–2290. doi: 10.1101/gr.1413403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauchet M., McEvoy B., Pearson L.N., Quillen E.E., Sarkisian T., Hovhannesyan K., Deka R., Bradley D.G., Shriver M.D. Measuring European population stratification with microarray genotype data. Am. J. Hum. Genet. 2007;80:948–956. doi: 10.1086/513477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellwood P. Prehistory of the Indo-Malaysian archipelago. University of Hawai'i Press; Honolulu, HI: 1997. [Google Scholar]
- Bulayeva K., Jorde L.B., Ostler C., Watkins S., Bulayev O., Harpending H. Genetics and population history of Caucasus populations. Hum. Biol. 2003;75:837–853. doi: 10.1353/hub.2004.0003. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza L.L., Edwards A.W.F. Analysis of human evolution. In: Geerts S.J., editor. Genetics today. Pergamon; New York: 1963. pp. 923–933. [Google Scholar]
- Cavalli-Sforza L.L., Menozzi P., Piazza A. The history and geography of human genes. Princeton University Press; Princeton, NJ: 1994. [Google Scholar]
- Chambers J.C., Elliott P., Zabaneh D., Zhang W., Li Y., Froguel P., Balding D., Scott J., Kooner J.S. Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat. Genet. 2008;40:716–718. doi: 10.1038/ng.156. [DOI] [PubMed] [Google Scholar]
- Chaubey G., Metspalu M., Kivisild T., Villems R. Peopling of South Asia: Investigating the caste–tribe continuum in India. Bioessays. 2007;29:91–100. doi: 10.1002/bies.20525. [DOI] [PubMed] [Google Scholar]
- Di X., Matsuzaki H., Webster T.A., Hubbell E., Liu G., Dong S., Bartell D., Huang J., Chiles R., Yang G., et al. Dynamic model based algorithms for screening and genotyping over 100 K SNPs on oligonucleotide microarrays. Bioinformatics. 2005;21:1958–1963. doi: 10.1093/bioinformatics/bti275. [DOI] [PubMed] [Google Scholar]
- Guthery S.L., Salisbury B.A., Pungliya M.S., Stephens J.C., Bamshad M. The structure of common genetic variation in United States populations. Am. J. Hum. Genet. 2007;81:1221–1231. doi: 10.1086/522239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C., Soares P., Mormina M., Macaulay V., Clarke D., Blumbach P.B., Vizuete-Forster M., Forster P., Bulbeck D., Oppenheimer S., et al. A mitochondrial stratigraphy for island Southeast Asia. Am. J. Hum. Genet. 2007;80:29–43. doi: 10.1086/510412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday E.G., Nyholt D.R., Tirupati S., John S., Ramachandran P., Ramamurti M., Ramadoss A.J., Jeyagurunathan A., Kottiswaran S., Smith H.J., et al. Strong evidence for a novel schizophrenia risk locus on chromosome 1p31.1 in homogeneous pedigrees from Tamil Nadu, India. Am. J. Psychiatry. 2008 doi: 10.1176/appi.ajp.2008.08030442. [DOI] [PubMed] [Google Scholar]
- Jakobsson M., Scholz S.W., Scheet P., Gibbs J.R., VanLiere J.M., Fung H.C., Szpiech Z.A., Degnan J.H., Wang K., Guerreiro R., et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- Jorde L.B. The genetic structure of subdivided human populations: A review. In: Mielke J.H., Crawford M.H., editors. Current developments in anthropological genetics, Vol. 1. Plenum Press; New York: 1980. pp. 135–208. [Google Scholar]
- Jorde L.B., Wooding S.P. Genetic variation, classification, and “race.”. Nat. Genet. 2004;36:S28–S33. doi: 10.1038/ng1435. [DOI] [PubMed] [Google Scholar]
- Jorde L.B., Bamshad M.J., Watkins W.S., Zenger R., Fraley A.E., Krakowiak P.A., Carpenter K.D., Soodyall H., Jenkins T., Rogers A.R. Origins and affinities of modern humans: A comparison of mitochondrial and nuclear genetic data. Am. J. Hum. Genet. 1995;57:523–538. doi: 10.1002/ajmg.1320570340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorde L.B., Rogers A.R., Bamshad M., Watkins W.S., Krakowiak P., Sung S., Kere J., Harpending H.C. Microsatellite diversity and the demographic history of modern humans. Proc. Natl. Acad. Sci. 1997;94:3100–3103. doi: 10.1073/pnas.94.7.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorde L.B., Watkins W.S., Bamshad M.J., Dixon M.E., Ricker C.E., Seielstad M.T., Batzer M.A. The distribution of human genetic diversity: A comparison of mitochondrial, autosomal, and Y chromosome data. Am. J. Hum. Genet. 2000;66:979–988. doi: 10.1086/302825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakar M.H. The Pacification of the Hazaras of Afghanistan. Afghanistan Council, Asia Society; New York: 1973. [Google Scholar]
- Li J.Z., Absher D.M., Tang H., Southwick A.M., Casto A.M., Ramachandran S., Cann H.M., Barsh G.S., Feldman M., Cavalli-Sforza L.L., et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- Marchani E.E., Watkins W.S., Bulayeva K., Harpending H.C., Jorde L.B. Culture creates genetic structure in the Caucasus: Autosomal, mitochondrial, and Y chromosomal variation in Daghestan. BMC Genet. 2008;9:47. doi: 10.1186/1471-2156-9-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountain J.L., Cavalli-Sforza L.L. Multilocus genotypes, a tree of individuals, and human evolutionary history. Am. J. Hum. Genet. 1997;61:705–718. doi: 10.1086/515510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M.R., Bryc K., King K.S., Indap A., Boyko A.R., Novembre J., Briley L.P., Maruyama Y., Waterworth D.M., Waeber G., et al. The Population Reference Sample, POPRES: A resource for population, disease, and pharmacological genetics research. Am. J. Hum. Genet. 2008;83:347–358. doi: 10.1016/j.ajhg.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novembre J., Johnson T., Bryc K., Kutalik Z., Boyko A.R., Auton A., Indap A., King K.S., Bergmann S., Nelson M.R., et al. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Price A.L., Butler J., Patterson N., Capelli C., Pascali V.L., Scarnicci F., Ruiz-Linares A., Groop L., Saetta A.A., Korkolopoulou P., et al. Discerning the ancestry of European Americans in genetic association studies. PLoS Genet. 2008;4:e236. doi: 10.1371/journal.pgen.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qamar R., Ayub Q., Mohyuddin A., Helgason A., Mazhar K., Mansoor A., Zerjal T., Tyler-Smith C., Mehdi S.Q. Y chromosomal DNA variation in Pakistan. Am. J. Hum. Genet. 2002;70:1107–1124. doi: 10.1086/339929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Race, Ethnicity, and Genetics Working Group. The use of racial, ethnic, and ancestral categories in human genetics research. Am. J. Hum. Genet. 2005;77:519–532. doi: 10.1086/491747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers A.R., Jorde L.B. Ascertainment bias in estimates of average heterozygosity. Am. J. Hum. Genet. 1996;58:1033–1041. [PMC free article] [PubMed] [Google Scholar]
- Rosenberg N.A. Standardized subsets of the HGDP-CEPH Human Genome Diversity Cell Line Panel, accounting for atypical and duplicated samples and pairs of close relatives. Ann. Hum. Genet. 2006;70:841–847. doi: 10.1111/j.1469-1809.2006.00285.x. [DOI] [PubMed] [Google Scholar]
- Rosenberg N.A., Pritchard J.K., Weber J.L., Cann H.M., Kidd K.K., Zhivotovsky L.A., Feldman M.W. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- Rosenberg N.A., Mahajan S., Ramachandran S., Zhao C., Pritchard J.K., Feldman M.W. Clines, clusters, and the effect of study design on the inference of human population structure. PLoS Genet. 2005;1:e70. doi: 10.1371/journal.pgen.0010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg N.A., Mahajan S., Gonzalez-Quevedo C., Blum M.G., Nino-Rosales L., Ninis V., Das P., Hegde M., Molinari L., Zapata G., et al. Low levels of genetic divergence across geographically and linguistically diverse populations from India. PLoS Genet. 2006;2:e215. doi: 10.1371/journal.pgen.0020215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaid D.J., Batzler A.J., Jenkins G.D., Hildebrandt M.A. Exact tests of Hardy–Weinberg equilibrium and homogeneity of disequilibrium across strata. Am. J. Hum. Genet. 2006;79:1071–1080. doi: 10.1086/510257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seldin M.F., Shigeta R., Villoslada P., Selmi C., Tuomilehto J., Silva G., Belmont J.W., Klareskog L., Gregersen P.K. European population substructure: Clustering of northern and southern populations. PLoS Genet. 2006;2:e143. doi: 10.1371/journal.pgen.0020143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares P., Trejaut J.A., Loo J.H., Hill C., Mormina M., Lee C.L., Chen Y.M., Hudjashov G., Forster P., Macaulay V., et al. Climate change and postglacial human dispersals in Southeast Asia. Mol. Biol. Evol. 2008;25:1209–1218. doi: 10.1093/molbev/msn068. [DOI] [PubMed] [Google Scholar]
- Stouffer S.A., Suchman E.A., DeVinney L.C., Star S.A., Williams R.M.J. The American soldier: Adjustment during army life. Princeton University Press; Princeton, NJ: 1949. [Google Scholar]
- Tang H., Quertermous T., Rodriguez B., Kardia S.L., Zhu X., Brown A., Pankow J.S., Province M.A., Hunt S.C., Boerwinkle E., et al. Genetic structure, self-identified race/ethnicity, and confounding in case-control association studies. Am. J. Hum. Genet. 2005;76:268–275. doi: 10.1086/427888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C., Plenge R.M., Ransom M., Lee A., Villoslada P., Selmi C., Klareskog L., Pulver A.E., Qi L., Gregersen P.K., et al. Analysis and application of European genetic substructure using 300 K SNP information. PLoS Genet. 2008;4:e4. doi: 10.1371/journal.pgen.0040004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff S.A., Verrelli B.C. Patterns of human genetic diversity: Implications for human evolutionary history and disease. Annu. Rev. Genomics Hum. Genet. 2003;4:293–340. doi: 10.1146/annurev.genom.4.070802.110226. [DOI] [PubMed] [Google Scholar]
- Vishwanathan H., Deepa E., Cordaux R., Stoneking M., Usha Rani M.V., Majumder P.P. Genetic structure and affinities among tribal populations of southern India: A study of 24 autosomal DNA markers. Ann. Hum. Genet. 2004;68:128–138. doi: 10.1046/j.1529-8817.2003.00083.x. [DOI] [PubMed] [Google Scholar]
- Wall J.D., Cox M.P., Mendez F.L., Woerner A., Severson T., Hammer M.F. A novel DNA sequence database for analyzing human demographic history. Genome Res. 2008;18:1354–1361. doi: 10.1101/gr.075630.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins W.S., Bamshad M., Dixon M.E., Bhaskara Rao B., Naidu J.M., Reddy P.G., Prasad B.V., Das P.K., Reddy P.C., Gai P.B., et al. Multiple origins of the mtDNA 9-bp deletion in populations of South India. Am. J. Phys. Anthropol. 1999;109:147–158. doi: 10.1002/(SICI)1096-8644(199906)109:2<147::AID-AJPA1>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Watkins W.S., Rogers A.R., Ostler C.T., Wooding S., Bamshad M.J., Brassington A.M., Carroll M.L., Nguyen S.V., Walker J.A., Prasad B.V., et al. Genetic variation among world populations: Inferences from 100 Alu insertion polymorphisms. Genome Res. 2003;13:1607–1618. doi: 10.1101/gr.894603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins W.S., Prasad B.V., Naidu J.M., Rao B.B., Bhanu B.A., Ramachandran B., Das P.K., Gai P.B., Reddy P.C., Reddy P.G., et al. Diversity and divergence among the tribal populations of India. Ann. Hum. Genet. 2005;69:680–692. doi: 10.1046/j.1529-8817.2005.00200.x. [DOI] [PubMed] [Google Scholar]
- Watkins W.S., Thara R., Mowry B.J., Zhang Y., Witherspoon D.J., Tolpinrud W., Bamshad M.J., Tiripati S., Padmavati R., Smith H., et al. Genetic variation in South Indian castes: Evidence from Y chromosome, mitochondrial, and autosomal polymorphisms. BMC Genet. 2008;9:86. doi: 10.1186/1471-2156-9-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir B.S., Cockerham C.C. Estimating F-statistics for the analysis of population structure. Evolution Int. J. Org. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells R.S., Yuldasheva N., Ruzibakiev R., Underhill P.A., Evseeva I., Blue-Smith J., Jin L., Su B., Pitchappan R., Shanmugalakshmi S., et al. The Eurasian heartland: A continental perspective on Y chromosome diversity. Proc. Natl. Acad. Sci. 2001;98:10244–10249. doi: 10.1073/pnas.171305098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witherspoon D.J., Marchani E.E., Watkins W.S., Ostler C.T., Wooding S.P., Anders B.A., Fowlkes J.D., Boissinot S., Furano A.V., Ray D.A., et al. Human population genetic structure and diversity inferred from polymorphic L1(LINE-1) and Alu insertions. Hum. Hered. 2006;62:30–46. doi: 10.1159/000095851. [DOI] [PubMed] [Google Scholar]
- Witherspoon D.J., Wooding S., Rogers A.R., Marchani E.E., Watkins W.S., Batzer M.A., Jorde L.B. Genetic similarities within and between human populations. Genetics. 2007;176:351–359. doi: 10.1534/genetics.106.067355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooding S., Ostler C., Prasad B.V., Watkins W.S., Sung S., Bamshad M., Jorde L.B. Directional migration in the Hindu castes: Inferences from mitochondrial, autosomal and Y chromosomal data. Hum. Genet. 2004;115:221–229. doi: 10.1007/s00439-004-1130-x. [DOI] [PubMed] [Google Scholar]
- Xing J., Watkins W.S., Zhang Y., Witherspoon D.J., Jorde L.B. High fidelity of whole-genome amplified DNA on high-density single nucleotide polymorphism arrays. Genomics. 2008;92:452–456. doi: 10.1016/j.ygeno.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N., Li H., Criswell L.A., Gregersen P.K., Alarcon-Riquelme M.E., Kittles R., Shigeta R., Silva G., Patel P.I., Belmont J.W., et al. Examination of ancestry and ethnic affiliation using highly informative diallelic DNA markers: Application to diverse and admixed populations and implications for clinical epidemiology and forensic medicine. Hum. Genet. 2005;118:382–392. doi: 10.1007/s00439-005-0012-1. [DOI] [PubMed] [Google Scholar]
- Yu N., Chen F.C., Ota S., Jorde L.B., Pamilo P., Patthy L., Ramsay M., Jenkins T., Shyue S.K., Li W.H. Larger genetic differences within Africans than between Africans and Eurasians. Genetics. 2002;161:269–274. doi: 10.1093/genetics/161.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]