Abstract
One and a half centuries after Charles Darwin and Alfred Russel Wallace outlined our current understanding of evolution, a new scientific era is dawning that enables direct observations of genetic variation. However, pure sequence-based molecular attempts to resolve the basal origin of placental mammals have so far resulted only in apparently conflicting hypotheses. By contrast, in the mammalian genomes where they were highly active, the insertion of retroelements and their comparative insertion patterns constitute a neutral, virtually homoplasy-free archive of evolutionary histories. The “presence” of a retroelement at an orthologous genomic position in two species indicates their common ancestry in contrast to its “absence” in more distant species. To resolve the placental origin controversy we extracted ∼2 million potentially phylogenetically informative, retroposon-containing loci from representatives of the major placental mammalian lineages and found highly significant evidence challenging all current single hypotheses of their basal origin. The Exafroplacentalia hypothesis (Afrotheria as the sister group to all remaining placentals) is significantly supported by five retroposon insertions, the Epitheria hypothesis (Xenarthra as the sister group to all remaining placentals) by nine insertion patterns, and the Atlantogenata hypothesis (a monophyletic clade comprising Xenarthra and Afrotheria as the sister group to Boreotheria comprising all remaining placentals) by eight insertion patterns. These findings provide significant support for a “soft” polytomy of the major mammalian clades. Ancestral successive hybridization events and/or incomplete lineage sorting associated with short speciation intervals are viable explanations for the mosaic retroposon insertion patterns of recent placental mammals and for the futile search for a clear root dichotomy.
Genomic variation is based on genetic convertibility and is amplified by gene flow and sexual recombination. While only a small fraction of genetic variation is directly exposed to the natural selection driving the evolution of species, the majority of changes are neutral (Kimura 1968). In the molecular Darwinian and post-genomic age when such variations are now directly observable in prodigious numbers, one still needs only compare the volumes of controversy in scientific reports to dispel the notion that all the mysteries of evolutionary history are now an open book. The basal node of the placental mammals is a prime example of such a controversy. After continuous morphological inconsistencies and a lack of resolution due to narrow temporal speciation events at the deeper divergences of the placental mammalian tree, the hope was that large-scale DNA sequencing and an increasing number of sampled species might resolve the higher-level mammalian relationships by reducing systematic biases. But the extraction and analysis of molecular traces of evolutionary history has been no less sensitive to misinterpretation than was the understanding of dusty specimens 150 years ago. The reliability of sequence data in genome-scale phylogenetic approaches is subject to unequal evolutionary rates among lineages, regional- or lineage-specific compositional biases, shifts in site-specific evolutionary rates over time and sequence regions (e.g., Nishihara et al. 2007), variable accuracies of multisequence alignments and analytical tools, and last but not least, the general liability of such data to homoplasy.
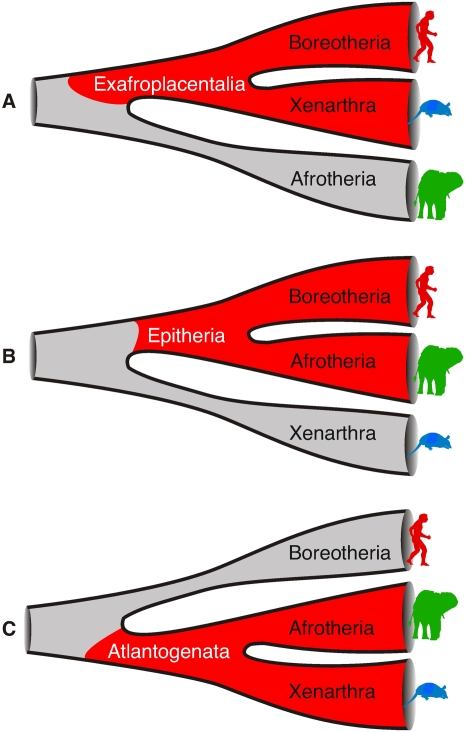
In a seminal work, Murphy et al. (2001a) investigated ∼10 kb of sequence information to substantiate the previously proposed (Waddell et al. 1999) major mammalian groups Afrotheria, Xenarthra, Laurasiatheria, and Euarchontoglires. Moderate support was found for Afrotheria as the basal split of placentals (Exafroplacentalia; Fig. 1A). A subsequent combination and expansion of previously published data (Madsen et al. 2001; Murphy et al. 2001a) found significant support for a basal split between Afrotheria and other placentals (Murphy et al. 2001b). Nikolaev et al. (2007) also found support for the Afrotheria as the first placental split, drawing on ∼200 kb of protein-coding sequences from the 1% of the human genome studied in the ENCODE pilot project (The ENCODE Project Consortium 2007). However, other studies of large-scale sequences (e.g., Hallström et al. 2007) and previous studies of smaller data sets could not significantly confirm an Afrotherian root (e.g., Madsen et al. 2001; Waddell et al. 2001; Delsuc et al. 2002; Amrine-Madsen et al. 2003). A sister-group relationship of Afrotheria and Xenarthra (Atlantogenata) also received significant support in analyses of sequences from ∼1700 conserved genome loci (Wildman et al. 2007), 2840 protein-coding genes (Hallström et al. 2007), and mitochondrial genomes (Kjer and Honeycutt 2007); and this was recently confirmed by large-scale analyses of 2.8 Mbp of protein-coding data (Hallström and Janke 2008) and large-scale genomic sequences (Prasad et al. 2008). Thus, despite many different molecular studies, the basal origin of placental mammals remains controversial and unresolved. Unfortunately, pure sequence data cannot resolve these apparent contradictions.
Figure 1.
Different hypotheses of the placental origin. (A) The Exafroplacentalia hypothesis proposes a mammalian clade merging Boreotheria (Supraprimates plus Laurasiathera) and Xenarthra, with Afrotheria as the sister group. (B) The Epitheria hypothesis merges Boreotheria and Afrotheria, with Xenarthra as the sister group. (C) The Atlantogenata hypothesis proposes Xenarthra and Afrotheria in one clade.
By contrast, the presence/absence patterns of inserted retroelements constitute a virtually homoplasy-free marker system with theoretically infinite character states (Steel and Penny 2000). Specific genomic insertions of such elements in the ancestor of two species reliably document their common ancestry. The very few known examples of discordance in retroelement presence/absence data can be explained by deletion via illegitimate recombination between perfect direct repeats flanking each insertion (van de Lagemaat et al. 2005), exact parallel insertions (Cantrell et al. 2001), or lineage sorting, a phenomenon related to incomplete allele fixation and frequent intervals of speciation events (Shedlock et al. 2004; Ray et al. 2006).
Analyses of a smaller number of retroelements as phylogenetic markers in mammals validated the four superordinal mammalian clades (Nishihara et al. 2005; Kriegs et al. 2006; Möller-Krull et al. 2007). However, in support of Shoshani and McKenna (1998), but in contradiction to most other molecular investigations, we found the first evidence, two L1MB5 retroelements, that placed Xenarthra (Epitheria hypothesis) (Fig. 1B) instead of Afrotheria at the base of the placental tree (Kriegs et al. 2006). Meanwhile, Murphy et al. (2007) presented two L1MB5 retroelements and four additional indels that favored a sister group relationship of Afrotheria and Xenarthra (Atlantogenata) (Fig. 1C) at the base of the placental tree. Thus, to date, even retroposon insertion patterns have not satisfactorily resolved the basal split of placental mammals.
With the intention of resolving the conflicting topologies of placental lineages, we aimed to examine a much larger sample of retroposed elements and in more species. For this study we took a slightly different tack and aligned whole genomes of three representative placental species, including additional species for informative loci to represent a maximum divergence within each of the groups, and scanned them for diagnostic retroelement insertions. In an analysis of ∼2 million potential phylogenetically informative loci, we found multiple retroposon insertion patterns significantly supporting all three placental speciation hypotheses, indicating a complex ancestral speciation scenario including successive early divergences in close temporal proximity and hybridization and/or lineage sorting events as viable sources for an effective “soft” polytomy. This very rare example of multiple retroposon incongruence goes a long way toward explaining the decades-long stream of apparently conflicting evidence for one or the other placental evolutionary history based on multiple marker systems including both morphological and molecular sequence evidence.
Results
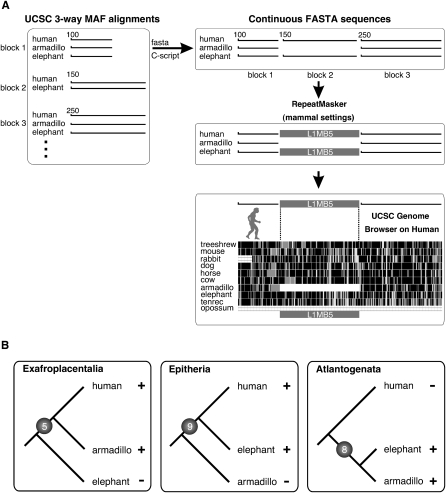
We focused our computational screening for phylogenetically informative markers on the insertion patterns of L1MB elements that were active during the critical period of early placental evolution ∼100 Mya (Kriegs et al. 2006; Murphy et al. 2007), and independently tested the three viable scenarios of placental origin: the Exafroplacentalia, Epitheria, and Atlantogenata hypotheses (Fig. 1). We screened the first of the two whole-genome three-way alignments (armadillo–elephant–human) for elements supporting the Exafroplacentalia (those present in armadillo and human but absent in elephant) and Atlantogenata (those present in armadillo and elephant but absent in human) hypotheses, and the second alignment (elephant–human–armadillo) for elements supporting the Epitheria hypothesis (those present in elephant and human but absent in armadillo; Fig. 2A). Using the criteria presented in the Methods section, we computationally preselected 1227 candidate loci and manually reinspected them using the University of California Santa Cruz (UCSC) sequence browser in combination with sequences from the trace archives. Manual screening of these loci revealed 22 conserved, potentially informative loci that we then realigned to verify the identities and boundaries of the inserted retroelements and to clearly characterize their respective flanking regions. To validate the presence/absence patterns of retroposed elements in these 22 loci in an expanded species sampling and to present as consistent a species representation for each marker as possible, we then retrieved orthologous loci in two distant supraprimates (human and mouse or guinea pig, rabbit), two laurasiatherians (dog and horse or cow), two Afrotheria (elephant and tenrec or hyrax), two Xenarthra (armadillo and sloth), and, as far as alignable, opossum as the outgroup. In some cases sufficient sequence data were not yet available from members of the Afrotheria clade (tenrec) or Xenarthra (sloth) to positively confirm the presence and or absence of all retroposons in a second representative species of the given clades. Some of those gaps were filled by experimental PCR amplification and sequencing of the corresponding loci in tenrec and sloth. Of the 22 phylogenetically informative markers analyzed in this study, 20 are highly reliable in that they provide recognizable information of potential direct repeats plus the corresponding unoccupied target site in the absence cases (see, e.g., Fig. 3). The remaining two markers are classified as “good,” with some restrictions regarding the recognition of clear direct repeats. Their broad distribution over 15 of the 23 human chromosomal pairs indicates the independent integration of the 22 phylogenetically informative elements (Supplemental Fig. S1).
Figure 2.
Computational strategy to extract phylogenetically informative retroposon presence/absence loci. (A) UCSC three-way MAF data were organized in conserved alignment blocks with chromosomal coordinates. Three adjacent blocks including an internal gap region in block 2 were transformed in FASTA format. Sequences above gap regions were analyzed for mammalian specific repeats using the RepeatMasker. Loci including such repeats were inspected in the UCSC Genome Browser. All available sequences of phylogenetically informative loci were transformed to FASTA sequences and manually realigned. (B) Five, nine, and eight loci were identified that support the three competing hypotheses of the origin of placental mammals, Exafroplacentalia, Epitheria, and Atlantogenata, respectively. (+) Presence of a diagnostic retroposon; (−) absence of a diagnostic retroposon.
Figure 3.
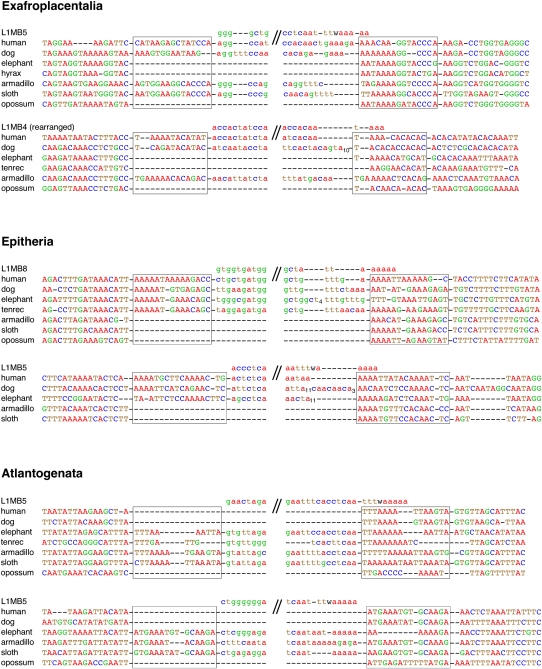
Sequence alignments of representative informative loci. Two representative, multispecies alignments of phylogenetically informative L1MB insertions supporting the Exafroplacentalia (markers 1b and 1d), Epitheria (markers 2a and 2c), and Atlantogenata (markers 3b and 3c) hypotheses. For full species sampling see Supplemental Material 1. Potential direct repeats are boxed. The sequences above each set of alignments represent the RepeatMasker consensus elements; w corresponds to a or t. Subscript numbers indicate serial identical nucleotides. Note that the 5′ ends of the L1MB elements are truncated. The 5′ and 3′ ends of the retroposed insertions are partially shown in lowercase letters.
In strong contrast to most of the mammalian branches that we and others investigated previously with this method (e.g., Nishihara et al. 2005, 2006; Huchon et al. 2007; Kriegs et al. 2007; Möller-Krull et al. 2007; Xing et al. 2007; Warren et al. 2008), we found a rare example of virtually equal support for three competing hypotheses (Fig. 2B; Supplemental Table S1; Kriegs 2007). We found five independent L1 insertions (2 L1MB4, 2 L1MB5, 1 L1MB7) shared by Boreotheria (supraprimates and laurasiatherians) and Xenarthra, but absent in Afrotheria, that would support the Exafroplacentalia hypothesis with Afrotheria as the sister group to all remaining placentals. Our search returned nine (including one previously reported by Kriegs et al. 2006) independent insertions of L1 elements (7 L1MB5, 1 L1MB8, 1 L1MB4) shared by Boreotheria and Afrotheria but absent in Xenarthra. Together with an additional independent marker reported by Kriegs et al. (2006), there are now ten retroposon markers that would support the Epitheria hypothesis. We also found eight (two of which were previously reported by Murphy et al. 2007) L1 insertions (7 L1MB5 and 1 L1MB8) that would support the Atlantogenata hypothesis, as they were present in Afrotheria and Xenarthra species and clearly absent in those of Boreotheria. Importantly, for each branching scenario we found 5, 9, and 8 clear markers that would individually translate to probabilities of P < 0.0001 that the individual hypotheses (by screening for markers in just one direction) are incorrect ([5 0 0], [9 0 0], [8 0 0]; Waddell et al. 2001).
Of all the phylogenetically informative mammalian markers investigated to date (Kriegs et al. 2006; Nishihara et al. 2006; Murphy et al. 2007), just the deepest split of the major placental clades has exhibited such extreme contradictions. The only hypothesis to best explain these apparently conflicting results is that there is no clear dichotomy at the base of placentals but rather a trifurcation.
Discussion
Comparative analyses of large-scale genome data afford a new era of phylogenetic inference combining the fields of evolution and genomics into one of phylogenomics. A completely objective history of the mammalian orders is one of the main visions of this new field of phylogenomics. Most of the interordinal mammalian branches have already been confirmed with convincing support from genome data, but problematic areas of the placental tree have also been recognized (Hallström and Janke 2008). As an example, narrow ancestral splitting events such as the early divergence of placentals have been contradictorily resolved by genome-wide analyses of protein-coding data (Hallström et al. 2007; Nikolaev et al. 2007). But, because nucleotide sequences are highly exposed to homoplasy, using them as phylogenetic markers cannot resolve these contradictions.
Our current data, with significant, nearly equivalent support for all three hypotheses in one study, could be explained by three possible scenarios: deletion via illegitimate recombination between perfect direct repeats flanking each insertion; exact parallel insertions; or lineage sorting, a phenomenon related to incomplete allele fixation and frequent intervals of speciation events (Shedlock et al. 2004; Ray et al. 2006). Precise deletion was observed in 0.5% of LINE1-mobilized SINE insertions in primates (van de Lagemaat et al. 2005). Statistically speaking, this could affect at most six of our 1227 candidate loci, and the chance of all six being in the 22 markers used for the final analysis is infinitesimally small. Moreover, the process of precise deletion is restricted to a very small period of time where both element flanking direct repeats must be identical to allow recombination. Van de Lagemaat et al. (2005) also did not consider the effects of lineage sorting or mosaic evolution in their calculation, hence the expected number of real precise deletions should be much lower than 0.5%. From a previous investigation (Ray et al. 2006), it is known that precise parallel insertion occurs in 0.05% of observed primate LINE1-mobilized SINE insertions. Considering our total of 1227 candidate loci, this is less than one. Furthermore, all previous investigations did not consider random truncations that are an essential, additional factor for establishing LINE element orthology.
Therefore, as the homoplasy of retroposon insertion patterns is extremely rare in mammals and was only described recently for one apparent conflicting marker challenging the newly described Pegasoferae clade (Nishihara et al. 2006), this leaves us with the last explanation, lineage sorting. We believe that the most parsimonious interpretation of the current data is that the ancestral placental populations were characterized by severe ancestral subdivisions and rejoinings, leading to a complex mosaic of phylogenetic relationships in recent species (Fig. 4). Effects of alternating divergence, hybridization, introgression, and incomplete lineage sorting might complicate our search for a clear dichotomy at the base of this tree and leave us with an indistinct, effective ‘soft’ polytomy, leading sometimes to one or the other solution depending on the size of the data set and the particular markers examined.
Figure 4.
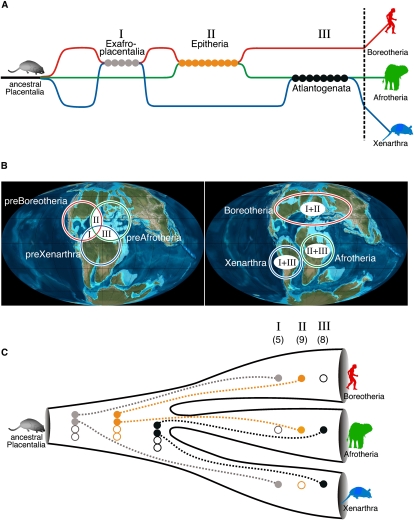
Hypothetical scenarios of interspersed ancestry of placentals. (A) Hybridization/introgression scenario: Initially, the three placental lineages diverged and separated from an Eomaia-like (Eomaia scansoria) common ancestor. Alternate gene flow between the lineages led to a mosaic pattern of relationships among the current species. The sequence of genetic flow leading to significant support for Exafroplacentalia, Epitheria, and Atlantogenata was chosen randomly and not meant to indicate a specific time course. The lineages leading to Boreotheria, Afrotheria, and Xenarthra are shown in red, green, and blue, respectively. Gray circles represent diagnostic retroposon insertions for Exafroplacentalia, orange circles for Epitheria, and black circles for Atlantogenata. (B) Geographic transitions at the time of separation of the three lineages during the Cretaceous. (Left terrestrial globe) Possible overlapping geographical distribution of the three placental lineages before continental drift, the expected period of genetic flow between the lineages; (right terrestrial globe) separation of the continents and divergence of placental lineages possibly prohibiting further gene flow (Murphy et al. 2001b; Wildman et al. 2007). Roman numerals indicate exchanges of genetic material between the species shown in A. (C) Randomly sorted ancestral alleles: Fixation of retroposon alleles was not completed in the ancestral population before the separation of lineages. After the proliferation of inserted retroposon alleles, the distributions to different lineages occurred randomly (broken lines). After fixation of the alleles in the terminal lineages, a mosaic pattern of relatedness remains. (I) Five orthologous markers represent Exafroplacentalia, (II) 9 markers (plus an additional marker previously described in Kriegs et al. 2006 that was undetected here) represent Epitheria, and (III) 8 markers represent Atlantogenata. Open circles represent alleles without retroelements.
Two different scenarios or a mixture of both might be responsible for the mosaic pattern that we found in our screening for informative retroposon insertions and that other studies found in phylogenetic investigations of subsets of genomic sequence data for these species. The earliest eutherian mammals (extant placentals plus closely related extinct mammals) originated about 125 Mya in the Early Cretaceous and based on appearance can probably be represented by the recently discovered and reconstructed skeletal remains of Eomaia scansoria (Ji et al. 2002). In the first scenario, the Eomaia-like ancestral population probably diverged into three distinct lineages of preAfrotheria, preXenarthra, and preBoreotheria. Limited gene flow and hybridization that occurred among these populations left behind mosaic signs of intermittent relationships that we identify about 125 million years later as a patchwork of Exafroplacentalia (I), Epitheria (II), and Atlantogenata (III) roots (Fig. 4A). This scenario requires temporary overlap of the three lineages or parts of their populations before final speciation occurred (Fig. 4B). A second scenario that could explain the results involves incomplete lineage sorting among the three populations (Fig. 4C). Because retroposon fixation requires possibly millions of years before being consistently represented in a population (Schmitz and Zischler 2002), alleles with and without certain elements surely coexisted temporarily, and, in the case of rapid speciation, were distributed randomly into different lineages. Such a process could contribute as well to the patchwork presentation of seemingly competing insertion patterns now present in recent species.
A mosaic of evolutionary signals, blueprinted in the human genome, was also recently described by Ebersberger et al. (2007). They reported that 23% of the genomic sequences (equal representations of genes and intergenic regions; 23,210 alignments, each ≥300 nucleotides [nt]) that they analyzed represented an incongruent genealogy in which chimpanzee was not the closest relative to human.
The majority of sequence-based investigations aimed at resolving the placental root, by examining numerous genes, sequence partition, and various analytic tools, also found almost equal support for one or the other of the various hypotheses (e.g., Madsen et al. 2001; Delsuc et al. 2002; Waddell and Shelley 2003), once more emphasizing the notion that early placental divergence constituted a patchwork evolution of the initial lineages.
That such conflicting results are so very rare requires some discussion of the actual markers that we recovered. The 22 phylogenetically informative markers providing significant support for a trifurcation at the basal node of placental mammals that were retrieved in the current screen included both of the previously described Atlantogenata markers (Murphy et al. 2007) but only one of the two previously described Epitheria markers (Kriegs et al. 2006). Although the undetected one was not present in the three-way multiple alignment format (MAF) alignments, we do consider it to be a reliable marker (Supplemental Fig. S2). A distinct advantage of the current expanded screening compared with previous searches is a more unbiased evaluation of equal amounts of data for all three hypothetical scenarios of placental origin based on whole-genome alignments. We based our previous strategies (Kriegs et al. 2006; Strategy II) on a random search of human intron data and screens of the limited amounts of trace sequences of elephant and armadillo available at the time, while the specifically designed, whole-genome, three-way alignments currently employed offer a much more comprehensive data source resulting in detection of a high number of potentially informative markers. However, the fact that we recovered only three of the four previously described retroposon markers for early placental splits indicates that the current limitations of an exhaustive and maximized unbiased search is the naturally imperfect and variable quality of available specific whole-genome alignments.
One critical point in using retroposon insertion data, especially in deep phylogenetic splits, is the continuous divergence of the elements and their flanking regions. Nevertheless, we recently demonstrated that retroposon data could provide reliable markers in deep mammalian phylogeny by supporting the more than 140-million-year-old therian split (Warren et al. 2008). As stated previously, 20 of the retroposons examined here had clearly recognizable information concerning their direct repeats and the other two, though recognizable, were not quite as clear.
Direct repeats generated by target site duplications in the process of retroposition are valuable landmarks of an orthologous insertion but are unlikely to remain unchanged throughout deep phylogenetic splits. Hence, there is an additional and more reliable orthology criterion available, particularly for the LINE-related retroposons such as L1MB elements. These elements insert almost exclusively as randomly truncated forms. Thus, as independent insertions of identically truncated LINE elements of the same subtype at the same genomic locus of two species is highly unlikely, determining the exact point of truncation of the observed orthologous elements will clearly trace them back to a common origin. A genome-wide screening for the 20,700 LINE1 (L1MB) truncations demonstrated a significant random distribution of truncations with no bias for specific truncation points (P < 0.002, Supplemental Fig. S3), and all investigated diagnostic LINE1 elements differ in their lengths (Supplemental Fig. S3). Thus, retroposon truncation points serve as an additional, powerful criterion for orthologous insertions.
The randomness of the insertions of LINE1-related elements is well demonstrated by Ray et al. (2006) with only a very few cases of observed conflicts. A genome-wide investigation of target site preferences indicates a slight preference for TT/AAAAA sites (Supplemental Fig. S4). This is well known as a kinkable DNA site (Jurka et al. 1998). However, the full target site duplication is much longer (8–30 nt) and rather individual. Nevertheless, the multiplicity of criteria, including (1) identical genomic insertion points, (2) element orientation, (3) identical LINE1 subtypes, (4) matching length and sequence of direct repeats, (5) concurrent truncation points, and (6) the complete consistency in diverse members of representative orders, make it improbable to have screened for artifacts.
Investigations of very young retroposon insertion events present another problem, lineage-specific exact or nearly exact deletions. For young retroposon insertions flanked by perfect identical direct repeats, precise removal might be possible via recombination involving the direct repeats (van de Lagemaat et al. 2005). Because direct repeats diversify quickly after insertion, this possibility should not be critical for deep phylogenetic splits (after altering of perfect direct repeats), but it is not completely impossible for such random perfect or nearly perfect deletions of retroelements to have accumulated, especially in deep divergences where exact comparisons of diverged direct repeats are difficult. However, the tendency of LINE1-related elements to integrate within A/T-rich sequence regions could promote such deletions. Comparing potential retroposon absence regions with a clear absence in an outgroup species is one way to diminish such potential misinterpretations. Unfortunately, for placentals the next outgroup are marsupials that diverged substantially. Because of the evolutionary distance between marsupials and placentals, even if possible, alignments between these species are, particularly for intergenic or intronic regions, only approximate and mostly not a critical proof of true absence. On the other hand, the outgroup comparison for informative L1MB elements is not essential, because L1MB elements are placental-specific and therefore necessarily absent in any outgroup species. Thus, the suggestion of Murphy et al. (2007) that one of our informative L1MB5 markers for Epitheria (Kriegs et al. 2006) is traceable in opossum may be based on a misalignment and misannotation of the corresponding BLAT browser location (Supplemental Fig. S2).
Conclusions
Comparing the genome-wide insertion patterns of retroelements is a powerful strategy to gain a virtually homoplasy-free picture of the evolutionary histories of species. Virtually homoplasy-free implies that this marker system is, as any other marker system, not insensitive to the effects of ancestral hybridization and incomplete lineage sorting combined with short splitting times. Because of the clear character polarity (presence of an element as the derived state) and the low probability of orthologous exact parallel insertions or deletions, the retroposon presence/absence marker system is exceptionally effective for detecting the ancestral exchange of genetic material among lineages and the lineage sorting effects that are difficult to pinpoint by the statistical evaluation of heterogeneous sequence data (Hallström and Janke 2008). Such effects are more difficult to recognize in deep phylogenetic splits and require maximized, unbiased, multidirectional screening of whole-genome phylogenetic signals. They must also be conducted with the notion in mind that in such rare cases, the minimum of three clear markers proposed by Waddell et al. (2001) is not absolutely sufficient as a significant clade support. The present example of apparently incongruent markers inherent in the early branching of placentals offers a seminal example of conflicting retroposon presence/absence patterns that, at the same time, shed new light on the decades-old, controversial scenarios of sequence-based phylogenies. As demonstrated, retroposon screening offers a powerful test of critical signals in narrow splits to detect nodes with more than two immediate descending branches. The basal rodent splits, the Euarchonta relationships, and the affiliations among laurasiatherians are only a few additional contentious examples that may possibly find elucidation by comparing the insertion histories of genome-wide, multidirectionally extracted, phylogenetically informative retroelements.
Methods
Genome alignments
To perform an exhaustive, genome-wide screening for phylogenetically informative retroposon insertions in placentals and to test the three viable phylogenetic hypotheses of the basal origin of placentals, we derived two different three-way (species) whole-genome sequence alignments in MAF format (UCSC): (1) armadillo–elephant–human (3050 gigabases) and (2) elephant–human–armadillo (2730 gigabases), the decisive difference between the two alignments being the full retroposon representation of the leading species that was used as the profile for the remaining species.
The three-way alignments were prepared using pairwise (armadillo–elephant and armadillo–human for the first three-way alignment and elephant–human and armadillo–elephant for the second three-way alignment) BLASTZ alignments (Chiaromonte et al. 2002; Kent et al. 2003; Schwartz et al. 2003) and the standard matrix used in the UCSC browser. The alignments were fed into axtChain with the parameters −chainMinScore = 3000 − chainLinearGap = medium, which organizes all alignments by chromosome (or scaffold) and creates a kd-tree from the gapless subsections (blocks) of the alignments. A dynamic program was then run over the kd-trees to find the maximally scoring chains of these blocks. Finally, MULTIZ (Blanchette et al. 2004) was used to generate the two three-way alignments referencing elephant and armadillo.
We used the first alignment to search for elements supporting the Atlantogenata [(armadillo + elephant) − human] and Exafroplacentalia [(armadillo + human) − elephant] hypotheses, and the second one to test the Epitheria [(elephant + human) − armadillo] hypothesis.
Presence/absence loci
The MAF alignments were organized in blocks of conservation including chromosomal coordinates (Fig. 2A). Using our own C-language script we extracted a total of 653,181 triple blocks, each composed of three continuous conserved regions, the middle one of which included sequence regions larger than 50 nt in two of the species that were missing in the third species (corresponding to the potential absence of the corresponding sequence in this species). All triple blocks were converted into FASTA format using another C-language script. In addition, using more relaxed conditions we screened all blocks (1,636,542) for orthologous retroposon regions in two species that corresponded to at least 80% of the gaps in the third species so as to also consider potentially informative but slightly misaligned regions.
Computational screening for potential informative retroposons
With the local RepeatMasker (www.repeatmasker.org) we screened the triple block sequences for mammalian-specific L1MB LINEs. We selected triple blocks in which at least 50% of block 2 was composed of an L1MB element (human + elephant 284 loci; human + armadillo 256 loci; elephant + armadillo 677 loci). In addition, to detect slightly misaligned but informative loci, all conserved blocks were independently screened for retroelements, and the corresponding regions were compared among all three species for potential absences (10 cases).
Visual screening for informative retroposons
Links to the Genome Browser (http://genome.ucsc.edu/cgi-bin/hgBlat) were generated to filter out loci with less than 70% sequence similarity between multiple species and retroposons that overlapped with blocks 1 and/or 3, leaving us with a total of 22 phylogenetically informative loci.
Experimental amplification to detect presence/absence of retroposons in sloth and tenrec
As sufficient sequence data were not yet available for all the phylogenetically informative loci in sloth and tenrec, we PCR-amplified some of those missing loci to provide a more complete presence/absence pattern. PCR reactions were performed using Phusion DNA Polymerase (New England BioLabs) for 30 sec at 98°C followed by 35 cycles of 10 sec at 98°C, 30 sec at primer-specific temperatures (Supplemental Table S3), and 30 sec at 72°C. Amplified PCR fragments were sequenced directly or purified on agarose gels, ligated into the pDrive Cloning Vector (Qiagen), and electroporated into TOP10 cells (Invitrogen). Sequencing was performed using the AmpliTaq FS Big Dye Terminator Kit (PE Biosystems). A list of the PCR primers used is shown in Supplemental Table S3.
Comparative analyses
For each locus identified in the MAF alignments, we compiled available sequence information of other selected mammalian species from trace or genomic data sequences from mouse, guinea pig, rabbit, dog, horse, cow, tenrec, hyrax, sloth, and as far as available, opossum. All inserted L1MB elements were aligned against the consensus Repbase sequences (www.girinst.org/repbase) (Kapitonov and Jurka 2008). Each insertion site was carefully checked to validate the orthology of the insertions based on the following criteria: (1) identical genomic insertion points; (2) element orientation; (3) identical LINE1 subtypes; (4) matching length and sequence of direct repeats; and (5) identical truncation points. We tried as much as was possible to compile a representative and consistent species sampling for each marker (Supplemental Table S1; Supplemental Material 1). This was not always possible because the genomic information from some species remains fragmented. For consistent comparisons for most markers we selected two Supraprimates from human, mouse, guinea pig, and rabbit; two Laurasiatheria from dog, horse, and cow; two Afrotheria from elephant, tenrec, and hyrax; two Xenarthra (armadillo and sloth); and if alignable, the opossum outgroup sequence.
Acknowledgments
We thank Marsha Bundman for editorial assistance and Ronald Blakey for providing the two terrestrial globes in Figure 4. Many thanks go to Webb Miller and Maria Nilsson for providing valuable comments on the paper. We thank Agencourt Biosciences, the Baylor Genome Sequencing Center, and the Washington University Genome Center for providing raw genomic sequences (trace archives). This work was supported by the Deutsche Forschungsgemeinschaft (SCHM1469/3-1).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article is online at http://www.genome.org/cgi/doi/10.1101/gr.090647.108.
References
- Amrine-Madsen H., Koepfli K.P., Wayne R.K., Springer M.S. A new phylogenetic marker, apolipoprotein B, provides compelling evidence for eutherian relationships. Mol. Phylogenet. Evol. 2003;28:225–240. doi: 10.1016/s1055-7903(03)00118-0. [DOI] [PubMed] [Google Scholar]
- Blanchette M., Kent W.J., Riemer C., Elnitski L., Smit A.F.A., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., et al. Aligning muliple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantrell M.A., Filanoski B.J., Ingermann A.R., Olsson K., DiLuglio N., Lister Z., Wichman H.A. An ancient retrovirus-like element contains hot spots for SINE insertion. Genetics. 2001;158:769–777. doi: 10.1093/genetics/158.2.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaromonte F., Yap V.B., Miller W. Scoring pairwise genomic sequence alignments. Pac. Symp. Biocomput. 2002;2002:115–126. doi: 10.1142/9789812799623_0012. [DOI] [PubMed] [Google Scholar]
- Delsuc F., Scally M., Madsen O., Stanhope M.J., de Jong W.W., Catzeflis F.M., Springer M.S., Douzery E.J. Molecular phylogeny of living xenarthrans and the impact of character and taxon sampling on the placental tree rooting. Mol. Biol. Evol. 2002;19:1656–1671. doi: 10.1093/oxfordjournals.molbev.a003989. [DOI] [PubMed] [Google Scholar]
- Ebersberger I., Galgoczy P., Taudien S., Taenzer S., Platzer M., von Haeseler A. Mapping human genetic ancestry. Mol. Biol. Evol. 2007;24:2266–2276. doi: 10.1093/molbev/msm156. [DOI] [PubMed] [Google Scholar]
- Hallström B.M., Janke A. Resolution among major placental mammal interordinal relationships with genome data imply that speciation influenced their earliest radiations. BMC Evol. Biol. 2008;8:162. doi: 10.1186/1471-2148-8-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallström B.M., Kullberg M., Nilsson M.A., Janke A. Phylogenomic data analyses provide evidence that Xenarthra and Afrotheria are sister groups. Mol. Biol. Evol. 2007;24:2059–2068. doi: 10.1093/molbev/msm136. [DOI] [PubMed] [Google Scholar]
- Huchon D., Chevret P., Jordan U., Kilpatrick C.W., Ranwez V., Jenkins P.D., Brosius J., Schmitz J. Multiple molecular evidences for a living mammalian fossil. Proc. Natl. Acad. Sci. 2007;104:7495–7499. doi: 10.1073/pnas.0701289104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Q., Luo Z.X., Yuan C.X., Wible J.R., Zhang J.P., Georgi J.A. The earliest known eutherian mammal. Nature. 2002;416:816–822. doi: 10.1038/416816a. [DOI] [PubMed] [Google Scholar]
- Jurka J., Klonowski P., Trifonov E.N. Mammalian retroposons integrate at kinkable DNA sites. J. Biomol. Struct. Dyn. 1998;15:717–721. doi: 10.1080/07391102.1998.10508987. [DOI] [PubMed] [Google Scholar]
- Kapitonov V.V., Jurka J. A universal classification of eukaryotic transposable elements implemented in Repbase. Nat. Rev. Genet. 2008;9:411–412. doi: 10.1038/nrg2165-c1. [DOI] [PubMed] [Google Scholar]
- Kent W.J., Baertsch R., Hinrichs A., Miller W., Haussler D. Evolution's cauldron: Duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl. Acad. Sci. 2003;100:11484–11489. doi: 10.1073/pnas.1932072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. Evolutionary rate at the molecular level. Nature. 1968;217:624–626. doi: 10.1038/217624a0. [DOI] [PubMed] [Google Scholar]
- Kjer K.M., Honeycutt R.L. Site specific rates of mitochondrial genomes and the phylogeny of eutheria. BMC Evol. Biol. 2007;7:8. doi: 10.1186/1471-2148-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegs J.O. Universität Münster; Germany: 2007. “Retroposed elements—Witnesses of the evolutionary history of placental mammals.”. PhD thesis, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegs J.O., Churakov G., Kiefmann M., Jordan U., Brosius J., Schmitz J. Retroposed elements as archives for the evolutionary history of placental mammals. PLoS Biol. 2006;4:e91. doi: 10.1371/journal.pbio.0040091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegs J.O., Churakov G., Jurka J., Brosius J., Schmitz J. Evolutionary history of 7SL RNA-derived SINEs in Supraprimates. Trends Genet. 2007;23:158–161. doi: 10.1016/j.tig.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Madsen O., Scally M., Douady C.J., Kao D.J., DeBry R.W., Adkins R., Amrine H.M., Stanhope M.J., de Jong W.W., Springer M.S. Parallel adaptive radiations in two major clades of placental mammals. Nature. 2001;409:610–614. doi: 10.1038/35054544. [DOI] [PubMed] [Google Scholar]
- Möller-Krull M., Delsuc F., Churakov G., Marker C., Superina M., Brosius J., Douzery E.J., Schmitz J. Retroposed elements and their flanking regions resolve the evolutionary history of xenarthran mammals (armadillos, anteaters, and sloths) Mol. Biol. Evol. 2007;24:2573–2582. doi: 10.1093/molbev/msm201. [DOI] [PubMed] [Google Scholar]
- Murphy W.J., Eizirik E., Johnson W.E., Zhang Y.P., Ryder O.A., O'Brien S.J. Molecular phylogenetics and the origins of placental mammals. Nature. 2001a;409:614–618. doi: 10.1038/35054550. [DOI] [PubMed] [Google Scholar]
- Murphy W.J., Eizirik E., O'Brien S.J., Madsen O., Scally M., Douady C.J., Teeling E., Ryder O.A., Stanhope M.J., de Jong W.W., et al. Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science. 2001b;294:2348–2351. doi: 10.1126/science.1067179. [DOI] [PubMed] [Google Scholar]
- Murphy W.J., Pringle T.H., Crider T.A., Springer M.S., Miller W. Using genomic data to unravel the root of the placental mammal phylogeny. Genome Res. 2007;17:413–421. doi: 10.1101/gr.5918807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev S., Montoya-Burgos J.I., Margulies E.H., Rougemont J., Nyffeler B., Antonarakis S.E. Early history of mammals is elucidated with the ENCODE multiple species sequencing data. PLoS Genet. 2007;3:e2. doi: 10.1371/journal.pgen.0030002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara H., Satta Y., Nikaido M., Thewissen J.G.M., Stanhope M.J., Okada N. A retroposon analysis of afrotherian phylogeny. Mol. Biol. Evol. 2005;22:1823–1833. doi: 10.1093/molbev/msi179. [DOI] [PubMed] [Google Scholar]
- Nishihara H., Hasegawa M., Okada N. Pegasoferae, an unexpected mammalian clade revealed by tracking ancient retroposon insertions. Proc. Natl. Acad. Sci. 2006;103:9929–9934. doi: 10.1073/pnas.0603797103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara H., Okada N., Hasegawa M. Rooting the eutherian tree: The power and pitfalls of phylogenomics. Genome Biol. 2007;8:R199. doi: 10.1186/gb-2007-8-9-r199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad A.B., Allard M.W., Green E.D. Confirming the phylogeny of mammals by use of large comparative sequence data sets. Mol. Biol. Evol. 2008;25:1795–1808. doi: 10.1093/molbev/msn104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D.A., Xing J., Salem A.H., Batzer M.A. SINEs of a nearly perfect character. Syst. Biol. 2006;55:928–935. doi: 10.1080/10635150600865419. [DOI] [PubMed] [Google Scholar]
- Schmitz J., Zischler H. Molecular cladistic markers and the infraordinal phylogenetic relationships of primates. In: Kay R.F., Ross C., editors. Anthropoid Origins: New Visions. Kluwer Academic Press; New York, NY: 2002. [Google Scholar]
- Schwartz S., Kent W.J., Smit A., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W. Human-mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shedlock A.M., Takahashi K., Okada N. SINEs of speciation: Tracking lineages with retroposons. Trends Ecol. Evol. 2004;19:545–553. doi: 10.1016/j.tree.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Shoshani J., McKenna M.C. Higher taxonomic relationships among extant mammals based on morphology, with selected comparisons of results from molecular data. Mol. Phylogenet. Evol. 1998;9:572–584. doi: 10.1006/mpev.1998.0520. [DOI] [PubMed] [Google Scholar]
- Steel M., Penny D. Parsimony, likelihood, and the role of models in molecular phylogenetics. Mol. Biol. Evol. 2000;17:839–850. doi: 10.1093/oxfordjournals.molbev.a026364. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Lagemaat L.N., Gagnier L., Medstrand P., Mager D.L. Genomic deletions and precise removal of transposable elements mediated by short identical DNA segments in primates. Genome Res. 2005;15:1243–1249. doi: 10.1101/gr.3910705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell P.J., Shelley S. Evaluating placental inter-ordinal phylogenies with novel sequences including RAG1, gamma-fibrinogen, ND6, and mt-tRNA, plus MCMC-driven nucleotide, amino acid, and codon models. Mol. Phylogenet. Evol. 2003;28:197–224. doi: 10.1016/s1055-7903(03)00115-5. [DOI] [PubMed] [Google Scholar]
- Waddell P.J., Cao Y., Hasegawa M., Mindell D.P. Assessing the Cretaceous superordinal divergence times within birds and placental mammals by using whole mitochondrial protein sequences and an extended statistical framework. Syst. Biol. 1999;48:119–137. doi: 10.1080/106351599260481. [DOI] [PubMed] [Google Scholar]
- Waddell P.J., Kishino H., Ota R. A phylogenetic foundation for comparative mammalian genomics. Genome Inform. 2001;12:141–154. [PubMed] [Google Scholar]
- Warren W.C., Hillier L.W., Marshall Graves J.A., Birney E., Ponting C.P., Grutzner F., Belov K., Miller W., Clarke L., Chinwalla A.T., et al. Genome analysis of the platypus reveals unique signatures of evolution. Nature. 2008;453:175–183. doi: 10.1038/nature06936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildman D.E., Uddin M., Opazo J.C., Liu G., Lefort V., Guindon S., Gascuel O., Grossman L.I., Romero R., Goodman M. Genomics, biogeography, and the diversification of placental mammals. Proc. Natl. Acad. Sci. 2007;104:14395–14400. doi: 10.1073/pnas.0704342104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J., Witherspoon D.J., Ray D.A., Batzer M.A., Jorde L.B. Mobile DNA elements in primate and human evolution. Am. J. Phys. Anthropol. 2007;Suppl. 45:2–19. doi: 10.1002/ajpa.20722. [DOI] [PubMed] [Google Scholar]