Abstract
Maf1 is a conserved repressor of transcription that functions at the downstream end of multiple nutrient and stress signaling pathways. How these different signaling pathways converge on Maf1 is not known. Previous work in yeast indicates that protein kinase A (PKA) regulates RNA polymerase (pol) III transcription, in part, by phosphorylating multiple sites in Maf1. Here we present additional evidence for this view and show that a parallel nutrient and stress-sensing pathway involving Sch9, an homologous kinase to metazoan S6 kinase, targets Maf1 at a subset of PKA sites. Using ATP analog-sensitive alleles of PKA and Sch9, we find that these two kinases account for the bulk of the phosphorylation on consensus PKA sites in Maf1. Deletion of Sch9 reduces RNA pol III transcription in a Maf1-dependent manner, yet the cells remain susceptible to further repression by rapamycin and other treatments. Because the rapamycin-sensitive kinase activity of the TORC1 complex is necessary for Sch9 function in vivo and in vitro, our results show that transcriptional regulation of RNA pol III and the coordinate control of ribosomal protein genes can be achieved by Sch9-dependent and -independent branches of the target of rapamycin (TOR) signaling pathway.
Maf1 is essential for repressing transcription by RNA polymerase (pol)2 III in yeast, and this function is conserved in mammals (1–4). In addition, human Maf1 is known to directly repress RNA pol II transcription of certain protein coding genes such as the TATA box-binding protein, TBP, and to indirectly repress transcription by RNA pol I (3). Repression by Maf1 occurs in response to a wide variety of nutritional and stress conditions, but the nature of the signaling pathways mediating these responses and how they converge on Maf1 is largely unknown (5).
In budding yeast, Maf1 is phosphorylated on consensus PKA sites under optimal growth conditions and is rapidly dephosphorylated under starvation or stress conditions that repress transcription (5, 6). Dephosphorylation of these PKA sites correlates with the relocation of Maf1 from the cytoplasm to the nucleus and is thought to regulate its inhibitory interaction with RNA pol III (5–9). Nuclear import of Maf1 is directed by two redundant nuclear localization sequences (NtNLS and CtNLS) that are differentially sensitive to PKA site phosphorylation. Specifically, the NtNLS is inhibited by phosphorylation as acidic substitutions at the PKA sites, which are normally fully functional, can prevent nuclear import and repression when the CtNLS is disabled (6). The localization of Maf1 in the cytoplasm serves to fine-tune its regulation but is not essential for preventing repression at inappropriate times. The accumulation of Maf1 in the nucleus, either by mutation of the protein or by deletion of the exportin of the protein, Msn5, does not cause repression in the absence of additional changes triggered by cellular signaling pathways (6, 10).
Numerous studies have implicated the RAS/PKA pathway and the rapamycin-sensitive TOR pathway as positive regulators of RNA pol III transcription (6, 11). However, it is not known how these pathways are integrated by Maf1 to affect the level of transcription. In this study, we show that PKA and another member of the AGC kinase family, Sch9, which is a direct downstream target of TOR (12), converge on Maf1 and together account for the bulk of the phosphorylation on consensus PKA sites. In addition, we find that the regulation of RNA pol III transcription and the co-regulated transcription of ribosomal protein (RP) genes involves bifurcation of the pathway downstream of TOR into Sch9-dependent and -independent branches.
EXPERIMENTAL PROCEDURES
Yeast Strains and Molecular Biology—All strains are derivatives of W303 unless otherwise noted and have been described previously (6, 9, 13) or were freshly generated (sch9Δ) by standard gene disruption techniques. Strains were grown in YPD or synthetic complete media as appropriate, and where indicated, treated with rapamycin (0.2 μg/ml) or C3-1′-naphthyl-methyl PP1 (1NM-PP1, 0.1 μm, Calbiochem). Procedures for Northern analysis have been reported previously (1) along with methods for site-directed mutagenesis of Maf1 and high resolution SDS-PAGE (6).
GST-Sch9 Purification and Kinase Assay—GST-Sch9 and the kinase-dead GST-Sch9 mutant (K441A) were expressed and purified from yeast strain BY4741 following a 10-min cycloheximide (25 μg/ml) treatment (12). Kinase reactions (20 μl) were performed for 30 min at 30 °C with [γ-32P]ATP as described previously (12) and contained 100 ng of full-length native recombinant yeast Maf1.3
Immunological Methods—Extracts from 4 × 108 cells were prepared by glass bead breakage into 0.5 ml of KBC100 breaking buffer containing protease and phosphatase inhibitors (9, 14). Maf1-Myc was immunoprecipitated and detected by Western blotting with Myc or phospho-PKA substrate-specific antibodies (6, 9).
RESULTS AND DISCUSSION
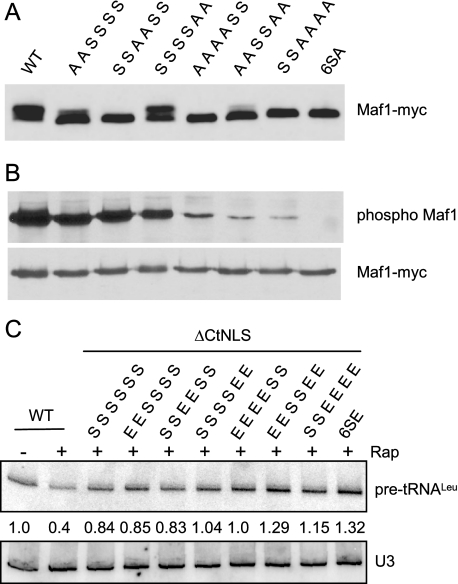
Regulation of Maf1 Involves Phosphorylation on Multiple Consensus PKA Sites—Saccharomyces cerevisiae Maf1 contains three pairs of clustered PKA consensus sites (serines 90, 101, 177, 178, 209, and 210). Phosphorylation of these sites negatively regulates the function of the NtNLS, one of two redundant NLSs in Maf1 (6). However, the importance of specific PKA sites in repression by Maf1 is unknown. To address this question, we first examined the phosphorylation of double, quadruple, and sextuple serine to alanine substitution mutants of the PKA consensus sites. Cell lysates prepared from log phase cultures of the various mutants were subjected to high resolution SDS-PAGE and Western blotting. As seen previously (6–8), rapidly growing wild-type cells have both slow migrating and fast migrating forms of Maf1 with predominantly more of the slow migrating hyperphosphorylated form (Fig. 1A). For each double mutant, we observed a decrease in the relative amount of the slow migrating form when compared with wild type. The changes were modest in the S209A,S210A mutant, pronounced in the S90A,S101A mutant, and complete in the S177A,S178A mutant, where only the fast migrating form was seen (Fig. 1A). In the S90A,S101A,S209A,S210A quadruple mutant, a further reduction in the amount of the slow migrating form of Maf1 was observed when compared with the parental double mutants. These data are consistent with the results of several mass spectrometry studies, which have detected phosphorylation of Maf1 at all six consensus PKA sites (15–17).3 Importantly, all mutants that included substitutions at S177,S178 showed only the fast migrating form of Maf1 (Fig. 1A). This result suggests that high resolving gels have a limited ability to report changes in Maf1 phosphorylation.
FIGURE 1.
Phosphorylation of multiple consensus PKA sites in Maf1. A, pairwise mutations (serine (S) to alanine (A)) of consensus PKA sites differentially affect the migration of Maf1 in high resolution SDS-PAGE. Lysates of a maf1Δ strain expressing plasmid-based MAF1-myc alleles were analyzed by Western blotting. The residues (Ser or Ala) at positions 90, 101, 177, 178, 209, and 210 are indicated, in order, above each lane. WT, wild type. B, Maf1-Myc proteins in panel A were immunoprecipitated, resolved by standard SDS-PAGE, and blotted with a phospho-PKA substrate-specific antibody (anti-RRXp(S/T), where X is any amino acid, Cell Signaling Technologies, upper panel) or anti-Myc antibody (lower panel). The phospho-Maf1 blot has been overexposed to show that there is no signal in the 6SA mutant. C, pairwise mutations (serine (S) to glutamic acid (E)) of consensus PKA sites (see above) were constructed in a Maf1 ΔCtNLS clone that contains a disabling mutation in the C-terminal NLS. Total RNA from log phase cells, treated or not with rapamycin (Rap) for 60 min, was analyzed by northern blotting with probes for pre-tRNALeu and U3 snRNA. The relative level of pre-tRNALeu, normalized for U3 snRNA loading is indicated. The average error associated with these measurements is ± 10%. In three independent experiments, the difference in the pre-tRNALeu signal between the ΔCtNLS (third lane from the left) and 6SEΔCtNLS mutants was 1.5 ± 0.1-fold.
To examine PKA site phosphorylation in Maf1 directly, the various mutants were immunoprecipitated from cell extracts, and phosphorylation was detected by Western blotting with a phospho-PKA substrate-specific antibody (6). Comparable amounts of Maf1 were detected in all samples (Fig. 1B, lower panel), and quantitative analysis of the blots showed that the total level of Maf1 phosphorylation (normalized for Maf1-Myc loading) was reduced to ∼77% (S90A,S101A), ∼65% (S177A,S178A), and ∼20% (S209A,S210A) of wild type in the double mutants. All quadruple mutants had less than 5% of the phosphorylation of wild-type Maf1, and no phosphorylation was detected in the 6SA mutant (simultaneous mutation of all six PKA phosphorylation sites to alanine). Notably, the S177A,S178A mutant, which retains around two-thirds of the total phosphate content on PKA sites shows only the fast migrating band in high resolving gels, whereas the S209A,S210A mutant with only one-fifth of the phosphate content of wild-type Maf1 has equivalent amounts of the slow and fast migrating species. Clearly, the migration of Maf1 in high resolving gels is especially sensitive to phosphorylation at Ser-177 and/or Ser-178. Perhaps more importantly, differences in the migration of Maf1 in high resolving gels may not always be correlated with the total level of phosphorylation.
Simultaneous mutation of all six PKA phosphorylation sites to glutamic acid (6SE) has no effect on repression by Maf1 (6). However, in the context of a mutation that cripples the CtNLS, the 6SE mutation renders Maf1 inactive for rapamycin-mediated repression (6). We used this assay to evaluate whether double or quadruple mutations to glutamic acid at different pairs of PKA phosphorylation sites compromise the function of Maf1. When compared with the parental ΔCtNLS mutant, which is partially defective in repression (6), the only double mutant that showed a significant decrease in Maf1 function was S209E,S210E (Fig. 1C). A comparable defect was caused by the S90E,S101E,S177E,S178E mutant. The other quadruple mutants were impaired to an extent that was not statistically different from the sextuple mutant (Fig. 1C). These data indicate that each pair of consensus PKA sites contributes, albeit modestly, to the regulation of Maf1 function.
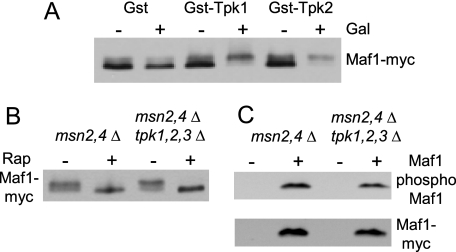
Phosphorylation of Maf1 by PKA in Yeast—Hyperactivation of PKA (by deletion of the Bcy1 regulatory subunit or an activating RAS2Val-19 mutation) promotes the phosphorylation of Maf1 and blocks repression of RNA pol III transcription under multiple conditions (6). Consistent with these observations, the Tpk1 isoform of PKA can phosphorylate recombinant Maf1 in vitro (6). However, carbon source-switching experiments show that although hyperactivation of PKA blocks repression of RNA pol III transcription upon transfer from fermentable glucose to non-fermentable glycerol, it does not prevent Maf1 dephosphorylation (18). This suggested that PKA targets proteins other than Maf1 in the regulation of RNA pol III transcription and raised questions about the glycerol-induced dephosphorylation of Maf1 in the hyperactive PKA strain. Accordingly, we felt that additional evidence was needed to strengthen the proposal that Maf1 is phosphorylated by PKA in vivo. As a first approach, we tested the effect of overexpressing different PKA catalytic subunits. Cells containing GST, GST-Tpk1, or GST-Tpk2 under the control of a GAL1 promoter were grown to log phase in raffinose-containing medium, and galactose was added to induce expression of the GST construct. Western blotting of Maf1 on high resolving gels showed that the protein is present predominantly in the fast migrating form prior to and after induction of the GST control (Fig. 2A). In contrast, induction of GST-Tpk1 or GST-Tpk2 led to a significant increase in the relative amount of the slow migrating form. These results are consistent with the idea that Tpk1 and Tpk2 can phosphorylate Maf1 in yeast cells.
FIGURE 2.
Phosphorylation of Maf1 upon over-expression of PKA. A, induction of GST-Tpk1 or GST-Tpk2 causes hyperphosphorylation of Maf1. GST expression plasmids were induced for 4 h by galactose addition to log phase cells growing in synthetic complete raffinose-uracil. Extract preparation and Western blotting of Maf1-Myc was performed as in Fig. 1A. B, deletion of PKA (tpk1,2,3Δ) does not affect the mobility of Maf1 in high resolving gels. Log phase cells containing a MAF1-myc plasmid were treated or not with rapamycin for 1 h. Extracts were blotted for Maf1-Myc as above. C, strains containing MAF1-myc or a control plasmid were grown to log phase for extract preparation, immunoprecipitation, and Western blotting as in Fig. 1B.
We next tested whether deletion of all three PKA catalytic subunits (tpk1,2,3Δ) affected the level of Maf1 phosphorylation in log phase cells grown on glucose. Because the tpk1,2,3Δ mutation is lethal in an otherwise wild-type background, these experiments were conducted in a strain that was also deleted for the stress-response transcription factors Msn2 and Msn4 to maintain strain viability (11). High resolving gel analysis showed Maf1 to be predominantly hyperphosphorylated in both the msn2,4Δ and the msn2,4Δ tpk1,2,3Δ strains but showed, as expected, only the fast migrating form of Maf1 after rapamycin treatment (Fig. 2B). However, when assayed by Maf1 immunoprecipitation and Western blotting, a modest but reproducible reduction in Maf1 phosphorylation was observed (to 70 ± 17% of wild type, n = 3, Fig. 2C). Along with other experiments described below, these data support our earlier conclusion that Maf1 is phosphorylated by PKA in yeast (6). In addition, we reasoned that the modest change in the level of PKA site phosphorylation of Maf1 in the tpk1,2,3Δ strain might be due to the compensatory effect of another kinase.
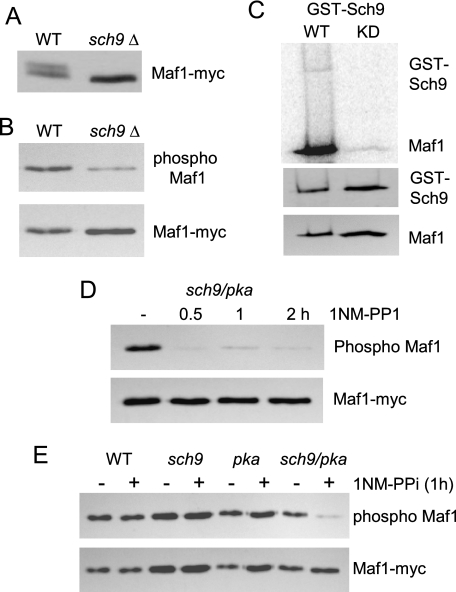
Sch9 Phosphorylates Maf1 on Consensus PKA Sites—In recent years, it has become increasingly apparent that Sch9 and PKA act in parallel pathways and regulate similar sets of functions (11). For example, nutrient-sensitive signaling by TORC1 kinase involves direct phosphorylation of Sch9 (12), and together with RAS/PKA signaling in response to the carbon source, determines the transcription of ribosome biosynthetic genes (11). Because RNA pol III transcription is coordinately regulated with the transcription of rDNA and RP genes (1), we considered that Sch9 might be a Maf1 kinase. To examine this possibility, extracts of log phase cells from wild-type and sch9Δ strains were analyzed on a high resolving gel. Notably, the sch9Δ extract contained only the fast migrating hypophosphorylated form of Maf1 (Fig. 3A). Moreover, in several independent experiments, immunoprecipitation and Western blotting showed that deletion of SCH9 reduced the level of phosphorylation at consensus PKA sites by ∼30 ± 13% (n = 3). These data indicate that Sch9 and PKA have overlapping target sites in Maf1. Next, we purified GST fusion proteins of wild-type Sch9 and a kinase-dead mutant form of the enzyme and performed in vitro kinase assays using purified recombinant S. cerevisiae Maf1 as a substrate. The wild-type enzyme exhibited autophosphorylation as reported previously (12) and readily phosphorylated recombinant Maf1 (Fig. 3C). In contrast, a slightly larger amount of kinase-dead Sch9 showed no detectable autophosphorylation, and the phosphorylation of Maf1 was dramatically reduced (<5% of the wild-type activity, normalized for Maf1 loading, Fig. 3C). Residual phosphorylation of Maf1 in this case presumably reflects the presence of contaminating kinase(s) in the preparation. Taken together, these data show that Sch9 is a Maf1 kinase.
FIGURE 3.
Sch9 phosphorylates an overlapping set of PKA consensus sites in Maf1. A, lysates of wild-type (WT) and sch9Δ strains were subjected to high resolution SDS-PAGE as in Fig. 1A. B, phosphorylation of PKA sites in Maf1 was analyzed in the indicated strains by immunoprecipitation and Western blotting as in Fig. 1B. C, in vitro phosphorylation of recombinant S. cerevisiae Maf1 by wild-type and kinase-dead (KD) GST-Sch9. After autoradiography (upper panel), the blot was probed with an antibody to GST and to Maf1 (lower panels). D, an analog-sensitive sch9 pka strain was treated for the indicated times with 1NM-PP1 before extract preparation and analysis of Maf1 phosphorylation as in Fig. 1B. E, wild-type and analog-sensitive strains were treated or not with 1NM-PP1 for 1 h before analysis of Maf1 as described above.
Finally, we tested the extent to which the preceding effects of Sch9 and PKA were due to separate, convergent actions of the kinases on Maf1 as opposed to potential cross-pathway effects by utilizing ATP analog-sensitive mutants of both enzymes. This strategy is broadly applicable to study protein kinase function and involves a single residue substitution in the conserved ATP-binding pocket of a kinase to confer chemical sensitivity to small molecule inhibitors (19). The strains used in our experiments either contain tpk1, tpk2, and tpk3 analog-sensitive mutations (designated pka) or an analog-sensitive sch9 mutation or combine all of these mutations (designated pka sch9). Previous work with these strains has demonstrated the cooperative role of Sch9 and PKA in regulating autophagy (13). A time course of treatment of the pka sch9 strain with the ATP analog 1NM-PP1 indicated that the phosphorylation of Maf1 on consensus PKA sites was largely eliminated after 30 min (Fig. 3D). This effect was specific to the pka sch9 strain because the wild-type, pka, and sch9 strains showed no effect on Maf1 phosphorylation after a 1-h treatment (Fig. 3E). This suggests that PKA and Sch9 independently target consensus PKA sites in Maf1.
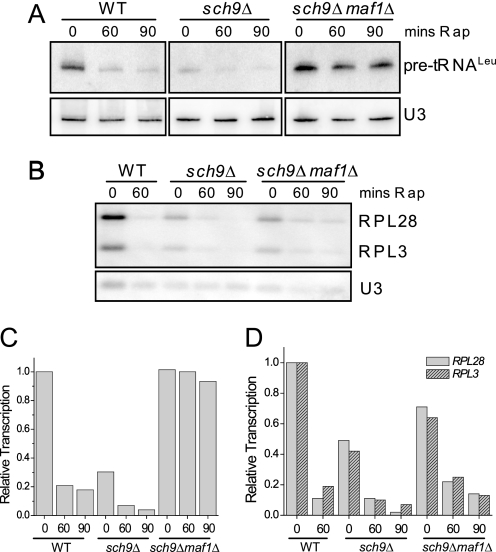
Deletion of Sch9 Causes a Maf1-dependent Reduction of RNA pol III Transcription—The functional overlap between the PKA and Sch9 pathways (11) together with the phosphorylation of Maf1 by these kinases and the effects of deregulating or deleting PKA on RNA pol III transcription (6) suggested that perturbing SCH9 would likely impact tRNA synthesis. Indeed, deletion of SCH9 reduced pre-tRNALeu synthesis to 24 ± 6% (n = 7) of the wild-type level under otherwise optimal growth conditions (Fig. 4, A and C). Importantly, this reduction in transcription is dependent on Maf1. Moreover, the reduced transcription in the sch9Δ strain is further diminished by rapamycin treatment (Fig. 4, A and C) and by all other repressing conditions that we have tested (including entry into stationary phase and treatments with tunicamycin, chlorpromazine, and methyl methane sulfonate).4 This result has important implications for TORC1 regulation of RNA pol III transcription. Rapamycin treatment inhibits TORC1 and TORC1-regulated Sch9 kinase activity (12). Therefore, if TORC1 acts exclusively through Sch9 to promote transcription by RNA pol III, then rapamycin should have an equivalent inhibitory effect in wild-type and sch9Δ strains. This is not the case (Fig. 4, A and C). The substantial further reduction of transcription in the rapamycin-treated sch9Δ strain indicates that TORC1 can also regulate RNA pol III transcription through an Sch9-independent pathway. Comparable results are seen for TORC1 regulation of RP gene transcription (Fig. 4, B and D) where a role for Sch9 has been known from numerous other studies (11). The reduced transcription of RP genes in sch9Δ cells is further inhibited by rapamycin. These effects on RP genes parallel other recent findings (20) and together support the conclusion that TOR regulation of ribosome and tRNA synthesis involves a bifurcation of the pathway downstream of TORC1 into minimally two branches, only one of which includes Sch9.
FIGURE 4.
Sch9 is required for high levels of tRNA and RP gene transcription. The indicated strains were treated with rapamycin for 0, 60, or 90 min, and total RNA was prepared for Northern analysis of pre-tRNALeu (A) or RP mRNAs (RPL28 and RPL3) (B). The data were quantified, normalized for loading relative to U3 snRNA, and plotted relative to the amount of transcript before rapamycin addition. WT, wild type; mins Rap, minutes of rapamycin. Pre-tRNA data are in C, and RP mRNA data are in D. The panels in A are non-adjacent lanes from the same gel.
Maf1 phosphorylation at consensus PKA sites can alter its distribution between the nucleus and the cytoplasm and provides one level of control over its function. However, as noted earlier, the localization of Maf1 in the cytoplasm is not an essential feature of its regulation because phospho-regulation of Maf1 can be accomplished when the protein is restricted to the nucleus by deletion of its exportin, Msn5 (10). Importantly, the role of both PKA and Sch9 in regulating Maf1 and its interactions with RNA pol III is compatible with the localization of Maf1 in either the nucleus or the cytoplasm. Tpk1 is localized to both compartments, whereas Tpk2 is primarily nuclear and is known to associate with the promoters of RP genes (21). Sch9 is predominantly localized to the vacuole membrane (12). A pool of TORC1 is also found at this location (22). However, a fraction of the cellular TORC1 and Sch9 is also present in the nucleus. TORC1 is physically associated with rDNA (23) in a rapamycin- and nutrient-sensitive manner, and Sch9 is known to associate with genes in response to osmotic stress (24). Thus, both functional and localization data are consistent with the ability of PKA and Sch9 to phosphorylate Maf1 in both the cytoplasm and the nucleus.
Conclusion—In the regulation of RNA pol III transcription, integration of the TOR and PKA pathways involves the phosphorylation of overlapping sites in Maf1 by Sch9 and PKA. These data, together with other studies (5, 12) allow the rapamycin-sensitive TOR signaling pathway to be delineated as a series of direct interactions between TORC1, Sch9, Maf1, and RNA pol III. Additional TORC1 regulation of RNA pol III transcription is achieved via an Sch9-independent pathway.
Acknowledgments
We thank Jim Broach for the analog-sensitive strains.
This work was supported, in whole or in part, by National Institutes of Health Grant GM42728. This work was also supported by funds from the Albert Einstein College of Medicine.
Footnotes
The abbreviations used are: pol, polymerase; PKA, protein kinase A; 1NM-PP1, C3-1′-naphthyl-methyl PP1; NLS, nuclear localization sequence; NtNLS, N-terminal NLS; CtNLS, C-terminal NLS; RP, ribosomal protein; GST, glutathione S-transferase; TOR, target of rapamycin.
H. Nika, J. Lee, I. M. Willis, and D. H. Hawke, manuscript in preparation.
J. Lee, R. D. Moir, and I. M. Willis, unpublished results.
References
- 1.Upadhya, R., Lee, J., and Willis, I. M. (2002) Mol. Cell 10 1489–1494 [DOI] [PubMed] [Google Scholar]
- 2.Reina, J. H., Azzouz, T. N., and Hernandez, N. (2006) PLoS ONE 1 e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson, S. S., Zhang, C., Fromm, J., Willis, I. M., and Johnson, D. L. (2007) Mol. Cell 26 367–379 [DOI] [PubMed] [Google Scholar]
- 4.Goodfellow, S. J., Graham, E. L., Kantidakis, T., Marshall, L., Coppins, B. A., Oficjalska-Pham, D., Gerard, M., Lefebvre, O., and White, R. J. (2008) J. Mol. Biol. 378 481–491 [DOI] [PubMed] [Google Scholar]
- 5.Willis, I. M., and Moir, R. D. (2007) Trends Biochem. Sci. 32 51–53 [DOI] [PubMed] [Google Scholar]
- 6.Moir, R. D., Lee, J., Haeusler, R. A., Desai, N., Engelke, D. R., and Willis, I. M. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 15044–15049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts, D. N., Wilson, B., Huff, J. T., Stewart, A. J., and Cairns, B. R. (2006) Mol. Cell 22 633–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oficjalska-Pham, D., Harismendy, O., Smagowicz, W. J., Gonzalez, d. P., Boguta, M., Sentenac, A., and Lefebvre, O. (2006) Mol. Cell 22 623–632 [DOI] [PubMed] [Google Scholar]
- 9.Desai, N., Lee, J., Upadhya, R., Chu, Y., Moir, R. D., and Willis, I. M. (2005) J. Biol. Chem. 280 6455–6462 [DOI] [PubMed] [Google Scholar]
- 10.Towpik, J., Graczyk, D., Gajda, A., Lefebvre, O., and Boguta, M. (2008) J. Biol. Chem. 283 17168–17174 [DOI] [PubMed] [Google Scholar]
- 11.Zaman, S., Lippman, S. I., Zhao, X., and Broach, J. R. (2008) Annu. Rev. Genet. 42 27–81 [DOI] [PubMed] [Google Scholar]
- 12.Urban, J., Soulard, A., Huber, A., Lippman, S., Mukhopadhyay, D., Deloche, O., Wanke, V., Anrather, D., Ammerer, G., Riezman, H., Broach, J. R., De Virgilio, C., Hall, M. N., and Loewith, R. (2007) Mol. Cell 26 663–674 [DOI] [PubMed] [Google Scholar]
- 13.Yorimitsu, T., Zaman, S., Broach, J. R., and Klionsky, D. J. (2007) Mol. Biol. Cell 18 4180–4189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shou, W., Verma, R., Annan, R. S., Huddleston, M. J., Chen, S. L., Carr, S. A., and Deshaies, R. J. (2002) Methods Enzymol. 351 279–296 [DOI] [PubMed] [Google Scholar]
- 15.Chi, A., Huttenhower, C., Geer, L. Y., Coon, J. J., Syka, J. E., Bai, D. L., Shabanowitz, J., Burke, D. J., Troyanskaya, O. G., and Hunt, D. F. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 2193–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, X., Gerber, S. A., Rudner, A. D., Beausoleil, S. A., Haas, W., Villen, J., Elias, J. E., and Gygi, S. P. (2007) J. Proteome. Res. 6 1190–1197 [DOI] [PubMed] [Google Scholar]
- 17.Albuquerque, C. P., Smolka, M. B., Payne, S. H., Bafna, V., Eng, J., and Zhou, H. (2008) Mol. Cell. Proteomics 7 1389–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ciesla, M., Towpik, J., Graczyk, D., Oficjalska-Pham, D., Harismendy, O., Suleau, A., Balicki, K., Conesa, C., Lefebvre, O., and Boguta, M. (2007) Mol. Cell Biol. 27 7693–7702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gregan, J., Zhang, C., Rumpf, C., Cipak, L., Li, Z., Uluocak, P., Nasmyth, K., and Shokat, K. M. (2007) Nat. Protoc. 2 2996–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smets, B., De Snijder, P., Engelen, K., Joossens, E., Ghillebert, R., Thevissen, K., Marchal, K., and Winderickx, J. (2008) FEMS Yeast Res. 8 1276–1288 [DOI] [PubMed] [Google Scholar]
- 21.Pokholok, D. K., Zeitlinger, J., Hannett, N. M., Reynolds, D. B., and Young, R. A. (2006) Science 313 533–536 [DOI] [PubMed] [Google Scholar]
- 22.De Virgilio, C., and Loewith, R. (2006) Oncogene 25 6392–6415 [DOI] [PubMed] [Google Scholar]
- 23.Li, H., Tsang, C. K., Watkins, M., Bertram, P. G., and Zheng, X. F. (2006) Nature 442 1058–1061 [DOI] [PubMed] [Google Scholar]
- 24.Pascual-Ahuir, A., and Proft, M. (2007) EMBO J. 26 3098–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]