Abstract
Viruses populate virtually every ecosystem on the planet, including the extreme acidic, thermal, and saline environments where archaeal organisms can dominate. For example, recent studies have identified crenarchaeal viruses in the hot springs of Yellowstone National Park and other high temperature environments worldwide. These viruses are often morphologically and genetically unique, with genomes that show little similarity to genes of known function, complicating efforts to understand their viral life cycles. Here, we review progress in understanding these fascinating viruses at the molecular level and the evolutionary insights coming from these studies.
The last decade has seen resurgent interest in the study of viruses that lie outside traditional agricultural and medical interests. One reason is the growing appreciation of the enormous abundance and impact of viruses on the greater biosphere. For example, the oceans are thought to contain ∼1031 viruses, a truly astronomical number (1), making viruses the most abundant biological entities in this ecosystem, where they catalyze turnover of 20% of the oceanic biomass per day (1). Remarkably, the virosphere has now been shown to extend to almost every known environment on earth, including the extreme acidic, thermal, and saline environments where archaeal organisms can be dominant. Thus, because of their abundance and variety, viruses are now thought to represent the greatest reservoir of genetic diversity on the planet (2).
A second reason to study archaeal viruses is a growing appreciation for the roles viruses play in evolution. Remarkably with >500 cellular genomes sequenced to date, most show a significant amount of viral or virus-like sequence within their genome, further evidence that viruses play a central role in horizontal gene transfer and help drive the evolution of their hosts. Roles for viruses in cellular evolution are also being considered. Current hypotheses contend that viruses have catalyzed several major evolutionary transitions, including the invention of DNA and DNA replication mechanisms (3), the origin of the eukaryotic nucleus (4), and thus a role in the formation of the three domains of life. In addition, there is also considerable interest in viral genesis and evolution in and of itself. To evaluate these hypotheses and to analyze evolutionary relationships among viruses, knowledge of viruses infecting the archaea is essential, yet these viruses are vastly understudied. Finally, interest in archaeal viruses stems also from the exceptional molecular insight viruses have traditionally provided into host processes; archaeal viruses are certain to provide new insights into the molecular biology of this poorly understood domain of life.
Pioneering studies by Wolfram Zillig et al. (5) identified the first archaeal viruses. Although initial studies suggested that viruses infecting the euryarchaea (principally halophiles and methanogens) were similar to head-tail bacteriophage, studies of viruses infecting the hyperthermophilic crenarchaea revealed morphologies suggesting new viral families. Indeed, work by several laboratories has led to the identification of seven new viral families infecting the crenarchaea, the Globuloviridae, Guttaviridae, Fuselloviridae, Bicaudaviridae, Ampullaviridae, Rudiviridae, and Lipothrixviridae (Fig. 1) (6, 7), with STIV3 (8) and STSV1 (9) awaiting assignment. All of these viruses contain double-stranded DNA genomes ranging in size from 13.7 to 75.3 kilobase pairs, encoding 31–74 ORFs. Although many package a circular genome, the filamentous Lipothrixviridae and rod-shaped Rudiviridae are notable exceptions and are the only viruses in any domain known to encapsidate linear double-stranded DNA. Although most crenarchaeal viruses are enveloped, the Rudiviridae are devoid of lipid, and with the exception of the Fuselloviridae, they employ a lytic life cycle, although only STIV and ATV (Bicaudaviridae) are known to cause cell lysis (11).4
FIGURE 1.
Morphological diversity in crenarchaeal viruses. A, clockwise, beginning at upper left: STIV (8), a PSV-like virus, Sulfolobus neozealandicus droplet-shaped virus (SNDV) (47), SSV1 (48), STSV1 (9), an ATV-like virus, an SIRV virus, and S. icelandicus filamentous virus (SIFV) (10). Micrographs of SIRV, PSV-like, and ATV-like viruses from Yellowstone National Park are the courtesy of M. J. Y. Other panels are reproduced, with permission, from Refs. 8–10, 47, and 48. B, cryoelectron microscopy reconstruction of the STIV particle (8) showing a cutaway view (20) of the T = 31 icosahedral capsid with turret-like projections that extend from each of the 5-fold vertices. Portions of the protein shell (blue) and inner lipid layer (yellow) have been removed to reveal the interior.
The exceptional morphology of these viruses has been reviewed (6, 7) and thus is only summarized here (Fig. 1). For the rod-shaped Rudiviridae, plugs are seen at both ends, from which three short tail fibers emanate, whereas the Lipothrixviridae show mop- or claw-like structures at both ends (6). Similarly, the non-tailed icosahedral viruses, STIV and euryarchaeal SH1, have large turrets or spikes that project from the surface (8, 12). In each case, these structures are thought to facilitate virus-host interactions. In contrast, other crenarchaeal viruses utilize a fusiform or lemon-shaped virion, a morphology unique to archaeal viruses. These fusiform viruses generally contain tail fibers or an extended tail on one end that is also involved in host recognition. For ATV, however, nascent particles are devoid of tails when released from the host (13). Remarkably, extended tails develop at both ends of the virion in an extracellular maturation process. Finally, Acidianus bottle-shaped virus (Ampullaviridae) shows an exceptional morphology that differs in its basic architecture from any known virus.
Genetic Diversity
These striking morphologies reflect the genetic diversity in these viral families. For example, while making a variety of new functional predictions, a recent comprehensive reanalysis concludes that these viral genomes are largely barren of recognizable features and that only a small pool of genes are shared by overlapping subsets of crenarchaeal viruses (11). The lack of sequence similarity to proteins with known function has, in turn, complicated efforts to elucidate viral life cycles, virus-host relationships, and the underlying genetics and biochemistry.
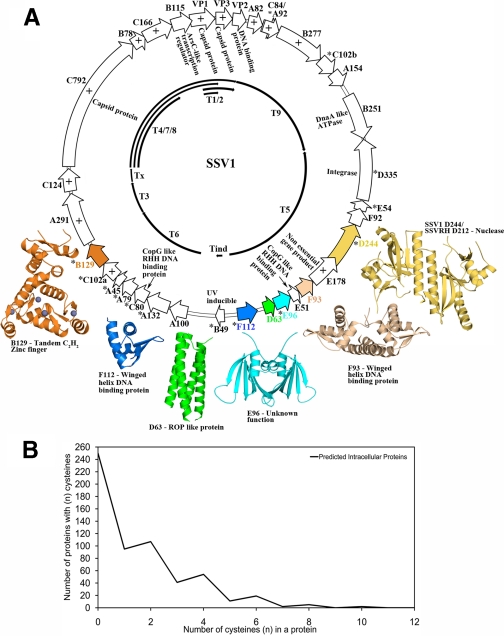
SSV1 is an excellent case in point. SSV1 is one of the best studied archaeal viruses and the type member of the Fuselloviridae, which are common in solfataric hot springs around the world. Its 60 × 90-nm fusiform virion (Fig. 1) packages a 15.5-kb circular double-stranded DNA genome. Palm et al. (15) reported the SSV1 genomic sequence in 1991, revealing 34 ORFs. Early analyses revealed only two ORFs similar to proteins of known function. Consistent with its lysogenic cycle, D335 is an integrase of the type I tyrosine recombinase family (15), although this activity is not essential (16), whereas B251 exhibits limited similarity to the ATP-binding domain of DnaA (17). With improved bioinformatic methods, additional similarities have been noted: E51 and C80 are potential CopG-like ribbon-helix-helix transcriptional regulators; C2H2 zinc-finger motifs are present in A45, A79, and B129; and B115 is annotated as a helix-turn-helix-type transcriptional regulator (11, 18, 19). However, 26 of 34 ORFs (∼75%) are not reliably identified by bioinformatic approaches. Thus, greater insight into these viral gene products is essential for a deeper comprehension of SSV1 and crenarchaeal viruses in general.
The Viral Particle
Purified virus can be analyzed by gel-based mass spectrometry or N-terminal sequencing to identify the structural and packaged proteins (6, 19–25). For example, N-terminal sequencing identified three SSV1 proteins, VP1, VP2, and VP3 (24). VP1 and VP3 are hydrophobic proteins embedded within the viral envelope, whereas VP2 is a packaged DNA-binding protein. Subsequent mass spectrometry analysis identified two additional proteins, C792 (a predicted membrane protein) and D244 (19). For STIV, a host protein packaged with the viral DNA was also identified (20). Lipid content can also be assessed using thin-layer chromatography (6, 21, 22, 25) or mass spectrometry (20, 25). Selective incorporation of host lipids into the viral envelope is observed for euryarchaeal SH1 (25) and for STIV (20), where complete viral assembly appears to take place within the cytosol in a membrane-independent process.4
High resolution cryoelectron microscopy single particle reconstructions have provided significant insight into archaeal viruses. For example, work with STIV has allowed the overall organization of the viral particle to be visualized in significant detail (8). The STIV reconstruction provided intricate details on the structure of the protruding turrets and clearly differentiated the internal lipid layer (Fig. 1B, yellow) from the external capsid proteins (blue). It also allowed the crystal structure of the major capsid protein to be placed within the T = 31 lattice of the viral particle, suggesting an electrostatic interaction between the negatively charged lipid layer and positively charged C terminus of the major capsid protein (26). Reconstruction of the euryarchaeal virus SH1 showed a clear structural relationship to STIV (12). The virus particle is built upon a T = 28 lattice that also includes an internal lipid layer and exterior surface turret-like projections. Notably, the structure of the SH1 major capsid protein is composed of a single β-barrel domain, providing an evolutionary link to the double β-barrel coat protein motif found in STIV.
Gene Expression
Insight into viral life cycles comes from transcriptome analysis during infection and viral production. Among temperate viruses, the Rudiviridae and Fuselloviridae are the best studied. For the Rudiviridae, a rather simple transcription pattern is found, where few genes exhibit temporal regulation (27). In addition, a host encoded transcription factor, Sta1, is capable of directing viral transcription (28). For SSV1, initial studies identified nine polycistronic transcripts (T1–T9) along with several regulatory elements, including a promoter for a UV-inducible transcript (T-ind) that lacks a TATA box (29, 30). Several transcripts share a common start site but result in transcripts of different lengths due to terminator read-through. These include the T1/2 and T4/7/8 transcripts. In the absence of UV induction, the transcription pattern for SSV1 appears relatively simple, with most transcripts produced constitutively (29). However, a recent microarray study (18) shows a chronological transcription cycle following UV irradiation, beginning with expression of T-ind. This is followed by early expression of T5 and T6 and then the shortly delayed expression of T9, whereas the T1/2, T3, and T4/7/8 transcripts are up-regulated at a later time point. These late stage transcripts encode at least four proteins found in the purified virion; VP1, VP2, and VP3 are encoded by T1/2, and C792 is encoded by the T4/7/8 transcripts. The T8 transcript also includes B115, a predicted helix-turn-helix-type transcriptional regulator that is the last gene to show significant up-regulation, suggesting that it may down-regulate SSV1 genes as viral replication is completed (18). These data also suggest that UV-enhanced transcription is not linked to an SOS-like response, as is common in bacteria (18).
STIV has also been studied with microarrays (31). Although transcript levels for all ORFs peak at 24 h post-infection, there is at least some temporal control. Transcripts for nine early genes were detected at 8 h post-infection. By 16 h, most of the viral genes were significantly expressed, including all of the known structural genes. However, three genes were not detected until 24 h post-infection, including the putative transcriptional regulator F93 (32). It will be interesting to determine whether F93 or the other late gene products down-regulate expression of the early and intermediate transcripts, as has been suggested for SSV1 B115, or play a role in directing cell lysis to release progeny virus. This study also identified 177 host genes that were differentially expressed upon infection with STIV. Of the annotated genes up-regulated 4-fold or more, many are associated with DNA replication or transcription. These include Sso7D (a 7-kDa DNA-binding protein found in purified STIV), cdc6-1 and cdc6-3 (associated with origins of replication in Sulfolobus solfataricus), a reverse gyrase, a transcription factor IIB homolog, and the M subunit of a DNA-directed RNA polymerase. This study begins to provide insight into virus-host interactions during a lytic infection.
Structural Annotation
Because sequence conservation is generally much weaker than structural conservation, structural studies may uncover functional and evolutionary relationships that are not apparent from the primary sequence (33). To this end, recent work includes structural analysis of the SSV1, STIV, and AFV3 gene products. Studies of SSV1 reveal structural similarity between D63 and ROP (repressor of primer) (34), an adaptor protein that serves to regulate ColE1 plasmid copy number in Escherichia coli (19). D63 may play a similar role in regulating replication of the viral genome. Similarly, structural and biochemical characterization has revealed a homodimeric winged-helix protein for F93 (35) and a monomeric winged-helix protein for F112 (19), suggesting they may serve as transcription factors, while Sulfolobus Spindle-shaped Virus Ragged Hills D212, a homolog of SSV1 D244, displays a nuclease fold (Protein Data Bank code 2w8m), and B129 reveals tandem C2H2 zinc-finger domains.5 However, even structural homology is not always found; the structure of SSV1 E96 shows little similarity to any protein with known function.5
Although not as advanced, the story is similar for STIV, where there is a clear structural homolog for three of four available structures. These include the major coat protein B345, which shows unmistakable similarity to viral capsid proteins in the bacterial and eukaryotic domains (26); A197, a putative glycosyltransferase (36); and F93, a putative transcriptional regulator (32). In contrast, the structure of B116 fails to reveal a structural homolog, although it did suggest a potential interaction with DNA, and a nonspecific interaction with DNA was subsequently demonstrated (37). Likewise, Keller et al. (38) determined the structure of AFV3-109, a B116 homolog from AFV3 that also interacts with DNA. In all, ∼80% of the STIV and SSV1 structures are yielding recognizable folds. In addition, the STIV B116 and AFV3-109 structures show that structural annotation is worthwhile, even when it fails to identify a homolog with known function.
Insight into Host Processes
Studies are also providing insight into critical host processes. For example, SSV1 was an important model system for pioneering studies of transcription in Archaea, contributing evidence toward acceptance of Archaea as a third domain of life (30). In addition, modifications to the SSV1 genome have provided the first shuttle vectors for the Sulfolobales (39).
More recent studies are providing insight into acquired viral resistance. Many bacterial genomes and all sequenced archaeal genomes contain arrays of CRISPRs that are separated by similarly sized non-repetitive spacers (2, 40). Remarkably, these “spacer” sequences are frequently derived from virus or other invading nucleic acid (2). Working in conjunction with the adjacent CRISPR-associated genes, the CRISPR sequences are now known to provide acquired resistance against bacteriophage (40). Several studies have begun to extend the work with bacteria and bacteriophage to the archaea and their viruses. For example, 126 repeat clusters containing 4005 spacer sequences from the available crenarchaeal genomes were analyzed for similarity to ORFs in four rudiviral genomes (41). At the nucleotide level, matches to 158 spacers were found, with an additional 148 matches at the protein level. Matches were appropriately restricted to spacer sequences from rudiviral host organisms, where ∼10% of the 3042 spacers yielded a positive match, consistent with the abundance of Rudiviridae in acidothermophilic environments.
Other investigators have turned to metagenomic analyses of acid mine drainages (42) and hyperthermal environments.6 In these studies, CRISPR spacer sequences were used to provide a remarkable record of the encounters between a host and its viruses, thus identifying specific virus-host interactions present within the larger metagenomic community. Furthermore, reads that exactly match short CRISPR spacers but lack the flanking CRISPR repeats are indicative of viral sequence and have been used to identify new archaeal viruses (42).6 These analyses also indicate rapid evolution of the CRISPR loci, suggesting that modulation of resistance levels occurs on a time scale of months. At the same time, resident viruses apparently adapt by extensive recombination, shuffling viral sequence motifs to evade the host CRISPR spacers (42), and new viruses migrate into the community (43).
The metagenomic data are valuable in other ways. Sequence matches to ORFans, proteins lacking significant sequence similarity to other proteins, are being found, resulting in the identification of new protein families. The data are also enlarging the size of existing protein families, facilitating identification of conserved residues. This is particularly useful when conserved residues are mapped to a representative structure. Metagenomic studies thus complement structural annotation.
Insight into host processes also comes from the structural studies themselves, where the structures of SSV1 B129 and SSV1 F112 (19) and STIV F93 (32) reveal a disulfide bond in a putative intracellular DNA-binding protein. In the case of F112 and F93, the disulfides have been shown to confer significant thermostability (19, 32). This is consistent with the observations of Yeates and co-workers (44, 45), who used sequence-structure mapping and proteomic approaches to conclude that disulfide bonds are common in the intracellular proteins of hyperthermophilic organisms. They also found evidence for intracellular disulfides in the genome of Pyrobaculum aerophilum, where they observed a preference for even numbers of cysteines in a size-restricted set of intracellular proteins (45). Similarly, analysis of cysteine distributions in crenarchaeal viral genomes has also revealed a clear preference for an even number of cysteines in the putative intracellular proteins of STIV, SIRV2, AFV1, PSV, and Thermoproteus tenax spherical virus 1 (19, 32). More importantly, this preference is also clear in a metagenome composed of 18 crenarchaeal viruses, where the larger sample size is of increased statistical significance (Fig. 2B). This is strong supporting evidence for an abundance of stabilizing intracellular disulfide bonds in hyperthermophilic organisms and extends the observation to the cellular proteins of their equally intriguing viruses.
FIGURE 2.
SSV1 genome. A, ORFs are shown as open arrows, whereas transcript identities are shown in the interior (black lines). Predicted membrane proteins are indicated (+), as are proteins containing cysteine (*). RHH, ribbon-helix-helix. B, shown is the distribution of cysteine in the hyperthermophilic viral metagenome. Eighteen crenarchaeal viral genomes were combined to create a viral metagenome (19). Similar to the analyses of P. aerophilum (45) and STIV (32), a genome enriched in intracellular proteins was produced by removing predicted extracellular proteins, membrane proteins, proteins exhibiting metal-binding motifs, and proteins found in purified viral particles. A preference for even numbers of cysteines in the predicted intracellular proteins of the metagenome is clearly seen, suggesting an abundance of intracellular disulfide bonds.
Evolutionary Insight
Despite the lack of obvious sequence similarity, the structure of the STIV major capsid protein reveals a fold common to the eukaryotic Paramecium bursaria Chlorella virus, the bacteriophage PRD1, and mammalian adenovirus (26). It has been suggested that fundamental structural aspects such as capsid architecture and possibly genome packaging machinery constitute a viral “self” that is inherited from a viral ancestor, whereas attributes such as host recognition and adaptation are more likely acquired from hosts via lateral gene transfer (46). This implies that the “STIV-adeno-PRD1” lineage may have evolved from a common ancestral virus, one that possibly predates divisions leading to the three domains of life (46).
Palm et al. (15) noted that the cysteine-containing gene products cluster in one-half of the SSV1 genome, suggesting that the Fuselloviridae arose from a genome fusion event (15). However, structural studies offer an alternative explanation (19, 32) that is consistent with the microarray work of Frols et al. (18). Specifically, the asymmetric cysteine distribution may simply reflect the operon-like organization of the genome, where early transcripts generally encode intracellular proteins that utilize their cysteine content to form stabilizing disulfide bonds (see above). In contrast, late transcripts encode predicted membrane proteins that might participate in later stages of viral assembly and most of the proteins incorporated in the viral particle. The decreased cysteine content in the late gene products may indicate these proteins gain little from incorporating disulfides, perhaps because the membrane provides a similar topological constraint or because disulfides are not stable to the external solfataric environment, where hydrogen sulfide and sulfite concentrations can be significant.
Concluding Remarks
Although our understanding of archaeal viruses has advanced significantly, much remains to be learned. For most if not all crenarchaeal viruses, we still lack a detailed understanding of many fundamental processes, including mechanisms of attachment, uptake, transcriptional regulation, genome replication, and viral assembly and release. The development of robust genetic systems will certainly aid in these endeavors, and their further development for the study of individual gene products is a priority. Nevertheless, studies to date are beginning to provide insight into the ecological roles these viruses play in their environments and their potential roles in viral evolution. Thus, although the field is still in its infancy, it is a rapidly advancing field that holds potential for significant discovery.
Supplementary Material
This work was supported by National Science Foundation Grants MCB-0236344, MCB-0132156, and MCB-0646499 and National Aeronautics and Space Administration Grant NAG5-8807. This minireview will be reprinted in the 2009 Minireview Compendium, which will be available in January, 2010.
Footnotes
The abbreviations used are: STIV, Sulfolobus turreted icosahedral virus; STSV1, Sulfolobus tengchongensis spindle-shaped virus 1; ORFs, open reading frames; ATV, Acidianus two-tailed virus; SSV1, Sulfolobus spindle-shaped virus 1; AFV3, Acidianus filamentous virus 3; CRISPR, clustered regularly interspaced short palindromic repeat; SIRV, Sulfolobus icelandicus rod-shaped virus; PSV, Pyrobaculum spherical virus.
S. K. Brumfield, A. C. Ortmann, P. Suci, T. Douglas, and M. J. Young, submitted for publication.
S. Menon, B. J. Eilers, and C. M. Lawrence, unpublished data.
A. C. Ortmann, M. Bateson, V. Ruigrok, F. F. Roberto, T. Douglas, and M. J. Young, submitted for publication.
References
- 1.Suttle, C. A. (2007) Nat. Rev. Microbiol. 5 801–812 [DOI] [PubMed] [Google Scholar]
- 2.Sorek, R., Kunin, V., and Hugenholtz, P. (2008) Nat. Rev. Microbiol. 6 181–186 [DOI] [PubMed] [Google Scholar]
- 3.Forterre, P. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 3669–3674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell, P. J. (2001) J. Mol. Evol. 53 251–256 [DOI] [PubMed] [Google Scholar]
- 5.Zillig, W., Prangishvilli, D., Schleper, C., Elferink, M., Holz, I., Albers, S., Janekovic, D., and Gotz, D. (1996) FEMS Microbiol. Rev. 18 225–236 [DOI] [PubMed] [Google Scholar]
- 6.Prangishvili, D., Forterre, P., and Garrett, R. A. (2006) Nat. Rev. Microbiol. 4 837–848 [DOI] [PubMed] [Google Scholar]
- 7.Ortmann, A. C., Wiedenheft, B., Douglas, T., and Young, M. (2006) Nat. Rev. Microbiol. 4 520–528 [DOI] [PubMed] [Google Scholar]
- 8.Rice, G., Tang, L., Stedman, K., Roberto, F., Spuhler, J., Gillitzer, E., Johnson, J. E., Douglas, T., and Young, M. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 7716–7720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiang, X., Chen, L., Huang, X., Luo, Y., She, Q., and Huang, L. (2005) J. Virol. 79 8677–8686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arnold, H. P., Zillig, W., Ziese, U., Holz, I., Crosby, M., Utterback, T., Weidmann, J. F., Kristjanson, J. K., Klenk, H. P., Nelson, K. E., and Fraser, C. M. (2000) Virology 267 252–266 [DOI] [PubMed] [Google Scholar]
- 11.Prangishvili, D., Vestergaard, G., Haring, M., Aramayo, R., Basta, T., Rachel, R., and Garrett, R. A. (2006) J. Mol. Biol. 359 1203–1216 [DOI] [PubMed] [Google Scholar]
- 12.Jaalinoja, H. T., Roine, E., Laurinmaki, P., Kivela, H. M., Bamford, D. H., and Butcher, S. J. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 8008–8013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haring, M., Vestergaard, G., Rachel, R., Chen, L., Garrett, R. A., and Prangishvili, D. (2005) Nature 436 1101–1102 [DOI] [PubMed] [Google Scholar]
- 14.Haring, M., Rachel, R., Peng, X., Garrett, R. A., and Prangishvili, D. (2005) J. Virol. 79 9904–9911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palm, P., Schleper, C., Grampp, B., Yeats, S., McWilliam, P., Reiter, W.-D., and Zillig, W. (1991) Virology 185 242–250 [DOI] [PubMed] [Google Scholar]
- 16.Clore, A. J., and Stedman, K. M. (2007) Virology 361 103–111 [DOI] [PubMed] [Google Scholar]
- 17.Koonin, E. V. (1992) Nucleic Acids Res. 20 1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frols, S., Gordon, P. M., Panlilio, M. A., Schleper, C., and Sensen, C. W. (2007) Virology 365 48–59 [DOI] [PubMed] [Google Scholar]
- 19.Menon, S. K., Maaty, W. S., Corn, G. J., Kwok, S. C., Eilers, B. J., Kraft, P., Gillitzer, E., Young, M. J., Bothner, B., and Lawrence, C. M. (2008) Virology 376 270–278 [DOI] [PubMed] [Google Scholar]
- 20.Maaty, W. S., Ortmann, A. C., Dlakic, M., Schulstad, K., Hilmer, J. K., Liepold, L., Weidenheft, B., Khayat, R., Douglas, T., Young, M. J., and Bothner, B. (2006) J. Virol. 80 7625–7635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bettstetter, M., Peng, X., Garrett, R. A., and Prangishvili, D. (2003) Virology 315 68–79 [DOI] [PubMed] [Google Scholar]
- 22.Vestergaard, G., Aramayo, R., Basta, T., Haring, M., Peng, X., Brugger, K., Chen, L., Rachel, R., Boisset, N., Garrett, R. A., and Prangishvili, D. (2008) J. Virol. 82 371–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahn, D. G., Kim, S. I., Rhee, J. K., Kim, K. P., Pan, J. G., and Oh, J. W. (2006) Virology 351 280–290 [DOI] [PubMed] [Google Scholar]
- 24.Reiter, W.-D., Palm, P., Henschen, A., Lottspeich, F., Zillig, W., and Grampp, B. (1987) Mol. Gen. Genet. 206 144–153 [Google Scholar]
- 25.Bamford, D. H., Ravantti, J. J., Ronnholm, G., Laurinavicius, S., Kukkaro, P., Dyall-Smith, M., Somerharju, P., Kalkkinen, N., and Bamford, J. K. (2005) J. Virol. 79 9097–9107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khayat, R., Tang, L., Larson, E. T., Lawrence, C. M., Young, M., and Johnson, J. E. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 18944–18949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kessler, A., Brinkman, A. B., van der Oost, J., and Prangishvili, D. (2004) J. Bacteriol. 186 7745–7753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kessler, A., Sezonov, G., Guijarro, J. I., Desnoues, N., Rose, T., Delepierre, M., Bell, S. D., and Prangishvili, D. (2006) Nucleic Acids Res. 34 4837–4845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reiter, W.-D., Palm, P., Yeats, S., and Zillig, W. (1987) Mol. Gen. Genet. 209 270–275 [DOI] [PubMed] [Google Scholar]
- 30.Reiter, W.-D., Palm, P., and Zillig, W. (1988) Nucleic Acids Res. 16 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ortmann, A. C., Brumfield, S. K., Walther, J., McInnerney, K., Brouns, S. J., van de Werken, H. J., Bothner, B., Douglas, T., van de Oost, J., and Young, M. J. (2008) J. Virol. 82 4874–4883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larson, E. T., Eilers, B., Menon, S., Reiter, D., Ortmann, A., Young, M. J., and Lawrence, C. M. (2007) Virology 368 249–261 [DOI] [PubMed] [Google Scholar]
- 33.Shapiro, L., and Harris, T. (2000) Curr. Opin. Biotechnol 11 31–35 [DOI] [PubMed] [Google Scholar]
- 34.Kraft, P., Kummel, D., Oeckinghaus, A., Gauss, G. H., Wiedenheft, B., Young, M., and Lawrence, C. M. (2004) J. Virol. 78 7438–7442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kraft, P., Oeckinghaus, A., Kummel, D., Gauss, G. H., Gilmore, J., Wiedenheft, B., Young, M., and Lawrence, C. M. (2004) J. Virol. 78 11544–11550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larson, E. T., Reiter, D., Young, M., and Lawrence, C. M. (2006) J. Virol. 80 7636–7644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larson, E. T., Eilers, B. J., Reiter, D., Ortmann, A. C., Young, M. J., and Lawrence, C. M. (2007) Virology 363 387–396 [DOI] [PubMed] [Google Scholar]
- 38.Keller, J., Leulliot, N., Cambillau, C., Campanacci, V., Porciero, S., Prangishvilli, D., Forterre, P., Cortez, D., Quevillon-Cheruel, S., and van Tilbeurgh, H. (2007) Virol. J., issue 4, article 12 [DOI] [PMC free article] [PubMed]
- 39.Jonuscheit, M., Martusewitsch, E., Stedman, K. M., and Schleper, C. (2003) Mol. Microbiol. 48 1241–1252 [DOI] [PubMed] [Google Scholar]
- 40.Brouns, S. J., Jore, M. M., Lundgren, M., Westra, E. R., Slijkhuis, R. J., Snijders, A. P., Dickman, M. J., Makarova, K. S., Koonin, E. V., and van der Oost, J. (2008) Science 321 960–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vestergaard, G., Shah, S. A., Bize, A., Reitberger, W., Reuter, M., Phan, H., Briegel, A., Rachel, R., Garrett, R. A., and Prangishvili, D. (2008) J. Bacteriol. 190 6837–6845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andersson, A. F., and Banfield, J. F. (2008) Science 320 1047–1050 [DOI] [PubMed] [Google Scholar]
- 43.Snyder, J. C., Wiedenheft, B., Lavin, M., Roberto, F. F., Spuhler, J., Ortmann, A. C., Douglas, T., and Young, M. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 19102–19107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beeby, M., Connor, B. D., Ryttersgaard, C., Boutz, D. R., Perry, L. J., and Yeates, T. O. (2005) PLoS Biol. 3 1549–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mallick, P., Boutz, D. R., Eisenberg, D., and Yeates, T. O. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 9679–9684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bamford, D. H., Burnett, R. M., and Stuart, D. I. (2002) Theor. Popul. Biol. 61 461–470 [DOI] [PubMed] [Google Scholar]
- 47.Arnold, H. P., Ziese, U., and Zillig, W. (2000) Virology 272 409–416 [DOI] [PubMed] [Google Scholar]
- 48.Martin, A., Yeats, S., Janekovic, D., Reiter, W.-D., Aicher, W., and Zillig, W. (1984) EMBO J. 3 2165–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.