Abstract
Several studies have found evidence for more positive selection on the chimpanzee lineage compared with the human lineage since the two species split. A potential concern, however, is that these findings may simply reflect artifacts of the data: inaccuracies in the underlying chimpanzee genome sequence, which is of lower quality than human. To test this hypothesis, we generated de novo genome assemblies of chimpanzee and macaque and aligned them with human. We also implemented a novel bioinformatic procedure for producing alignments of closely related species that uses synteny information to remove misassembled and misaligned regions, and sequence quality scores to remove nucleotides that are less reliable. We applied this procedure to re-examine 59 genes recently identified as candidates for positive selection in chimpanzees. The great majority of these signals disappear after application of our new bioinformatic procedure. We also carried out laboratory-based resequencing of 10 of the regions in multiple chimpanzees and humans, and found that our alignments were correct wherever there was a conflict with the published results. These findings throw into question previous findings that there has been more positive selection in chimpanzees than in humans since the two species diverged. Our study also highlights the challenges of searching the extreme tails of distributions for signals of natural selection. Inaccuracies in the genome sequence at even a tiny fraction of genes can produce false-positive signals, which make it difficult to identify loci that have genuinely been targets of selection.
A powerful approach for finding genes affected by positive selection is to align the coding sequences of closely related species (for example human and chimpanzee) and more distantly related out-groups (for example macaque), and to screen these alignments for loci, where on one lineage there is a much higher rate of protein coding changes than is observed on other lineages (Hughes and Nei 1988; Nielsen et al. 2005; Bakewell et al. 2007; Rhesus Macaque Genome Sequencing and Analysis Consortium 2007). This test has been formalized as the study of the ratio of the rate of nonsynonymous substitutions per site that could harbor a nonsynonymous mutation (dN), to the rate of synonymous substitutions per site that could harbor a synonymous mutation (dS). If the value of ω = dN/dS is significantly greater than 1 in specific codons or on a specific lineage, the observation is interpreted as evidence of a history of positive selection (Nielsen 2001).
The macaque genome (Rhesus Macaque Genome Sequencing and Analysis Consortium 2007) provides a valuable reference for studies comparing the human and chimpanzee genomes (The Chimpanzee Sequencing and Analysis Consortium 2005), both by making it possible to determine the lineage on which a mutation occurred and by providing a way to estimate the degree of sequence conservation at each codon averaged over primate evolutionary history. Two recent analyses have scanned the genome to identify lists of putative positively selected genes (PSGs) in which there is statistically significant evidence of an acceleration in the rate of amino acid changes on the human or chimpanzee lineages since the two species diverged (Bakewell et al. 2007; Rhesus Macaque Genome Sequencing and Analysis Consortium 2007). Intriguingly, of the genes that met thresholds for being PSGs in human or chimpanzee, but not both, the studies found a significant excess on the chimpanzee side. For example, 59 of 61 genes in the study by Bakewell et al. (2007) that met a false discovery rate (FDR) threshold of <5% showed evidence of positive selection in chimpanzees; we call this set of genes “test set 1.” Similarly, 13 of the 14 genes in a second analysis that met a P-value threshold of <0.001 showed evidence of positive selection in chimpanzees (Rhesus Macaque Genome Sequencing and Analysis Consortium 2007); we call this set of genes “test set 2.” (Of concern, however, the lists of the most significant chimpanzee PSGs in the two studies did not overlap.) A third study aligned human, chimpanzee, mouse, rat, and dog genes and also found evidence for accelerated positive selection in chimpanzees (Arbiza et al. 2006).
A potential concern for dN/dS-based tests for positive selection, when applied on a genome-wide scale, is that they can be confounded by a small error rate in the data. Even if a great majority of bases are correctly determined, if there are a handful affected by errors, and especially if these errors are clustered within particular codons, a statistical signal can be generated that will cause these genes to artifactually appear as PSGs. A genome scan examines many thousands of genes, so that even if the overall error rate is low (<<1%), enough genes with false clusters of mutations could be observed to make it difficult to distinguish true signals. The concern is particularly acute for a comparison of human and chimpanzee. Due to the lower quality of the chimpanzee than that of the human genome sequence, more false-positive mutations are expected in the chimpanzee. The errors in the chimpanzee sequence can produce an artifactual signal of accelerated evolution on the chimpanzee lineage if they appear to reflect multiple nonsynonymous changes specific to the chimpanzee lineage. Moreover, such artifactual signals can be statistically significant in light of the low average divergence between these closely related species. This could provide a trivial explanation for the signal of accelerated chimpanzee evolution that has been suggested by several recent studies (Arbiza et al. 2006; Bakewell et al. 2007; Rhesus Macaque Genome Sequencing and Analysis Consortium 2007).
The two analyses that compared human, chimpanzee, and macaque genes applied multiple filters to increase the quality of their alignments and to minimize errors. Bakewell et al (2007) (who primarily analyzed the 4× chimpanzee assembly; panTro1), repeated their analyses in data sets in which they only analyzed nucleotides with chimpanzee sequence quality scores of at least Q0, Q10, and Q20 (corresponding to estimated error rates of <1, <0.1, and <0.01 per base pair) (Ewing et al. 1998). They found that the dN/dS ratio averaged across the genome achieved an asymptote with the most stringent of these filters. However, this method for assessing the efficacy of quality filtering may not be sufficient, as false-positive signals are expected to arise from the extreme tail of the statistical distribution, and genome averages are not very sensitive to the behavior of the extreme tail. Quality score filtering also cannot eliminate errors arising from misassembly of the chimpanzee genome or inaccuracies in multiple sequence alignment. The Rhesus Macaque Genome Sequencing and Analysis Consortium (2007) applied a different set of filters to their alignments using the more complete 6× chimpanzee assembly (panTro2). The most novel of these filters were synteny and frame-shift filters. The latter filter prohibited insertion/deletion changes (indels) that produced a frame shift in the alignment that was not compensated within 15 bases.
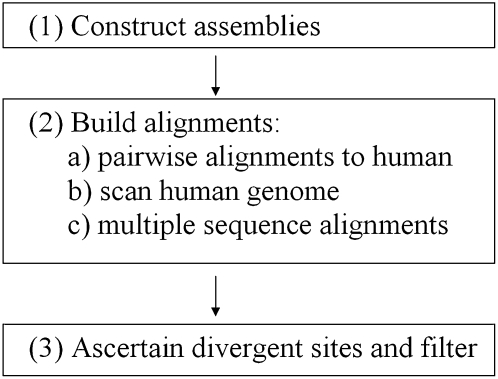
Here we reanalyzed genes that were highlighted as positively selected in chimpanzees in both test set 1 and test set 2 (see Methods). We implemented a bioinformatics procedure (Fig. 1) whose goal was to generate aligned bases of high reliability, even at the expense of a loss of some exon coverage. The procedure had three steps:
(1) We used the ARACHNE genome assembler to generate a de novo genome assembly of chimpanzee, corresponding to about 7× coverage of the genome since it used approximately the same raw data as the panTro2 6× assembly, but also included an additional ∼7 million sequencing reads that became available in public databases after the preparation of that assembly. We also generated a de novo assembly of macaque, which included about 6× coverage and corresponded to approximately the same raw data as the rheMac2 assembly (Jaffe et al. 2003; S. Gnerre, E. Lander, K. Lindblad-Toh, and D. Jaffe, in prep.). We modified ARACHNE so that we did not automatically set heterozygous sites within the sequenced genomes (single nucleotide polymorphisms [SNPs]) to be of low quality as is done in many current assemblies including chimpanzee. Instead, if we could identify a SNP with confidence, we picked one of the bases and allowed its score to be high (see Methods). A particular benefit of our bioinformatic procedure was that the genome assembly for each species was compared with human in a way that generated a syntenic map between that species and human, reducing the rate of misalignment.
(2) We generated alignments of each of the genomes with human, breaking long alignments into a series of small alignment problems that can be more reliably processed using conventional aligners (we used ClustalW, version 1.83; Larkin et al. 2007). The position of each of the smaller alignments was guided by the synteny map built during our reassembly of chimpanzee and macaque. This acted as a filter to prevent possible alignments to paralogous regions, in contrast to the more common reciprocal BLAST approach (Nembaware et al. 2002). There was an advantage in using our own assemblies, as it allowed us to customize the generation of consensus sequence in each species.
(3) We applied a series of filters to remove problematic regions. This included short alignments (<100 base pairs [bp]), regions near the ends of alignments, and near insertion/deletion polymorphisms (Methods). Alignments of genes could then be obtained by stripping out introns. We identified divergent sites only at nucleotides that passed a set of aggressive base quality filters. We required the quality score of every nucleotide used in analysis to be at least Q30, all bases within five nucleotides to have a quality score of at least Q20, and no base to be in a hypermutable CpG dinucleotide.
Figure 1.
Alignment pipeline. Flowchart of our bioinformatic procedure for generating multiple sequence alignments. For non-human species in step 1, publicly available traces are turned into genome assemblies using ARACHNE (Jaffe et al. 2003). This allows us to construct a synteny map and to use assembly information to guide the positioning of the non-human sequence on the reference (human) genome. In step 2, pairwise alignments of non-human sequence with its human counterpart are constructed using synteny information and information on the uniqueness of the alignment to filter out spurious alignments and regions of duplication. BLASTZ (Schwartz et al. 2003) is used to generate local alignments that are then combined to create a nonoverlapping pairwise alignment, allowing for the possibility of local inversions. The human genome is scanned to determine regions that have alignments to all the non-human species. Multiple sequence alignments are constructed using ClustalW (Larkin et al. 2007). In step 3, alignments are scanned to determine divergent sites, after which aggressive filters are applied (see Methods).
We applied this procedure to 49 of the chimpanzee PSGs from test set 1 and 10 of the chimpanzee PSGs from test set 2, corresponding to all the genes for which we obtained enough coverage in our alignments (after filtering) to permit useful comparison. If these genes genuinely reflect accelerated evolution on the chimpanzee lineage since the split from humans, we would expect to confirm a signal of accelerated evolution in chimpanzees at these genes by “branch-site” tests of evolution similar to the tests that the authors applied. We only replicated 1 of the 49 signals of accelerated evolution on the chimpanzee lineage that we were able to reanalyze from test set 1, and 5 of the 10 signals of accelerated evolution that we were able to reanalyze from test set 2. We also experimentally resequenced 10 of the regions where previous analyses had reported a signal of selection, while our reanalysis had not, and confirmed that our alignments were correct wherever a direct comparison could be made.
Results
We analyzed two data sets (sets of genes) for which putative chimpanzee PSGs had been identified (Bakewell et al. 2007; Rhesus Macaque Genome Sequencing and Analysis Consortium 2007). For each data set, we generated human–chimpanzee–macaque alignments using our bioinformatics pipeline (with macaque as the out-group) and identified nonsynonymous and synonymous divergent sites after applying all filters (see Methods).
Filtering eliminates almost all signals of accelerated chimpanzee evolution from the genes in test set 1
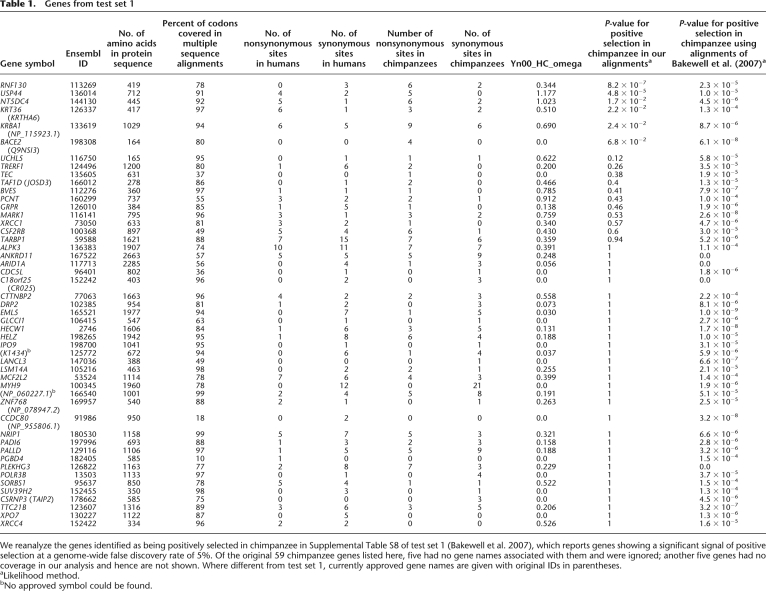
Table 1 shows our analysis of the putative chimpanzee PSGs from test set 1. From the 59 genes that the authors identified as chimpanzee PSGs, we obtained multiple sequence alignments for 49, and >80% amino acid coverage for over 30. Totaling the counts of nonsynonymous and synonymous sites across genes, we observed a higher ratio of nonsynonymous to synonymous sites in chimpanzees (0.86) than in humans (0.55). This is expected from ascertainment bias, since these genes were chosen from a set of thousands of genes as showing a signal of accelerated evolution on the chimpanzee lineage. However, we found that only one gene (RNF130) with nominally significant evidence for an excess of chimpanzee over human nonsynonymous sites (P = 0.02 in a binomial test for asymmetry of nonsynonymous sites, not corrected for multiple hypothesis testing).
Table 1.
Genes from test set 1
We reanalyze the genes identified as being positively selected in chimpanzee in Supplemental Table S8 of test set 1 (Bakewell et al. 2007), which reports genes showing a significant signal of positive selection at a genome-wide false discovery rate of 5%. Of the original 59 chimpanzee genes listed here, five had no gene names associated with them and were ignored; another five genes had no coverage in our analysis and hence are not shown. Where different from test set 1, currently approved gene names are given with original IDs in parentheses.
aLikelihood method.
bNo approved symbol could be found.
To powerfully detect signals of positive selection on the chimpanzee lineage, we applied test 2 of the “improved branch-site likelihood method” developed by Zhang and colleagues (Yang and Nielsen 2000, 2002; Zhang et al. 2005) using “Test 2” in the PAML software package version 4 (Yang 1997), which was also used to search for PSGs in Bakewell et al. (2007). Genes that show a strong signal or positive selection in chimpanzees, by this test, tend to be ones where multiple amino acid changes specific to the chimpanzee are observed in the same codon, a pattern that is unlikely to occur by chance since genuine chimpanzee-specific divergent sites only occur about once per 250 bp on average (and even less often in coding regions). To detect positive selection in chimpanzees, the improved branch-site likelihood method splits the genealogy into foreground (chimpanzee) and background (human and macaque) branches. It then calculates a likelihood ratio for the data being fit better by a null model in which all species have the same ω at each codon, or a selection model in which the foreground branch is allowed to have a class of codons with a higher ω. A likelihood ratio test (the difference between two times the log-likelihood of the data under the selection model to the analogous quantity under the null model) then provides a statistic that is χ2 distributed with one degree of freedom, which can be translated into a P-value (see Methods).
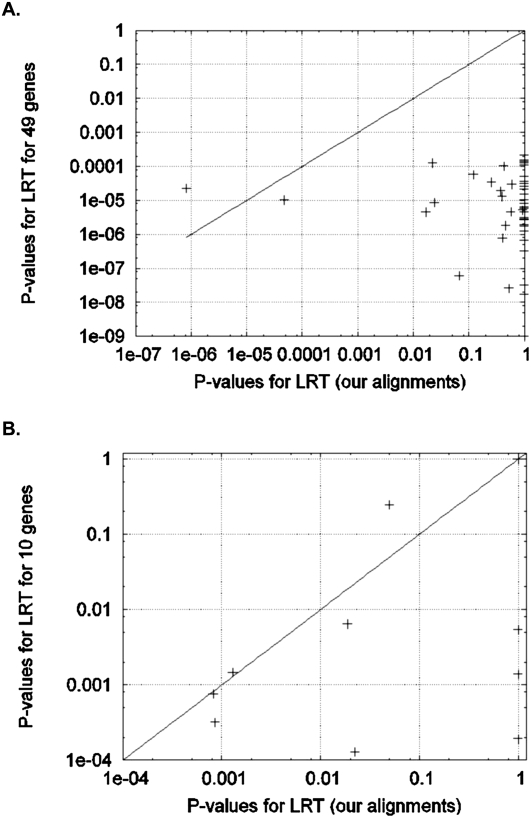
We performed the improved branch site likelihood test on the original alignments of test set 1 as well as our realignments of 49 genes from that study. P-values for positive selection in our analysis of the alignments of test set 1 are all very statistically significant (P << 0.05), as the authors reported previously (Bakewell et al. 2007). However, in the analysis of our new alignments, only two genes continue to produce significant signals: RNF130 (P = 8.2 × 10−7) and USP44 (P = 4.8 × 10−5) (Fig. 2A; Table 1). The signal at RNF130 is supported by six nonsynonymous chimpanzee-specific mutations in a region where many nucleotides were removed by our filters. This indicates that even our own filtering has probably not been aggressive enough at RNF130, and it may be a false positive in both our analysis and that of test set 1. The signal for positive selection at USP44 remains striking at P = 4.8 × 10−5 in our reanalysis. However, after applying a Bonferroni correction to account for the 13,888 genes scanned to obtain test set 1, this gene is not unambiguously flagged as a target of selection (P = 0.49).
Figure 2.
Positive selection in the chimpanzee lineage for each gene from each test set (see Methods). (A) Likelihood ratio test analysis of genes comparing our alignments with the alignments of test set 1: The majority of the 49 genes that we reanalyzed from test set 1 showed significant P-values (<0.0001) for positive selection in chimpanzee when we analyzed them using the alignments provided to us by the authors. However, our alignments indicate no positive selection in chimpanzee (P > 0.05) at all genes except for RNF130 and USP44. As described in the text, there is evidence that RNF130 is a false positive that emerges both from our analysis pipeline and that of test set 1. (B) The same analysis finds that five of the 10 signals of chimpanzee PSGs that we reanalyzed from test set 2 do not replicate after our realignment.
Detailed examination of the signals of positive selection from test set 1
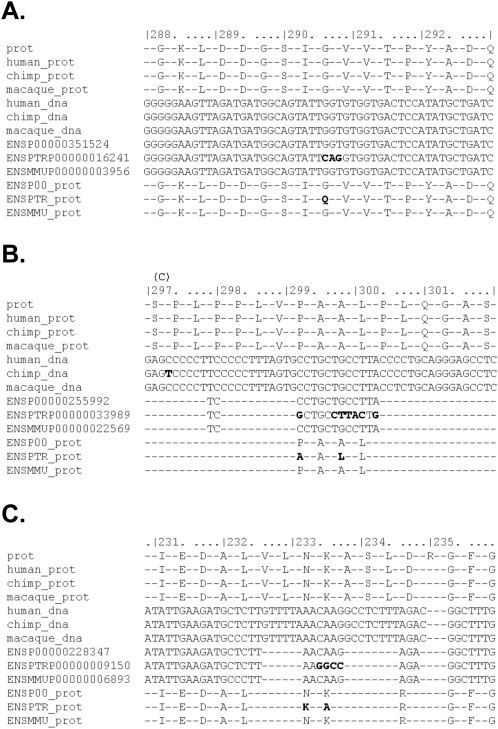
To understand the reason why many of the signals of accelerated evolution in chimpanzees disappear in our reanalysis, we created meta-alignments for each of the 49 genes that we reanalyzed (see Methods). These allowed us to compare the alignments in the published analyses to our own alignments that did not produce the same signals of selection (Fig. 3), and to diagnose the reasons for the discrepancies. The visualizations of the meta-alignments for all genes are available at our website, http://genepath.med.harvard.edu/~reich/Data%20Sets.htm.
Figure 3.
Sequence error revealed by alignments. Genome sequence errors in the template chimpanzee genome sequence used in test set 1 dominate the signal for positive selection in chimpanzee. We generated meta-alignments of each of the 49 genes that we reanalyzed for this study, which compare our alignments (with the new bioinformatics pipeline) with the alignments originally provided by the authors of the studies of test set 1 (Bakewell et al. 2007) (see Methods). Three examples from test set 1 are presented, where clusters of chimpanzee-specific divergent sites within a codon appear to be causing a false-positive signal of a chimpanzee PSG: (A) HELZ, (B) KRBA1 (NP_115923.1), and (C) POLR3B. In our realignment, these clusters of divergent sites disappear. There are 13 lines in each meta-alignment. Line 1 is the reference Ensembl human protein sequence for the gene. Lines 2–4 show the protein translations of our DNA alignments for human, chimpanzee, and macaque, respectively, and lines 5–7 show the DNA alignments themselves. Lines 8–10 show the corresponding alignment published in test set 1 (Bakewell et al. 2007). Lines 11–13 show translations of the DNA from test set 1. ENSP (lines 8,11) and ENSPTR (lines 9,12) refer to human and chimpanzee, respectively. Positions within the protein alignment that do not match the protein consensus are highlighted. Sequence differences are highlighted above the alignment. “C” indicates a synonymous chimpanzee divergent site. Macaque divergent sites are not highlighted.
To understand why there was an excess of signals of positive selection in test set 1 that we did not replicate in our reanalysis, we recall that the main signal that is tested by the improved branch site model is at least two chimpanzee-specific divergent sites within the same codon, which is not expected to occur by chance on a short lineage such as chimpanzee in the absence of selection. However, this kind of pattern can arise due to sequence errors, misassembly, or misalignment. Figure 3 shows three examples of genes that appear to be chimpanzee PSGs in test set 1, but where the signal disappears in our reanalysis. The discrepancies appear to be due to clusters of chimpanzee-specific mutations in the panTro1 4× assembly that do not replicate in our assembly (nucleotides 2906–2908 in HELZ, 2992–3002 in KRBA1 [NP_115923.1], and 2334–2337 in POLR3B; Fig. 3).
A clue about why we failed to replicate most of the 59 strongest signals of positive chimpanzee evolution from test set 1 comes from the observation that many of the sites contributing to the signals map to nucleotides of relatively low sequence quality in the panTro 4× assembly. When we raised the sequence quality filter to the Q30 minimum from our study (and at least Q20 for five bases in either direction), 49% of the 76 codons with two chimpanzee-specific nonsynonymous changes were removed. (This filter only removed 5% of the panTro1 4× genome as a whole.) However, a more stringent sequence quality filter is not by itself sufficient to account for the excess of chimpanzee PSGs observed in test set 1. To show this, the authors of test set 1 examined 233 genes that were nominally significant chimpanzee PSGs (P < 0.05) when analyzed using the panTro1 4× chimpanzee assembly, and repeated their analysis using the panTro2 6× assembly. The signal of positive selection replicated in 89% of these genes.
Having shown that the signals of accelerated evolution in test set 1 are enriched in less reliable sequences, we manually examined the 302 chimpanzee-specific divergent sites that were present in those alignments but not ours. We focused on codons where two chimpanzee-specific nonsynonymous changes were observed in test set 1, but not replicated in our alignments (Table 2):
(1) We found that 23% of these codons occurred at nucleotides where the underlying genome sequence differs between the 4× chimpanzee assembly used in test set 1 and our 7× assembly. The average quality score of these nucleotides was Q32 in panTro1, whereas only 3.7% of nucleotides in panTro1 as a whole have quality scores this low (Table 2). Encouragingly, 60% of these codons would have been removed from test set 1 simply by replacing their Q20 filter with our more stringent filter (Table 2, row a).
(2,3) We found that 14% of these codons occurred in regions that we filtered out during genome assembly (Table 2, row b) or post-processing (Table 2, row c). These sites are likely to mostly reflect errors caused by aligning nonorthologous sequence, and their impact could be reduced by more stringent filtering of assemblies and alignments.
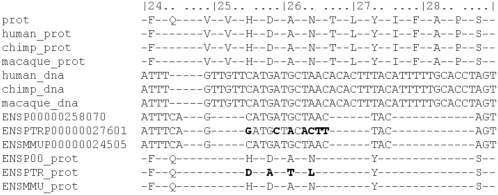
(4) We found that 30% of these codons mapped to regions where the local alignments used by the two studies are different. Based on manual inspection, these problems almost always occur near gaps in the raw input data used in test set 1 (Fig. 4). A potential solution to this class of false positives is to require very complete input sequences from all species, and to throw out alignments that show insertion/deletion polymorphisms (indels) within a minimum distance of divergent sites (Table 2, row d).
(5) We found that 33% of these codons occurred where different transcript definitions were used in the two studies, or where we were unable to confidently map the nucleotide in our alignment to that of test set 1. Even stringent filters cannot correct such ambiguities (Table 2, row e).
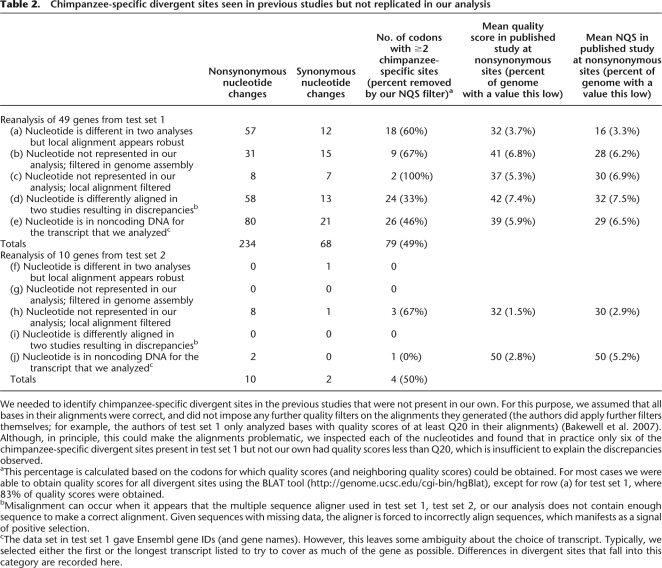
Table 2.
Chimpanzee-specific divergent sites seen in previous studies but not replicated in our analysis
We needed to identify chimpanzee-specific divergent sites in the previous studies that were not present in our own. For this purpose, we assumed that all bases in their alignments were correct, and did not impose any further quality filters on the alignments they generated (the authors did apply further filters themselves; for example, the authors of test set 1 only analyzed bases with quality scores of at least Q20 in their alignments) (Bakewell et al. 2007). Although, in principle, this could make the alignments problematic, we inspected each of the nucleotides and found that in practice only six of the chimpanzee-specific divergent sites present in test set 1 but not our own had quality scores less than Q20, which is insufficient to explain the discrepancies observed.
aThis percentage is calculated based on the codons for which quality scores (and neighboring quality scores) could be obtained. For most cases we were able to obtain quality scores for all divergent sites using the BLAT tool (http://genome.ucsc.edu/cgi-bin/hgBlat), except for row (a) for test set 1, where 83% of quality scores were obtained.
bMisalignment can occur when it appears that the multiple sequence aligner used in test set 1, test set 2, or our analysis does not contain enough sequence to make a correct alignment. Given sequences with missing data, the aligner is forced to incorrectly align sequences, which manifests as a signal of positive selection.
cThe data set in test set 1 gave Ensembl gene IDs (and gene names). However, this leaves some ambiguity about the choice of transcript. Typically, we selected either the first or the longest transcript listed to try to cover as much of the gene as possible. Differences in divergent sites that fall into this category are recorded here.
Figure 4.
Example of different alignment between test set 1 and our analysis (TEC). Different alignments between the analysis of test set 1 and our reanalysis are one of the largest contributors to the signal of chimpanzee-specific PSGs, accounting for 30% of codons with at least two chimpanzee-specific changes. By visual inspection of the alignments where these problems occur, we concluded that a major problem appears to be missing sequence in the input data used in the published study. This example shows the TEC gene, where the alignment based on the panTro1 4× assembly (prefixed by ENSPTRP) gives a strong signal of positive selection. However, our reanalysis (shown in lines “human_dna,” “chimp_dna,” and “macaque_dna”) show that there is an alignment solution without evidence for positive selection. A description of each line is given in the legend for Figure 3.
Examination of the signals of positive selection of test set 2
In the study of test set 2, the authors found that of 14 genes that they identified as PSGs on the chimpanzee or human lineage, 13 were on the chimpanzee side (one of these genes was identified as a PSG in both chimpanzee and human) (Rhesus Macaque Genome Sequencing and Analysis Consortium 2007). Of the chimpanzee PSGs, one was in duplication (MAGEB6) and was thus excluded by our bioinformatic procedure, and there were two for which we did not have adequate coverage for other reasons. Our reanalysis of the remaining genes is presented in Table 3. Applying the improved branch site likelihood method to these data to maximize comparability to test set 1, we found that only five genes were nominally significant and none was significant after applying a Bonferroni correction, correcting for 10,376 alignments tested. Thus, the evidence of an excess of chimpanzee, compared with human, PSGs is attenuated by our reanalysis of test set 2 just as in test set 1. However, the total number of discrepancies between the previous study and ours is reduced for this test set.
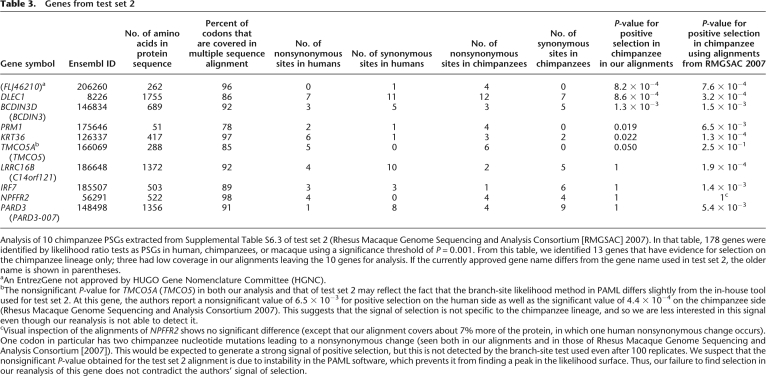
Table 3.
Genes from test set 2
Analysis of 10 chimpanzee PSGs extracted from Supplemental Table S6.3 of test set 2 (Rhesus Macaque Genome Sequencing and Analysis Consortium [RMGSAC] 2007). In that table, 178 genes were identified by likelihood ratio tests as PSGs in human, chimpanzees, or macaque using a significance threshold of P = 0.001. From this table, we identified 13 genes that have evidence for selection on the chimpanzee lineage only; three had low coverage in our alignments leaving the 10 genes for analysis. If the currently approved gene name differs from the gene name used in test set 2, the older name is shown in parentheses.
aAn EntrezGene not approved by HUGO Gene Nomenclature Committee (HGNC).
bThe nonsignificant P-value for TMCO5A (TMCO5) in both our analysis and that of test set 2 may reflect the fact that the branch-site likelihood method in PAML differs slightly from the in-house tool used for test set 2. At this gene, the authors report a nonsignificant value of 6.5 × 10−3 for positive selection on the human side as well as the significant value of 4.4 × 10−4 on the chimpanzee side (Rhesus Macaque Genome Sequencing and Analysis Consortium 2007). This suggests that the signal of selection is not specific to the chimpanzee lineage, and so we are less interested in this signal even though our reanalysis is not able to detect it.
cVisual inspection of the alignments of NPFFR2 shows no significant difference (except that our alignment covers about 7% more of the protein, in which one human nonsynonymous change occurs). One codon in particular has two chimpanzee nucleotide mutations leading to a nonsynonymous change (seen both in our alignments and in those of Rhesus Macaque Genome Sequencing and Analysis Consortium [2007]). This would be expected to generate a strong signal of positive selection, but this is not detected by the branch-site test used even after 100 replicates. We suspect that the nonsignificant P-value obtained for the test set 2 alignment is due to instability in the PAML software, which prevents it from finding a peak in the likelihood surface. Thus, our failure to find selection in our reanalysis of this gene does not contradict the authors' signal of selection.
To demonstrate how our filtering removes at least some of the signal of chimpanzee PSGs in test set 2, Figure 5 shows meta-alignments (see Methods) for the two genes in Table 3 where the P-values are significant in test set 2, but not significant in our realignments despite high exon coverage. At IRF7, the P-value changes from P = 1.4 × 10−3 to P = 1, and at LRRC16B (C14orf121) from P = 1.9 × 10−4 to P = 1. For both of these genes, the divergent sites that are contributing most strongly to the signal are adjacent to a break in the genomic alignments, which suggests that they may be less reliably aligned (in our bioinformatic pipeline, we apply a filter to remove bases within five positions of the end of a genomic alignment). Indeed, in Table 2 (row h), chimpanzee-specific divergent sites near breaks in the alignments are an important contributor to discrepancies: they account for most of the nonsynonymous changes in nucleotides that are present in the alignments of test set 2, but not in our reanalysis.
Figure 5.
Signals of selection from test set 2 removed by our filtering. Two genes, where strong signals of selection from test set 2, disappear in our reanalysis. For both of these genes, the chimpanzee-specific divergent sites in the alignments of test set 2 are adjacent to a break in the genomic alignments (Rhesus Macaque Genome Sequencing and Analysis Consortium 2007). In our filtering, we remove nucleotides within five positions of the end of a genomic alignment, which appears to abolish these signals. Table 2 shows that 80% of nonsynonymous chimpanzee-specific divergent sites that are present in test set 2, but not our reanalysis, are screened out by our bioinformatic procedure because they are close to the ends of alignments. There are 13 lines in each meta-alignment. Lines 1–7 are as described in Figure 3, but for test set 2. An asterisk indicates that the position is filtered. Macaque divergent sites are not highlighted.
Resequencing to test previously reported chimpanzee PSGs
As an experimental check on these results, we also attempted to resequence 17 loci where one of the previous studies found evidence for multiple nonsynonymous substitutions in the same codon on the chimpanzee lineage, providing strong evidence for positive selection in chimpanzees. We carried out PCR amplification and bi-directional sequencing on an ABI 3730 sequencer on a panel of eight humans and eight chimpanzees at these sites (including Clint, the chimpanzee used for the chimpanzee reference sequence). We obtained clear results at 10 loci (Table 4). Meta-alignments comparing our resequencing to previously reported data are presented at http://genepath.med.harvard.edu/~reich/Data%20Sets.htm.
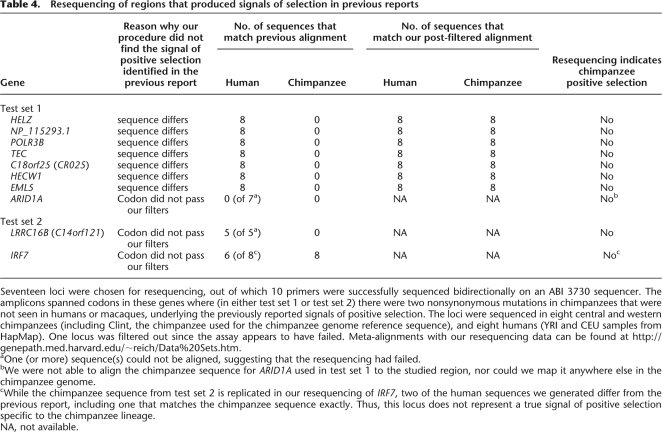
Table 4.
Resequencing of regions that produced signals of selection in previous reports
Seventeen loci were chosen for resequencing, out of which 10 primers were successfully sequenced bidirectionally on an ABI 3730 sequencer. The amplicons spanned codons in these genes where (in either test set 1 or test set 2) there were two nonsynonymous mutations in chimpanzees that were not seen in humans or macaques, underlying the previously reported signals of positive selection. The loci were sequenced in eight central and western chimpanzees (including Clint, the chimpanzee used for the chimpanzee genome reference sequence), and eight humans (YRI and CEU samples from HapMap). One locus was filtered out since the assay appears to have failed. Meta-alignments with our resequencing data can be found at http://genepath.med.harvard.edu/~reich/Data%20Sets.htm.
aOne (or more) sequence(s) could not be aligned, suggesting that the resequencing had failed.
bWe were not able to align the chimpanzee sequence for ARID1A used in test set 1 to the studied region, nor could we map it anywhere else in the chimpanzee genome.
cWhile the chimpanzee sequence from test set 2 is replicated in our resequencing of IRF7, two of the human sequences we generated differ from the previous report, including one that matches the chimpanzee sequence exactly. Thus, this locus does not represent a true signal of positive selection specific to the chimpanzee lineage.
NA, not available.
Our resequencing demonstrates that at seven codons with a direct discrepancy between our alignment and the previously reported alignment, our alignment was correct in every case (Table 4). In addition, there were three codons where a previous study had found a signal of selection that we had not replicated in our study because we had filtered out the codon as being in a region of unreliable sequence. Our resequencing showed that the signals of selection at these codons were problematic as well. Two of the codons (in ARID1A from test set 1 and LRRC16B [C14orf121] from test set 2) were clear false positives in the sense that our resequencing showed that the chimpanzee sequence always matched the human reference sequence (Table 4). The third codon (in IRF7) was a special case in that our resequencing data for all eight chimpanzees matched for the chimpanzee sequence reported in test set 2. However, the codon was polymorphic in humans, with one of the human alleles exactly matching the chimpanzee. Thus, this locus does not clearly harbor a signal of positive selection specific to chimpanzees (Table 4).
Discussion
Several analyses have identified an excess of genes that appear to have been positively selected in chimpanzees but not humans (Arbiza et al. 2006; Bakewell et al. 2007; Rhesus Macaque Genome Sequencing and Analysis Consortium 2007), which led Bakewell et al. (2007) to hypothesize that positive selection may have been more effective in chimpanzees than in humans, since we evolved from our common great ape ancestor. However, a concern is that these analyses are sensitive to errors in DNA sequence and alignment. If sequence and alignment errors are not removed by stringent filters, genome scans may artifactually detect an excess of signals of positive selection in the genome of lower quality.
In this analysis we analyzed genes that were previously identified as candidates for positive selection. We reassessed the signal of selection at these genes by building novel genome assemblies, constructing novel gene alignments, and then analyzing these for signals of selection. Any true signal should be robust to this procedure.
The genes identified as chimpanzee PSGs in the data set of test set 1 were obtained using the draft chimpanzee assembly (panTro1, which had about 4× coverage), whereas our reanalysis of these genes had the advantage of using more genomic data (about 7× coverage). Our re-evaluation of 49 of the genes with the strongest signals confirms only one as having a nominally significant evidence of being a chimpanzee PSG. While the majority of the strongest signals that the authors identified disappeared after our reanalysis, our results do not disprove the hypothesis of the authors of test set 1, that chimpanzees have experienced more positive selection than humans since the two species split (Bakewell et al. 2007). It is possible that a signal of accelerated evolution could still be found even after applying more aggressive filtering to the set of 233 genes that the authors of test set 1 identified by their P < 0.05 threshold. In light of our results, however, future studies should apply more stringent filters to the underlying sequence data and to multiple sequence alignments, in order to provide convincing evidence for the hypothesis that some species experienced more positive selection than others
The authors of test set 1 recognized that a higher rate of errors in the chimpanzee sequence is likely to be contributing to some of their signals. However, they also argued that it was not sufficient to explain their entire signal. To support this inference, they reported that the ratio of chimpanzee-to-human PSGs (considering all genes with P < 0.05 as PSGs for this analysis) decreased from 2.57 when using all bases with phred quality score Q ≥ 10 in the 4× assembly, to 1.51 when using all bases with Phred quality score Q ≥ 20, to 1.38 when focusing on the same set of genes but now using the higher quality 6× assembly. They pointed out that an excess of chimpanzee PSGs is still inferred when they increase the stringency of their filters, but this result is also concerning, as the ratio seems to be falling substantially for each increase in filter stringency that is applied.
Our reanalysis of the chimpanzee PSGs identified in test set 2 identified a smaller proportion of disagreements between our analysis and the previous report, which in part reflects the more reliable 6× panTro2 chimpanzee assembly that was used for this test set (Rhesus Macaque Genome Sequencing and Analysis Consortium 2007). Nevertheless, only 5 of the 10 genes that we were able to reanalyze were confirmed as being positively selected at nominal significance (P < 0.05), and we showed that one of these signals at LRRC16B (C14orf121) is a false positive when we sequenced the nucleotides underlying the signal in a panel of eight chimpanzees and eight humans. Thus, the evidence of accelerated chimpanzee evolution attenuates in this data set as well. A related observation was made by some of the same authors in an updated analysis of six-way human–chimpanzee–macaque–dog–mouse–rat (Kosiol et al. 2008), which found a more modest excess of chimpanzee, compared with human, PSGs. We conclude that there is no consistent evidence of more positive evolution on the chimpanzee than the human lineage since the two species split. However, there are interesting suggestions of this signal, and the question should be investigated in further analyses.
Methodologically, our manuscript is also interesting in that it highlights a general challenge to scans for selection in the age of whole genome sequences. Genome-wide scans search tens of thousands of genes for unusual patterns, and then focus on the extreme tail of outlying genes as candidates for selection. However, this strategy can be confounded by even a tiny rate of error in the underlying data, if the error can masquerade as the signal that is being sought. A small error rate can produce enough genes with apparently unusual signals, to outnumber true signals. A similar problem arises in genome-wide association studies to find disease genes, and medical geneticists routinely take rigorous measures to address these problems. However, the problem is not as well appreciated in studies of evolution.
It is important to point out that our alignment method may not be any better overall than that used to produce test set 1 or test set 2, and it is possible that if we applied our bioinformatic procedure to the entire genome, we would find our own extreme tail of chimpanzee PSGs. The goal of the present study is simply to highlight that genes that appear extremely unusual in a genome scan may often be artifacts of rare errors in the underlying data. Our analysis has demonstrated that it is important to verify input data, and to assess the robustness of results to sequence quality scores and alignment stringency. Finally, it is important to confirm a subset of loci in an independent resequencing study to verify the bioinformatics procedure and to confirm the strongest signals of selection.
Methods
Construction of primate alignments
We developed a novel bioinformatic procedure to generate and filter primate alignments, consisting of three major components: (1) We generate de novo genome assemblies for all of the compared species except for human (for human, we use the Build 36 reference sequence and assume that it is correct). Each assembly is then “assisted” by comparing to the human genome, which allows for the construction of larger supercontigs. Finally, consensus sequence is generated for each genome. (2) We use synteny information to remove problematic regions. Pairwise alignments are then constructed between each assembly and human, before building multiple sequence alignments. (3) We aggressively filter the data, using sequence quality scores to remove erroneous sites. While we lose a substantial fraction of our data, what we have left is more reliable.
De novo genome assemblies
We generated genome assemblies for all species except for human, using publicly available paired reads downloaded from the NCBI trace archive, and the ARACHNE genome assembly software (Jaffe et al. 2003).
Assisted assembly
We used the assisted assembly methodology (S. Gnerre, E. Lander, K. Lindblad-Toh, and D. Jaffe, in prep.) to improve the chimpanzee and macaque assemblies based on knowledge of the human genome sequence (Build 36). The idea is to use synteny information with a closely related species to improve the long-range connectedness of a de novo assembly that was made only using data from the species itself. To build an assisted assembly, we carry out a de novo assembly using ARACHNE, and then independently align the read pairs that form the raw material for the assembly to the reference genome (human). Because we know the position of each read in the supercontig, we can use the relative positioning of the reads on the reference genome to connect together supercontigs, for which there is only weak evidence of connectedness in the de novo assembly. For example, to avoid false joins in an assembly, which is commonly caused by chimeric read-pairs, a de novo ARACHNE assembly requires the presence of at least two links across a gap in a sequence before joining two supercontigs. For an assisted assembly, we can join two supercontigs based on the evidence from only a single-read pair spanning the gap, as long as the distance spacing of the read pair on the reference genome is compatible with the expected insert size (specified by the mean and standard deviation for the clones from the library). Thus, the alignments to another species are used to confirm that an insert is not chimeric.
New consensus code
The assemblies were generated with an improved version of the ARACHNE assembler (Jaffe et al. 2003). In the original version of ARACHNE, the genome was assumed to be haploid (the assembler had been designed to deal with inbred genomes), and the quality scores of all sites in which more than one haplotype was observed were set to zero (this happens, for example, for within-species SNPs).
The new consensus code allows for polymorphic genomes. It selects one of the two haplotypes, and uses a rigorously defined quality score to represent the consensus on a diploid genome interval. Specifically, the new consensus code is based on a two-step algorithm. The first step consists of generating an initial approximation for the bases of the consensus, by selecting a very small set of overlapping reads provided by the layout algorithm. This defines the backbone for the consensus. In the second step, all the reads in the consensus are realigned to the backbone. A sliding window of 12 bp (or more, in regions where reads match perfectly with each other) is examined. For each sliding window, the reads contained within the window are separated into groups, in which each group (ideally) is consistent with a single haplotype. Low quality regions of the reads are ignored, and the consensus is assigned low quality scores inside windows that appear to contain more than two groups (these are probably over-collapsed repeats). The highest scoring group is then used to define the consensus, both in terms of bases and quality scores. The advantages of this approach over existing assembly algorithms are that (1) SNPs within diploid genomes are assigned a realistic quality score that can, in fact, be very high rather than simply scoring zero (eliminating SNPs could bias evolutionary analyses) and (2) the base called at the SNP is more reliable.
Synteny information and construction of alignments
We chose the human genome assembly as a reference since this assembly is the most complete of the three species for which we have data. We defined an “anchor” for our synteny analysis as a pair of sequencing reads that belong to the same clone, and where both reads align uniquely, validly, and in opposite orientations to the reference (human) genome. This filters out regions of the genome that have duplications and could thus lead to spurious alignments. Each anchor read can be used to attach the supercontig to which the read belongs to the reference genome. If all anchors place and orient the supercontig coherently onto the reference, then the supercontig is anchored onto the reference in a syntenic manner. This synteny information then guides the pairwise alignment of a read onto the human reference, since its positioning is anchored by its supercontig. The synteny information removes the need to perform a reciprocal BLAST analysis (Moreno-Hagelsieb and Latimer 2008) to identify orthologous genes across species. The problem of such analyses is that they may be complicated by ambiguous alignments. The aggressive use of synteny information in our alignments is a considerable advantage of our procedure of reassembling each non-human genome and aligning it to the human reference sequence.
Comparison of chimpanzee and macaque assemblies with publicly available assemblies
We compared our genome assemblies of chimpanzee and macaque to public assemblies according to two metrics: (1) total coverage (i.e., the average number of times a particular nucleotide is represented) and (2) the N50 contig length, which is a length-weighted average of contig size, such that the average nucleotide in an assembly will appear in a contig of N50 size or greater. The 7.3× chimpanzee assembly constructed for this work has an N50 contig length of 43.5 kb (total contig length of 2.83 Gb). This compares with the publicly available panTro2 6× assembly, with an N50 contig length of 29 kb (total contig length of 2.97 Gb). The 6.2× macaque assembly has an N50 contig length of 34.9 kb (total contig length of 2.9 Gb). This compares with the publicly available 5.1× rheMac2 assembly with an N50 contig length of 25.7 kb (total contig length of 2.87 Gb). Further details about our assemblies are provided at our website, http://genepath.med.harvard.edu/~reich/Data%20Sets.htm.
To summarize, we found that there were several advantages in constructing our own assemblies: (1) We were able to incorporate reads that were deposited into the trace archives since the publication of the main assemblies. (2) We were able to use the ARACHNE assembly format, allowing us to use the analysis modules in the ARACHNE toolset (Jaffe et al. 2003). (3) We were able to generate consensus sequence that does not automatically set the quality score for heterozygous sites (SNPs) to zero, which greatly reduces bias that might be associated with analyzing heterozygous alleles in the diploid genome. (4) We were able to take advantage of a synteny-based approach for aligning supercontigs rather than reads, thereby reducing misalignments. The full de novo assemblies that we generated for chimpanzee and macaque are available on request.
Construction of pairwise alignments
Once assemblies were built, we identified sections of the genome on which to generate multiple sequence alignments. For consensus sequence in the non-human assemblies, we built pairwise alignments to the corresponding positions in the human assembly (using the positioning information of the sequence on its supercontig, and synteny information to determine where the supercontig aligns to the human assembly). We also required that this alignment generated a unique mapping between query and target bases, and also allowed local inversions. We could not find existing tools that fulfilled these criteria. In particular, SLagan (Brudno et al. 2003) generated overlapping alignments, and was not sufficiently fast to apply on a genome-wide scale. We constructed a tool that assimilates BLASTZ (Schwartz et al. 2003) alignments using a greedy approach. We resolved conflicts between overlaps in alignments by scoring the overlapping section using both flanking alignments and choosing the alignment with the better score.
Construction of multiple sequence alignments
Each pairwise alignment generates a series of breakpoints on the human reference genome sequence. Regions flanked by breakpoints where we obtained at least one sequence for each organism were considered fully populated and were used to define a region for multiple sequence alignment. If there were multiple sequences for a single organism, these were all retained for the alignment. RepeatMasker was used to filter low complexity regions and short tandem repeats. We then generated alignments with ClustalW (version 1.83) (Larkin et al. 2007).
Filtering
The alignments were scanned for divergent sites (ignoring alignments shorter than 100 bp) and classified according to divergent site class. In an attempt to avoid arbitrary filtering thresholds, we determined our filter thresholds based on the analysis of five-way alignments of human (H), chimpanzee (C), orangutan (O), gorilla (G), and macaque (M). We studied the behavior of five quantities: (1) the sum of divergent sites that clustered human and gorilla (HG) and chimpanzee and gorilla (CG) to the exclusion of all other species, (2) HC divergent sites, (3) the sum of H and C divergent sites, (4) G divergent sites, and (5) divergent sites not consistent with a genealogical tree. Each was plotted as a function of the following filters: (1) minimum quality score at the position of the divergent site, (2) minimum quality score in a window around the divergent site whose size we varied, (3) distance of divergent site from the nearest indel (insertion/deletion polymorphism), (4) distance of divergent site from the nearest other divergent site, and (5) distance of divergent site from the end of the alignment. Each quantity was normalized by the counts for the sum of O and M divergent sites. Thresholds for each filter were then set based on the point at which the quantities stabilized. The thresholds we eventually chose for analysis were: (1) a minimum quality score of 30 at a divergent site, (2) a minimum quality score of 20 for all positions within five bases of a divergent site, (3) a minimum distance to an indel of 10 bases, (4) no adjacent divergent sites, and (5) a minimum distance to the end of an alignment of five bases. These thresholds were judged to be valuable in cleaning data from the five-species alignments, and we then applied them to the three-species alignments of human, chimpanzee, and macaque.
Detecting positive selection using the improved branch-site likelihood method
We used the codeml program in PAML (version 4) (Yang 1997). The branch-site test that we use aims to detect positive selection that is sensitive to the presence of many changes within the same codon restricted to a particular lineage, which, if real, is a strong signal of positive selection (Zhang et al. 2005). The test is conservative overall but exhibits better power than branch-based tests. It requires that branches of the phylogeny are divided a priori into foreground (where selection may have occurred, in this case in chimpanzee) and background lineages (human and macaque). The likelihood ratio test then compares a model with selection on a particular branch with a null model where no selection has occurred in the foreground lineage, integrating the analysis over sites (codons). We used a χ2 distribution (with one degree of freedom) as suggested to obtain a P-value (Zhang et al. 2005). This is the same test as was used to detect positive selection in test set 1, and is similar to the test used in test set 2 (the software used there is not publicly available) (Bakewell et al. 2007; Rhesus Macaque Genome Sequencing and Analysis Consortium 2007). We ran both modelA and modelAnull for at least 10 replicates to attempt to find the global optimum in each of the likelihood surfaces. The maximum likelihood observed in these replicates was used for calculating a P-value.
Constructing genic alignments from genomic alignments
For a given gene, the Ensembl database (version 49) (Hubbard et al. 2007) was used to (1) obtain the position of the gene in the human assembly (Build 36), (2) to obtain the positions and sequence for the exons for a transcript, and (3) to obtain the resulting protein translation. Typically, a number of different transcripts were available. Either the first transcript was used, or, if there was a significant difference in the lengths of the transcripts, the longest was chosen. To extract the exonic regions from our genomic alignments, the exons were translated into all six possible reading frames and aligned to the protein sequence, and trimmed where necessary. This ensured that each section extracted from the genomic alignments exactly corresponds to the protein sequence (for human).
Generating meta-alignments between an alignment in a previous analysis and our own
To facilitate a comparison between an alignment of a set of sequences in one analysis and an alignment of a set of sequences in another analysis, we generated meta-alignments; that is, “alignments of alignments.” To do this, we took the human DNA sequence from our alignments and the previously published alignments, either test set 1 or test set 2, and used dynamic programming (needle from EMBOSS; Rice et al. 2000) to globally align them. This is used as a guide to stitch the two alignments together. Examples of the alignments are shown in Figure 3, and alignments for all 59 genes that we analyzed are available in http://genepath.med.harvard.edu/~reich/Data%20Sets.htm. Additionally, three-way comparisons of our alignments, those of test set 1 or test set 2, and newly available independent alignments from Ensembl (version 52) (Hubbard et al. 2007) are also here. The meta-alignments highlight differences between the published alignments and our realignments. The visual comparison allows us to diagnose the reason for the previously reported signal of accelerated evolution on the chimpanzee lineage.
Resequencing of regions to compare our study to previous reports
We chose 17 loci for resequencing where either test set 1 or test set 2 found multiple nonsynonymous changes in a codon in the chimpanzee lineage that were not observed in the human or macaque lineages, and that were not replicated by our own bioinformatic procedure (Bakewell et al. 2007; Rhesus Macaque Genome Sequencing and Analysis Consortium 2007). We obtained consensus sequence between our human and chimpanzee reference sequence for each of these loci, marking an “N” for each site that was discrepant between human and chimpanzee, and designed primers using the Primer3 software using all the default settings (Rozen and Skaletsky 2000). We carried out PCR and sequenced the amplicons bidirectionally using an ABI 3730 sequencing on a panel of eight chimpanzees and eight humans. The eight chimpanzees included Clint, the western chimpanzee used for the chimpanzee genome reference sequence, four other western chimpanzees (Gina, Yvonne, NA03448, and NA03450), and three central chimpanzees (Clara, Masuku, and Noemie). The eight humans were from HapMap (The International HapMap Consortium 2007) and included four YRI West Africans (NA18502, NA18870, NA19201, and NA19238) and four CEU of North European ancestry (NA07348, NA10831, NA10863, and NA12753). Six amplicons were not successfully sequenced, and one appeared to fail and hence we did not consider the locus. The resulting 10 loci were aligned to the meta-alignments of our data and the previously reported studies.
Acknowledgments
We thank M. Bakewell, G. Zhang, and A. Siepel for sharing with us the sequence alignments used in our study. We thank M. Bakewell, M. Clamp, M. Garber, A. Keinan, N. Patterson, A. Price, T. Sharpe, and G. Zhang for helpful comments and discussions. This work was supported by a Burroughs Wellcome Career Development Award in the Biomedical Sciences to D.R., a SPARC award from the Broad Institute of Harvard and MIT to D.R., and NIH grant U01-HG004168 to D.R.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GenBank (http://www.ncbi.nlm.nih.gov/Genbank/) under accession nos. FJ821202–FJ821288.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.086512.108.
References
- Arbiza L., Dopazo J., Dopazo H. Positive selection, relaxation, and acceleration in the evolution of the human and chimpanzee genome. PLoS Comput. Biol. 2006;2:288–300. doi: 10.1371/journal.pcbi.0020038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakewell M., Shi P., Zhang J. More genes underwent positive selection in chimpanzee evolution than in human evolution. Proc. Natl. Acad. Sci. 2007;104:7489–7494. doi: 10.1073/pnas.0701705104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brudno M., Do C., Cooper G., Kim M.F., Davydov E., Green E.D., Sidow A., Batzoglou S. LAGAN and Multi-LAGAN: Efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 2003;13:721–731. doi: 10.1101/gr.926603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Ewing B., Hillier L., Wendl M., Green P. Basecalling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Hubbard T.J.P., Aken B.L., Beal K., Ballester B., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., et al. Ensembl 2007. Nucleic Acids Res. 2007;35:610–617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A.L., Nei M. Patterns of nucleotide substitution at major histocompatibility class I loci reveals overdominant selection. Nature. 1988;335:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe D.B., Butler J., Gnerre S., Mauceli E., Lindblad-Toh K., Mesirov J.P., Zody M.C., Lander E.S. Whole-genome sequence assembly for mammalian genomes: ARACHNE 2. Genome Res. 2003;13:91–96. doi: 10.1101/gr.828403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosiol C., Vinar T., da Fonseca R.R., Hubisz M.J., Bustamante C.D., Nielsen R., Siepel A. Patterns of positive selection in six mammalian genomes. PLoS Genet. 2008;4:1–17. doi: 10.1371/journal.pgen.1000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. ClustalW and ClustalX version 2. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Moreno-Hagelsieb G., Latimer K. Choosing BLAST options for better detection of orthologs as reciprocal best hits. Bioinformatics. 2008;24:319–324. doi: 10.1093/bioinformatics/btm585. [DOI] [PubMed] [Google Scholar]
- Nembaware V., Crum K., Kelso J., Seioghe C. Impact of the presence of paralogs on sequence divergence in a set of mouse-human orthologs. Genome Res. 2002;12:1370–1376. doi: 10.1101/gr.270902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R. Statistical test of selection neutrality in the age of genomes. Heredity. 2001;86:641–647. doi: 10.1046/j.1365-2540.2001.00895.x. [DOI] [PubMed] [Google Scholar]
- Nielsen R., Bustamante C., Clark A.G., Glanowski S., Sackton T.B., Hubisz M.J., Fledel-Alon A., Tanenbaum D.M., Civello D., White T.J., et al. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 2005;3:70. doi: 10.1371/journal.pbio.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhesus Macaque Genome Sequencing and Analysis Consortium. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Rice P., Longden I., Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S., Misener S., editors. Bioinformatics methods and protocols: Methods in molecular biology. Humana; Totowa, NJ: 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Schwartz S., Kent J.W., Smit A., Zhang Z., Baertsch R., Hardison R.H., Haussler D., Miller W. Human-mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yang Z., Nielsen R. Estimating synonymous and non-synonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- Yang Z., Nielsen R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 2002;19:908–917. doi: 10.1093/oxfordjournals.molbev.a004148. [DOI] [PubMed] [Google Scholar]
- Zhang J., Nielsen R., Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]