Abstract
Identifying conserved alternative splicing (AS) events among evolutionarily distant species can prioritize AS events for functional characterization and help uncover relevant cis- and trans-regulatory factors. A genome-wide search for conserved cassette exon AS events in higher plants revealed the exonization of 5S ribosomal RNA (5S rRNA) within the gene of its own transcription regulator, TFIIIA (transcription factor for polymerase III A). The 5S rRNA-derived exon in TFIIIA gene exists in all representative land plant species but not in green algae and nonplant species, suggesting it is specific to land plants. TFIIIA is essential for RNA polymerase III-based transcription of 5S rRNA in eukaryotes. Integrating comparative genomics and molecular biology revealed that the conserved cassette exon derived from 5S rRNA is coupled with nonsense-mediated mRNA decay. Utilizing multiple independent Arabidopsis overexpressing TFIIIA transgenic lines under osmotic and salt stress, strong accordance between phenotypic and molecular evidence reveals the biological relevance of AS of the exonized 5S rRNA in quantitative autoregulation of TFIIIA homeostasis. Most significantly, this study provides the first evidence of ancient exaptation of 5S rRNA in plants, suggesting a novel gene regulation model mediated by the AS of an anciently exonized noncoding element.
Recent genome-wide estimates of the alternative splicing (AS) rate of plant genes indicate that AS is more prevalent than originally expected (Reddy 2007), although substantially fewer plant genes undergo AS than has been observed for human genes. Approximately 20% of multiexon genes in several plant species, including Arabidopsis, rice, moss, and maize, are predicted to be alternatively spliced based on EST/cDNA evidence (Campbell et al. 2006; Wang and Brendel 2006; Barbazuk et al. 2008). The functional significance of almost all known and predicted plant AS events has yet to be determined; however, the results of a few functional analyses conducted demonstrate roles for AS in important plant processes such as photosynthesis, defense response, flowering, and cereal grain quality (Reddy 2007), as well as some metabolic pathways (Gorlach et al. 1995) and mRNA processing (Golovkin and Reddy 1996; Kalyna et al. 2006). Identification of conserved AS events among multiple evolutionarily distant species will prioritize AS events for functional characterization and help identify relevant cis- and trans-regulatory factors (Xing and Lee 2006; Reddy 2007; Barbazuk et al. 2008).
AS events that occur within the translated regions of mRNAs may contribute to the diversity of the proteome (Stamm et al. 2005). Some splice isoforms containing a premature stop codon (PTC) are often not translated. Instead, they are targeted for nonsense-mediated mRNA decay (NMD), which is an RNA surveillance system that recognizes PTC-containing mRNAs and targets them for degradation (Maquat 2004). Lewis et al. (2003) proposed that coupled AS and NMD, also termed RUST (regulated unproductive splicing and translation), may function to regulate protein expression by generating NMD-targeted isoforms. Recently, ultraconserved elements in mammals associated with cassette exons in some splicing activator SR (serine-arginine rich) proteins have been discovered, and transcript isoforms that include these conserved sequences contain PTCs and are subjected to NMD. The conserved nature of these elements in SR proteins suggests that their unproductive splicing is functionally important for the autoregulation or homeostatic control of these splicing regulators (Lareau et al. 2007; Ni et al. 2007). Conserved AS–NMD events have also been reported within SR proteins in plants (Kalyna et al. 2006).
A genome-wide comparative analysis for AS events conserved between Arabidopsis and rice (Oryza sativa sp. Nipponbare), which diverged 140–150 Mya (Chaw et al. 2004), identified a conserved cassette exon splicing event associated with highly conserved sequences within the TFIIIA gene (transcription factor for polymerase III A). In eukaryotes, 5S ribosomal RNA (5S rRNA) is transcribed by RNA polymerase III (Pol III) and is required for the assembly of the large ribosomal subunit (Szymanski et al. 2003). TFIIIA is essential for the Pol III-based transcription of 5S rRNA. TFIIIA was the first zinc finger (ZF) protein characterized and the first protein found to bind both 5S rDNA and 5S rRNA (Engelke et al. 1980; Pelham and Brown 1980). The dual functions of TFIIIA suggest a negative feedback autoregulation model for 5S rRNA transcription (Cassiday and Maher III 2002). A few cis- and trans-factors have been implicated in the developmental regulation of the TFIIIA in Xenopus (Pfaff and Taylor 1992, 1998; Griffin et al. 2003; Penberthy et al. 2003); however, the transcriptional regulation of TFIIIA is not well understood. Post-transcriptional regulation of TFIIIA has never been reported in any species.
The current analysis provides evidence of post-transcriptional autoregulation of TFIIIA in plants mediated by coupling a cassette exon AS event in TFIIIA pre-mRNA with NMD. This cassette exon is highly conserved in land plants and exhibits striking sequence and predicted secondary structure similarity to 5S rRNA. Similar to exonization of Alu elements in mammalian genomes (Sorek et al. 2002), the cassette exon may have derived from a 5S rDNA integration into the TFIIIA DNA and acquired a role in regulating TFIIIA homeostasis.
Results
Identification of a conserved cassette exon AS event within the TFIIIA gene of plants
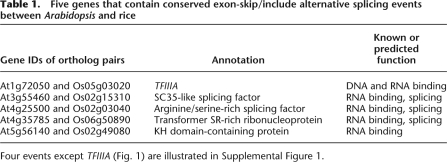
Five cassette exon AS events conserved between A. thaliana and O. sativa were discovered by comparative sequence analysis (Table 1; Fig. 1; Supplemental Fig. 1). All of the genes associated with these five events potentially encode RNA-binding proteins. Four of the genes are known or predicted to code splicing factors (Wang and Brendel 2004). The fifth, TFIIIA (At1g72050 and Os05g03020), has not been reported to be involved in RNA splicing, nor is there evidence of alternative splicing occurring in animal TFIIIA. The exon-skipping (ES) mRNA isoform of TFIIIA encodes the fully functional peptide containing nine ZFs in both AtTFIIIA and OsTFIIIA (Fig. 1A). Expression of the exon-including (EI) mRNA isoform of AtTFIIIA would possibly result in a truncated peptide consisting of the first two ZFs due to a PTC introduced by the cassette exon. This cassette exon is flanked by the exons encoding the second and third ZFs of AtTFIIIA (Fig. 1A). Sequence comparison suggests that the cassette exon in AtTFIIIA (Exon3) has been duplicated in rice. The duplicated exons (Exon4 and Exon5) also function as cassette exons during the processing of OsTFIIIA pre-mRNA (Fig. 1B; Supplemental Table 1), confirming that the cassette exon event is conserved. The inclusion of both exons or Exon5 only in OsTFIIIA mRNA results in a PTC. In both species, the PTC is located >50 nt upstream of an exon–exon junction, suggesting the EI isoform is a potential target for the NMD pathway (Hori and Watanabe 2007).
Table 1.
Five genes that contain conserved exon-skip/include alternative splicing events between Arabidopsis and rice
Four events except TFIIIA (Fig. 1) are illustrated in Supplemental Figure 1.
Figure 1.
Identification of a conserved cassette exon event associated with the TFIIIA gene in Arabidopsis and rice. (A) Exon-inclusion (EI) and exon-skipping (ES) isoforms of TFIIIA transcripts in Arabidopsis and rice (red boxes represent cDNA/EST alignments indicating exon locations; green boxes represent predicted gene models; orange boxes represent predicted coding regions). (B) A conserved ES event associated with AtTFIIIA (At1G72050) and OsTFIIIA (Os05G03020) and the sequence alignment (BLAST parameters: −W 7 –F F) between orthologous exon skipped/included (gray box) and the flanking constitute orthologous exons (green box, the second and third C2H2 zinc finger). The inclusion of the skipped/included exon introduces an in-frame stop codon (red triangle in the top panel and red text in the alignments of the bottom panel). OsTFIIIA Exon5 has higher similarity to AtTFIIIA cassette exon (Exon3) than OsTFIIIA Exon4 (81% vs. 74%).
Identification of a 5S-rRNA-like cis-element that coincides with the conserved cassette exon AS event
The available angiosperm EST data sets provide evidence for the EI isoform of TFIIIA in many plant species (Supplemental Table 1). Cross-species EST alignment and gene structure prediction suggest the existence of the cassette exon in Selaginella moellendorffii, an early vascular plant lacking true leaves and roots that last shared a common ancestor with angiosperms over 400 Mya (Stewart and Rothwell 1993). No obvious TFIIIA ortholog was found in the green algae Chlamydomonas genome. Sequence comparison of TFIIIA orthologs suggests the occurrence of the cassette exon and its AS in all land plant species surveyed (Supplemental Tables 1 and 2). Using reverse transcription polymerase chain reaction (RT-PCR), we also isolated both EI and ES isoforms from the tomato and the bryophyte Physcomitrella patens (a moss), which diverged from vascular plants ∼450 Mya (Rensing et al. 2008).
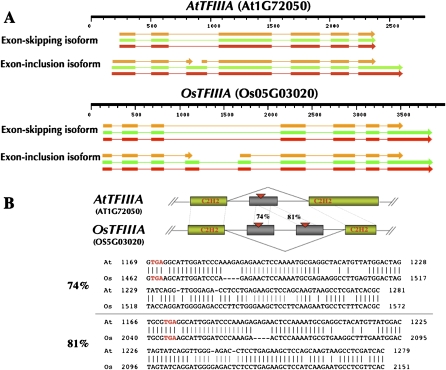
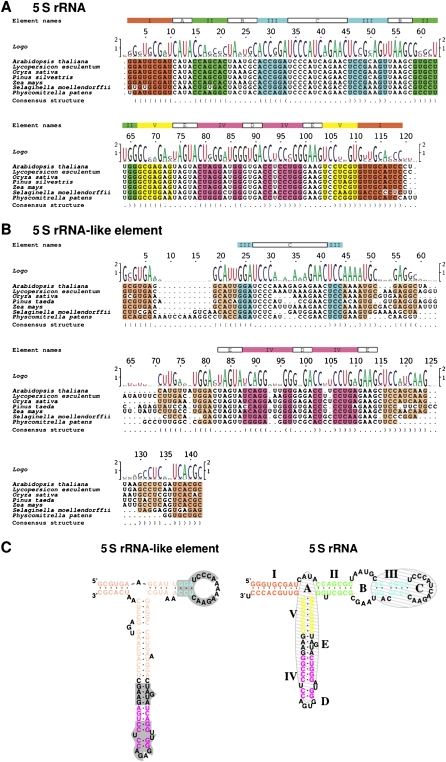
Multiple RNA sequence alignment of all 52 available TFIIIA orthologs identified an evolutionarily conserved region (ECR) in the TFIIIA gene (Supplemental Fig. 2) that corresponds to the cassette exon of AtTFIIIA (Exon3) and OsTFIIIA (Exon4 and Exon5). It is noteworthy that this ECR exhibits greater sequence conservation than any other coding region in TFIIIA. The predicted secondary structure of the ECR bears considerable similarity to the secondary structure of plant 5S rRNA (Fig. 2). In addition to a common Y-shaped structure exhibited by both elements, the 5S-rRNA-like element in TFIIIA contains the structural signatures of 5S rRNA: (1) Helix III and Loop D, which are required for the binding of ribosomal protein L5 and the import of the 5S rRNA–protein complex into the nucleus; (2) Helix IV and Loop E, which can be bound by TFIIIA during export of 5S rRNA from the nucleus; and (3) the thermodynamically stable Loop D (Szymanski et al. 2003). The predicted secondary structure of the 5S-rRNA-like element of TFIIIA contains a longer Helix V with a different Loop A and no Helix II or Loop B (Fig. 2C).
Figure 2.
Identification of a 5S-rRNA-like cis-element that coincides with the conserved cassette exon AS event. For clarity, only those sequences corresponding to the ECR in seven plant species representing evolutionarily significant nodes from Bryophyta to Magnoliophyta were displayed (for complete alignments see Supplemental Fig. 2). Alignments are based on 49 5S rRNA (A) and 52 5S-rRNA-like element sequences (B). (C) Similarity between the predicted secondary structure of the 5S-rRNA-like element of AtTFIIIA (left) and 5S rRNA (right). Element names are given according to the standard 5S rRNA nomenclature: Helices are designated by roman numbers (I, II, III, IV, and V) in colored boxes; loop regions are designated by letters (A, B, C, D, and E) in open boxes. Helical regions, which belong together, are colored identically in the element line, in the alignment, and in the structure models; similar colors are used to emphasize features similar in 5S rRNA and the 5S-rRNA-like element. In lines labeled “Logo,” the size of nucleotide characters is proportional to the frequency of occurrence of this nucleotide at this position; the height of the character stack signifies the information content (in bits) of the sequences at that position (Beitz 2000). The last line of the alignments shows the predicted consensus structure in bracket-dot notation (Wilm et al. 2008). In the structure model of 5S rRNA, binding sites of TFIIIA and ribosomal protein L5 are shaded in gray. In the structure model of the 5S-rRNA-like element, sequence regions identical in 5S rRNA and the 5S-rRNA-like element are highlighted in gray.
Phylogenetic analyses of 5S rRNAs and 5S-rRNA-like elements
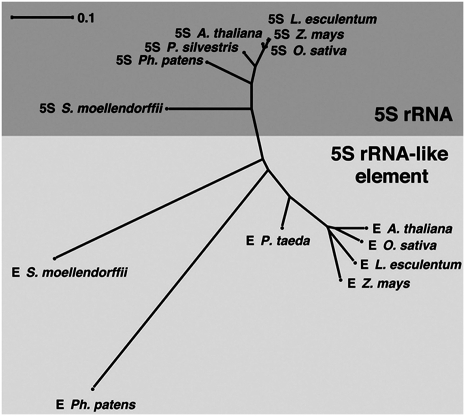
Sequence alignment (Supplemental Fig. 2C) and subsequent phylogenetic analysis shows a clear separation between the groups of 5S rRNAs and 5S-rRNA-like elements, with the sequences from P. patens and S. moellendorffii placed close to the center of the tree (Fig. 3). These results suggest that the 5S-rRNA-like element was derived from a 5S rRNA inserted into the TFIIIA gene of the common ancestor genome of all land plants.
Figure 3.
Phylogenetic relationship of plant 5S rRNAs (dark gray) and 5S-rRNA-like elements (light gray). The tree is based on the program SplitsTree4 (Huson and Bryant 2006), LogDet distance (calculated on basis of the alignment of 5S rRNAs and 5S-rRNA-like elements shown in Supplemental Fig. 2C, but after gap removal) and neighbor joining. Only the leaves with the sequences given in Figure 2 are labeled.
Coupling of the cassette exon AS event in TFIIIA mRNA and NMD
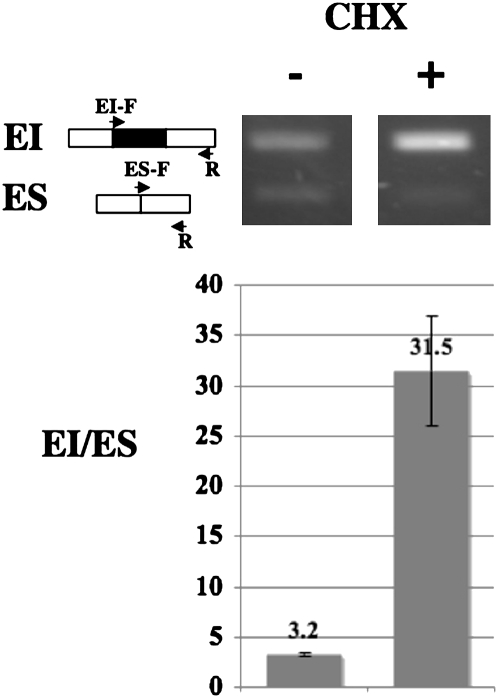
The EI mRNA isoforms of AtTFIIIA and OsTFIIIA harbor a PTC within the cassette exon. Cycloheximide (CHX), a known inhibitor of the NMD pathway (Beelman and Parker 1994), was applied to Arabidopsis seedlings and its effect on the abundance ratio of the EI to ES mRNA isoforms was examined. CHX treatment resulted in an increase in the ratio of EI to ES mRNA by almost tenfold (Fig. 4), which suggests that selective inclusion of the PTC-containing cassette exon may function to regulate TFIIIA protein expression by generating NMD-targeted isoforms. Furthermore, the conserved cassette exon is coincidental with a PTC in all land plants with available ESTs/cDNAs supporting the EI isoform (Supplemental Table 1), which indicates further that regulation of TFIIIA protein expression by coupling the nonproductive splicing of the PTC-containing cassette exon with the NMD pathway is also conserved in land plants.
Figure 4.
RT-PCR validation of accumulation of EI mRNA isoform after blocking NMD with cycloheximide (CHX) treatment for 4 h.
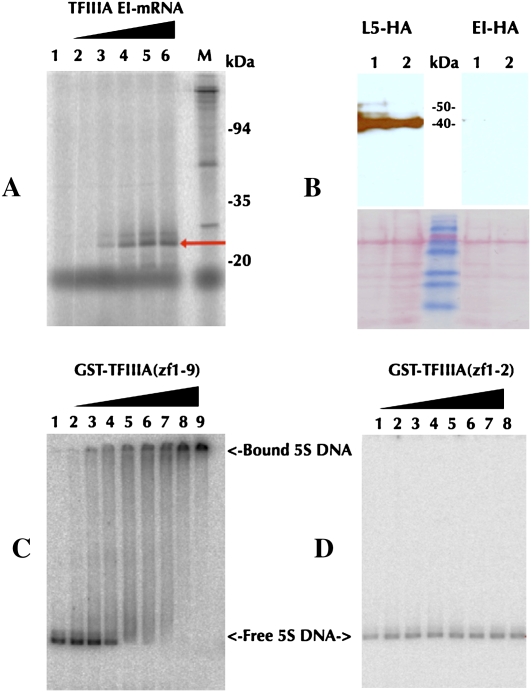
Multiple independent lines of experimental evidence further support RUST of the EI mRNA isoform. In vitro translation analysis indicated that the Arabidopsis EI mRNA isoform can produce a peptide of ∼15 kDa that may correspond to a truncated TFIIIA containing only zf1-2, which is expected as a result of the PTC. The longer product that would correspond to TFIIIA(zf3-9) through an alternate translation re-initiation process (Wang and Wessler 1998) could not be detected in vitro (Fig. 5A) or in vivo using Arabidopsis transgenic plants overexpressing a construct containing EI cDNA fused to a C-terminal HA-tag (Fig. 5B). Electrophoretic mobility shift assays demonstrated that the full-length recombinant GST-LeTFIIIA protein binds to 5S rDNA in subnanomolar concentrations (Fig. 5C), which is consistent with a previous report on GST-AtTFIIIA (Mathieu et al. 2003). In contrast, the recombinant protein of truncated tomato TFIIIA (GST-LeTFIIIA(zf1-2)) did not exhibit any binding activity to 5S rDNA (Fig. 5D).
Figure 5.
Characterization of the EI mRNA isoform of TFIIIA. (A) In vitro translation of Arabidopsis EI mRNA. Lanes (1–6) 0, 0.5, 1, 2, 4, and 8 μg EI mRNA, respectively, added in a cell-free wheat germ extract. (B) In vivo translation of EI. Western blot using anti-HA antibody (top) and SDS-PAGE analysis (bottom) of the overexpression lines using L5-cDNA (left) and EI-cDNA (right) fused with HA-tag. Electrophoretic mobility shift assay using GST-LeTFIIIA(zf1-9) (C) and GST-LeTFIIIA(zf1-2) (D). 5S DNA was titrated with increasing concentrations of GST-LeTFIIIA(zf1-9) (lanes 1–9: 0, 0.2, 0.4, 0.9, 1.8, 3.5, 7, 14, and 28 nM, respectively) or GST-LeTFIIIA(zf1-2) (lanes 1–8: 0, 14, 28, 55, 110, 220, 440, and 880 nM, respectively).
Molecular and physiological phenotypes of TFIIIA overexpression lines
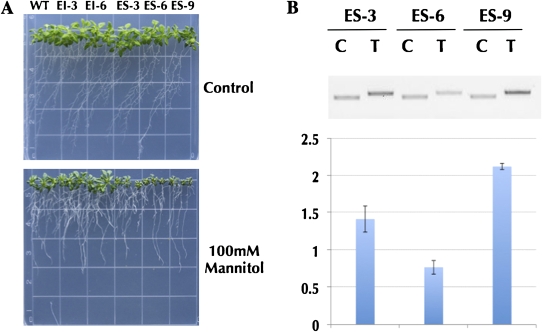
The common origin as well as the conserved alternative splicing pattern of TFIIIA genes in land plants may have important implications in both gene regulation and impact on plant fitness. To determine possible molecular and physiological consequences of perturbing the TFIIIA gene, we generated transgenic Arabidopsis plants overexpressing EI or ES isoforms of TFIIIA and tested their ability to withstand abiotic stresses. Consistent with the hypothesis that EI mRNA undergoes unproductive splicing, the EI-overexpressing lines (EI-3 and EI-6) have no obvious phenotype compared with wild-type plants under normal, osmotic, or salt stress conditions (Fig. 6A). Therefore, in vivo and in vitro studies indicate that the TFIIIA EI transcript isoform has no detectable biological function under the conditions tested, lending further support to the hypothesis that it is degraded through the NMD pathway and may not contribute to protein accumulation.
Figure 6.
Phenotypes of transgenic Arabidopsis lines overexpressing EI and ES isoforms of TFIIIA under osmotic stress. (A) Overexpressing TFIIIA-EI lines (EI-3,6) and TFIIIA-ES lines (ES-3, 6, 9) and Col-0 as wild type (WT) without (top) and with (bottom) treatment of 100 mM mannitol. (B) Relative accumulation levels of TFIIIA transgene mRNA (T) in ES-3, ES-6, and ES-9 using Tubulin 8 (TUB8) as a reference gene (C).
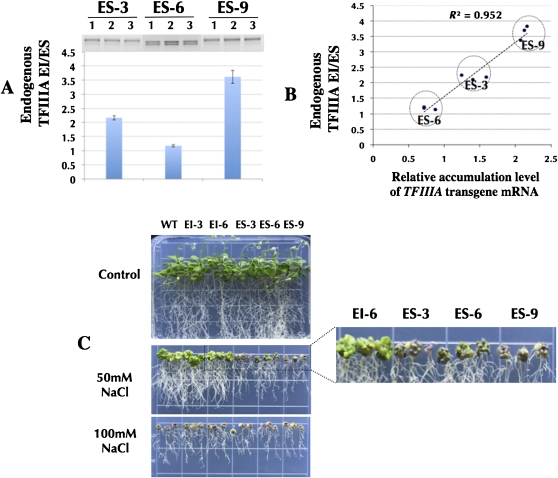
Among the independent transgenic lines overexpressing TFIIIA (ES isoform), three representative homozygote lines (ES-3, ES-6, ES-9) with different expression levels of the AtTFIIIA transgene were chosen for further investigation. The expression level of the transgene was the highest in ES-9 and lowest in ES-6 (Fig. 6B). Semiquantitative RT-PCR was used to examine the ratio of EI to ES mRNA, the splicing products of the endogenous TFIIIA pre-mRNA, in these overexpression lines. The endogenous TFIIIA EI/ES ratios were strongly positively correlated with the levels of the mRNA accumulation of TFIIIA transgenes in these lines (R2 = 0.952; Fig. 7). These results demonstrate that the increased expression of TFIIIA (and presumably the amount of TFIIIA protein) results in an increased accumulation of processed EI isoform of the endogenous TFIIIA mRNA. Thus, TFIIIA may affect the splicing of its own pre-mRNA by increasing the proportion of NMD-targeted mRNA isoforms in response to abundant TFIIIA. Similar to the regulation of expression of the mammalian SR protein gene SC35 by a proposed negative feedback loop mediated by RUST (Lareau et al. 2007), high levels of TFIIIA cause a greater fraction of the TFIIIA transcripts to be spliced into the unproductive isoform and then degraded through NMD, thereby down-regulating TFIIIA production.
Figure 7.
Strong accordance between molecular and phenotypic evidences in multiple independent transgenic Arabidopsis lines overexpressing TFIIIA under salt stress. (A) mRNA accumulation ratio of endogenous TFIIIA EI to ES isoform in ES-3, ES-6, and ES-9 lines. Quantification (bottom) with three replicates for each line. (B) The high positive correlation (R2 = 0.952) between the relative accumulation level of TFIIIA transgene mRNA and mRNA accumulation ratio of endogenous TFIIIA EI to ES isoform in ES-3, ES-6, and ES-9 lines with three replicates for each line. (C) Overexpression lines without (top) and with treatments of 50 mM NaCl (middle) and 100 mM NaCl (bottom).
Three Arabidopsis transgenic lines with different expression levels of transgenic AtTFIIIA provide an opportunity to study the functional significance of the homeostasis of TFIIIA. Under salt stress (50 mM NaCl; Fig. 7C), seedlings of the transgenic TFIIIA overexpression line with the highest accumulation of TFIIIA transgene mRNA (ES-9) exhibited the most severe stress phenotypes (i.e., the least tolerance) whereas seedlings of the transgenic TFIIIA overexpression line with the lowest accumulation of TFIIIA transgene mRNA (ES-6) exhibited the least severe stress phenotypes. The correlation between the salt stress phenotypes and the levels of TFIIIA transgene mRNA accumulation suggests that TFIIIA homeostasis is required to maintain the function of plant cells under salt stress.
Discussion
Autoregulation of TFIIIA expression by coupling alternative splicing of the 5S rRNA-derived exon with NMD
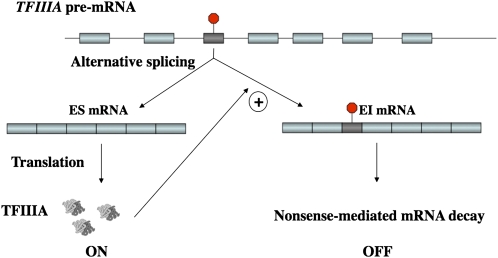
RUST has been proposed to be critical for autoregulation or homeostatic control of some splicing regulators (Lareau et al. 2007; Ni et al. 2007). In some members of the human family of splicing regulators, highly or ultraconserved elements are alternatively spliced and a strong coupling of AS and NMD in these genes is observed (Lareau et al. 2007; Ni et al. 2007). Comparative genomic analysis reveals that the most evolutionarily conserved regions in TFIIIA pre-mRNA are the 5S-rRNA-derived exonic regions, suggesting the presence of regulatory elements. We hypothesized that inclusion of a 5S-rRNA-derived exon in TFIIIA mRNA can trigger NMD (Figs. 4 and 5). Consistent with our hypothesis, the level of EI mRNA of AtTFIIIA is decreased in lines overexpressing UPF1 (an RNA helicase known to be involved in NMD) but is amplified 17-fold in upf1 mutants compared with wild-type plants (Yoine et al. 2006). The coupling of the cassette exon AS event in AtTFIIIA mRNA with NMD resembles the mechanism of regulation of mammalian SR proteins through nonproductive alternative splicing coupled with NMD proposed by Lareau et al. (2007). Although we do not know exactly how TFIIIA regulates itself, all currently available evidence supports a post-transcriptional negative feedback autoregulation model for plant TFIIIA genes, as illustrated in Figure 8. According to this model, when in abundance, TFIIIA interacts with its own pre-mRNA via the 5S-rRNA-like structural elements helix IV and loop E, thereby activating the inclusion of exon 3 during the splicing process and triggering NMD to inhibit synthesis of new TFIIIA. When TFIIIA protein levels are low, the PTC-containing exon is skipped, which results in an increase in the ES isoform and a consequent increase in production of new TFIIIA protein. While this model suggests that TFIIIA interacts directly with TFIIIA pre-mRNA, it is possible that other factors in addition to TFIIIA are involved in TFIIIA and 5S rRNA regulation. For example, 5S rRNA synthesis is also closely linked to ribosomal protein L5 in Xenopus oocytes (Pittman et al. 1999). Since the 5S-rRNA-like element also has the binding site for ribosomal protein L5, a contribution to TFIIIA regulation by this protein cannot be excluded but remains to be demonstrated. Another well-studied example of post-transcriptional regulation via negative feedback represents the RNA-binding protein AtGRP7 that is subject to circadian oscillations in Arabidopsis (Staiger et al. 2003). Overexpression of AtGRP7 promotes the use of a cryptic intronic 5′ splice site resulting in a PTC-containing AtGRP7 transcript that can be rapidly degraded, presumably by NMD (Schoning et al. 2007).
Figure 8.
Autoregulatory model for homeostatic maintenance of the level of TFIIIA in the plant cell via conserved coupling of the alternative splicing and NMD of TFIIIA gene.
The possibility that TFIIIA regulates its own splicing is very intriguing. Pairwise coexpression analysis of TFIIIA using large-scale microarray data across different tissues (http://bar.utoronto.ca/ntools/cgi-bin/ntools_expression_angler.cgi) (Toufighi et al. 2005) indicates that TFIIIA expression is highly correlated with the expression of CypRS64 (At4g32420) (R2 = 0.82) and CPSF160 (At5g51660) (R2 = 0.81; Supplemental Fig. 3). CypRS64 is a protein consisting of an N-terminal peptidyl–prolyl cis/trans isomerase domain and a C-terminal domain with many SR/SP dipeptides. CypRS64 has been demonstrated to interact with the splicing machinery in Arabidopsis and suggested to be functional in early steps of spliceosomal assembly (Lorkovic et al. 2004). CPSF160, a cleavage and polyadenylation specificity factor, has been shown to interact with U2 snRNA and provides evidence for the coupling of pre-mRNA 3′ processing and splicing (Kyburz et al. 2006). While it is possible that the observed correlation of expression of TFIIIA, CypRS64, and CPSF160 may be purely circumstantial, the notion that TFIIIA itself may play some role in the splicing processes, consistent with its observed role in influencing the processing of its own pre-mRNA, warrants further investigation.
The origin of 5S rRNA-derived exon within the TFIIIA gene in plants
Phylogenetic analysis of 5S rRNAs and 5S-rRNA-like elements in TFIIIA (Fig. 3) suggests that the 5S-rRNA-like element may have originated from a 5S rRNA element inserted into an intron of the TFIIIA gene. The occurrence of the 5S-rRNA-like element in the TFIIIA intron in a bryophyte (moss) and all vascular plants investigated including a lycophyte (S. moellendorffii), but not in green algae or other nonplant genomes, suggests that the origin of this insertion occurred after the separation of land plants and unicellular green algae and before the divergence of nonvascular and vascular plants in the Silurian (over 400 Mya) (Rensing et al. 2008). Widespread, highly conserved 5S-rRNA-like elements in TFIIIA genes among land plant species indicate that this insertion has been favored. The observation that the region containing the 5S-rRNA-like element shows better conservation than any other coding regions within the TFIIIA gene demonstrates the occurrence of the directional selection pressure on this element.
In all angiosperms investigated, the cassette exon of TFIIIA contains the whole 5S-rRNA-like element (Supplemental Table 1). This suggests that some cis-element(s) within the 5S-rRNA-like element may function as exonic splicing enhancers (Tian and Maniatis 1993). The moss TFIIIA EI isoform includes only 80% of the 5S-rRNA-like element present within the TFIIIA gene. This is presumably a result of a different splice site selection, which reflects the divergent evolution of exonized 5S-rRNA within TFIIIA genes during land plant evolution. To our knowledge, this is the first reported observation of 5S rRNA, a noncoding gene, being exonized into genic sequence in plant genomes. More interestingly, the exonization of 5S rRNA occurred within its own transcription factor (TFIIIA) with high conservation among all land plants investigated. Our concerted evolutionary and functional analyses indicate that this 5S-rRNA-derived exon might have been alternatively spliced before the divergence of nonvascular and vascular plants.
The conservation of 5S-rRNA-like elements as well as the alternative splicing pattern of TFIIIA genes in land plants may have played a role in the successful colonization of land by these organisms. Since water deficit and the associated osmotic stress are a major challenge for land plants, we tested the fitness of the homozygous transgenic Arabidopsis plants under osmotic stress by mannitol or salt. Indeed, a highly significant positive correlation between the molecular phenotypes and physiological phenotypes of AtTFIIIA overexpression lines was evident (Figs. 6 and 7), which reveals the functional importance of TFIIIA homeostasis for plants under abiotic stress via RUST. This may have contributed to the adaptation of these plants to the prevailing stress when their ancestors invaded land. To our knowledge, this is the first report of molecular exaptation, a preciously adapted sequence co-opted to serve a different function, associated with exonization in plants. In mammals, most cases of exonization–exaptation are derived from Alu elements and short interspersed repetitive elements (Bejerano et al. 2006; Sorek 2007). 5S rRNA exonization is clearly distinct from those cases. A recent report that Cassandra retrotransposons carry conserved 5S rRNA sequences in both long terminal repeats (LTRs) in widespread vascular plants (Kalendar et al. 2008) leads to the speculation that the ancient insertion of 5S rRNA could be mediated by retroposition of Cassandra retrotransposons. With more available plant genomes, deeper transcriptome sequencing and the power of comparative genomics, we expect more conserved AS events and more cases of coexonization–exaptation to be reported in plants.
Methods
Genome-wide identification of conserved cassette exon AS events
The pseudochromosomes of Arabidopsis (Version 5) and Oryza (Version 4) were downloaded from TAIR (www.arabidopsis.org) and TIGR (www.tigr.org/tdb/e2k1/osa1). The cDNA/EST data for both species were retrieved from GenBank on August 31, 2006, according to Wang and Brendel (2006). The PASA (Campbell et al. 2006) and GMAP software (Wu and Watanabe 2005) were used to identify cassette exon events. Reciprocal best BLAST matches were used to determine the orthologous gene pairs that both contain ES events. Only the event containing the orthologous cassette exon(s) is deemed as the conserved ES event (illustrated in Fig. 1).
Collection and gene structure prediction of TFIIIA orthologs
The protein and nucleotide sequences of AtTFIIIA and OsTFIIIA were used to BLAST against plant genome databases at www.phytozome.net and NCBI plant EST databases to identify genomic sequences for TFIIIA orthologs. Reciprocal best BLAST matches assured the orthology of TFIIIA if possible. We also cloned and sequenced the TFIIIA genomic and cDNA sequences in tomato (DQ882178-DQ882180), Arabidopsis, and Physcomitrella patens (moss) (EU574733–EU574735). GeneSeqer (Usuka et al. 2000) was used to seek the EST supporting evidence of gene structures and AS events for TFIIIA orthologs.
Phylogenetic analyses of 5S rRNA and 5S-rRNA-like elements
Alignments of 5S rRNA sequences (see Fig. 2A and Supplemental Fig. 2A) and of 5S-rRNA-like sequences (see Fig. 2B and Supplemental Fig. 2B) were generated with mafft (ginsi version 6.240) (Katoh et al. 2005) and hand-optimized with respect to a consensus secondary structure using ConStruct (Wilm et al. 2008). We created a common alignment by inserting gap-only columns into the individual alignments (shown in Supplemental Fig. 2A,B) to compensate for the unequal length of structural elements similar in both RNAs. The tree shown in Figure 3 was drawn using SplitsTree4 (Huson and Bryant 2006) with LogDet distances and neighbor joining. Nearly identical trees were obtained from the PHYLIP package (Felsenstein 1989) using DNADIST with Kimura's two-parameter model. Sequence logos were generated as described by Schneider and Stephens (1990).
In vitro translation of EI-mRNA of AtTFIIIA
Full-length EI AtTFIIIA mRNA (GenBank AC: AY054225) was transcribed in vitro according to the RiboMAX Protocol (Promega). In vitro translation in a cell-free wheat germ extract (Promega) was performed according to the manufacturer's instructions. De novo synthesized proteins were labeled with 35S-L-Methionine and subjected to SDS-PAGE. The dried gels were exposed on PhosphorImager screens, and bands were visualized on a Bioimager FAS 3000 (Fuji).
Cloning, expression, and purification of GST-LeTFIIIA and GST-LeTFIIIA(zf1-2)
The cDNAs of LeTFIIIA and LeTFIIIA(zf1-2) were cloned into pGEX-4T-3 GST-fusion expression vector (Amersham). The constructs were transformed in Escherichia coli Rosetta2 cells (Novagen) for expression. A modified protocol from Frangioni and Neel (1993) was used to improve the solubility of active GST fusion proteins and purification. After binding of the GST fusion proteins to 50% glutathione sepharose 4B bead suspension (Amersham), the proteins were eluted upon addition of elution buffer (50 mM Tris/HCl at pH 8.8, 10 mM reduced glutathione, 0.1% Triton X, 1 mM β-mercaptoethanol, 100 μM ZnCl2).
Electrophoretic mobility shift assay
GST-LeTFIIIA zf(1-2) and full-length GST-LeTFIIIA were mixed with a 32P-labeled 330-bp DNA fragment comprising the 5S rRNA (GenBank accession no. X55697), respectively. The reaction mixture (containing 20 mM Tris-HCl at pH 8.0, 100 mM KCl, 1 mM MgCl2, 0.1 mM ZnCl2, 0.004% BPB, 0.008% 2-Mercaptoethanol, 3% glycerin) was preincubated for 30 min at room temperature. Samples were cooled on ice and loaded onto a 5% polyacrylamide gel (43:1 acrylamide/bisacrylamide) containing 20 mM NaOAc, 8 mM Tris, pH 8.3 adjusted with glacial acetic acid. Electrophoresis was performed in Hoefer SE600 apparatus at 50 V/cm and 4°C for 1 h. Gels were dried and exposed on PhosphorImager screens.
Overexpression constructs and plant transformation
The cDNAs for EI- and ES-mRNA isoforms of AtTFIIIA were amplified by RT-PCR, and the PCR products were cloned into the pCR8-TOPO vector (Invitrogen) and then cloned into the pMDC32 through LR clonase recombination (Invitrogen). The cDNAs of ribosomal protein L5 (GenBank accession no. AY186611) and EI isoform with the elimination of stop codons were also cloned into p35S-BAR-FH vector with an HA-tag at the C-terminal. Plant transformation was performed with Agrobacterium tumefaciens GV3101. Hygromycin- and Basta-resistant plants transformed by the pMDC32- and p35S-BAR-FH-derived constructs were validated by RT-PCR, respectively. Further screening identified T2 homozygote plants for phenotyping and quantitative RT-PCR.
Plant material and growth conditions
Arabidopsis Col-0 was used as control for all experiments. Two-week-old seedlings were treated with 10 μM cycloheximide for 4 h. For mannitol and NaCl treatment, one-week-old seedlings grown on vertically incubated one-half MS agar plates were transferred to one-half MS supplemented with different concentrations of mannitol or NaCl. Pictures were taken three (for mannitol) or four (for NaCl) weeks after the transfer.
RNA preparation and quantitative RT-PCRs
RNA was isolated from two-week-old seedlings using RNeasy Plant mini Kit (QIAGEN), and three seedlings were pooled as each of three biological replicates. SuperScript III Reverse Transcriptase and oligo(dT) primer (Invitrogen) were used for synthesis of the first-strand cDNA. Combined with the same reverse primer (TGCAGACAACCTGCTTCTGA), forward primers CATTCGCTCGAGAGATCTTTTAC and CGCTCGAGGAAATAAATAGCC were used to amplify both isoforms of AtTFIIIA. TUB8 was used as reference gene in semiquantitative RT-PCR (TUB8 primers: CGTGGATCACAGCAATACAGAGCC and CCTCCTGCACTTCCACTTCGTCTTC). An Alphaimager 2200 gel doc system (Alpha Innotech) and ImageJ software (http://rsbweb.nih.gov/ij/) were used for PCR product (band) quantification. The final quantification was obtained after subtracting background intensity.
Acknowledgments
We thank Joseph Cho and Ralph S. Quatrano at Washington University (WU) for moss RNA isolation, and Kevin Lutke and Hao Peng at Donald Danforth Plant Science Center (DDPSC) for their assistance on plant transformation and screening of transgenic plants. We are indebted to Elke Reinartz for purification of recombinant proteins. We also thank Craig Pikaard at WU, Ruth Davenport at DDPSC, Detlev Riesner at Heinrich-Heine-Universität, Karen M. McGinnis at Florida State University, and Gordon Burleigh and David Oppenheimer at the University of Florida for their discussion and comments. This work was supported by awards from the National Science Foundations Plant Genome Program (DBI-0501758), The National Research Initiative (NRI) Plant Genome Program of the USDA Cooperative State Research, Education and Extension Service (CSREES) to W.B.B. (20078-35300-18460), and funds from the Donald Danforth Plant Science Center.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GenBank (http://www.ncbi.nlm.nih.gov/Genbank/) under accession nos. DQ882178–DQ882180, EU574733–EU574735, EU924184, EU918200, EU919737.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.086876.108.
References
- Barbazuk W.B., Fu Y., McGinnis K.M. Genome-wide analyses of alternative splicing in plants: Opportunities and challenges. Genome Res. 2008;18:1381–1392. doi: 10.1101/gr.053678.106. [DOI] [PubMed] [Google Scholar]
- Beelman C.A., Parker R. Differential effects of translational inhibition in cis and in trans on the decay of the unstable yeast MFA2 mRNA. J. Biol. Chem. 1994;269:9687–9692. [PubMed] [Google Scholar]
- Beitz E. TeXshade: Shading and labeling of multiple sequence alignments using LaTeX2ε. Bioinformatics. 2000;16:135–139. doi: 10.1093/bioinformatics/16.2.135. [DOI] [PubMed] [Google Scholar]
- Bejerano G., Lowe C.B., Ahituv N., King B., Siepel A., Salama S.R., Rubin E.M., Kent W.J., Haussler D. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature. 2006;441:87–90. doi: 10.1038/nature04696. [DOI] [PubMed] [Google Scholar]
- Campbell M.A., Haas B.J., Hamilton J.P., Mount S.M., Buell C.R. Comprehensive analysis of alternative splicing in rice and comparative analyses with Arabidopsis. BMC Genomics. 2006;7:327. doi: 10.1186/1471-2164-7-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassiday L.A., Maher L.J., III Having it both ways: Transcription factors that bind DNA and RNA. Nucleic Acids Res. 2002;30:4118–4126. doi: 10.1093/nar/gkf512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaw S.M., Chang C.C., Chen H.L., Li W.H. Dating the monocot-dicot divergence and the origin of core eudicots using whole chloroplast genomes. J. Mol. Evol. 2004;58:424–441. doi: 10.1007/s00239-003-2564-9. [DOI] [PubMed] [Google Scholar]
- Engelke D.R., Ng S.Y., Shastry B.S., Roeder R.G. Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell. 1980;19:717–728. doi: 10.1016/s0092-8674(80)80048-1. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Mathematics vs. evolution: Mathematical evolutionary theory. Science. 1989;246:941–942. doi: 10.1126/science.246.4932.941. [DOI] [PubMed] [Google Scholar]
- Frangioni J.V., Neel B.G. Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal. Biochem. 1993;210:179–187. doi: 10.1006/abio.1993.1170. [DOI] [PubMed] [Google Scholar]
- Golovkin M., Reddy A.S. Structure and expression of a plant U1 snRNP 70K gene: Alternative splicing of U1 snRNP 70K pre-mRNAs produces two different transcripts. Plant Cell. 1996;8:1421–1435. doi: 10.1105/tpc.8.8.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlach J., Raesecke H.R., Abel G., Wehrli R., Amrhein N., Schmid J. Organ-specific differences in the ratio of alternatively spliced chorismate synthase (LeCS2) transcripts in tomato. Plant J. 1995;8:451–456. doi: 10.1046/j.1365-313x.1995.08030451.x. [DOI] [PubMed] [Google Scholar]
- Griffin D., Penberthy W.T., Lum H., Stein R.W., Taylor W.L. Isolation of the B3 transcription factor of the Xenopus TFIIIA gene. Gene. 2003;313:179–188. doi: 10.1016/s0378-1119(03)00678-4. [DOI] [PubMed] [Google Scholar]
- Hori K., Watanabe Y. Context analysis of termination codons in mRNA that are recognized by plant NMD. Plant Cell Physiol. 2007;48:1072–1078. doi: 10.1093/pcp/pcm075. [DOI] [PubMed] [Google Scholar]
- Huson D.H., Bryant D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Kalendar R., Tanskanen J., Chang W., Antonius K., Sela H., Peleg O., Schulman A.H. Cassandra retrotransposons carry independently transcribed 5S RNA. Proc. Natl. Acad. Sci. 2008;105:5833–5838. doi: 10.1073/pnas.0709698105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyna M., Lopato S., Voronin V., Barta A. Evolutionary conservation and regulation of particular alternative splicing events in plant SR proteins. Nucleic Acids Res. 2006;34:4395–4405. doi: 10.1093/nar/gkl570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Kuma K., Toh H., Miyata T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyburz A., Friedlein A., Langen H., Keller W. Direct interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 3′ end processing and splicing. Mol. Cell. 2006;23:195–205. doi: 10.1016/j.molcel.2006.05.037. [DOI] [PubMed] [Google Scholar]
- Lareau L.F., Inada M., Green R.E., Wengrod J.C., Brenner S.E. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Green R.E., Brenner S.E. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl. Acad. Sci. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorkovic Z.J., Lopato S., Pexa M., Lehner R., Barta A. Interactions of Arabidopsis RS domain containing cyclophilins with SR proteins and U1 and U11 small nuclear ribonucleoprotein-specific proteins suggest their involvement in pre-mRNA splicing. J. Biol. Chem. 2004;279:33890–33898. doi: 10.1074/jbc.M400270200. [DOI] [PubMed] [Google Scholar]
- Maquat L.E. Nonsense-mediated mRNA decay: Splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell Biol. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- Mathieu O., Yukawa Y., Prieto J.L., Vaillant I., Sugiura M., Tourmente S. Identification and characterization of transcription factor IIIA and ribosomal protein L5 from Arabidopsis thaliana. Nucleic Acids Res. 2003;31:2424–2433. doi: 10.1093/nar/gkg335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J.Z., Grate L., Donohue J.P., Preston C., Nobida N., O'Brien G., Shiue L., Clark T.A., Blume J.E., Ares M., Jr Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes & Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham H.R., Brown D.D. A specific transcription factor that can bind either the 5S RNA gene or 5S RNA. Proc. Natl. Acad. Sci. 1980;77:4170–4174. doi: 10.1073/pnas.77.7.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penberthy W.T., Griffin D., Hall R.K., Taylor W.L. The Xenopus B2 factor involved in TFIIIA gene regulation is closely related to Sp1 and interacts in a complex with USF. Gene. 2003;305:205–215. doi: 10.1016/s0378-1119(03)00384-6. [DOI] [PubMed] [Google Scholar]
- Pfaff S.L., Taylor W.L. Characterization of a Xenopus oocyte factor that binds to a developmentally regulated cis-element in the TFIIIA gene. Dev. Biol. 1992;151:306–316. doi: 10.1016/0012-1606(92)90235-9. [DOI] [PubMed] [Google Scholar]
- Pfaff S.L., Taylor W.L. Xenopus TFIIIA gene transcription is dependent on cis-element positioning and chromatin structure. Mol. Cell. Biol. 1998;18:3811–3818. doi: 10.1128/mcb.18.7.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman R.H., Andrews M.T., Setzer D.R. A feedback loop coupling 5 S rRNA synthesis to accumulation of a ribosomal protein. J. Biol. Chem. 1999;274:33198–33201. doi: 10.1074/jbc.274.47.33198. [DOI] [PubMed] [Google Scholar]
- Reddy A.S. Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu. Rev. Plant Biol. 2007;58:267–294. doi: 10.1146/annurev.arplant.58.032806.103754. [DOI] [PubMed] [Google Scholar]
- Rensing S.A., Lang D., Zimmer A.D., Terry A., Salamov A., Shapiro H., Nishiyama T., Perroud P.F., Lindquist E.A., Kamisugi Y., et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- Schneider T.D., Stephens R.M. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoning J.C., Streitner C., Page D.R., Hennig S., Uchida K., Wolf E., Furuya M., Staiger D. Auto-regulation of the circadian slave oscillator component AtGRP7 and regulation of its targets is impaired by a single RNA recognition motif point mutation. Plant J. 2007;52:1119–1130. doi: 10.1111/j.1365-313X.2007.03302.x. [DOI] [PubMed] [Google Scholar]
- Sorek R. The birth of new exons: Mechanisms and evolutionary consequences. RNA. 2007;13:1603–1608. doi: 10.1261/rna.682507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R., Ast G., Graur D. Alu-containing exons are alternatively spliced. Genome Res. 2002;12:1060–1067. doi: 10.1101/gr.229302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger D., Zecca L., Wieczorek Kirk D.A., Apel K., Eckstein L. The circadian clock regulated RNA-binding protein AtGRP7 autoregulates its expression by influencing alternative splicing of its own pre-mRNA. Plant J. 2003;33:361–371. doi: 10.1046/j.1365-313x.2003.01629.x. [DOI] [PubMed] [Google Scholar]
- Stamm S., Ben-Ari S., Rafalska I., Tang Y., Zhang Z., Toiber D., Thanaraj T.A., Soreq H. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- Stewart W.N., Rothwell G.W. Paleobotany and the Evolution of Plants. Cambridge University Press; New York: 1993. [Google Scholar]
- Szymanski M., Barciszewska M.Z., Erdmann V.A., Barciszewski J. 5 S rRNA: Structure and interactions. Biochem. J. 2003;371:641–651. doi: 10.1042/BJ20020872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M., Maniatis T. A splicing enhancer complex controls alternative splicing of doublesex pre-mRNA. Cell. 1993;74:105–114. doi: 10.1016/0092-8674(93)90298-5. [DOI] [PubMed] [Google Scholar]
- Toufighi K., Brady S.M., Austin R., Ly E., Provart N.J. The botany array resource: e-Northerns, expression angling, and promoter analyses. Plant J. 2005;43:153–163. doi: 10.1111/j.1365-313X.2005.02437.x. [DOI] [PubMed] [Google Scholar]
- Usuka J., Zhu W., Brendel V. Optimal spliced alignment of homologous cDNA to a genomic DNA template. Bioinformatics. 2000;16:203–211. doi: 10.1093/bioinformatics/16.3.203. [DOI] [PubMed] [Google Scholar]
- Wang B.B., Brendel V. The ASRG database: Identification and survey of Arabidopsis thaliana genes involved in pre-mRNA splicing. Genome Biol. 2004;5:R102. doi: 10.1186/gb-2004-5-12-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B.B., Brendel V. Genome-wide comparative analysis of alternative splicing in plants. Proc. Natl. Acad. Sci. 2006;103:7175–7180. doi: 10.1073/pnas.0602039103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wessler S.R. Inefficient reinitiation is responsible for upstream open reading frame-mediated translational repression of the maize R gene. Plant Cell. 1998;10:1733–1746. doi: 10.1105/tpc.10.10.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilm A., Linnenbrink K., Steger G. ConStruct: Improved construction of RNA consensus structures. BMC Bioinformatics. 2008;9:219. doi: 10.1186/1471-2105-9-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T.D., Watanabe C.K. GMAP: A genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005;21:1859–1875. doi: 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- Xing Y., Lee C. Alternative splicing and RNA selection pressure–evolutionary consequences for eukaryotic genomes. Nat. Rev. Genet. 2006;7:499–509. doi: 10.1038/nrg1896. [DOI] [PubMed] [Google Scholar]
- Yoine M., Ohto M.A., Onai K., Mita S., Nakamura K. The lba1 mutation of UPF1 RNA helicase involved in nonsense-mediated mRNA decay causes pleiotropic phenotypic changes and altered sugar signalling in Arabidopsis. Plant J. 2006;47:49–62. doi: 10.1111/j.1365-313X.2006.02771.x. [DOI] [PubMed] [Google Scholar]