Abstract
Using bacteria artificial chromosome (BAC) end sequences (16.9 Mb) and high-quality alignments of genomic sequences (17.4 Mb), we performed a global assessment of the divergence distributions, phylogenies, and consensus sequences for Alu elements in primates including lemur, marmoset, macaque, baboon, and chimpanzee as compared to human. We found that in lemurs, Alu elements show a broader and more symmetric sequence divergence distribution, suggesting a steady rate of Alu retrotransposition activity among prosimians. In contrast, Alu elements in anthropoids show a skewed distribution shifted toward more ancient elements with continual declining rates in recent Alu activity along the hominoid lineage of evolution. Using an integrated approach combining mutation profile and insertion/deletion analyses, we identified nine novel lineage-specific Alu subfamilies in lemur (seven), marmoset (one), and baboon/macaque (one) containing multiple diagnostic mutations distinct from their human counterparts—Alu J, S, and Y subfamilies, respectively. Among these primates, we show that that the lemur has the lowest density of Alu repeats (55 repeats/Mb), while marmoset has the greatest abundance (188 repeats/Mb). We estimate that ∼70% of lemur and 16% of marmoset Alu elements belong to lineage-specific subfamilies. Our analysis has provided an evolutionary framework for further classification and refinement of the Alu repeat phylogeny. The differences in the distribution and rates of Alu activity have played an important role in subtly reshaping the structure of primate genomes. The functional consequences of these changes among the diverse primate lineages over such short periods of evolutionary time are an important area of future investigation.
Alu repeats are primate-specific short interspersed sequence elements (SINEs), ∼300 nt in length, propagating within a genome through retrotransposition (Schmid 1996). They are the most abundant repeat sequences found in humans, with more than 1.1 million copies accounting for ∼10% of the human genome sequence (Lander et al. 2001). Recent work increasingly recognizes that Alu elements have a greater impact than expected on phenotypic change, diseases, and evolution. Alu elements were demonstrated to mediate insertion mutagenesis, “exonization” by alternative splicing, genomic rearrangements, segmental duplication, and expression regulation causing disorders like Hunter syndrome, hemophilia A, and Sly syndrome (Batzer and Deininger 2002). The oldest Alu elements were estimated to emerge either coincident with or immediately after the radiation of primates. Based on Alu subfamily sequence diversity, a major burst in Alu amplification was estimated to have occurred 25–50 million years ago (Mya) (Shen et al. 1991). Younger Alu repeat elements have emerged in the hominoid, although the rate of more recent retrotransposition events has declined (Batzer and Deininger 2002). Owing to their unidirectional mode of evolution, SINE insertions have been used as largely homoplasy-free character states in cladistic analyses of primates (Schmitz et al. 2001; Roos et al. 2004). Alu insertion loci have also been used to clarify relationships among New World monkeys (NWM), Old World monkeys (OWM), and the human–chimpanzee–gorilla trichotomy (Salem et al. 2003; Ray and Batzer 2005; Ray et al. 2005; Xing et al. 2005).
Alu elements in human lineage have been extensively characterized (Batzer and Deininger 2002). They are divided into subfamilies based on the extent of sequence diversity and diagnostic mutations (Britten et al. 1988; Jurka and Smith 1988). The monomeric repeats (such as FAM, FRAM, and FLAM) are the oldest Alu-related elements derived from the 7SL RNA gene. The more recent dimeric Alu elements consist of two similar but not identical monomers with a short adenine-rich linker between the two monomers and a longer and more variable A-rich region at the 3′-end. The various dimeric Alu subfamilies have been identified in different evolutionary ages with overlap. AluJo and AluJb are the most ancient Alu dimeric subfamilies. AluS represents the major burst of Alu elements, which contains subfamilies such as Sx, Sp, Sq, Sg, and Sc, with Sx being the most common. AluY is the youngest subfamily in the hominoid lineage, which continues to retrotranspose, and is subsequently polymorphic in the population. Pevzner and colleagues identified 213 human Alu subfamilies at a much finer resolution using a novel method (Alucode) (Price et al. 2004). This method first split Alu subfamilies based on “biprofiles,” that is, linkage of pairs of nucleotide values, and then used the calibration of Alu mutation rates to split subfamilies containing overrepresented individual mutations. These observations generally support the master-gene hypothesis for Alu amplifications, i.e., Alu subfamilies originated through successive waves of fixation from sequential small subsets of master elements (Batzer and Deininger 2002).
To date, genome-wide characterization of Alu repeats in nonhuman primates has been limited to chimpanzee and macaque (The Chimpanzee Sequencing and Analysis Consortium 2005; Gibbs et al. 2007). Most chimpanzee-specific elements belong to a subfamily (AluYc1) that is very similar to the source gene in the human–chimpanzee last common ancestor. In macaque, Alu elements have evolved into four currently active lineages: AluYRa1-4, AluYRb1-4, AluYRc1-2, and AluYRd1-4 (Han et al. 2007). Currently, there are three macaque consensus sequences: AluMacYa3, AluMacYb2, and AluMAcYb4 in Repbase (Version 13.5). For other primate genomes, most studies have been based on PCR cross-amplification among diverse primate taxa and, therefore, are potentially biased to either conserved regions or limited to closely related species. Ray and Batzer (2005) recovered 48 NWM-specific Alu elements using a combination of PCR and computational approaches and reported three NWM-specific subfamilies: AluTa7, AluTa10, and AluTa1. In another publication, Herke et al. (2007) reported a few loci (such as DQ822065) from the lemur derived from PCR display. Initial comparative analysis based on small samples of primate genomic sequences demonstrated that the fixation rates of retroelements (especially SINE/Alu) vary radically in different primate lineages (Liu et al. 2003; Hedges et al. 2004). In this study, we analyze Alu elements in randomly sampled BAC end sequences (BES) and finished genomic sequence alignments (ALN) from five nonhuman primates—lemur, marmoset, macaque, baboon, and chimpanzee—using two distinct approaches combining mutation profile and insertion/deletion analysis. The five species, including great apes (chimpanzee), OWM (baboon and macaque), NWM (marmoset), and prosimians (lemur), are estimated to have diverged from humans at distant time points, ∼6, 25, 25, 35, and 55 Mya, respectively (Goodman 1999). Thus, this spectrum of the taxa provides a vista of Alu-element changes at different nodes during primate evolution.
Results
Alu repeat identification
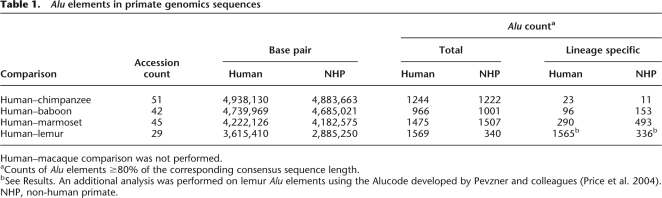
We used RepeatMasker (Smit 1999) to initially identify and extract Alu elements for primate genomic sequence. We analyzed two different sources, namely, 16.9 Mb of end-sequence data generated from randomly selected large-insert BAC clones from different primate species (Supplemental Table S1) and 17.4 Mb of orthologous sequence alignments of finished nonhuman primate BAC sequences aligned to the human reference genome (Table 1). Alu elements in nonhuman primates, especially those lineage-specific Alu elements and/or those in more distantly related species like marmoset and lemur, may differ significantly from human consensus sequences; therefore, they may be difficult to recognize by RepeatMasker. To eliminate this bias and exclude the possibility of incomplete annotation, we separately analyzed all indels (insertions or deletions >100 bp) based on human–marmoset and human–lemur genomic sequence alignments using previously described methods (Liu et al. 2003). In total, we identified 1475 human and 1507 marmoset Alu elements from human–marmoset sequence alignments; 1569 human and 340 lemur Alu elements were identified from human–lemur alignments. No additional Alu repeats were identified based on our independent analysis of indels (>100 bp).
Table 1.
Alu elements in primate genomics sequences
Human–macaque comparison was not performed.
aCounts of Alu elements ≥80% of the corresponding consensus sequence length.
bSee Results. An additional analysis was performed on lemur Alu elements using the Alucode developed by Pevzner and colleagues (Price et al. 2004). NHP, non-human primate.
Pairwise sequence divergence distribution
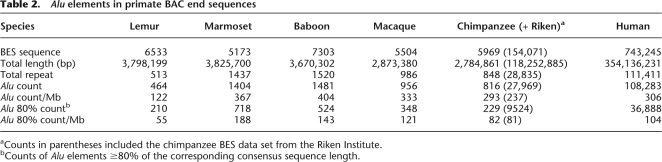
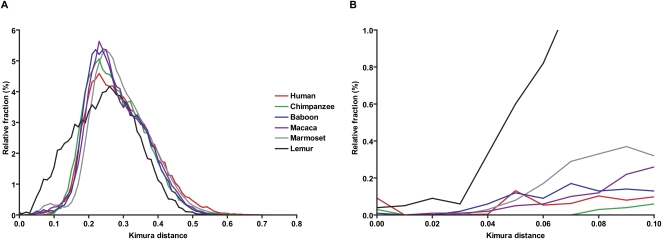
In order to provide an unbiased assessment of Alu repeat sequence properties, we generated BAC end sequence data from more than 2500 randomly selected genomic clones from five nonhuman primate species (Supplemental Table S1). We identified all Alu repeat elements whose insert length was ≥80% of the corresponding consensus sequence length (Table 2). Compared to all other primates analyzed in this study, the marmoset genome shows the greatest density of Alu repeats (188 repeats/Mb), while the lemur genome shows the least (55 repeats/Mb) (Table 2). In human BES, the density of Alu repeats is 104 repeats/Mb, which is lower than the genome-wide density of human Alu repeats at 315 repeats/Mb, mainly because of the short length of BES. We performed an all-by-all pairwise sequence divergence analysis of all available Alu elements within each species (210–718 Alu repeats) and computed the genetic distance among all alignments using the Kimura two-parameter model. We plotted the distribution of pairwise divergences within each species (Fig. 1, bin size = 0.01, termed “K-plots”) as a function of genetic distance. Notable differences among the K-plots were observed when lemur was compared to other primates. All anthropoids including human, great apes (chimpanzee), OWM (baboon, macaque), and NWM (marmoset) show a similar asymmetric divergence profile with a mode at 0.23 substitutions/per site and a relative small fraction of high-identity Alu repeat elements. In contrast, the lemur shows a broader, more symmetric distribution with a much greater abundance of highly identical (potentially evolutionarily “young”) Alu repeats when compared to other primates. A detailed inspection of the most identical Alu repeats (Fig. 1B, with Kimura distance <0.10) also provides evidence of a slight increase in the fraction of most identical Alu repeats (<0.01) in human as compared to chimpanzee, consistent with previous observations (Liu et al. 2003; Hedges et al. 2004; Watanabe et al. 2004; The Chimpanzee Sequencing and Analysis Consortium 2005). Similar K-plots were obtained for Alu elements derived from finished primate genomic sequences (data not shown).
Table 2.
Alu elements in primate BAC end sequences
aCounts in parentheses included the chimpanzee BES data set from the Riken Institute.
bCounts of Alu elements ≥80% of the corresponding consensus sequence length.
Figure 1.
(A) Sequence divergences of Alu elements. (B) An enlarged view for Kimura Distances <0.10.
Characterization of lineage-specific Alu repeat elements from BAC end sequences
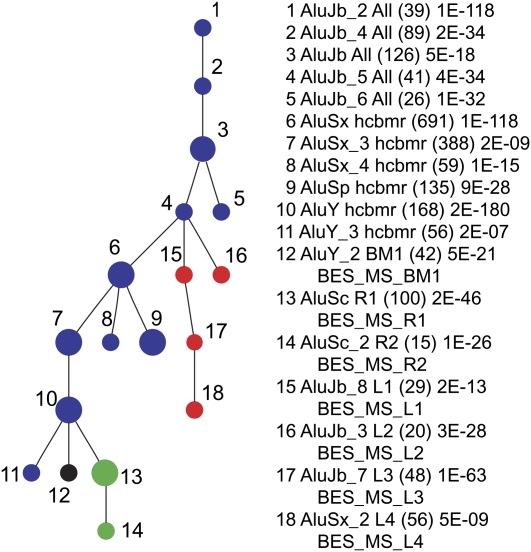
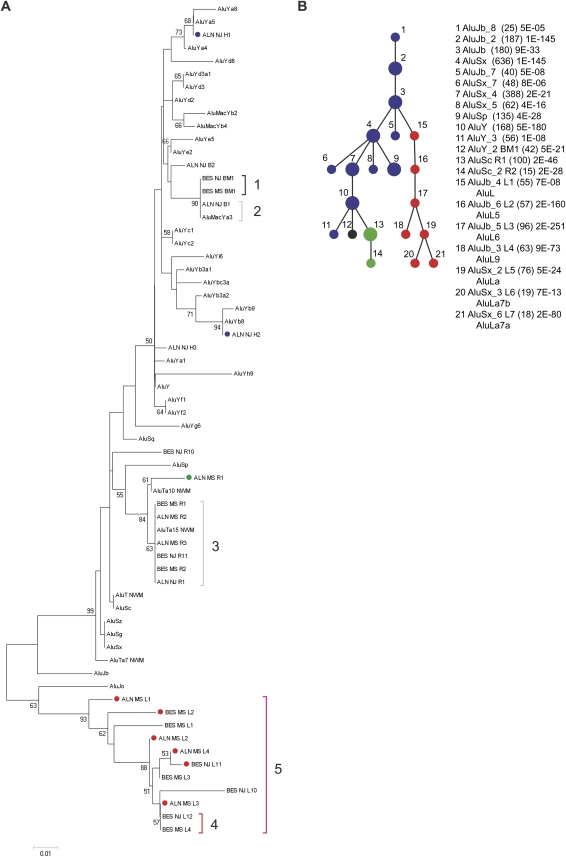
We used two distinct approaches to study lineage-specific Alu subfamilies. First, we categorized Alu subfamilies using the program Alucode (Price et al. 2004). Based on our analysis of 2128 Alu repeats from six primate species, we identified 18 distinct subfamilies: subfamily composition ranges from 15 to 691 with most subfamilies containing 50–100 elements (P-value for subfamily partition ranges from 2 × 10−180 to 2 × 10−7) (see Price et al. 2004 for the P-value definition and calculation). We next constructed a minimum spanning (MS) tree for these 18 Alu subfamilies to summarize their evolutionary relationship (Fig. 2). We identified 11 subfamilies shared among different species (Nodes 1–11) and seven putative lineage-specific subfamilies (Nodes 12–18, named BES_MS_BM1, BES_MS_R1-2, and BES_MS_L1-4).
Figure 2.
The minimum spanning tree of 18 Alu subfamilies. The tree is based on an Alucode analysis of 2128 Alu repeats extracted from primate BES data. (Blue) Eleven families were shared among human and at least one nonhuman primate species while seven were lineage specific: (black) baboon–macaque; (green) marmoset; (red) lemur. The number of Alu elements (in parentheses) and the P-value within each group are indicated.
As a second method, we constructed Alu neighbor-joining (NJ) trees independently for genomic sequences from lemur (Supplemental Fig. S3) and marmoset (Supplemental Fig. S4) as well as from all six primate species including human (Supplemental Fig. S5). We used the tree topology to cluster related Alu elements into groups. The groups were named as follows: lemur (BES_NJ_L1–12), marmoset (BES_NJ_R1–11), and baboon/macaque (BES_NJ_BM1). The analysis clearly identified monophyletic clades that appear lineage specific with modest bootstrap support (Supplemental Fig. S5). These six putative lineage-specific subfamilies are lemur's BES_NJ_L10–12 (green, labeled as “Lemur AluJ”), marmoset's BES_NJ_R10–11 (purple, labeled as “Marmoset AluS”), and baboon/macaque's BES_NJ_BM1 elements (red, labeled as “Baboon/macaque AluY”). Based on the majority rule, Alu consensus sequences were derived from each group. We constructed a NJ tree using all derived Alu consensus sequences with known primate Alu consensus sequences (Supplemental Fig. S6).
Characterization of lineage-specific Alu repeat elements from orthologous sequence alignments
As a second source of data, we constructed optimal global sequence alignments between finished nonhuman primate genomic BAC clones and the human genome reference sequence using previously described methods (Liu et al. 2003; She et al. 2006). We generated a total of 51 human–chimpanzee, 42 human–baboon, 45 human–marmoset, and 29 human–lemur genomic alignments (Table 1; http://bfgl.anri.barc.usda.gov/Alusite). Based on these alignments, we classified all Alu elements into two categories (lineage specific or shared) based on the presence or absence of an ∼300-bp insertion deletion event within the alignment. We limited our analysis to full-length Alu repeats that are not chimeric (single subfamily designation) and show flanking target site duplications. We assume that the majority of 300-bp insertions arise as a result of new retrotransposition events as opposed to precise deletion of the repeat. The term “lineage specific” is relative only to the two species being compared. We constructed NJ trees based on multiple sequence alignments of these lineage-specific Alu repeat elements (Fig. 3A,B) and Alu subfamily consensus sequences (Repbase).
Figure 3.
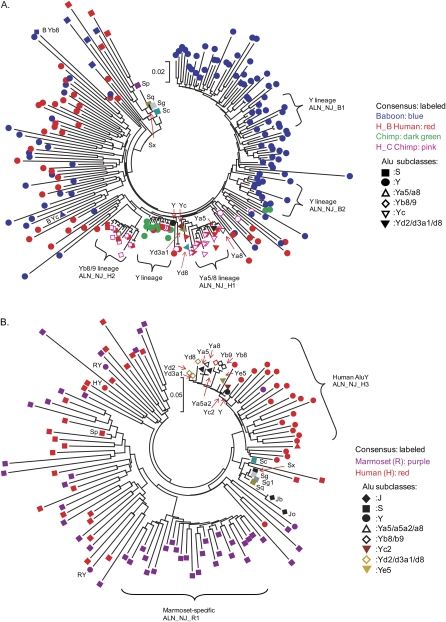
Neighbor-joining trees of lineage-specific Alu elements derived from genomic sequence alignments. All lineages with brackets were supported by the bootstrap values >50% with n = 1000 replicates. Clades with significant bootstrap support include two human-specific (Yb8/9, Ya5/8), a human/chimpanzee-specific AluY, and two baboon-specific AluY repeat families (A) and one human-specific AluY and one marmoset-specific AluY repeat families (B).
The phylogenetic analysis of lineage-specific Alu repeats derived from human–baboon and human–chimpanzee orthologous sequence alignments reveals three different categories of repeat (Fig. 3A): (1) an interleaved set of divergent human- and baboon-specific copies that are equivalent in number between the two species; (2) a monophyletic set of chimpanzee- and human-specific repeats with high sequence similarity to recently active AluY (Y lineage), Ya5/8 (ALN_NJ_H1), and Yb8/9 (ALN_NJ_H2) subfamilies; and (3) a more abundant set of baboon-specific AluY elements (ALN_NJ_B1 and ALN_NJ_B2) including both ancestral and young elements. There have been 60% more baboon-specific Alu retrotransposition events as a result of the expansion of the third category (Table 1).
A similar topology was obtained from Alu phylogenetic trees constructed from human and marmoset orthologous sequence alignments (Fig. 3B): We identified (1) an interleaved group of divergent human and marmoset repeats that are related to AluS consensus sequences; (2) a monophyletic marmoset-specific AluS/Sc lineage (ALN_NJ_R1); and (3) a human-specific AluY set (human AluY, ALN_NJ_H3). The last two lineages showed significant bootstrap support. By count, once again, marmoset-specific elements were 70% more abundant than human-specific elements (Table 1).
Although human–lemur genomic sequence alignments are complicated by greater sequence divergence between the two genomes, we identified only four pairs of Alu repeats as orthologous from a total of 1569 human and 340 lemur annotated Alu repeats. These data suggest that the anthropoid lineage (represented by human) has experienced a 4.6-fold increase in Alu activity when compared to prosimians (Table 1). Finally, we generated a minimal spanning tree using Alu elements derived from human–lemur, human–marmoset genomic sequences. Similar to the BES analysis (Fig. 2), we identified three marmoset- and four lemur-specific Alu subfamilies with statistical significance (named ALN_MS_R1-3, ALN_MS_L1-4 in Supplemental Fig. S7A,B), respectively.
Subfamily consensus sequences and phylogeny
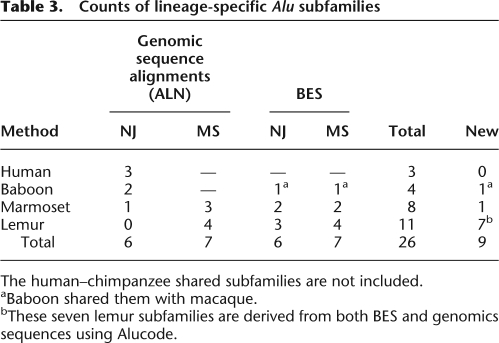
Table 3 summarizes all 26 putative lineage-specific Alu subfamilies identified using four combinations of data (ALN vs. BES) and methods (NJ vs. MS) in lemur, marmoset, baboon/macaque, and human. We performed phylogenetic analyses (NJ and MS) on these 26 consensus sequences with 34 known primate Alu subfamilies. In the NJ tree shown in Figure 4A (the cladogram of this tree is in Supplemental Fig. S8B), the accepted relationship among known primate Alu consensus sequences was recovered as expected. Several subfamilies confirmed known primate Alu trees, including two of human ALN_NJ_H1 and ALN_NJ_H2 subfamilies (blue dots) that closely cluster with human AluYa5/8 and AluYb8/9, respectively. This confirmed the earlier observation that most human-specific Alu elements belong to AluYa5 and AluYb8 subfamilies that have evolved since the chimpanzee–human divergence and differ substantially from the ancestral source gene (Hedges et al. 2004). Baboon/macaque ALN_NJ_B1 subfamilies (gray bracket 2) grouped with AluMacYa3. Marmoset consensus sequences BES_MS_R1, ALN_MS_R2, ALN_MS_R3, BES_NJ_R11, BES_MS_R2, and ALN_NJ_R1 grouped with AluTa15 in NWM (gray bracket 3).
Table 3.
Counts of lineage-specific Alu subfamilies
The human–chimpanzee shared subfamilies are not included.
aBaboon shared them with macaque.
bThese seven lemur subfamilies are derived from both BES and genomics sequences using Alucode.
Figure 4.
Phylogenetic trees of primate lineage-specific Alu consensus elements. (A) Neighbor-joining tree: All branches are labeled with the bootstrap values (>50%) with n = 1000 replicates. (B) Minimum spanning tree. The color and label schemes are as described in Figure 2.
In spite of the above-mentioned ancestry sharing, multiple lineage-specific consensus sequences were discovered corresponding to distinct clades such as BES_NJ_BM1 and BES_MS_BM1 of baboon/macaque (black bracket 1) and ALN_MS_R1 and BES_MS_R1 of marmoset (green dots). Lemur Alu subfamilies share ancestry from the human J subfamilies but have their own trajectory of evolution since divergence. This clade (pink brackets 5) includes ALN_MS_L1, BES_MS_L2, ALN_MS_L2, ALN_MS_L4, BES_NJ_L11, ALN_MS_L3 (red dots), BES_NJ_L12, and BES_MS_L4 (red bracket 4). To further classify lemur Alu subfamilies, we combined the BES data (210 lemur Alu elements in Fig. 2) with the 340 lemur Alu repeats from genomic sequence alignments (ALN) and then rebuilt a MS tree using Alucode. The new MS tree (Fig. 4B) agrees well with the other NJ, MS trees (Figs. 2 and 4A; Supplemental Fig. S7). In Figure 4B, we identified seven statistically significant lemur lineage-specific Alu subfamilies (Nodes 15–21, named AluL, AluLL5, AluL6, AluL9, AluLa, AluLa7a, and AluLa7b). Therefore, we concluded that similar results were obtained irrespective of data source or method. In conclusion, we generated nine new Alu consensus sequences (Table 3; Supplemental Table S4). We further estimated that ∼70% (384/550) of lemur and 16% (115/718) of marmoset Alu elements belong to lineage-specific subfamilies (Fig. 4B). All Alu subfamilies' consensus sequences and selected multiple alignments can be found in Additional Supplemental File (Supplemental Figs. S9–S14; Supplemental File S15).
Subfamily diagnostic mutations
We inspected the diagnostic nucleotide features of lemur, marmoset, baboon/macaque, and human lineage-specific Alu consensus sequences as compared to known primate Alu consensus sequences (Fig. 5; Supplemental Figs. S10–S14; Supplemental Table S2). In Figure 5, we compare lemur consensus sequences with human AluJo and AluJb. These lemur consensus sequences are distinct at approximately 25–30 positions. AluLa, AluLa7a, and AluLa7b also have the distinctive poly(A) linker of 35–37 nt between the left and right monomer (alignment position 132–168). Depending on the presence of this linker or not, we divided seven lemur subfamilies into two Alu consensus sequences: AluL and AluLa. Using the Alu naming convention (Batzer et al. 1996), we tentatively identify these seven subfamilies as AluL, AluL5, AluL6, AluL9, AluLa, AluLa7a, and AluLa7b (Supplemental Table S2) based on these diagnostic nucleotide differences from their consensus sequences (Supplemental Figs. S10 and S11). Although grouped with human AluSc and Sp, marmoset Alu subfamilies have 14–18 distinct nucleotide changes and an insertion of 3–6 nt between positions 264 and 269 (Supplemental Fig. S12). As discussed above, six marmoset subfamilies (Fig. 4A, gray bracket 3) are essentially the same as AluTa15 sharing almost all its diagnostic nucleotides (Ray and Batzer 2005). One marmoset Alu subfamily (ALN_MS_R1) is related to AluTa10 with a few more mutations and can be assigned as AluTa14 (Supplemental Table S2). BES_NJ_BM1/BES_MS_BM1 consensus is close to human Ye2/5 subfamilies. It is identical to AluMacYa3 with the exception of a transition from “G” to “A” at the position 205 (Supplemental Fig. S13). Thus, it can be assigned as AluMacYa4.
Figure 5.
Aligned consensus sequences of lemur-specific Alu subfamilies. The consensus sequences of two human Alu subfamilies (AluJo and AluJb) are from Repbase. The seven lemur Alu subfamilies we identified include AluL, AluL5, AluL6, AluL9, AluLa, AluLa7a, and AluLa7b.
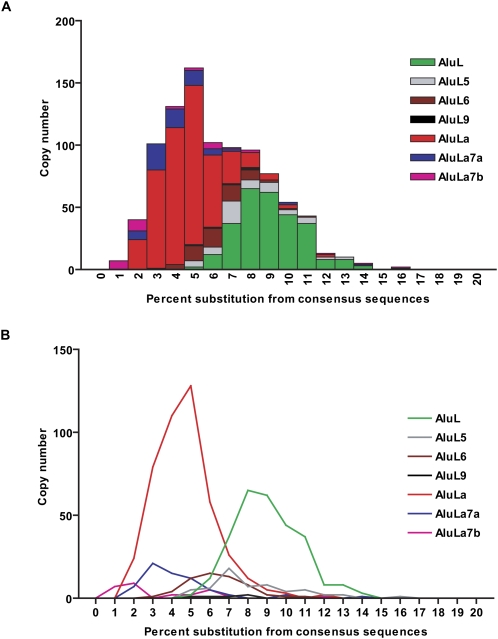
We also performed an age/divergence distribution analysis of all currently available lemur sequences using these seven lineage-specific Alu consensus sequences (Lander et al. 2001). The divergence levels reported by RepeatMasker were corrected by the CpG content of each repeat. We plotted the divergence distribution either by summing all seven subfamilies or separately for each subfamily (Fig. 6, bin size = 0.01). In the stacking plot (Fig. 6A), two bursts in Alu amplification can be detected (around 0.05 and 0.08 substitutions/site) and estimated to occur ∼20 and 32 Mya assuming a substitution rate of 2.5 × 10−9 substitutions/site per year (Price et al. 2004). Notable differences among the distributions are observed when each subfamily is considered: AluL and AluLa subfamilies are the major divergence profiles that are likely responsible for the two bursts; other minor profiles include AluL5, AluL6, and AluL9, which derived from AluL, while AluLa7a and AluLa7b, which are the youngest subfamilies, derived from AluLa. These results generally agree well with the MS trees in terms of age and fractions (Fig. 4B) and verified the relationship among these seven subfamilies. However, the multiple modes of these distribution profiles suggest that these seven subfamilies may still represent a mixed population and could be further divided into distinct subfamilies when more sequences are available.
Figure 6.
Sequence divergence distribution of lemur-specific Alu subfamilies. The sequence divergence distributions are plotted in bins corresponding to 0.01 increments after correction for the CpG content by (A) summing all seven subfamilies or (B) separately for each subfamily.
Discussion
In this project, we performed a global characterization of Alu elements in diverse primate genomes using an integrated approach combining phylogenetic (NJ and MS trees) and insertion/deletion analysis of orthologous genomic alignments. Our analyses were based on two independent data sets: BES data and finished genomic sequences. BAC end sequences were randomly sampled from primate genomes. Compared to PCR cross-species amplification, the approach is potentially less biased capturing a broader spectrum of repeat diversity. High-quality finished genomic sequences (150–170 kb) offer the advantage that Alu retrotransposition events can be classified as shared or lineage specific in the context of orthologous sequence alignments. We found that Alu subfamilies derived from independent analyses (MS and NJ trees) of both BES and finished genomic sequences are in close agreement. Our analysis supports a model in which a burst of Alu activities occurred during the emergence of anthropoids (35 Mya) but after the divergences of prosimians (55 Mya). Divergence analyses support the master-gene hypothesis for Alu amplifications within individual primate lineages. With respect to human, chimpanzee, and macaque, our sampling has confirmed previous analysis (The Chimpanzee Sequencing and Analysis Consortium 2005; Gibbs et al. 2007) as well as provided new insights especially with respect to more divergent primate genomes.
Our analysis reveals a fundamentally distinct sequence divergence distribution profile between prosimians and anthropoids. We find that prosimian Alu repeats have a >10-fold increase in the relative fraction of high-identity Alu repeats when compared to anthropoids (Fig. 2). Our analysis of lemur suggests that this is a combination of the anthropoid burst in AluSc retrotransposition (>35 Mya) and a subsequent, continual decline in retrotransposition activity among various anthropoid lineages. Both marmoset and macaque show a significant excess of lineage-specific events when compared to chimpanzee and humans (Fisher's exact test P-value of 5 × 10−16), although we note a previously reported trend for the doubling of lineage-specific events in human when compared to chimpanzee (Liu et al. 2003). The broader sequence divergence spectrum in prosimians may reflect a more steady state of Alu retrotransposition as opposed to the anthropoid burst and decline.
Several molecular and cellular mechanisms may account for lineage-specific changes in Alu consensus sequences and their differential activity, including changes in insertion site availability, competence of active parental (master) elements, and efficiency of reverse transcription (Liu et al. 2003; Ohshima et al. 2003). Additionally, we speculate that the lineage-specific changes in Alu activity could also be due to the changes in the host primates and their environment during their 60 million years of evolution. A similar situation was found that lineage-specific expansions of retroviral inserted within the genomes of African great apes but not in humans and orangutans (Yohn et al. 2005).
A few exceptions in our phylogenetic analyses shed further insight on the evolutionary forces that shaped Alu elements. We observed, for example, a small subset of lineage-specific events that share diagnostic mutational differences with more ancient Alu repeat elements. Such elements may represent perfect deletions of more ancient elements, perhaps as a result of non-allelic homologous recombination, gene conversion events between Alu events, or the low levels of recent activity of the older subfamilies. We characterized nine new lineage-specific Alu consensus sequences in more diverse primate genomes: seven subfamilies in lemur: AluL, AluL5, AluL6, AluL9, AluLa, AluLa7a, and AluLa7b; one in marmoset: AluTa14; and one in baboon/macaque: AluMacYa4. The phylogenetic clustering of these Alu subfamilies according to species support that they were lineage-specific master genes for Alu amplification in these nonhuman primates. The nine new lineage-specific Alu subfamilies expand our understanding of Alu evolution and their impact on primate genome architecture.
Earlier studies using PCR and bioinformatics strategies also confirmed our discoveries. Our results showed that recent lemur-specific Alu consensus sequences (AluLa) contain a distinct poly(A) linker between the left and right Alu monomers. It agreed with previous data using Alu PCR amplification from lemur, sifaka, and galago (Zietkiewicz et al. 1998). Deininger and colleagues also had similar observations for active galago Alu elements (Daniels and Deininger 1983, 1991). However, it is difficult to associate those limited individual lemur loci (such as DQ822065 amplified by Herke et al. [2007]) with our lemur-specific consensus sequences at this stage. More prosimian sequence data are needed to make a meaningful comparison possible. A comparison with Ray and Batzer (2005) demonstrated that multiple subfamilies identified in NWM are essentially identical to AluTa15 sharing most of its diagnostic mutations. Our results derived from a larger subset of Alu elements (446 sequences from both BES and genomic sequences) further confirmed that the AluTa15 subfamily expanded later in NWM evolution and may have arisen from AluTa7 or AluTa10 (177 sequences).
In summary, our analysis has provided an evolutionary framework for further classification and refinement of the Alu repeat phylogeny. The differences in the distribution and rates of Alu activity have played an important role in subtly reshaping the structure of primate genomes (Bailey et al. 2003). The functional consequences of these changes among the diverse primate lineages over such short periods of evolutionary time are an important area of future investigation.
Methods
Genomic sequence alignment and analyses
BAC libraries were constructed in Peter de Jong's laboratory at Children's Hospital Oakland Research Institute, Oakland, CA (http://www.chori.org/bacpac/) for the common chimpanzee (Pan troglodytes CH251), the (olive) baboon (Papio anubis RP41), the rhesus macaque (Macaque mulatta CH250), and the common marmoset (Callithrix jacchus CH259), while the lemur BAC library (Lemur catta LB2) was constructed by Jan-Fang Cheng's laboratory at Lawrence Berkeley National Laboratory. Large genomic sequences (>50 kb in length) from chimpanzee (RP43), baboon (RP41), marmoset (CH259), and lemur (LB2) were retrieved from GenBank. Orthologous sequence relationships were identified, and optimal global alignments were constructed and validated as described previously (Liu et al. 2003). In total, we examined 51 loci (5.0 Mb) for human–chimpanzee, 42 loci (5.0 Mb) for human–baboon, 45 loci (4.0 Mb) for human–marmoset, and 29 loci (2.8 Mb) for human–lemur genomic sequence alignments (She et al. 2006). Large gaps (>100 bp) in these pairwise alignments were subdivided into one of two categories based on their association with a repeat sequence as described previously (Liu et al. 2003). Briefly, we classified an indel as a retrotransposition if at least 80% of the indel contained one predominant repeat (LINE, SINE, LTR). We considered the known interspersed repeat phylogeny based on the established repeat subfamilies (Smit 1999). For L1 and Alu elements, insertion sequences were examined for the presence of target-site duplications and a polyadenylation tail at the site of integration. The directionality of these retrotransposition events were unambiguously assigned to a specific lineage.
BAC end sequencing
We generated 24,513 BAC end sequences from 12,200 randomly sampled clones as part of an effort to randomly sample sequence from a diversity panel of primate genomes (BES originally generated at The Institute for Genomic Research, Supplemental Table S1; sequence and quality data are downloadable at http://bfgl.anri.barc.usda.gov/Alusite/). DNA sequence was isolated from single-colony-derived templates and prepared as described previously (Zhao et al. 2000). With the exception of the marmoset, the average Q20 length was 433.5 bp (Supplemental Table S1). Marmoset BES of higher quality were produced with improved sequencing techniques, as described previously (Zhao et al. 2001). Table 2 includes extra chimpanzee BES from the Riken Institute (Fujiyama et al. 2002) and extra human BES (Lander et al. 2001). For Figure 6, besides the BES generated in this study, we also included 10,101 lemur (BES and whole-genome shotgun) reads and 43 lemur accessions assembled from 116,761 shotgun reads.
Alu-element identification and phylogenetic analyses
We initially detected Alu repeat elements using the slow search option (-s of RepeatMasker version 2002/07/13) with Repbase (http://www.girinst.org/, version 9.04). Owing to the variable lengths of poly(A) tails (Batzer and Deininger 2002), the default human consensus sequences were trimmed at their 3′ poly(A) until only five bases of adenine remained. We selected all Alu repeats with at least 80% length of the consensus repeat. We then examined those indels that were not captured by RepeatMasker. None of these indels displayed any grouping or any Alu distinct features based on either length (∼300 bp) or sequence identity (including diagnostic mutations). Therefore, we were convinced that the default human consensus library is sufficiently robust to identify Alu elements in other primates.
Pairwise sequence alignments and divergences of Alu elements were computed by Multipair, a ClustalW-like program (Thompson et al. 1994) that aligns all possible sequence pairs using Smith-Waterman algorithm (Myers and Miller 1988) and estimates the genetic distances according to the Kimura two-parameter model. Sequence divergences of Alu elements from the consensus sequences were computed by RepeatMasker. Divergence levels reported by RepeatMasker were corrected for the CpG content of each repeat by DCpG = D/(1 + 9FCpG). Distribution histograms were plotted using a 0.01 bin size. For major branches within phylogenetic trees, multiple sequence alignments were performed with ClustalW at the default setting. The consensus sequences were derived using the simple majority rule. Degenerated nucleotides were defined according to the standard IUPAC codes. MEGA (Kumar et al. 2001) was used to construct NJ trees using the Kimura two-parameter model. The minimum spanning trees of primate Alu subfamilies, that is, the trees with Alu subfamilies as nodes that minimize the sum of edge distances, were constructed using Alucode. Under the null hypothesis of uniformity, the P-value for the linkage was calculated using the nonparametric computation as described by Price et al. (2004). Since Alucode can run on a wide range of resolutions, it can split a small Alu population into large numbers of subfamilies. Based on the size of our data, we chose MINCOUNT = 15 with all other default parameters. Under this setting, Alucode created similar numbers of Alu subfamilies as the conventional NJ method.
Acknowledgments
This work was supported in part by CRIS Project No. 1265-31000-090-00D from USDA and by National Institutes of Health grant GM058815 to E.E.E. E.E.E. is an investigator of the Howard Hughes Medical Institute.
Footnotes
[Supplemental material is available online at www.genome.org and at http://bfgl.anri.barc.usda.gov/Alusite.]
Article is online at http://www.genome.org/cgi/doi/10.1101/gr.083972.108.
References
- Bailey J.A., Liu G., Eichler E.E. An Alu transposition model for the origin and expansion of human segmental duplications. Am. J. Hum. Genet. 2003;73:823–834. doi: 10.1086/378594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer M.A., Deininger P.L. Alu repeats and human genomic diversity. Nat. Rev. Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- Batzer M.A., Deininger P.L., Hellmann-Blumberg U., Jurka J., Labuda D., Rubin C.M., Schmid C.W., Zietkiewicz E., Zuckerkandl E. Standardized nomenclature for Alu repeats. J. Mol. Evol. 1996;42:3–6. doi: 10.1007/BF00163204. [DOI] [PubMed] [Google Scholar]
- Britten R.J., Baron W.F., Stout D.B., Davidson E.H. Sources and evolution of human Alu repeated sequences. Proc. Natl. Acad. Sci. 1988;85:4770–4774. doi: 10.1073/pnas.85.13.4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Daniels G.R., Deininger P.L. A second major class of Alu family repeated DNA sequences in a primate genome. Nucleic Acids Res. 1983;11:7595–7610. doi: 10.1093/nar/11.21.7595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels G.R., Deininger P.L. Characterization of a third major SINE family of repetitive sequences in the galago genome. Nucleic Acids Res. 1991;19:1649–1656. doi: 10.1093/nar/19.7.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiyama A., Watanabe H., Toyoda A., Taylor T.D., Itoh T., Tsai S.F., Park H.S., Yaspo M.L., Lehrach H., Chen Z., et al. Construction and analysis of a human-chimpanzee comparative clone map. Science. 2002;295:131–134. doi: 10.1126/science.1065199. [DOI] [PubMed] [Google Scholar]
- Gibbs R.A., Rogers J., Katze M.G., Bumgarner R., Weinstock G.M., Mardis E.R., Remington K.A., Strausberg R.L., Venter J.C., Wilson R.K., et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Goodman M. The genomic record of Humankind's evolutionary roots. Am. J. Hum. Genet. 1999;64:31–39. doi: 10.1086/302218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K., Konkel M.K., Xing J., Wang H., Lee J., Meyer T.J., Huang C.T., Sandifer E., Hebert K., Barnes E.W., et al. Mobile DNA in Old World monkeys: A glimpse through the rhesus macaque genome. Science. 2007;316:238–240. doi: 10.1126/science.1139462. [DOI] [PubMed] [Google Scholar]
- Hedges D.J., Callinan P.A., Cordaux R., Xing J., Barnes E., Batzer M.A., Salem A.H., Kilroy G.E., Watkins W.S., Schienman J.E., et al. Differential Alu mobilization and polymorphism among the human and chimpanzee lineages. Genome Res. 2004;14:1068–1075. doi: 10.1101/gr.2530404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herke S.W., Xing J., Ray D.A., Zimmerman J.W., Cordaux R., Batzer M.A. A SINE-based dichotomous key for primate identification. Gene. 2007;390:39–51. doi: 10.1016/j.gene.2006.08.015. [DOI] [PubMed] [Google Scholar]
- Jurka J., Smith T. A fundamental division in the Alu family of repeated sequences. Proc. Natl. Acad. Sci. 1988;85:4775–4778. doi: 10.1073/pnas.85.13.4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Jakobsen I.B., Nei M. MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Liu G., Zhao S., Bailey J.A., Sahinalp S.C., Alkan C., Tuzun E., Green E.D., Eichler E.E. Analysis of primate genomic variation reveals a repeat-driven expansion of the human genome. Genome Res. 2003;13:358–368. doi: 10.1101/gr.923303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers E.W., Miller W. Optimal alignments in linear space. Comput. Appl. Biosci. 1988;4:11–17. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- Ohshima K., Hattori M., Yada T., Gojobori T., Sakaki Y., Okada N. Whole-genome screening indicates a possible burst of formation of processed pseudogenes and Alu repeats by particular L1 subfamilies in ancestral primates. Genome Biol. 2003;4:R74. doi: 10.1186/gb-2003-4-11-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A.L., Eskin E., Pevzner P.A. Whole-genome analysis of Alu repeat elements reveals complex evolutionary history. Genome Res. 2004;14:2245–2252. doi: 10.1101/gr.2693004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D.A., Batzer M.A. Tracking Alu evolution in New World primates. BMC Evol. Biol. 2005;5:51. doi: 10.1186/1471-2148-5-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D.A., Xing J., Hedges D.J., Hall M.A., Laborde M.E., Anders B.A., White B.R., Stoilova N., Fowlkes J.D., Landry K.E., et al. Alu insertion loci and platyrrhine primate phylogeny. Mol. Phylogenet. Evol. 2005;35:117–126. doi: 10.1016/j.ympev.2004.10.023. [DOI] [PubMed] [Google Scholar]
- Roos C., Schmitz J., Zischler H. Primate jumping genes elucidate strepsirrhine phylogeny. Proc. Natl. Acad. Sci. 2004;101:10650–10654. doi: 10.1073/pnas.0403852101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem A.H., Ray D.A., Xing J., Callinan P.A., Myers J.S., Hedges D.J., Garber R.K., Witherspoon D.J., Jorde L.B., Batzer M.A. Alu elements and hominid phylogenetics. Proc. Natl. Acad. Sci. 2003;100:12787–12791. doi: 10.1073/pnas.2133766100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid C.W. Alu: Structure, origin, evolution, significance and function of one-tenth of human DNA. Prog. Nucleic Acid Res. Mol. Biol. 1996;53:283–319. doi: 10.1016/s0079-6603(08)60148-8. [DOI] [PubMed] [Google Scholar]
- Schmitz J., Ohme M., Zischler H. SINE insertions in cladistic analyses and the phylogenetic affiliations of Tarsius bancanus to other primates. Genetics. 2001;157:777–784. doi: 10.1093/genetics/157.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She X., Liu G., Ventura M., Zhao S., Misceo D., Roberto R., Cardone M.F., Rocchi M., Green E.D., Archidiacano N., et al. A preliminary comparative analysis of primate segmental duplications shows elevated substitution rates and a great-ape expansion of intrachromosomal duplications. Genome Res. 2006;16:576–583. doi: 10.1101/gr.4949406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M.R., Batzer M.A., Deininger P.L. Evolution of the master Alu gene(s) J. Mol. Evol. 1991;33:311–320. doi: 10.1007/BF02102862. [DOI] [PubMed] [Google Scholar]
- Smit A.F. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. ClustalW: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H., Fujiyama A., Hattori M., Taylor T.D., Toyoda A., Kuroki Y., Noguchi H., BenKahla A., Lehrach H., Sudbrak R., et al. DNA sequence and comparative analysis of chimpanzee chromosome 22. Nature. 2004;429:382–388. doi: 10.1038/nature02564. [DOI] [PubMed] [Google Scholar]
- Xing J., Wang H., Han K., Ray D.A., Huang C.H., Chemnick L.G., Stewart C.B., Disotell T.R., Ryder O.A., Batzer M.A. A mobile element based phylogeny of Old World monkeys. Mol. Phylogenet. Evol. 2005;37:872–880. doi: 10.1016/j.ympev.2005.04.015. [DOI] [PubMed] [Google Scholar]
- Yohn C.T., Jiang Z., McGrath S.D., Hayden K.E., Khaitovich P., Johnson M.E., Eichler M.Y., McPherson J.D., Zhao S., Paabo S., et al. Lineage-specific expansions of retroviral insertions within the genomes of African great apes but not humans and orangutans. PLoS Biol. 2005;3:e110. doi: 10.1371/journal.pbio.0030110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Malek J., Mahairas G., Fu L., Nierman W., Venter J.C., Adams M.D. Human BAC ends quality assessment and sequence analyses. Genomics. 2000;63:321–332. doi: 10.1006/geno.1999.6082. [DOI] [PubMed] [Google Scholar]
- Zhao S., Shatsman S., Ayodeji B., Geer K., Tsegaye G., Krol M., Gebregeorgis E., Shvartsbeyn A., Russell D., Overton L., et al. Mouse BAC ends quality assessment and sequence analyses. Genome Res. 2001;11:1736–1745. doi: 10.1101/gr.179201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zietkiewicz E., Richer C., Sinnett D., Labuda D. Monophyletic origin of Alu elements in primates. J. Mol. Evol. 1998;47:172–182. doi: 10.1007/pl00006374. [DOI] [PubMed] [Google Scholar]