Abstract
The mitochondrial (mt) genomes of animals typically consist of a single circular chromosome that is ∼16-kb long and has 37 genes. Our analyses of the sequence reads from the Human Body Louse Genome Project and the patterns of gel electrophoresis and Southern hybridization revealed a novel type of mt genome in the sucking louse, Pediculus humanus. Instead of having all mt genes on a single chromosome, the 37 mt genes of this louse are on 18 minicircular chromosomes. Each minicircular chromosome is 3–4 kb long and has one to three genes. Minicircular mt chromosomes are also present in the four other species of sucking lice that we investigated, but not in chewing lice nor in the Psocoptera, to which sucking lice are most closely related. We also report unequivocal evidence for recombination between minicircular mt chromosomes in P. humanus and for sequence variation in mt genes generated by recombination. The advantages of a fragmented mt genome, if any, are currently unknown. Fragmentation of mt genome, however, has coevolved with blood feeding in the sucking lice. It will be of interest to explore whether or not life history features are associated with the evolution of fragmented chromosomes.
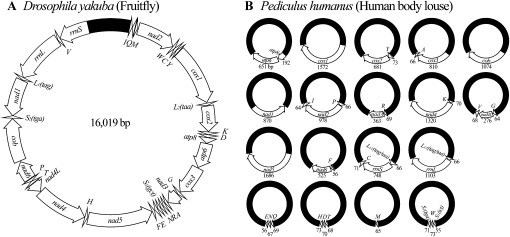
Changes in the number and structure of chromosomes have long been thought to play a role in the evolution of organisms (Williams 1966; White 1973, 1978), although the mechanisms involved are not well understood (Sites and Moritz 1987; Rieseberg 2001). It is well known that eukaryotes and prokaryotes have very different numbers and structures of chromosomes. Eukaryotes have two types of chromosomes: one in nuclei and the other in mitochondria and chloroplasts. The nuclear chromosomes of eukaryotes are linear and vary greatly in number, even among species that are closely related. At one extreme, the jumper ant, Myrmecia pilosula, has only one pair of nuclear chromosomes; at the other extreme, the Adder's Tongue fern, Ophioglossum reticulatum, has 630 pairs of nuclear chromosomes. The Indian muntjac deer, Muntiacus muntjak, has three pairs of nuclear chromosomes, whereas the Chinese muntjac deer, Muntiacus reevesi, has 23 pairs of nuclear chromosomes (Wurster and Benirschke 1970). In contrast to eukaryotes, most prokaryotes have only one circular chromosome, but a number of bacteria have linear or multiple chromosomes (Jumas-Bilak et al. 1998; Prozorov 2008). The mitochondria and chloroplasts of eukaryotes evolved from endosymbiotic bacteria; thus, the chromosomes in these organelles have many characteristics of bacterial chromosomes (Gray 1999). The mitochondrial and chloroplast chromosomes of most eukaryotes are circular and most eukaryotes have only one mitochondrial and/or chloroplast chromosome (Gray 1999). Linear and multiple mitochondrial and chloroplast chromosomes, however, have been found in protists (including algae, ciliates, flagellates, slime molds, and ichthyosporeans), oomycetous fungi, yeasts, and cnidarians (Nosek et al. 1998; Zhang et al. 1999; Burger et al. 2003). In animals (Metazoa), the mitochondrial chromosomes are typically circular, ∼16-kb long and have 37 genes (Boore 1999; Fig. 1A).
Figure 1.
(A) The typical mitochondrial (mt) genome of animals, represented here by a fruitfly, Drosophila yakuba; (B) the mt genome of the human body louse, Pediculus humanus. The mt genome of Drosophila yakuba consists of a single circular chromosome that is 16,019-bp long and has 37 genes (GenBank accession no. NC_001322). The mt genome of P. humanus, however, consists of 18 minicircular chromosomes; each minicircular chromosome is 3000–4000-bp long and has one to three genes. Genes are represented as boxes and are drawn to scale. The length of each gene of P. humanus is indicated near the box that represents the gene. Arrows indicate the direction of transcription. Protein-coding genes are abbreviated as atp6 and atp8 (for ATP synthase subunits 6 and 8), cox1-3 (for cytochrome c oxidase subunits 1 to 3), cob (for cytochrome b), and nad1-6 and 4L (for NADH dehydrogenase subunits 1–6 and 4L). rrnL and rrnS are for large and small rRNA subunits. tRNA genes are shown with the single-letter abbreviations of their corresponding amino acids. The noncoding regions are in black. The 18 minichromosomes of P. humanus are in alphabetical order according to the names of their protein-coding and rRNA genes; those with only tRNA genes are in the last row.
Lice (order Phthiraptera; class Insecta) are obligate ectoparasites of birds and mammals. The Phthiraptera has four suborders: Anoplura, Amblycera, Ischnocera, and Rhyncophthirina (Barker 1994). The Anoplura are sucking lice, whereas the other three suborders are chewing lice. Sucking lice parasitize placental mammals (Durden and Musser 1994) and feed exclusively on blood. Chewing lice infest birds and mammals and feed primarily on epidermal features, such as hairs and feathers (Price et al. 2003). The mitochondrial (mt) genomes of three chewing lice have been sequenced entirely: a wallaby louse, Heterodoxus macropus (Amblycera), a pigeon louse, Campanulotes bidentatus (Ischnocera), and a screamer louse, Bothriometopus macrocnemis (Ischnocera) (Shao et al. 2001b; Covacin et al. 2006; Cameron et al. 2007). Like most other animals, these three chewing lice have their 37 mt genes on a single circular chromosome, 14,670-, 14,804-, and 15,564-bp long, respectively. Over several years we attempted, but were unable, to amplify the entire mt genomes of any sucking lice by either polymerase chain reaction (PCR) (Nelson et al. 1996) or rolling circle amplification (RCA) (Simison et al. 2006), although these techniques have successfully amplified the entire mt genomes of many animals. We suspected that the structure of mt genomes of sucking lice differed substantially from those of chewing lice and other animals. The Human Body Louse Genome Project allowed us to investigate this possibility.
In this study we report that the single mt chromosome typical of animals has evolved into 18 minichromosomes in the human body louse, Pediculus humanus (a sucking louse). This novel type of mt genome is present in the other sucking lice that we investigated but not in chewing lice nor the Psocoptera, to which sucking lice are most closely related. We compared the structure of mt genomes and the life history of sucking lice with chewing lice and the Psocoptera. It appears that blood feeding has coevolved with minicircular mt chromosomes in the lineage leading to sucking lice. We also report evidence for frequent recombination between mt minichromosomes in P. humanus. Recombination between mt minichromosomes could increase sequence variation in mt genes, and increased sequence variation might have been an advantage to the ancestor of sucking lice when they were adapting to a new diet, blood, and new hosts, the eutherian mammals.
Results
The mt genome of the human body louse, P. humanus, consists of 18 minicircular chromosomes; each minicircular chromosome is 3–4 kb long and has one to three genes
The Human Body Louse Genome Project used a whole-genome shotgun sequencing strategy and generated 1,480,551 nt sequence reads from the nuclear genome and the mt genome of P. humanus, and the genomes of the endosymbiotic bacteria of this louse (Pittendrigh et al. 2006). Notably, 99.84% (37,084) of the 37,144 sequence reads that contain mt genes are from the library that has inserts of 4 ± 0.8 kb, although this library accounts for only 53.12% of the total sequence reads. The extremely biased distribution of the sequence reads that contain mt genes toward the 4 ± 0.8-kb library indicates that the mt genome of P. humanus consists of multiple chromosomes that are 4 ± 0.8 kb in size, rather than a single chromosome that is ∼16 kb. The presence of multiple mt chromosomes was confirmed by the assembly of sequence reads and by our analyses of the patterns of gel electrophoresis and Southern hybridization (see below).
Of the 37,144 sequence reads that had mt genes, 36,615 were assembled into 18 contigs. Each contig has a different mt gene, or a cluster of two or three mt genes, flanked by noncoding sequences. These genes and gene clusters range from 65 (trnM gene) to 1686 bp (nad5 gene) (Fig. 1B). Together, the 18 contigs contain all of the 37 mt genes typical of animals. The structure of these contigs indicates that the mt genome of P. humanus consists of 18 minichromosomes; each of these minichromosomes has one to three genes (Fig. 1B).
To confirm the sizes of the mt minichromosomes of P. humanus and to find out whether these minichromosomes were circular or linear, we examined the patterns of agarose gel electrophoresis and Southern hybridization of undigested and ApaI-digested mtDNA of P. humanus. Each of the 18 contigs (above) has one, and only one, ApaI cut site (G∧GGCC∨C) near the 3′-end of each gene or gene cluster. If the mt minichromosomes of P. humanus are 4 ± 0.8 kb in size and are circular, then the ApaI digestion of minichromosomes will generate DNA fragments of 4 ± 0.8 kb. On the other hand, if the mt minichromosomes are 4 ± 0.8 kb in size and are linear, then the ApaI digestion of minichromosomes will generate DNA fragments of two different sizes; each size will be smaller than 4 ± 0.8 kb and the sum of the two sizes will be 4 ± 0.8 kb. If the mt minichromosomes are 4 ± 0.8 kb in size and are in both circular and linear configurations, then the ApaI digestion of minichromosomes will generate DNA fragments of three different sizes: one size will be 4 ± 0.8 kb, whereas the sum of the other two sizes will be 4 ± 0.8 kb.
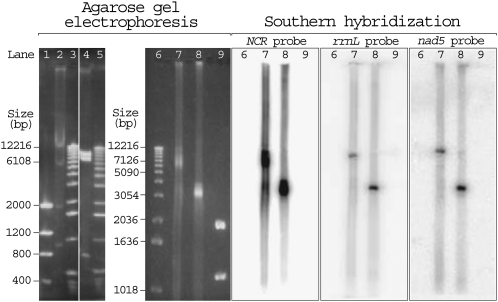
Our agarose gel electrophoresis showed that the undigested mtDNA of P. humanus was a smear with a predominant size of 6–9 kb, whereas the ApaI-digested mtDNA of P. humanus was a block of fragments that vary in size from 3 to 4 kb (Fig. 2, lanes 7,8, left). The agarose gel electrophoresis results were confirmed by Southern hybridization with three probes: (1) an NCR probe, which was designed from a segment of the noncoding region that was highly conserved among all of the 18 mt minichromosomes (see below); (2) an rrnL probe, which was specific to the mt minichromosomes that contained the rrnL gene; and (3) an nad5 probe, which was specific to the mt minichromosomes that contained the nad5 gene (Fig. 2, right). The patterns of agarose gel electrophoresis and Southern hybridization of undigested and ApaI-digested mtDNA of P. humanus can only be explained by the 18 mt minichromosomes of P. humanus being circles that vary in size from 3 to 4 kb. The variation in size among the 18 minichromosomes is due to the variation in the size of both the coding regions (see above) and the noncoding regions (see below). The smear from 6 to 9 kb in the undigested mtDNA of P. humanus (Fig. 2, lane 7) is most likely nicked circular DNA, 3–4 kb in size. This apparent size difference between undigested and digested circular DNA was also observed in our control, the circular plasmids of similar size (data not shown).
Figure 2.
Agarose gel electrophoresis (left) and Southern hybridization (right) of the mitochondrial (mt) DNA of a fruitfly, D. melanogaster; a mouse, Mus musculus; and the human body louse, P. humanus. (Lanes 1,9) Two microliters low mass ladder DNA marker (Invitrogen); (lane 2) EcoRI-digested mtDNA extracted from182 mg of fruitflies; (lanes 3,5,6) 2 μL DNA molecular weight marker X (Roche); (lane 4) BamHI-digested mtDNA extracted from 36 mg of mouse liver tissues; (lane 7) undigested mtDNA extracted from 212 mg of human body lice; (lane 8) ApaI-digested mtDNA extracted from 212 mg of human body lice. The fruitfly and the mouse were experimental controls. As expected, the 19,517-bp mt chromosome of the fruitfly was cut into four fragments by EcoRI: ∼12,000, ∼5500, ∼1700, and ∼900 bp (lane 2); and the 16,300-bp mt chromosome of the mouse was cut into three fragments by BamHI: ∼8600, ∼7000, and ∼700 bp (lane 4). The undigested mtDNA of P. humanus migrated as a smear with a predominant size of 6000–9000 bp (lane 7), whereas the ApaI-digested mtDNA of P. humanus appeared as a block that ranges from 3000 to 4000 bp (lane 8).
From the evidence above, we conclude that the mt genome of P. humanus consists of 18 minicircular chromosomes that are 3–4 kb in size (Fig. 1B). Each minicircular chromosome has a coding region and a noncoding region. Five of the 18 minicircular chromosomes have only one gene: cob, cox1, nad1, nad5, and trnM, respectively. Seven minicircular chromosomes have two genes: atp8-atp6, trnY-cox2, cox3-trnA, trnR-nad3, trnK-nad4, trnF-nad6, and trnL-rrnL. The other six minicircular chromosomes have three genes: trnP-nad2-trnI, trnG-nad4L-trnV, trnL-rrnS-trnC, trnQ-trnN-trnE, trnT-trnD-trnH, and trnS1-trnW-trnS2 (Note: hyphens link neighboring genes on the same minicircular chromosome). Where there are two or three genes in a minichromosome, these genes are on the same strand and are either immediately next to one another, or overlap by 1–2 bp, or have 1–4 bp of noncoding sequences in between.
The noncoding regions of the minicircular mitochondrial chromosomes of P. humanus are highly polymorphic but share three blocks of highly conserved nucleotide sequence
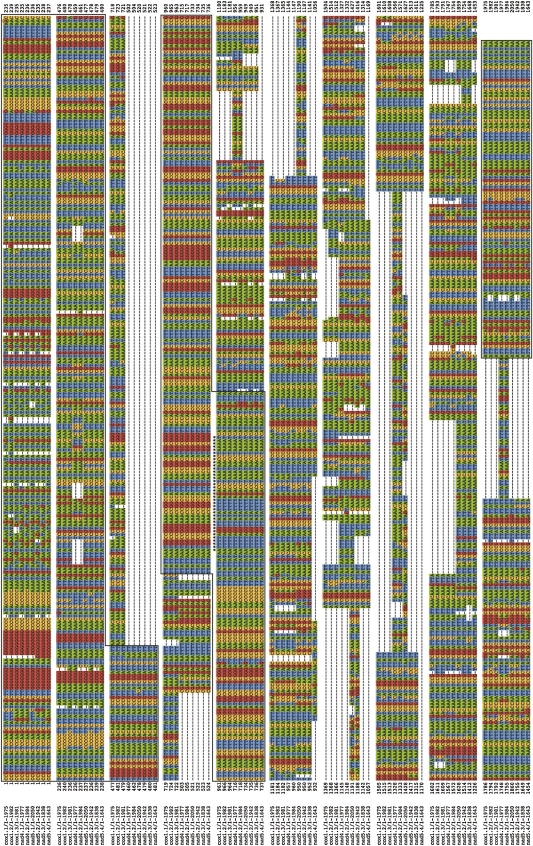
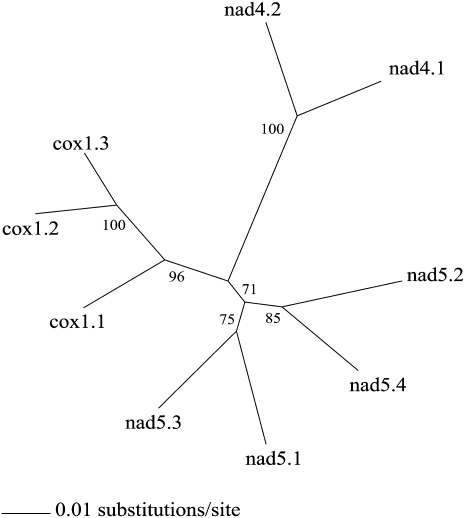
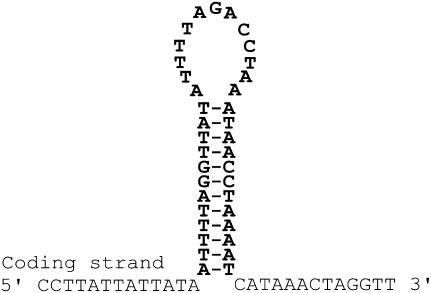
To obtain full-length sequences of noncoding regions, we sequenced nine plasmid clones entirely: three clones with inserts of the cox1 minicircular chromosome, two clones with inserts of the nad4 minicircular chromosome, and four clones with inserts of the nad5 minicircular chromosome (Fig. 3). The sequence identity among the noncoding regions of different clones of the same type of minicircular chromosome is higher (92%–97%) than that among the noncoding regions of different minicircular chromosomes (77%–87%); an unrooted neighbor-joining (NJ) tree has all of the noncoding regions of the same minicircular chromosomes together (Fig. 4). The noncoding regions are highly polymorphic, even among clones of the same minicircular chromosome (Fig. 3). These polymorphisms are mainly within repetitive motifs that vary in sequence, length (11–373 bp) and copy number (2–7). The nine plasmid clones that we sequenced were unlikely from the same individual louse because the libraries in the Human Body Louse Genome Project were constructed from hundreds of lice (Pittendrigh et al. 2006). So, the polymorphisms in the noncoding regions we revealed in this study are more likely among different individuals of P. humanus than within an individual louse. The noncoding regions of the minicircular chromosomes, on the other hand, have three blocks of highly conserved sequences (Fig. 3). A block of sequence at the start and another at the end of the noncoding regions, 523- and 101-bp long, respectively, have 90% and 99% nucleotide identity, respectively. An internal block (302 bp) is almost perfectly conserved and part of this block has the potential to form a stable stem loop (Fig. 5). Stem loops are the initiation sites of replication and transcription of the mt genomes of several animals (Wong and Clayton 1985; Clayton 1991; L'Abbe et al. 1991).
Figure 3.
Alignment of the nucleotide sequences of the noncoding regions from plasmid clones with the cox1 (three clones), nad4 (two clones), and nad5 (four clones) minicircular mitochondrial chromosomes. Dashes indicate gaps. The three highly conserved blocks of sequences at the start, middle, and end of the noncoding region are boxed. The sequence that can form a stem-loop is marked with asterisks (see also Fig. 5). The coding region (data not shown) of each clone is located between the end and start of these noncoding regions.
Figure 4.
An unrooted neighbor-joining (NJ) tree inferred from the nucleotide sequences of the noncoding regions of cox1, nad4, and nad5 minicircular mitochondrial chromosomes (see also Fig. 3). Gaps in sequence alignment were excluded. Bootstrap support in percentage (1000 replicates) is shown near each branch. Noncoding regions (e.g., cox1.1) are named after the minicircular chromosome and the plasmid clone number.
Figure 5.
A putative stem-loop in the 302-bp highly conserved segment of the noncoding regions of the minicircular mitochondrial chromosomes of the human body louse, P. humanus.
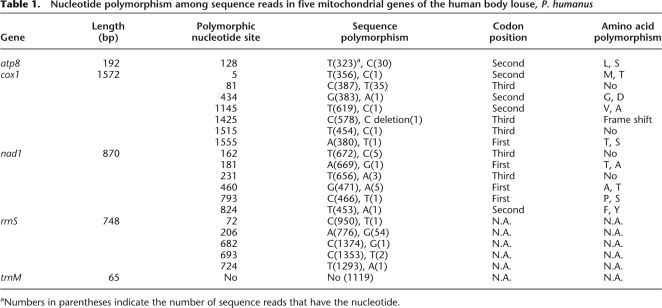
Compared with the noncoding regions, the coding regions of the minicircular chromosomes are much less polymorphic. We analyzed the sequence variation in five mitochondrial genes: three protein-coding genes, atp8, cox1, and nad1, one rRNA gene, rrnS, and one tRNA gene, trnM. We found a single nucleotide polymorphism (SNP) in atp8, seven SNPs in cox1, six SNPs in nad1, five SNPs in rrnS, but no SNPs in trnM (Table 1). The numbers of SNPs at the first, second, and third codon positions of the three protein-coding genes are almost equal. This is unexpected, because SNPs at the first and second codon positions may cause changes in amino acid sequence more frequently than SNPs at the third codon position. It is possible that some of the SNPs in the three protein-coding genes, especially those that appear only once in sequence reads (Table 1), occurred recently and thus have not been eliminated by selection. Alternatively, the amino acid changes caused by SNPs at the first and the second codon position may have only minor effects on the function of the proteins encoded by these genes.
Table 1.
Nucleotide polymorphism among sequence reads in five mitochondrial genes of the human body louse, P. humanus
aNumbers in parentheses indicate the number of sequence reads that have the nucleotide.
Mitochondrial minicircular chromosomes are present in other sucking lice but not in chewing lice and the Psocoptera
We tested by PCR and sequenced part of the mt genomes of the human head louse, Pediculus capitis; the human crab louse, Pthirus pubis; the chimpanzee louse, Pediculus schaeffi; and the langur louse, Pedicinus ancoratus. We found that at least 12 minicircular mt chromosomes were present in the human head louse, at least nine in the human crab louse, at least five in the chimpanzee louse, and at least one in the langur louse (Fig. 6). These lice represent all three of the families of sucking lice (suborder Anoplura) of primates: Pediculidae, Pthiridae, and Pedicinidae (Durden and Musser 1994). Thus, minicircular mt chromosomes likely evolved in or before the most recent common ancestor of the sucking lice of primates, which apparently lived ∼22.5 Mya (Reed et al. 2004).
Figure 6.
Evolution of minicircular mitochondrial (mt) chromosomes in the sucking lice of primates. The phylogeny of the sucking lice of primates and the estimate of the date of their most recent common ancestor (22.5 Mya) are after Barker et al. (2003) and Reed et al. (2004). The preferred habitats of lice on humans (i.e., body, head, and pubis) are in parentheses. The 18 minicircular mt chromosomes of the human body louse were sequenced entirely; the minicircular mt chromosomes of other lice were partially sequenced or identified by PCR tests (underlined). Hyphens link neighboring genes on the same minicircular chromosome.
Sucking lice are most closely related to chewing lice and the Psocoptera (Barker 1994; Johnson et al. 2004). The mt genomes of three species of chewing lice and one species of the Psocoptera, i.e., a wallaby louse, Heterodoxus macropus (suborder Amblycera); a pigeon louse, Campanulotes bidentatus (suborder Ischnocera); a screamer louse, B. macrocnemis (Ischnocera); and Lepidopsocid sp. (Psocoptera), have been sequenced entirely (Shao et al. 2001b, 2003; Covacin et al. 2006; Cameron et al. 2007). Furthermore, seven species of chewing lice and two species of Psocoptera have been sequenced partially (Shao et al. 2001a, b; Covacin et al. 2006; Cameron et al. 2007). There is no evidence for minicircular mt chromosomes in any of these chewing lice nor the Psocoptera.
Discussion
Sucking lice are the first group of animals known in which fragmented mt chromosomes likely replaced the typical mt chromosomes
The multiple mt minicircular chromosomes of the sucking lice we reported in this study evolved, with little doubt, from a single circular mt chromosome typical of animals. Chromosomal fragmentation has been found previously in the nuclear genomes of ciliates (Prescott 1994), the chloroplast genomes of dinoflagaellates (Zhang et al. 1999), and the mitochondrial genomes of an ichthyosporean, Amoebidium parasiticum (Burger et al. 2003), a diplonemid flagellate, Diplonema papillatum (Marande et al. 2005) and a mesozoan, Dicyema misakiense (Watanabe et al. 1999). The nature of the chromosomal fragmentation in these eukaryotes, however, differs in: (1) the form (i.e., linear or circular), the number (from less than 10 to more than 24,000), and the replication capability of the fragmented chromosomes; (2) whether or not the genes on each fragmented chromosome are full length or partial; and (3) the presence or absence of the typical chromosomes. The ciliates have two sets of nuclear chromosomes; one set is in the germ-line micronucleus and has the typical linear structure with many genes on each chromosome, whereas the other set is in the somatic macronucleus and has hundreds or even thousands of linear, gene-sized chromosomes (Prescott 1994). The genes on the fragmented chromosomes of the ciliates are full length and are apparently able to replicate. The dinoflagaellates have up to 14 minicircular chloroplast chromosomes; each has one or a few full-length genes and is likely able to replicate (Zhang et al. 1999). There is no evidence for typical chloroplast chromosomes in the dinoflagaellates. The diplonemid flagellate, D. papillatum, may have ∼56 minicircular mt chromosomes; each has only part (<300 bp) of a gene (Marande et al. 2005). There is no evidence for typical mt chromosomes in D. papillatum, and it is not clear whether the fragmented chromosomes of this flagellate are able to replicate or not. Three mt genes (cox1, cox2, and cox3) of the mesozoan, D. misakiense, were found in three separate DNA minicircles (Watanabe et al. 1999). These DNA minicircles, however, have apparently been generated from typical mt chromosomes, and do not appear to be able to replicate (Awata et al. 2005). Each DNA minicircle of D. misakiense has a few full-length mt genes. In the present study, we did not find any evidence for typical mt chromosomes in P. humanus. The sucking lice, thus, are the second lineage of animals known (in addition to dicyemids) to have fragmented mt chromosomes and, further, the only lineage of animals known in which fragmented mt chromosomes apparently replaced the typical mt chromosomes.
Life history may be associated with the evolution of fragmented chromosomes
According to a model by Smith and Szathmary (1993), there is positive selection for genes to be linked together on a chromosome rather than being separated from one another. Having genes linked together on a chromosome, such as the typical mt chromosome of animals, ensures that each daughter organelle/cell will receive all of the genes on the chromosome and the ratios of these genes are not changed after segregation. Although chromosomes with many genes may take longer to replicate than chromosomes with one or a few genes, this disadvantage may be less than the advantage of having genes linked together on one chromosome. The model of Smith and Szathmary (1993) seems to explain well why the nuclear and organelle chromosomes of the vast majority of the eukaryotes whose chromosomes have been studied have many genes, whereas only a small number of eukaryotes (listed above) have chromosomes with one or a few genes.
If chromosomes with many genes were selected for, why did the chromosomes of the exceptional eukaryotes we listed above become fragmented and thus have only one or a few genes? What drove the evolution of fragmentation of chromosomes in these eukaryotes? Chromosomes with one or a few genes may replicate faster than typical chromosomes that have many genes. Replication advantage alone, however, does not appear to be sufficient to lead to chromosomal fragmentation. Thus, other factors may be more important than replication advantage in driving the chromosomal fragmentation. In the case of sucking lice, the fragmented mt chromosomes have coevolved with blood feeding. Chewing lice feed primarily on the hairs and feathers of their mammal and bird hosts; the Psocoptera feed on algae, fungi, and fragments of plant and insect tissues. Sucking lice, however, feed exclusively on the blood of their mammalian hosts. It will be of interest to explore the relationship between these two derived features of sucking lice.
Did the fragmentation of mitochondrial chromosomes have a role in the evolution of sucking lice?
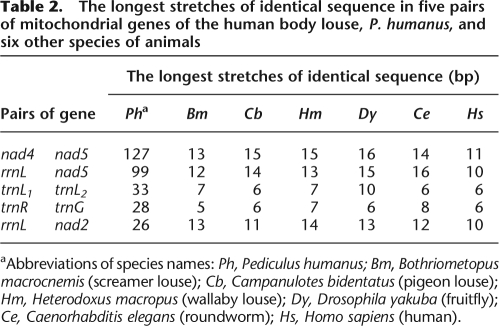
Changes in the number and structure of chromosomes have long been thought to play a role in the evolution of organisms (Williams 1966; White 1973, 1978). It is still not well understood, however, how chromosome changes may interact with organism evolution (Sites and Moritz 1987; Rieseberg 2001). Could mt chromosomal fragmentation have a role in the evolution of sucking lice? If yes, what could that role be? We note that several mt genes of P. humanus have stretches of identical sequences that are much longer than expected. Any two mt protein-coding and rRNA genes of an animal can be expected by chance to have identical sequences that are up to 16-bp long, and any two mt tRNA genes to have identical sequences that are up to 10-bp long (Table 2). Five pairs of mt genes of P. humanus, nad4 and nad5, rrnL and nad5, trnL1 and trnL2, trnR and trnG, and rrnL and nad2, however, have identical sequences that are 127, 99, 33, 28, and 26 bp, respectively. These are eight, six, three, three, and two times longer than expected. Why do so many mt genes of P. humanus, which are on different minichromosomes, have identical sequences that are much longer than expected? The only plausible explanation is that the minichromosomes of P. humanus recombine with each other and recombine often. Since increased chromosome numbers in nuclear genomes tend to cause increased genetic recombination (Qumsiyeh 1994), we propose that the recombination between minichromosomes is likely facilitated by the fact that the mt genome of P. humanus comprises 18 minichromosomes. The extreme sequence variation in the noncoding regions of the minicircular chromosomes of P. humanus is also consistent with our proposal of frequent recombination between minichromosomes. Mitochondrial minichromosomes may have been selected for during the evolution of sucking lice, possibly because recombination between minichromosomes increases the genetic variation in mt genes. This might have been an advantage to the sucking lice when they were adapting to a new diet, blood, and new hosts, the eutherian mammals.
Table 2.
The longest stretches of identical sequence in five pairs of mitochondrial genes of the human body louse, P. humanus, and six other species of animals
aAbbreviations of species names: Ph, Pediculus humanus; Bm, Bothriometopus macrocnemis (screamer louse); Cb, Campanulotes bidentatus (pigeon louse); Hm, Heterodoxus macropus (wallaby louse); Dy, Drosophila yakuba (fruitfly); Ce, Caenorhabditis elegans (roundworm); Hs, Homo sapiens (human).
How did a single typical mitochondrial chromosome fragment into many minichromosomes?
It is unlikely that all of the minicircular mt chromosomes in a sucking louse were generated from a single “big bang” event that fragmented a typical mt chromosome into many minichromosomes. A “big bang” event would not generate multiple minicircular chromosomes that all have the sequences needed for replication and transcription, because there is only one set of such sequences in each typical mt chromosome (see Taanman 1999). Rather, the multiple minicircular mt chromosomes of the sucking lice were more likely generated from a series of events that involved excision and rejoining of fragments of mt chromosome over a long period of time. In each event, a fragment that contained the noncoding region and one or a few genes near the noncoding region had been excised from a typical mt chromosome. The two ends of this fragment apparently joined together to form a functional minicircular mt chromosome. If this new mt minicircular chromosome had a selective advantage over the typical mt chromosome (above), then the genes on this minicircular mt chromosome would eventually take over the function of their counterparts on the typical mt chromosome, and thus make their counterparts on the typical mt chromosome redundant. The deletion of the redundant genes from the typical mt chromosome would move other genes closer to the noncoding region. The next event of excision-and-rejoining would generate another new minicircular mt chromosome. Eventually, all of the genes on a typical mt chromosome would be on minicircular chromosomes.
In summary, we discovered that the single mt chromosome that is typical of animals has evolved into 18 minichromosomes in the human body louse, P. humanus. This novel type of mt chromosome is also present in the other sucking lice whose mt genomes have been studied, but not in chewing lice nor the Psocoptera. Blood feeding appears to have coevolved with minicircular mt chromosomes in sucking lice. Whether this is a cause–effect association or chance association remains to be seen. Having multiple mt chromosomes may facilitate recombination between minichromosomes in sucking lice, and thus increases sequence variation in mt genes. Increased sequence variation in mt genes may have been an advantage to the sucking lice when they were adapting to a new diet, blood, and new hosts, the eutherian mammals. It remains to be elucidated, however, how changes in life history traits, such as feeding style, may interact with changes in mt genomic structure.
Methods
Assembly of the mitochondrial genome of the human body louse, P. humanus
Initial shotgun assembly of the sequence reads from the Human Body Louse Genome Project yielded six ∼3500-bp contigs that contained seven mt genes of P. humanus, whose sequences were in GenBank (cob, cox1, cox3-trnA, nad4, rrnL, and rrnS). These six contigs could not be linked to each other by paired sequence reads from the same plasmid clones; this was unexpected. We inspected these contigs and found two blocks of highly conserved noncoding sequences that flanked the coding sequence of each contig. These two blocks are 523- and 101-bp long and share 90% and 99% nucleotide identity, respectively (Fig. 3). These conserved noncoding sequences were used subsequently to identify, by Mega BLAST (Zhang et al. 2000), 37,144 sequence reads that contained the mt genes of P. humanus. Assembly of these sequence reads yielded an additional 12 contigs that contained the other 30 mt genes of P. humanus. The 18 contigs, together, have all of the 37 genes that are found commonly in the mt genomes of animals (Fig. 1A). The average sequence coverage for the coding regions of the 18 contigs is 916×, and ranges from 434× (nad5) to 1773× (trnL/rrnL).
Isolation, digestion, and hybridization of mitochondrial DNA
The mtDNAs of the fruitfly, Drosophila melanogaster (Oregon strain); the mouse, Mus musculus (Quackenbush strain); and the human body louse, P. humanus (Orlando strain) were isolated with an alkaline lysis method (Tamura and Aotsuka 1988) and digested with EcoRI, BamHI, and ApaI (Roche), respectively. The mt genomes of M. musculus and D. melanogaster have been sequenced entirely (GenBank accession nos. DQ874614 and U37541) and were used as experimental controls. The mtDNAs were resolved by agarose gel (0.9%) electrophoresis. The gel was stained with ethidium bromide, run for 1 h at 100 V and 60 mA, visualized with a UV transilluminator, and then photographed. The mtDNA of P. humanus on the agarose gel was transferred to three nylon membranes and hybridized with NCR, rrnL, and nad5 probes, respectively. The NCR probe was amplified by PCR from the 302-bp internal segment of the noncoding region that is almost perfectly conserved among all of the 18 minicircular mt chromosomes of P. humanus. The rrnL probe was amplified by PCR from a section of rrnL (nt 616–1064) and was specific to the rrnL minicircular mt chromosome. The nad5 probe was amplified by PCR from a section of nad5 (nt 1226–1453) and was specific to the nad5 minicircular mt chromosome. The probes were labeled with Megaprime DNA Labeling System (Amersham) and dCTP [α-32P]EasyTide Lead (PerkinElmer) and were hybridized with the DNA fixed on the nylon membranes for 16 h. Unincorporated probes were then washed off. The nylon membranes were exposed to phosphor-storage screens for 12 h and hybridization images were taken with a Typhoon 9400 scanner.
PCR amplification and sequencing of mitochondrial genomes
Total DNA was extracted with the DNeasy Tissue Kit (QIAGEN) from: (1) the human head louse, Pediculus capitis, from Brisbane, Australia; (2) the human crab louse, Pthirus pubis, from Brisbane, Australia; (3) the chimpanzee louse, Pediculus schaeffi, from the Tacugama Chimpanzee Sanctuary, Sierra Leone; and (4) the langur louse, Pedicinus ancoratus, from the Cuc Phuong National Park, Vietnam. The sequences of the genes on the 18 minicircular mt chromosomes of the human body louse, P. humanus, were aligned with the sequences of their counterpart genes in the wallaby louse, H. macropus (Shao et al. 2001b), and the pigeon louse, C. bidentatus (Covacin et al. 2006). The two most conserved regions in the gene/gene cluster of each minicircular mt chromosome of the human body louse were identified, and two primers, one forward and one reverse, were designed from each of these two regions. In total, 72 primers were designed from the 18 minicircular mt chromosomes of the human body louse. The total DNA and these primers were used in PCR tests to discover whether or not any of the 18 minicircular mt chromosomes that occurred in the human body louse were present in other sucking lice.
Two PCRs were set up to test for the presence of each minicircular mt chromosome in a species of louse: One PCR targeted a small section, 200–500 bp, of the coding region of the minicircular chromosome, whereas the other PCR targeted the rest of the minicircular chromosome, 2500–3100 bp. If both PCRs produce amplicons of expected sizes, then, and only then, the minicircular chromosome was concluded to be present. Otherwise, conclusions could not be drawn about the presence or absence of that minicircular chromosome. To ensure the reliability of PCR tests, both positive and negative controls were executed with each PCR test. Further, 13 of the 27 minicircular mt chromosomes identified by PCR tests in the human head louse, the human crab louse, the chimpanzee louse, and the langur louse were confirmed by sequencing their corresponding PCR amplicons (Fig. 6).
Elongase Enzyme Mix (GIBCO) and Takara La Taq (Takara) were used in PCR amplification, following the instructions of the manufacturers. PCR amplicons were checked by agarose gel (1%) electrophoresis. The sizes of PCR amplicons were estimated by comparison with molecular markers. PCR amplicons used for sequencing were purified with Wizard SV Gel/PCR Clean-up System (Promega) and cloned with pGEM-T Easy Vector System (Promega). Sequencing reactions were with BigDye Terminator Kit (ABI) and resolved by capillary separation at the Australian Genome Research Facilities in Brisbane.
Acknowledgments
We thank Bao Tran and John Gill for transposon-mediated sequencing of plasmid clones; Natalie Leo, Cath Covacin, Mike Cranfield, Asami Kabasawa, Rosa Garriga, Margaret Mobbs, Tilo Nadler, Cynthia Pollard, and Chris Whittier for louse samples; Markus Riegler for fly samples; and Maryam Ashrafi and Michael Moore for mouse samples: We thank Wen Jun Liu, Yvette Emmanuel, Carl Morrow, and James Fraser for help with Southern hybridization. We also thank Nick Campbell, and three reviewers for comments that greatly improved this manuscript. This research was partly funded by an Australian Postdoctoral Fellowship from the Australian Research Council and an Early Career Researcher grant from The University of Queensland to R.S.
Footnotes
[The nucleotide sequences of the minicircular mitochondrial chromosomes reported in this study have been submitted to GenBank (http://www.ncbi.nlm.nih.gov/Genbank/) under accession nos. EU219983–EU219995, FJ499473–FJ499490, and FJ514591–FJ514599.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.083188.108.
References
- Awata H., Noto T., Endoh H. Differentiation of somatic mitochondria and the structural changes in mtDNA during development of the dicyemid Dicyema japonicum (Mesozoa) Mol. Genet. Genomics. 2005;273:441–449. doi: 10.1007/s00438-005-1157-2. [DOI] [PubMed] [Google Scholar]
- Barker S.C. Phylogeny and classification, origins, and evolution of host associations of lice. Int. J. Parasitol. 1994;24:1285–1291. doi: 10.1016/0020-7519(94)90195-3. [DOI] [PubMed] [Google Scholar]
- Barker S.C., Whiting M., Johnson K.P., Murrell A. Phylogeny of the lice (Insecta, Phthiraptera) inferred from small subunit rRNA. Zool. Scr. 2003;32:407–414. [Google Scholar]
- Boore J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999;27:1767–1780. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G., Forget L., Zhu Y., Gray M.W., Lang B.F. Unique mitochondrial genome architecture in unicellular relatives of animals. Proc. Natl. Acad. Sci. 2003;100:892–897. doi: 10.1073/pnas.0336115100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron S.L., Johnson K.P., Whiting M.F. The mitochondrial genome of the screamer louse Bothriometopus (Phthiraptera: Ischnocera): Effects of extensive gene rearrangements on the evolution of the genome. J. Mol. Evol. 2007;65:589–604. doi: 10.1007/s00239-007-9042-8. [DOI] [PubMed] [Google Scholar]
- Clayton D.A. Replication and transcription of vertebrate mitochondrial DNA. Annu. Rev. Cell Biol. 1991;7:453–478. doi: 10.1146/annurev.cb.07.110191.002321. [DOI] [PubMed] [Google Scholar]
- Covacin C., Shao R., Cameron S., Barker S.C. Extraordinary number of gene rearrangements in the mitochondrial genomes of lice (Phthiraptera: Insecta) Insect Mol. Biol. 2006;15:63–68. doi: 10.1111/j.1365-2583.2005.00608.x. [DOI] [PubMed] [Google Scholar]
- Durden L.A., Musser G.G. The sucking lice (Insecta, Anoplura) of the world —a taxonomic checklist with records of mammalian hosts and geographical distributions. Bull. Am. Mus. Nat. Hist. 1994;218:1–90. [Google Scholar]
- Gray M.W. Evolution of organellar genomes. Curr. Opin. Genet. Dev. 1999;9:678–687. doi: 10.1016/s0959-437x(99)00030-1. [DOI] [PubMed] [Google Scholar]
- Johnson K.P., Yoshizawa K., Smith V.S. Multiple origins of parasitism in lice. Proc. Biol. Sci. 2004;271:1771–1776. doi: 10.1098/rspb.2004.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumas-Bilak E., Michaux-Charachon S., Bourg G., Ramuz M., Allardet-Servent A. Unconventional genomic organization in the alpha subgroup of the Proteobacteria. J. Bacteriol. 1998;180:2749–2755. doi: 10.1128/jb.180.10.2749-2755.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Abbe D., Duhaime J.F., Lang B.F., Morais R. The transcription of DNA in chicken mitochondria initiates from one major bidirectional promoter. J. Biol. Chem. 1991;266:10844–10850. [PubMed] [Google Scholar]
- Marande W., Lukes J., Burger G. Unique mitochondrial genome structure in diplonemids, the sister group of kinetoplastids. Eukaryot. Cell. 2005;4:1137–1146. doi: 10.1128/EC.4.6.1137-1146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson W.S., Prodohl P.A., Avise J.C. Development and application of long-PCR for the assay of full-length animal mitochondrial DNA. Mol. Ecol. 1996;5:807–810. doi: 10.1111/j.1365-294x.1996.tb00376.x. [DOI] [PubMed] [Google Scholar]
- Nosek J., Tomaska L., Fukuhara H., Suyama Y., Kovac L. Linear mitochondrial genomes: 30 Years down the line. Trends Genet. 1998;14:184–188. doi: 10.1016/s0168-9525(98)01443-7. [DOI] [PubMed] [Google Scholar]
- Pittendrigh B.R., Clark J.M., Johnston J.S., Lee S.H., Romero-Severson J., Dasch G.A. Sequencing of a new target genome: The Pediculus humanus humanus (Phthiraptera: Pediculidae) genome project. J. Med. Entomol. 2006;43:1103–1111. doi: 10.1603/0022-2585(2006)43[1103:soantg]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Prescott D.M. The DNA of ciliated protozoa. Microbiol. Rev. 1994;58:233–267. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price R.D., Hellenthal R.A., Palma R.L., Johnson K.P., Clayton D.H. World checklist of chewing lice with host associations and keys to families and genera. In: Price RD R.D., et al., editors. The chewing lice: World checklist and biological overview. INHS special publication 24; Illinois Natural History Survey, IL: 2003. [Google Scholar]
- Prozorov A.A. Additional chromosomes in bacteria: Properties and origin. Microbiology. 2008;77:385–394. [PubMed] [Google Scholar]
- Qumsiyeh M.B. Evolution of number and morphology of mammalian chromosomes. J. Hered. 1994;85:455–465. doi: 10.1093/oxfordjournals.jhered.a111501. [DOI] [PubMed] [Google Scholar]
- Reed D.L., Smith V.S., Hammond S.L., Rogers A.R., Clayton D.H. Genetic analysis of lice supports direct contact between modern and archaic humans. PLoS Biol. 2004;2:e340. doi: 10.1371/journal.pbio.0020340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg L.H. Chromosomal rearrangements and speciation. Trends Ecol. Evol. 2001;16:351–358. doi: 10.1016/s0169-5347(01)02187-5. [DOI] [PubMed] [Google Scholar]
- Shao R., Campbell N.J., Schmidt E.R., Barker S.C. Increased rate of gene rearrangement in the mitochondrial genomes of three orders of hemipteroid insects. Mol. Biol. Evol. 2001a;18:1828–1832. doi: 10.1093/oxfordjournals.molbev.a003970. [DOI] [PubMed] [Google Scholar]
- Shao R., Campbell N.J.H., Barker S.C. Numerous gene rearrangements in the mitochondrial genome of the wallaby louse, Heterodoxus macropus (Phthiraptera) Mol. Biol. Evol. 2001b;18:858–865. doi: 10.1093/oxfordjournals.molbev.a003867. [DOI] [PubMed] [Google Scholar]
- Shao R., Dowton M., Murrell A., Barker S.C. Rates of gene rearrangement and nucleotide substitution are correlated in the mitochondrial genomes of insects. Mol. Biol. Evol. 2003;20:1612–1619. doi: 10.1093/molbev/msg176. [DOI] [PubMed] [Google Scholar]
- Simison W.B., Lindberg D.R., Boore J.L. Rolling circle amplification of metazoan mitochondrial genomes. Mol. Phylogenet. Evol. 2006;39:562–567. doi: 10.1016/j.ympev.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Sites J.W., Moritz C. Chromosomal evolution and speciation revisited. Syst. Zool. 1987;36:153–174. [Google Scholar]
- Smith J.M., Szathmary E. The origin of chromosomes. 1. Selection for linkage. J. Theor. Biol. 1993;164:437–446. doi: 10.1006/jtbi.1993.1165. [DOI] [PubMed] [Google Scholar]
- Taanman J.W. The mitochondrial genome: Structure, transcription, translation and replication. Biochim. Biophys. Acta. 1999;1410:103–123. doi: 10.1016/s0005-2728(98)00161-3. [DOI] [PubMed] [Google Scholar]
- Tamura K., Aotsuka T. Rapid isolation method of animal mitochondrial DNA by the alkaline lysis procedure. Biochem. Genet. 1988;26:815–819. doi: 10.1007/BF02395525. [DOI] [PubMed] [Google Scholar]
- Watanabe K.I., Bessho Y., Kawasaki M., Hori H. Mitochondrial genes are found on minicircle DNA molecules in the mesozoan animal Dicyema. J. Mol. Biol. 1999;286:645–650. doi: 10.1006/jmbi.1998.2523. [DOI] [PubMed] [Google Scholar]
- White M.J.D. Animal cytology and evolution. University Press; Cambridge, UK: 1973. [Google Scholar]
- White M.J.D. Modes of speciation. W.H. Freeman; San Francisco, CA: 1978. [Google Scholar]
- Williams G.C. Adaptation and natural selection: A critique of some current evolutionary thought. Princeton University Press; Princeton, NJ: 1966. [Google Scholar]
- Wong T.W., Clayton D.A. In vitro replication of human mitochondrial DNA: Accurate initiation at the origin of light-strand synthesis. Cell. 1985;42:951–958. doi: 10.1016/0092-8674(85)90291-0. [DOI] [PubMed] [Google Scholar]
- Wurster D.H., Benirschke K. Indian muntjac, Muntiacus muntjak: A deer with a low diploid chromosome number. Science. 1970;168:1364–1366. doi: 10.1126/science.168.3937.1364. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Green B.R., Cavalier-Smith T. Single gene circles in dinoflagellate chloroplast genomes. Nature. 1999;400:155–159. doi: 10.1038/22099. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Schwartz S., Wagner L., Miller W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]