Abstract
Metal-activated transcription factor 1 (MTF1) mediates the induction of metallothioneins I and II by zinc and stress signals. The mechanism of MTF1 activation has not been well understood. We analyzed the interaction between arsenic (As3+) and MTF1 for Mt1 induction. As3+ potently induces Mt1 mRNA expression in mouse hepa1c1c7 cells. Induction is dependent upon functional MTF1 as induction is lost in Mtf1 knockout cells but is restored upon reconstitution with Mtf1; moreover, As3+ induces the binding of MTF1 to the metal response elements of endogenous Mt1. Induction is not affected by modulating zinc concentrations but is markedly enhanced by cycloheximide. Phenylarsine oxide (PAO), which covalently binds to vicinal protein cysteine thiol groups, induces Mt1 with a magnitude of higher potency than that of As3+. PAO affinity beads effectively pulls down the carboxyl half of MTF1 (MTF1321–675) by binding to a cluster of five cysteine residues near the terminus. Preincubation with As3+, Cd2+, Co2+, Ni2+, Ag+, Hg2+, and Bi3+ blocks pulldown of MTF1321–675 by PAO beads in vitro and in vivo, indicating that binding of the metal inducers to the same C-terminal cysteine cluster as PAO occurs. Deletion of the C-terminal cysteine cluster or mutation of the cysteine residues abolishes or markedly reduces the transcription activation activity of MTF1 and the ability of MTF1 to restore Mt1 induction in Mtf1 knockout cells. The findings demonstrate a critical role of the C-terminal cysteine cluster of MTF1 in arsenic sensing and gene transcription via arsenic-cysteine thiol interaction.
Humans are constantly exposed to a wide variety of metals from dietary, environmental, and occupational sources. Trace elements, such as copper, iron, and zinc, are required for certain biological functions under physiological concentrations. However, most metals are toxic and cause a wide range of pathological conditions, including cancer, toxicity, and chronic diseases in humans (1, 2). Induction of metallothioneins (MT I and II),2 metal transporters, and antioxidative proteins represent a major means of metal detoxification in the body to maintain trace elements within a physiological range or to protect the body from the damage by metal overload. MTs are small, cysteine-rich, metal-binding proteins expressed in all eukaryotes and some prokaryotes. MTs I and II are highly inducible by heavy metals (such as Zn2+, Cd2+, Co2+, Ni2+, Ag+, Hg2+, and Bi3+), alkylating agents (e.g. iodoacetate), antioxidants, glucocorticoids, and inflammatory signals (such as lipopolysaccharide) (3–5). MTs regulate metal homeostasis by sequestering metals in protein-bound forms, providing a zinc reserve, and serving as a scavenger to quench ROS and other free radicals. On the other hand, metal transporters export metals out of the cells, and antioxidant proteins/enzymes antagonize metal-induced oxidative stress.
Transcriptional regulation of MTs, metal transporters, and certain antioxidant proteins is coordinated by metal-activated transcription factor 1 (MTF1), a member of the zinc finger family (6–10). MTF1 appears to be evolutionally conserved from Drosophila, to pufferfish Fugu rubripes, mouse, and human both structurally and functionally, reflecting a critical role of MTF1 in maintaining metal homeostasis across species (6, 9–13). MTF1 contains six pairs of Cys2His2 type of zinc fingers at the amino half as the signature DNA-binding domain (DBD) and three separable regions (acidic, proline-rich, and serine/threonine-rich) in the carboxyl half as the transcription activation domain (14).
In Drosophila, MTF1 (dMTF1) plays dual roles in copper homeostasis: at limiting copper concentrations, it induces the Ctr1B copper importer gene to increase cellular copper uptake; whereas at high copper concentrations, it induces MTs to chelate excess copper. Induction of both genes requires the binding of dMTF1 to the metal-responsive elements (MREs) located in the enhancers of the genes (15, 16). In mammalian species, MTF1 may have adapted into more complex biological functions. Disruption of the Mtf1 gene in mice impairs liver development at gestation day 14 resulting in liver decay, generalized edema, and embryonic death (17), revealing a critical role of MTF1 in the regulation of liver-specific, developmental genes. This function of MTF1 may not involve Mt1 and Mt2, because embryonic lethality is not observed in a double deletion of Mt1 and Mt2 (18, 19). Mice with conditional knockout of Mtf1 mature to adulthood but are highly sensitive to the toxicity of metals, such as Cd2+ (20). Adult Mtf1 null mice also exhibit relative leucopenia, in particular lymphocyte reduction (20), suggesting a role of MTF1 in lymphocyte development. The target genes of MTF1 responsible to its function in liver and lymphocyte development remain elusive. In addition, linkage analysis and haplotype mapping identified Mtf1 as a lymphoma susceptibility gene in mice, because a mutation in MTF1 (S424P) predisposes mice to ionizing radiation-induced thymic lymphomas, presumably by up-regulating radiation-resistant genes to promote tumor growth (21). In mammalian cells, coordinated regulation of MTs and zinc transporter 1 by MTF1 plays a major role in protection against the toxicity of zinc and other metals (22).
Induction of Mt1 in the liver serves as a prototype of MTF1 action in response to metals. Quiescent MTF1 resides in the cytoplasm; MTF1 is activated by metals and is translocated into the nucleus, where it binds to multiple copies of MREs in the enhancer of Mt1 to mediate gene transcription. A critical question on MTF1 action is how it senses metals. In one model, Zn2+ plays a central role: Zn2+ activates MTF1 by binding to a few or all of the zinc fingers of the protein leading to nuclear translocation and MRE-binding; other metals and stress signals, such as Cd2+ and ROS, mobilize Zn2+ from protein-bound Zn2+ pools to increase the concentration of intracellular free Zn2+, which is then available to MTF1 zinc fingers for activation of the protein (23–26). This postulate is supported by the observations that Zn2+ binds to MTF1 zinc fingers and stabilizes the MTF1·MRE complex both in vitro and in chromatin (27, 28). On the other hand, Cd2+ does not appear to bind to the fingers in a way to preserve the canonical ββα-structure of the zinc fingers for optimal binding to DNA (29). Antioxidants, such as tBHQ, indeed increase intracellular free Zn2+ concentration (30); however, Cd2+ does not appear to affect the Zn2+ pool at 10 μm at which it significantly induces Mt1 (31). Activation of MTF1 may also involve the removal of one or more inhibitory proteins, because inhibition of protein synthesis or exposing cells to heat shock synergizes with metal inducers to give rise to “superinduction” of MTs (7, 31, 32). However, the nature of the inhibitory protein(s) remains unclear. Other inducers, such as glucocorticoids and alkylating agents, may also interact with MTF1-associated proteins to activate MTF1, because these inducers are unlikely to interact with the zinc fingers directly. Phosphorylation of MTF1 was observed and may contribute to nuclear translocation of activated MTF1 (33–35).
Arsenic (As3+) is a human carcinogen and a pleiotropic toxic metalloid that causes cancer (particularly of the skin, lung, bladder, and liver), neuropathy, cardiovascular lesions, ovarian dysfunction, aberrant embryonic development, and postnatal growth retardation in humans (36–42). As3+ is ubiquitous in the environment and has become a major public health concern worldwide, because millions of people are at risk of drinking water contaminated with arsenic (43). On the other hand, As3+ has been effectively used as therapeutics, such as in the treatment of acute promyelocytic leukemia (44). The molecular targets of As3+ for many of its biological effects remain unidentified. As3+ appears to induce Mt1 in rodents, but the mechanism of induction has not been addressed. Given the importance of MTF1 and MTs in metal detoxification, we examined the role and mechanism of action of MTF1 in the induction of Mt1 by As3+. The findings reveal that MTF1 mediates the induction of Mt1 by As3+ and As3+ binds directly to the C-terminal cysteine thiol groups of MTF1 to induce the genes. The findings demonstrate a critical role of the C-terminal cysteine cluster of MTF1 in arsenic sensing and gene transcription by MTF1 in vivo.
EXPERIMENTAL PROCEDURES
Cell Culture—Mouse hepa1c1c7 cells were provided by Dr. J. P. Whitlock, Jr. (Stanford University, Stanford, CA). The cells were cultured in α-minimal essential medium with 10% fetal bovine serum (FBS) and 5% CO2. Mouse Nrf2 wild-type (Nrf2 WT) and Nrf2 knockout (Nrf2 KO) embryonic fibroblast cells were derived from wild-type and Nrf2 null mice, respectively. The cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% FBS as described previously (45). MTF1 WT and MTF1 KO (dko7) fibroblast cells were kind gifts from Dr. W. Schaffner (Universitat Zurich, Zurich, Switzerland). The cells were cultured in DMEM supplemented with 10% FBS. The MRE-βGeo reporter cell line, BHK3038, was kindly provided by Dr. R. D. Palmiter (University of Washington, Seattle, WA). The cells were cultured in DMEM with 10% FBS medium. HEK293T cells were purchased from ATCC (Manassas, VA) and were cultured in DMEM with 10% FBS.
Chemical Reagents—Arsenic chloride (AsCl3), bismuth ammonium citrate (C12H22BiN3O14), cadmium chloride (CdCl2), cobaltous chloride (Cl2Co), mercury chloride (HgCl2), nickel chloride (NiCl2), phenylarsine oxide (PAO), potassium dichromate (K2Cr2O7), silver nitrate (AgNO3), tert-butylhydroquinone (tBHQ), and zinc chloride (ZnCl2) were purchased from Sigma-Aldrich. Affi-Gel 10 gel was from Bio-Rad (Hercules, CA). 4-Amino-phenylarsine oxide (p-aminophenyl arsenoxide, dihydrate) was purchased from Toronto Research Chemicals, Inc. (Toronto, Ontario, Canada).
Plasmid Construction, Point Mutations, and Cell Transfection—The full-length coding region of mouse MTF1 cDNA (amino acid residues 1–675, aa1–675) was amplified by reverse transcription and PCR from total RNA of hepa1c1c7 cells and was verified by sequencing. MTF1 was subcloned into the pcDNA3.1HA.his vector (Invitrogen) to generate pMTF11–675. A dominant-negative MTF1 (MTF11–320) was generated by cloning the DNA binding domain (aa1–320) of MTF1 into pcDNA3.1HA.his. pMTF1321–675 was constructed by cloning the carboxyl half (aa321–675) of MTF1 into the pcDNA3.1HA.his. pMTF1321–631 encodes the carboxyl half of MTF1 without the C-terminal 44 amino acid residues that contain the C-terminal five cysteine cluster. pMTF11–631 was generated by cloning the aa1–631 fragment of MTF1 into pcDNA3.1HA.his. The pGMTF1 fusion constructs were generated by cloning the Gal41–147 and a corresponding MTF1 fragment into pcDNA3.1HA.His to obtain pGMTF11–675, pGMTF11–320, pGMTF1321–675, and pGMTF1321–631. pGVP16 was generated by cloning the VP16 transcription activation domain (aa411–456) into pGal41–147. The pGMTF11–320VP16 was constructed by inserting MTF11–320 into the pGVP16. MTF1 C-terminal cysteine point mutation constructs were obtained by using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The cysteine mutants include pGMTF1321–675C632FC634F, pGMTF1321–675C636FC638F, pGMTF1321–675C653F, pGMTF1321–675C636FC638FC653F, pGMTF1321–6755CysMut, pMTF11–6755CysMut, and pMTF1321–6755CysMut; 5CysMut = C632FC634FC636FC638FC653F. All plasmid DNA constructs and point mutations were confirmed by sequencing. The sequences of the primers used for plasmid construction and mutations are available upon request. The plasmids were transfected into cultured cells using the Lipofectamine plus reagents from Invitrogen.
RNA Preparation and Northern Blotting—Total RNA was isolated from cells using the Qiagen total RNA isolation kit. Total RNA of 3 μg each was fractionated in a 1.2% formaldehyde agarose gel, transferred to a super-charged nylon membrane, and blotted with the digoxigenin-labeled riboprobe prepared with the digoxigenin-labeling reagents (Roche Applied Science, Indianapolis, IN). Plasmid constructs for riboprobes of mouse Mt1, Ho1, β-Gal, actin, and Gapdh were verified by sequencing. Northern signals were visualized by chemiluminescence using a digoxigenin RNA detection kit with CDP Star as a substrate (Roche Applied Science). actin and Gapdh were probed as loading control.
Measurement of β-Galactosidase Activity—β-Galactosidase (β-gal, encoded by β-Geo) was measured by using the β-galactosidase assay kit (Promega, Madison, WI). In a typical assay, BHK3038 cells were cultured in a 12-well plate and were treated with tBHQ (30 μm) or As3+ (5 μm) for 5 h. The cells were lysed in 100 μl of 1× reporter lysis buffer (Promega) by shaking for 10 min at room temperature, followed by vortex for 15 s. Cell debris was removed by spinning in a microcentrifuge at the maximal speed for 2 min. Twenty-five μl of supernatant was transferred to a 96-well plate containing 25 μl of the 1× lysis buffer, and 50 μl of the 2× assay buffer was then added. The 96-well plate was covered with a plastic wrapper and was cultured at 37 °C for 30 min to 3 h (until yellow color is visible). A stop solution (1 m sodium carbonate) of 150 μl per well was added for 5 min. Absorbance at 420 nm (A420) was measured on a 96-well plate reader. Each sample was repeated three to six times.
Luciferase Assay—Hepa1c1c7 cells were transiently transfected with appropriate plasmid DNA constructs with the Renilla internal control plasmid (Promega) by using Lipofectamine plus reagent. Cells were lysed with 1× passive lysis buffer (Promega) after 36–48 h of transfection. The plate was shaken for 10 min, and the lysate was collected and mixed by vortex for 15 s. Cell debris was removed by centrifugation for 2 min at 4 °C. Twenty microliters of cell lysate was mixed with 100 μl of the luciferase assay reagent in a luminometer tube and the dual-luciferase activity was measured using the TD20/20 luminometer (Sunnyvale, CA); luciferase activity was normalized to both the protein concentration and Renilla luciferase activity of each sample.
Immunoblotting—Cells were lysed on ice with a radioimmune precipitation assay buffer containing protease and phosphatase inhibitors for 30 min. The cell lysate was sonicated briefly and was centrifuged at 14,000 × g for 20 min to remove cell debris. Cell lysate (10–20 μg each) was fractionated in 10% SDS-PAGE, transferred to polyvinylidene difluoride membranes (Bio-Rad), and blocked with 5% nonfat milk in PBST (phosphate-buffered saline (PBS) plus 0.05% Tween 20). The membrane was blotted with primary antibody at 4 °C for overnight with shaking, followed by incubation with horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature. Protein bands were visualized using enhanced chemiluminescence detection reagents from Amersham Biosciences. Actin was blotted as loading control.
In Vitro Transcription and Translation—The TnT quick-coupled transcription/translation system (Promega) was used for in vitro transcription and translation of proteins. The reaction was incorporated with or without biotin tRNA for detection with streptavidin-horseradish peroxidase. Reaction products (1–5 μl) were separated on 10% SDS-PAGE, transferred onto a polyvinylidene difluoride membrane, and immunoblotted with an antibody or with streptavidin-horseradish peroxidase, followed by detection with chemiluminescence.
PAO Beads Conjugation—10 mg of 4-aminophenylarsene oxide was dissolved into 3.05 ml of methanol and was mixed with 1.22 ml of Affi-Gel (50% slurry, Bio-Rad) for 2 h at room temperature. 100 μl of aminoethanol was then added to block the remaining active groups of Affi-Gel. The mixture was washed three times with methanol and three times with PBS, followed by resuspension with 0.6 ml of PBS. An Affi-Gel control was prepared with 1.22 ml of Affi-Gel (50% slurry) mixed with 3.05 ml of methanol and 1 ml of aminoethanol with shaking at room temperature for 2 h.
Measurement of Protein Free Thiol Groups—Protein free thiol groups were measured as described previously with modifications (46, 47). Purified MT I protein (Sigma, 10 μg) in 100 μl was mixed with arsenic or PAO for 30 min. The protein was precipitated by adding 10 μg of bovine serum albumin and 10% ice-cold trichloroacetic acid containing 1 mm dithiothreitol. The suspension was centrifuged at 14,000 × g for 5 min at 4 °C. The precipitate was suspended in 400 μl of Ellman's reagent (DTNB buffer, composed of, 0.5 m potassium phosphate, pH 7.4, containing 0.2 mm 5, 5′-dithiobis(2-nitrobenzoic acid) and 5 mm EDTA). The mixture was incubated at 4 °C for 30 min. Insoluble materials were removed by centrifugation for 5 min at 14,000 × g at 4 °C, and the absorbance was measured at 412 nm.
ChIP Assay—Hepa1c1c7 cells were grown in a 10-cm dish to reach 90% confluence. The cells were treated with As3+ for 5 h. ChIP assay was performed as described previously with modifications (48). Briefly, DNA-proteins were cross-linked by incubation of cells with 1% formaldehyde at 37 °C for 10 min. Excess formaldehyde was quenched with 0.125 m glycine. Cells were collected in 1 ml of a lysis buffer (5 mm Pipes, pH 8.0, 85 mm KCl, and 0.5% IGEPAL CA-630) with proteinase inhibitors. Nuclei were pelleted by centrifugation. The nuclei were washed and resuspended in nuclei lysis buffer. DNA was sonicated to an average size of 200–1000 bp. Sheared chromatin was diluted in an immunoprecipitation dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA, and 16.7 mm Tris/HCl, pH 8.0, and 167 mm NaCl) precleared with protein G and salmon sperm DNA; immunoprecipitation was performed with rabbit anti-mouse MTF1 antibodies or a normal rabbit IgG (as a control). The immunocomplex was reverse cross-linked by incubation with 5 m NaCl at 65 °C overnight. The DNA samples were purified and amplified by real-time PCR following standard procedures. Threshold cycles (CT values) were determined using iCycler IQ software (Bio-Rad). Real-time PCR results were normalized using CT values with 1% of input as an internal control. Relative DNA amounts were calculated from CT values for each sample by interpolating into the standard curve obtained using a series dilution of standard DNA samples run under the same conditions. Primer set for amplifying the MRE of Mt1 was as follows: forward, CGGAGGCCATTGTATTGTCT and reverse, GCTGGGTTGGTCCGATACTA.
Data Quantification and Statistical Analysis—Quantification of RNA or protein bands were performed with Image-Quant software (Molecular Dynamics, San Jose, CA). Statistical analysis was performed with one-way analysis of variance followed by t test using Prism (GraphPad Software, San Diego, CA).
RESULTS
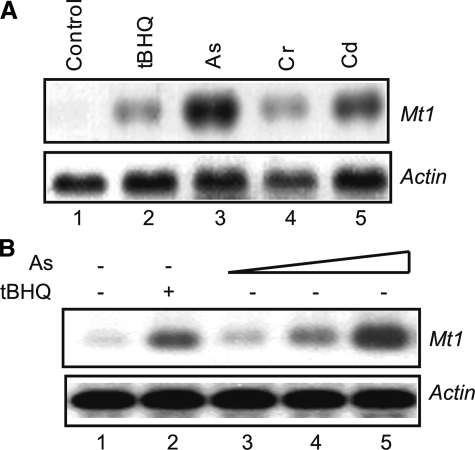
Arsenic Induces Mt1 via MTF1—To investigate the mechanism of Mt1 induction by As3+, we first characterized induction in hepa1c1c7 cells, a mouse hepatoma cell line highly responsive to toxic metals. Induction of Mt1 mRNA by As3+ (10 μm, 5 h) was compared with those by several known inducers of Mt1, including the phenolic antioxidant tBHQ (30 μm), and carcinogenic metals Cr6+ (10 μm) and Cd2+ (10 μm). Mt1 mRNA was nearly undetectable by Northern in the cells in the absence of the inducers (Fig. 1A, lane 1). tBHQ, Cr6+, and Cd2+ induced Mt1 as expected (lanes 2, 4, and 5). Notably, As3+ induced Mt1 expression to a higher level than other inducers at an equal (for Cr6+ and Cd2+) or lower concentration (for tBHQ) (Fig. 1A, compare lane 3 with lanes 2, 4, and 5). Induction by As3+ is concentration-dependent; induction was clearly observed at 2 μm and reached >10-fold higher at 10 μm (Fig. 1B).
FIGURE 1.
Induction of Mt1 gene expression by arsenic. A, comparison of induction by metals and phenolic antioxidant. Hepa1c1c7 cells were treated with tBHQ (30 μm), As3+ (10 μm), Cr6+ (10 μm), or Cd2+ (10 μm) for 5 h. Total RNA was analyzed by Northern blotting. Actin was used as a loading control. B, concentration dependence. Cells were treated with tBHQ (30 μm) or As3+ at 1, 5, and 10 μm for 5 h. Total RNA was blotted with Mt1 and actin riboprobes.
The enhancer of the mouse Mt1 gene contains several copies of MREs and an antioxidant responsive element (ARE) that overlaps with an upstream stimulatory factor-binding site (8, 49). Because As3+ is known to induce a number of cytoprotective genes, such as Nqo1, via ARE (50), we first examined if induction of Mt1 by arsenic requires Nrf2, the principal transcription factor for ARE-dependent gene induction (Fig. 2A). tBHQ was used as a positive control, because it is known to induce Nqo1 through ARE and Mt1 through MRE, respectively (30, 45). As expected, tBHQ induced Nqo1 in Nrf2 WT but not Nrf2 KO cells (Fig. 2A, middle panel, compare lanes 2 and 7); on the other hand, tBHQ induced Mt1 in both Nrf2 WT and KO cells (upper panel, lanes 2 and 7). As2+ induced both Mt1 and Nqo1 in Nrf2 WT cells (Fig. 2A, upper and middle panels, compare lane 1 with lanes 3–5). Whereas induction of Nqo1 was lost in the Nrf2 KO cells (Fig. 2A, middle panel, lanes 6 and 8–10), induction of Mt1 was unaffected in Nrf2 KO cells (upper panel, compare lane 6 with lanes 8–10), indicating that Nrf2 is required for induction of Nqo1 but not Mt1. Together, the data revealed that induction of Mt1 by As3+ is unlikely to be mediated through ARE and Nrf2.
FIGURE 2.
Induction of Mt1 by arsenic requires MTF1. A, Nrf2 WT and KO cells were treated with tBHQ (30 μm), or As3+ (1, 5 and 10 μm) for 5 h. Total RNA was blotted with Mt1, Nqo1, and actin riboprobes. B, MTF1 WT, MTF1 KO, and MTF1 KO cells stably transfected with pMTF11–675 were treated with As3+ (10 μm) for 5 h. Total RNA was blotted for Mt1, Ho1, and actin mRNA expression.
The role of MRE and MTF1 in Mt1 induction by As3+ was examined. As3+ induced Mt1 in MTF1 WT fibroblasts but not MTF1 KO cells (Fig. 2B, upper panel, lanes 1–4). Moreover, reconstitution of MTF1 expression in MTF1 KO cells (KO/MTF1) restored the induction (Fig. 2B, upper panel, lanes 5 and 6). Ho1, which does not depend on MRE for induction, was induced by As3+ in all three cell types independently of MTF1 (Fig. 2B, middle panel). The results indicate that induction of Mt1 by As3+ requires MTF1.
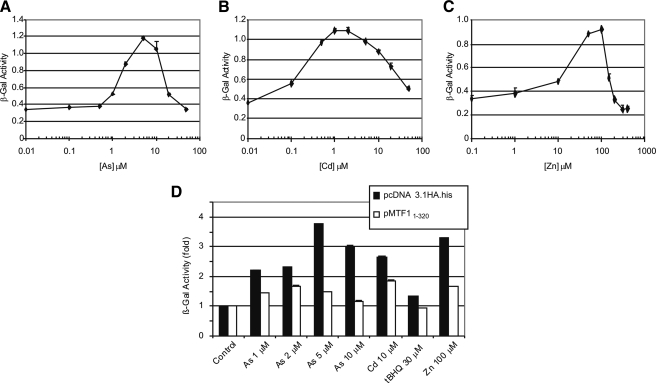
As3+ stimulated β-gal reporter expression under the control of the MREs of Mt1 in BHK3038 cells that express β-gal through the MTF1/MRE pathway (Fig. 3A). Induction was observed in a concentration range of 0.5–5 μm, above which the β-gal activity decreased concentration-dependently due to increased toxicity of As3+ at higher concentrations. Both the concentration range and the EC50 (∼1.5 μm) of β-gal induction by As3+ were lower than those for the induction of the Mt1 gene, possibly due to a higher sensitivity of reporter induction. Induction of the reporter by As3+ is similar to those by Cd2+ and Zn2+ in the overall shape of concentration-response curves (Fig. 3, B and C). Induction potency is in the order of Cd2+ > As3+ > Zn2+ with estimated EC50 values of 0.25, 1.5, and 22 μm, respectively. To further ascertain the role of MTF1/MRE in Mt1 induction, a dominant-negative form of MTF1 (MTF11–320) containing the DBD but lacking of the transcription activation domain was expressed in the BHK3038 cells. BHK3038 transfected with the empty vector served as a negative control. The β-gal reporter was induced by treatments with As3+ (from 1 to 10 μm), Cd2+ (10 μm), tBHQ (30 μm), or Zn2+ (100 μm) as expected. Induction was significantly inhibited in all cells expressing the dominant negative MTF1 compared with control (Fig. 3D), further supporting that MTF1 mediates Mt1 induction by As3+.
FIGURE 3.
Induction of MRE-β-gal reporter by arsenic. BHK3038 were treated with metals for 5 h. Total cell lysate was measured forβ-gal activity as described under “Experimental Procedures.” A–C, concentration curves of β-gal induction by As3+, Cd2+, or Zn2+, respectively. D, BHK3038 cells were transfected with pcDNA3.1HA.his (vector control) or pMTF11–320 (dominant negative MTF1) for 48 h and were treated with As3+ (1, 2, 5, and 10 μm), Cd2+ (10 μm), tBHQ (30 μm), or Zn2+ (100 μm) for 5 h. β-gal activity was measured.
To examine if MTF1 directly mediates the induction of endogenous Mt1 by As3+, chromatin immunoprecipitation (ChIP) was performed. A polyclonal antibody preparation recognized the endogenous MTF1, in vitro expressed MTF1, and transiently expressed MTF1, all having an estimated apparent molecular mass of ∼110 kDa (Fig. 4A) (31). MTF1 binds to the endogenous Mt1 MREs constitutively at a low but detectable level; binding was markedly increased upon treatment with As3+ (Fig. 4B). Taken together, the results established that MTF1 mediates the induction of Mt1 by As3+ through binding to its MREs. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay indicated that MTF1 WT cells were resistant to the toxicity of arsenic at concentrations of up to 50 μm, whereas the MTF1 KO cells had reduced survival in an arsenic concentration-dependent manner (data not shown). Moreover, KO cells reconstituted with MTF1 were resistant to arsenic toxicity similarly to the WT cells. Thus, MTF1 is required for defense against arsenic toxicity, which is, at least in part, attributable to MTF1-mediated induction of the Mt genes.
FIGURE 4.
Recruiting MTF1 to endogenous MRE by As3+. A, immunoblotting of MTF1. Cell lysate from hepa1c1c7, in vitro transcription/translation-produced MTF1 (TnT MTF1), and cell lysate from HEK293T cells transfected with pMTF11–675 were blotted with anti-MTF1 antibodies. B, ChIP assay. Hepa1c1c7 cells were treated with tBHQ (30 μm) or As3+ (10 μm) for 5 h. ChIP was performed and precipitated DNA was amplified by real time PCR with MRE primers. Quantitative data represent means and standard deviations from three experiments. Normal rabbit IgG was used as a negative control.
Role of Intracellular Free Zinc, ROS, and Labile Repressor—Some MTF1 inducers, such as tBHQ, activate MTF1 by mobilizing intracellular zinc from protein-bound forms to free zinc (30); increased intracellular free zinc can then bind and activate MTF1. Thus, modulating the concentration of intracellular free zinc may perturb induction of Mt1 by tBHQ through MTF1. To examine if As3+ induces Mt1 via zinc, hepa1c1c7 cells were cultured in a zinc-depleted medium to reduce the intracellular zinc concentration (30); induction of Mt1 in the cells was compared with that of normal culture (Fig. 5). As3+ induced Mt1 in MTF1 WT, but not KO cells, and stable expression of MTF-1 in KO restored the induction when the cells were cultured in a normal medium as expected (Fig. 5A, upper panels). A similar induction pattern was observed in the cells that were cultured in a zinc-depleted medium for a prolonged period of time (Fig. 5A, lower panels), indicating that depletion of extracellular zinc, which reduces the intracellular free zinc pool, does not appear to interfere with the induction of Mt1 by As3+. In Fig. 5B, induction of MRE-β-gal reporter was measured in BHK3038 cells that were cultured in a normal or zinc-depleted medium. Depletion of zinc in the medium did not decrease but increased reporter induction by either As3+ or Zn2+, indicating that reduction of intracellular free zinc did not reduce induction but may have sensitized cells to MRE-inducers to give rise to increased induction.
FIGURE 5.
Induction of Mt1 by arsenic does not depend on zinc. A, effect of zinc depletion. MTF1 WT, MTF1 KO, and MTF1 KO cells stably transfected with pMTF11–675 were cultured in a normal medium (DMEM plus 10% FBS) or a zinc-depleted medium (DMEM plus 10% of Chelex 100-treated FBS) for 48 h. The cells were treated with arsenic (10 μm) for 5 h. Total RNA was blotted for Mt1 and actin. B, MRE-βGal induction. BHK3038 cells were cultured in a normal medium or a zinc-depleted medium for 48 h and were treated with arsenic (5 and 10 μm) or Zn2+ (100 μm) for 5 h. Cell lysate was assayed for β-gal activities. C, effect of EDTA. BHK3038 cells were treated with EDTA plus Cd2+ (10 μm), As3+ (2, 5, or 10 μm), or Zn2+ (100 μm) for 5 h, or the cells were pretreated with EDTA for 2 h, followed by washing with fresh medium and treating with Cd2+ 10 μm or As3+ at 2, 5, and 10 μm for 5 h. β-gal induction was measured.
EDTA chelates bivalent cations. Co-treatment of BHK3038 cells with EDTA and Cd2+, As3+, or Zn2+, respectively, blocked the induction of β-gal by the metal inducers. However, treatment of BHK3038 cells with EDTA for 2 h followed by washing away of EDTA and replacing with a fresh medium did not apparently affect induction of β-gal expression by either As3+ or Cd2+ (Fig. 5C), suggesting a direct effect of As3+ and Cd2+ on the induction. Together, the data imply that arsenic directly induces Mt1 independently of Zn2+ concentrations in the culture medium or within the cells under the experimental conditions.
As3+ promotes the production of ROS in many cell types. A potential role of As3+-induced oxidative stress in MTF1 activation was examined using the antioxidant, N-acetylcysteine (NAC), which inhibits ROS production. NAC itself did not affect the MRE-β-gal expression in a concentration range from 10 μm to 10 mm in BHK3038 cells (Fig. 6). Co-treatment with As3+ (5 μm) and NAC from 5 μm to 1 mm only slightly reduced reporter induction at high concentrations in which NAC was 100 to 200 times in excess of As3+ in comparison with treatment of As3+ alone. Only at a concentration of 2000 times excess (i.e. 10 mm) did NAC block the induction by As3+. The apparently low efficiency of NAC in inhibiting the MTF1/MRE-dependent transcription of Mt1 negates a direct role of ROS in MTF1 activation by As3+.
FIGURE 6.
Effect of NAC on MRE reporter induction. BHK3038 cells were treated with NAC at 10 μm, 1 mm, and 10 mm alone or at 5 μm, 10 μm, 20 μm, 50 μm, 1 mm, and 10 mm plus arsenic (5 μm) for 5 h. β-gal was measured.
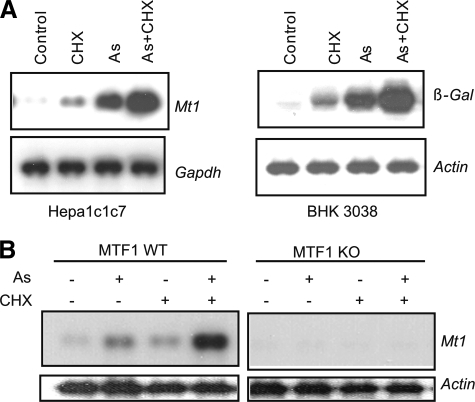
Inhibition of protein synthesis induces Mt1 and superinduces the gene in the presence of an activator of MTF1, such as Cd2+ and tBHQ, implicating a labile repressor in MTF1-mediated gene transcription (7, 31). The labile factor may suppress MTF1 as an MTF1-binding protein to sequester MTF1 in a dormant form (7) or as a nuclear factor that promotes the degradation of activated MTF1 in the nucleus (31). We examined if induction of Mt1 by As3+ is also regulated by this labile repressor. In Fig. 7A, cycloheximide (CHX), a potent inhibitor of protein synthesis, induced Mt1 in hepa1c1c7 cells or MRE-controlled β-gal in BHK3038 cells. As3+ induced both genes to a higher level than CHX. Co-treatment with arsenic and CHX increased the induction of the genes to much higher levels (i.e. superinduction). Furthermore, both the induction by CHX and superinduction by CHX plus As3+ required MTF1, because both activities were lost in the MTF1 KO cells (Fig. 7B). Thus, induction of Mt1 by arsenic is likely regulated by the same labile repressor that suppresses MTF1 function constitutively and in the induction by Cd2+ and tBHQ.
FIGURE 7.
Superinduction of Mt1 by As3+ and CHX. A, Hepa1c1c7 or BHK3038 cells were treated with CHX (10 μg/ml), arsenic (10 μm), or both for 5 h. Total RNA was blotted for mRNA expression of Mt1 or β-Gal. Gapdh and actin were measured as loading controls. B, MTF1 WT or KO cells were treated with arsenic (10 μm), CHX (10 μg/ml), or both for 5 h. Expression of Mt1 and actin mRNAs was measured by Northern blotting.
As3+ and Other Metal Inducers Bind to MTF1 C-terminal Cysteine Residues in Vitro and in Vivo—The observation that induction of Mt1 by As3+ does not depend on Zn2+ and ROS suggests a direct interaction of As3+ with MTF1 or an MTF1-associated protein. Because As3+ is reactive with protein thiol groups, we examined the possibility of As-thiol interaction in the activation of MTF1. Mouse MTF1 contains two clusters of cysteine residues (Fig. 8A). The N-terminal cluster includes the six pairs of cysteine residues within the six zinc finger structures plus two additional cysteine residues N-terminal to the zinc fingers. The C-terminal cluster contains five cysteine residues near the C-terminal end of MTF1. To investigate the potential role of the thiol clusters in MTF1 activation by As3+, PAO, which forms stable covalent binding with two vicinal cysteine thiols of a protein, was used. We first showed that PAO potently induced Mt1 in the concentration range of 0.01 to 1 μm (Fig. 8B, lanes 2–7); induction was reduced at 5 μm, which is attributable to increased toxicity of PAO at concentrations of >1 μm (data not shown). The potency of PAO for Mt1 induction was estimated to be ∼10 times higher than that of As3+ (EC50 of 0.5 μm versus 5 μm). Furthermore, both As3+ and PAO were shown to bind the free thiol groups of proteins, as revealed by reduction in the contents of free thiol groups of purified MT1 protein after incubation with PAO or As3+ (Fig. 8C).
FIGURE 8.
Characterization of PAO. A, schematic presentation of MTF1 and cysteine residues. B, induction of Mt1 by PAO and arsenic. Hepa1c1c7 cells were treated with tBHQ at 30 μm, PAO at 0.01, 0.05, 0.1, 0.5, 1, or 5 μm, or arsenic at 1, 5, or 10 μm for 5 h. Total RNA was blotted for Mt1 and actin mRNA expression. C, direct binding of arsenic and PAO to the thiol groups of MT1. Purified MT1 protein was incubated with arsenic or PAO for 30 min; the mixture was precipitated with trichloroacetic acid, and free thiol groups were measured by using Ellman reagent with absorbance at 412 nm.
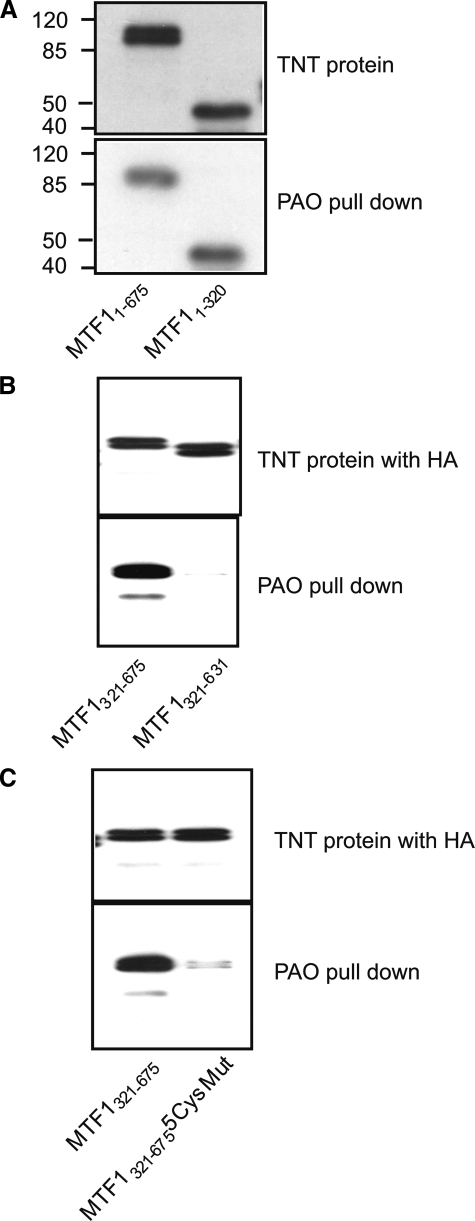
Binding of the N- or C-terminal cysteine clusters of MTF1 to As2+ was directly tested using PAO affinity matrix pulldown. MTF1 and deletion fragments were produced by in vitro transcription and translation (Fig. 9, A and B, upper panels). Both the full-length MTF1 (MTF11–675) and the DBD (MTF11–320) that comprises most of the six zinc fingers were precipitated by PAO pulldown as expected (Fig. 9A, lower panel). The C-terminal half (MTF1321–675) and its deletion mutant that lacks the C-terminal cysteine cluster (MTF1321–631) were examined in Fig. 9B. The PAO affinity matrix pulled down the C-terminal half of MTF1 but not the C-terminal deletion mutant (lower panel), indicating that PAO binds to the C-terminal cysteine cluster. To further test the biding of PAO to the cysteine residues, the C-terminal cysteine residues were mutated to phenylalanine (MTF1321–6755CysMut). PAO beads efficiently pulled down MTF1321–675 but failed to precipitate MTF1321–6755CysMut, even though both proteins were expressed at similar levels (Fig. 9C). The results demonstrate a direct binding of PAO with the C-terminal thiol cluster.
FIGURE 9.
PAO directly binds to the C-terminal cysteine cluster of MTF1. A, biotin-labeled MTF11–675 and MTF11–320 was produced in vitro in TnT reticulocyte lysate and detected with streptavidin (upper panel). The proteins were pulled down with PAO matrix and detected with streptavidin (lower panel). B, MTF1321–675 and MTF1321–631 with a HA tag were produced in vitro and detected with an anti-HA antibody (upper panel). The proteins were pulled down with PAO matrix and detected with anti-HA (lower panel). C, HA-MTF1321–675 and HA-MTF1321–675 with all five cysteine residues mutated to phenylalanine were produced in vitro. The proteins were pulled down with PAO and detected with anti-HA. “5CysMut” = C632FC634FC636FC638FC653F.
Binding of As3+ and other metal inducers with the C-terminal cysteine cluster was examined in Fig. 10. MTF1321–675 expressed in vitro was incubated with As3+ (10 μm, 2 h) prior to PAO pulldown. Fig. 10A shows that preincubation with As3+ effectively abolished binding of PAO with the protein. Similarly, treatment of cells expressing MTF1321–675 with As3+ (10 μm, 2 h) blocked pulldown of the peptide by PAO beads (Fig. 10B). Furthermore, most other metal inducers, Cd2+ (10 μm), Co2+ (150 μm), Ni2+ (150 μm), Ag+ (10 μm), Hg2+ (10 μm), or Bi3+ (50 μm), but not Zn2+ (150 μm), blocked or significantly inhibited PAO pulldown of MTF1321–675 expressed in cells, which were treated with the inducers for 2 h prior to PAO pulldown (Fig. 10C). Taken together, these results demonstrated that As3+ and most other metal inducers bind directly to the C-terminal cysteine residues of MTF1 both in vitro and in vivo. The failure of Zn2+ to inhibit PAO pulldown could be due to the much lower affinity of Zn2+ to cysteine thiols than that of PAO.
FIGURE 10.
Metal inducers of Mt1 bind to MTF1 C-terminal cysteine residues. A, binding with As3+ in vitro. TnT HA-MTF1321–675 was incubated with As3+ (10 μm) for 2 h and then pulled down with PAO and blotted with anti-HA. B, binding with As3+ in vivo. MTF1 KO cells were transfected with pMTF1321–675 HA.His and treated with As3+ at 10 μm for 2 h. MTF1321–675 was pulled down with PAO and blotted with anti-HA. C, binding with other metal inducers in vivo. MTF KO cells transfected with pMTF1321–675 HA.His were treated with Zn2+ (150 μm), Cd2+ (10 μm), Co2+ (150 μm), Ni2+ (150 μm), Ag+ (10 μm), Hg2+ (10 μm), or Bi3+ (50 μm) for 2 h. MTF1321–675 was pulled down with PAO and blotted with anti-HA.
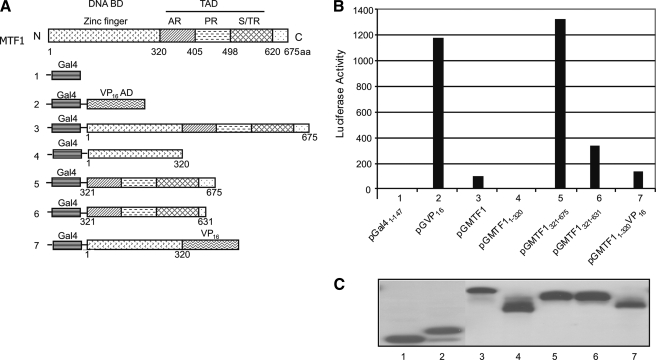
MTF1 C-terminal Cysteine Residues Are Required for Transcription Activation by MTF1 and for Induction of Endogenous Mt1—We asked whether binding of As3+ to the C-terminal cysteine cluster plays one or more functional roles in Mt1 induction. Because MTF1 C-terminal half comprises mostly the TA domain, we examined the impact of the cysteine residues on the TA activity of MTF1. Gal4-MTF1 chimeric proteins, in which the Gal4-DNA binding domain (Gal4) and MTF1 or its deletion fragments were fused together (Fig. 11A), were expressed in cells to examine the TA activities of the fragments. Gal4 DBD (pGal41–147), which lacks a TA domain, was used as a negative control and showed little activity (Fig. 11B, lane 1), whereas Gal4-Vp16 (pGVP, lane 2), a positive control, activated reporter expression markedly. Full-length MTF1 (pGMTF1) exhibited a very low TA activity (lane 3) and MTF1 DBD (pGMTF11–320) showed no TA activity (lane 4). On the other hand, the C-terminal half (pGMTF1321–675) had a high TA activity comparable to that of Vp16 (compare lane 5 with 2) and thus, is a strong TA domain. However, deletion of the C-terminal cysteine cluster (pGMTF1321–631, lane 6) largely reduced the TA activity (to ∼23% of pGMTF1321–675), revealing a critical role of the C-terminal cysteine residues in MTF1 transcription activation.
FIGURE 11.
MTF1 C-terminal cysteine cluster is required for transcription activation. A, schematic presentation of MTF1 domains fused to the Gal4 DNA-binding domain (Gal4). B, Hepa1c1c7 cells were co-transfected with pG5IL2Luc and each construct shown above. 48 h later, the cells were lysed and measured for luciferase expression. C, protein expression of chimeric Gal4-MTF1 and Gal4-MTF1 deletion mutants.
The remarkably low activity of the full-length MTF1 compared with the C-terminal half suggest an inhibitory activity of the N-terminal fragment of MTF1. To test this notion, MTF1 DBD was inserted between Gal4 DBD and VP16. Remarkably, MTF1 DBD totally blocked the TA activity of VP16, demonstrating a potent inhibitory function of MTF1 DBD on a heterologous TA activity (Fig. 11B, compare lane 7 with 2). All Gal4 fusion proteins were expressed at comparable levels (Fig. 11C), excluding variable protein expression and stability, as contributing factors to the large differences in the TA activities observed from the fusion proteins. The results revealed that the C-terminal half of MTF1 contains a strong TA function that requires the C-terminal cysteine cluster, whereas the MTF1 DBD is a strong inhibitor of MTF1 or heterologous VP16 TA activities in addition to its DNA binding function.
Point mutation was performed to further dissect the role of the C-terminal cysteine residues in MTF1 transcription activation (Fig. 12). Replacing cysteine residues 632 and 634 with phenylalanine (pGMTF1321–675C632FC634F) significantly reduced the TA activity compared with the wild-type (pGMTF1321–675), but the reduction was only ∼52%, much less dramatic than that of pGMTF1321–631 (Fig. 12A, compare lane 4 with lanes 2 and 3). Double mutations at C636 and C638 (pGMTF1321–675C636FC638F) resulted in a 33% reduction in the TA activity (compare lane 5 with 2). On the other hand, a single mutation at C653 (pGMTF1321–675C653F) resulted in >80% inhibition. A triple mutation (pGMTF1321–675 C636FC638FC653F) or five cysteine mutation (pGMTF1321–675 5CysMut) caused further reduction in TA activity to a similar level to that of pGMTF1321–631. The point mutant proteins were expressed at comparable levels (Fig. 12B). The findings revealed that most of the cysteine residues at the C terminus of MTF1 play a role in its TA function and may do so through a synergistic action among the residues; however, of the five cysteines, Cys-653 is likely the most critical.
FIGURE 12.
Point mutations of MTF1 C-terminal cysteine residues. A, point mutations of MTF1 C-terminal cysteine residues (cysteine to phenylalanine), C632FC634F, C636FC638F, C653F, C636FC638FC653F, and 5CysMut (C632FC634FC636FC638FC653F), were generated with pGMTF1321–675 as the template, respectively. Co-transfection of pG5IL2Luc with one of the point mutation plasmids was performed, and luciferase activities were measured 48 h after transfection. B, protein expression of chimeric Gal4-MTF1 and Gal4-MTF1 cysteine mutants.
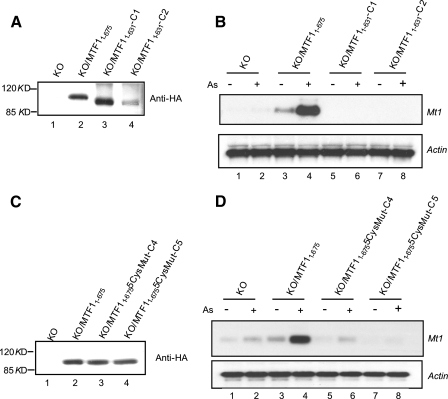
Lastly, the role of the C-terminal cysteine cluster in the induction of endogenous Mt1 was examined by comparing the abilities of MTF1 and the deletion mutant (MTF11–631) or five cysteine mutant (MTF11–6755CysMut) to reconstitute Mt1 induction in MTF1 KO cells (Fig. 13). MTF11–675 and MTF11–631 were stably expressed in MTF1 KO cells, respectively, as detected by immunoblotting with an anti-HA tag antibody (Fig. 13A). The MTF1 KO cells stably expressing MTF1 (KO/MTF11–675) showed a detectable basal expression and a strong induction of Mt1 by As3+ compared with the KO cells (Fig. 13B, compare lanes 3 and 4 with lanes 1 and 2). Remarkably, two MTF1 KO clones expressing MTF11–631 (KO/MTF11–631-C1 and KO/MTF11–631-C2) did not show either the basal expression or induction of Mt1 in the presence of As3+ (Fig. 13B, compare lanes 5–8 with lanes 1 and 2). In a separate experiment, MTF11–675 and MTF11–6755CysMut were expressed in KO cells (Fig. 13C). Cells expressing MTF11–675 are fully functional for induction of Mt1. On the contrary, one mutant clone (MTF11–6755CysMut-C4) showed only slight induction that is similar to that in KO cells (compare lanes 6 and 5 with lanes 1 and 2), and the other clone (MTF11–6755CysMut-C5) did not show detectable expression or induction of the gene (lanes 7 and 8). Together, the findings strongly support a critical role of the C-terminal cysteine clusters of MTF1 in the induction of Mt1 by As3+ via As3+-MTF1 C-terminal cysteine thiol interaction.
FIGURE 13.
MTF1 C-terminal cysteine residues are required for induction of endogenous Mt1. A, MTF1 KO cells were stably transfected with pMTF11–675 or pMTF11–631. Cell lysate was blotted with anti-HA to show expression of MTF1 or the deletion mutant. B, cell clones stably expressing MTF11–675 or MTF11–631 (C1 and C2) were treated with arsenic (5 μm) for 5 h. Total RNA was blotted for Mt1 or actin. C, MTF1 KO cells were stably transfected with pMTF11–675 or pMTF11–6755CysMut in which all five C-terminal cysteine residues were mutated to phenylalanine. Cell lysate was blotted with anti-HA to show expressed MTF1 or the cysteine mutant. D, cell clones stably expressing MTF11–675 or MTF11–6755CysMut (C4 and C5) were treated with arsenic (5 μm) for 5 h. Total RNA was blotted for Mt1 or actin.
DISCUSSION
MTF1 Mediates the Induction of Mt1 by As3+—As3+ elicits pleiotropic effects in animals and humans causing a wide range of lesions in multiple organs; moreover, arsenic is highly potent in many of the responses, suggesting multiple specific molecular targets and signaling pathways in arsenic responses. As3+ potently induces Mt1 with EC50 values of 5 μm for induction of Mt1 and 1.5 μm for induction of MRE reporter, which are similar to those by Cd2+ and a magnitude higher than those by Zn2+. The pathway mediating Mt1 induction by As3+ has not been analyzed.
The enhancer of Mt1 contains a putative ARE that overlaps with a binding site of the upstream stimulatory factor (8). We have previously shown that As3+ activates Nrf2, a cap `n' collar basic region leucine zipper transcription factor, to mediate induction of ARE-dependent phase II detoxification enzymes, such as NAD(P)H:quinone oxidoreductase 1 and glutathione S-transferase 1A, as well as several antioxidative enzymes/proteins, such as heme oxygenase 1, thioredoxin, and thioredoxin reductase 1 (50). Induction of the genes via Nrf2 by As3+ and other toxic metals is critical for protection against oxidative stress induced by the metals (51, 52). However, Nrf2 does not appear to be required for induction of Mt1 by As3+, because induction remains unaltered in the absence of functional Nrf2, even though an ARE is present in the enhancer of mouse Mt1.
In this study, we provided several lines of convincing evidence demonstrating that MTF1 mediates the induction of Mt1 by As3+. First, As3+ induces Mt1 in MTF1 WT but not KO cells, whereas reconstitution of the KO cells with full-length MTF1 restores the induction. Second, As3+ induces the MRE-driven β-gal expression similarly to Cd2+ and Zn2+. Third, As3+ induces the binding of MTF1 to the MREs of the endogenous chromatin Mt1 in cells. Fourth, induction by As3+ is susceptible to inhibition by a labile repressor; inhibition of protein synthesis by cycloheximide in the presence of As3+ results in the superinduction of Mt1, which is characteristic of Mt1 induction by a number of inducers through the MTF1/MRE pathway (31). Lastly, loss of MTF1 increases the sensitivity of cells to arsenic-induced toxicity, supporting MTF1 as a key mediator of the adaptive response for survival in the presence of As3+. We conclude that MTF1 is the critical mediator of Mt1 induction by As3+.
MTF1 Is an Arsenic Sensor through the C-terminal Cysteine Residues—A central issue that remains elusive in understanding the regulation of metal homeostasis is how the body senses changes in the concentrations of metals. In light of the importance of MTF1 and MTs in metal response across species, the MTF1/MRE pathway provides an opportunity for unraveling the molecular events of metal sensing. Much of the attention in the research of MTF1 activation by metals in the past has been focused on the zinc fingers of MTF1 in which six pairs of cysteine residues can bind a range of metals (9, 24, 26). Binding with Zn2+ stabilizes MTF1-MRE interaction. However, binding of the fingers with other metals, such as Cd2+, does not appear to induce conformations favored for DNA binding (9). It was proposed that Zn2+ plays a central role in MTF1 activation by interacting with the zinc fingers; other inducers, including metals and antioxidants, may mobilize Zn2+ in cells to increase free Zn2+, which binds to the zinc fingers and activates MTF1. One alternative hypothesis of MTF1 activation involves dissociation of MTF1 from one or more inhibitory proteins, because inhibition of protein synthesis or exposing cells to heat shock significantly increases the induction of Mt1 (e.g. superinduction) (7, 31, 32). A third postulate of MTF1 activation suggests that a four-cysteine residue stretch at the C terminus of human MTF1 plays a role in metal-induced transcription of an MRE reporter gene, because the cysteine residues are conserved among the human, mouse, pufferfish, and Drosophila proteins and deletion of the cysteine cluster diminished the ability of MTF1 to support the induction of the reporter (53).
Our analysis of the mechanism of Mt1 induction by As3+ does not support a central role of Zn2+ in arsenic sensing by MTF1, because modulating zinc concentrations did not affect the induction of Mt1 or MRE β-gal reporter expression by As3+. In fact, depletion of intracellular free Zn2+ did not decrease but consistently enhanced the induction of the MRE reporter. This result is consistent with our previous report that treatment with tBHQ but not Cd2+ elevates cytoplasmic free Zn2+ concentrations (30, 31). Therefore, it is plausible to postulate that some inducers, such as tBHQ and ROS, activate MTF1 via intracellular free Zn2+, whereas metal inducers, such as As3+, Cd2+, Co2+, Ni2+, Ag+, Hg2+, and Bi3+, do not rely on Zn2+ for Mt1 induction. In the latter scenario, Zn2+ may still be necessary for maintaining the zinc fingers in a conformation favorable for DNA binding; however, additional critical signaling events by As3+ and other inducers lead to the activation of MTF1 as discussed below.
Our study demonstrates a critical role of binding of metal inducers to the five-cysteine cluster at the C terminus of MTF1 for Mt1 induction. First, PAO affinity beads efficiently bind and pull down the C-terminal half of MTF1 (MTF1321–675) that comprises most of the transcription activation domain and contains the terminal five-cysteine cluster. Deletion or mutation of the cysteine residues abolishes the binding, thus providing the first direct evidence for the binding of an arsenic inducer to the C-terminal cysteine cluster of mouse MTF1. Second, a number of metal inducers, including As3+, Cd2+, Co2+, Ni2+, Ag+, Hg2+, and Bi3+, bind to the same cysteine cluster as PAO does both in vitro and in vivo, because the metal inducers effectively block PAO pulldown of MTF1321–675. Third, deletion of the cysteine cluster abolishes the TA activity of the MTF1 TA domain; mutation of Cys-632, Cys-634, Cys-636, and Cys-638 of the cysteine cluster significantly reduces the TA activity, whereas mutation of Cys-653 largely diminishes and mutation of all five cysteine residues completely blocks the TA activity. Therefore, at a molecular level, the C-terminal cysteine cluster is critical for MTF1 transcription activation. In this regard, a synergistic action among the cysteine residues is likely, and Cys-653 appears to be essential. Finally, we showed that the C-terminal cysteine cluster is required for induction of endogenous Mt1 in intact cells, because deletion of the cysteine cluster or mutation of the five cysteine residues totally repressed the ability of MTF1 to restore Mt1 induction in MTF1 KO cells. Taken together, these findings strongly support the notion that interaction of As3+ and other metal inducers with the C-terminal cysteine cluster is critical for MTF1 activation and Mt1 induction by the metal inducers. This metal-sensing function of the C-terminal cysteine cluster appears to be independent of the zinc fingers located in the amino half of MTF1.
A New Function of MTF1 Zinc Fingers: Direct Regulation of Transcription Activation—Examination of the transcription activation activity of MTF1 domains reveals a novel regulatory role of the zinc fingers of MTF1. Specifically, the DBD represses the transcription activation activity of MTF1 to give rise to a very low TA activity of the full-length MTF1, whereas deletion of the DBD results in the expression of a strong MTF1 TA activity that is comparable to that of the TA domain of VP16. Moreover, MTF1 zinc fingers totally block the heterologous VP16 TA activity, indicating that the inhibitory activity of the zinc fingers is not dependent on the sequence context of MTF1 TA domain. Therefore, MTF1 zinc fingers appear to play multiple regulatory roles in MTF1 activation. In addition to mediating DNA binding and nuclear translocation, which is mediated through the nuclear import and export signaling sequences flanking the zinc fingers, our study demonstrates a new function of the zinc fingers, i.e. direct inhibition of the transcription activation activity of the C-terminal TA domain. We envision that repressing the TA function of MTF1 by the zinc fingers is necessary to keep MTF1 in a quiescent state in the absence of an inducer. The zinc fingers may directly interact with the TA domains to mediate suppression. In this model, a specific interface between the zinc fingers and the TA domain is required for effective interaction. Alternatively, the zinc fingers may interact with a repressor protein that in turn interacts with and inhibits the TA domains. Thirdly, the DNA-binding domain may take a conformation that hinders the interaction between the TA domain and transcription co-activators in the absence of an inducer. Because the zinc fingers also inhibit the TA activity of heterologous VP16 TA domain, the second and third hypotheses are more likely than the first postulate.
Footnotes
The abbreviations used are: MT, metallothionein; ARE, antioxidant-responsive element; β-gal, β-galactosidase; ChIP, chromatin immunoprecipitation; CHX, cycloheximide; DBD, DNA-binding domain; HO1, heme oxygenase 1; MRE, metal-responsive element; MTF1, metal-activated transcription factor 1; NAC, N-acetylcysteine; NQO1, NAD(P)H:quinone oxidoreductase; Nrf2, Nuclear factor erythroid 2-related factor 2; PAO, phenylarsine oxide; tBHQ, tert-butylhydroquinone; FBS, fetal bovine serum; DMEM, Dulbecco's modified Eagles medium; WT, wild type; KO, knockout; aa, amino acid(s); PBS, phosphate-buffered saline; HA, hemagglutinin; TA, transcription activation.
References
- 1.Liu, J., Goyer, R. A., and Waalkes, M. P. (2008) in Casarett and Doull's Toxicology. The Basic Science of Poisons (Klaassen, C. D., ed) 7th Ed., pp. 931–979, McGraw-Hill Medical, New York
- 2.Sarkar, B. (1983) Biological Aspects of Metals and Metal-related Diseases, pp. 1–318, Raven Press, New York
- 3.Palmiter, R. D. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 8428–8430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lichtlen, P., and Schaffner, W. (2001) Bioessays 23 1010–1017 [DOI] [PubMed] [Google Scholar]
- 5.Klaassen, C. D., Liu, J., and Choudhuri, S. (1999) Annu. Rev. Pharmacol. Toxicol. 39 267–294 [DOI] [PubMed] [Google Scholar]
- 6.Radtke, F., Heuchel, R., Georgiev, O., Hergersberg, M., Gariglio, M., Dembic, Z., and Schaffner, W. (1993) EMBO J. 12 1355–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palmiter, R. D. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 1219–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andrews, G. K. (2000) Biochem. Pharmacol. 59 95–104 [DOI] [PubMed] [Google Scholar]
- 9.Giedroc, D. P., Chen, X., and Apuy, J. L. (2001) Antioxid. Redox Signal. 3 577–596 [DOI] [PubMed] [Google Scholar]
- 10.Ma, Q. (2008) Chem. Res. Toxicol. 21 1651–1671 [DOI] [PubMed] [Google Scholar]
- 11.Auf der Maur, A., Belser, T., Elgar, G., Georgiev, O., and Schaffner, W. (1999) Biol. Chem. 380 175–185 [DOI] [PubMed] [Google Scholar]
- 12.Balamurugan, K., Egli, D., Selvaraj, A., Zhang, B., Georgiev, O., and Schaffner, W. (2004) Biol. Chem. 385 597–603 [DOI] [PubMed] [Google Scholar]
- 13.Brugnera, E., Georgiev, O., Radtke, F., Heuchel, R., Baker, E., Sutherland, G. R., and Schaffner, W. (1994) Nucleic Acids Res. 22 3167–3173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Radtke, F., Georgiev, O., Muller, H. P., Brugnera, E., and Schaffner, W. (1995) Nucleic Acids Res. 23 2277–2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Selvaraj, A., Balamurugan, K., Yepiskoposyan, H., Zhou, H., Egli, D., Georgiev, O., Thiele, D. J., and Schaffner, W. (2005) Genes Dev. 19 891–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balamurugan, K., Egli, D., Hua, H., Rajaram, R., Seisenbacher, G., Georgiev, O., and Schaffner, W. (2007) EMBO J. 26 1035–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gunes, C., Heuchel, R., Georgiev, O., Muller, K. H., Lichtlen, P., Bluthmann, H., Marino, S., Aguzzi, A., and Schaffner, W. (1998) EMBO J. 17 2846–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masters, B. A., Kelly, E. J., Quaife, C. J., Brinster, R. L., and Palmiter, R. D. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 584–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michalska, A. E., and Choo, K. H. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 8088–8092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang, Y., Wimmer, U., Lichtlen, P., Inderbitzin, D., Stieger, B., Meier, P. J., Hunziker, L., Stallmach, T., Forrer, R., Rulicke, T., Georgiev, O., and Schaffner, W. (2004) FASEB J. 18 1071–1079 [DOI] [PubMed] [Google Scholar]
- 21.Tamura, Y., Maruyama, M., Mishima, Y., Fujisawa, H., Obata, M., Kodama, Y., Yoshikai, Y., Aoyagi, Y., Niwa, O., Schaffner, W., and Kominami, R. (2005) Oncogene 24 399–406 [DOI] [PubMed] [Google Scholar]
- 22.Palmiter, R. D. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 4918–4923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andrews, G. K. (2001) Biometals 14 223–237 [DOI] [PubMed] [Google Scholar]
- 24.Li, Y., Kimura, T., Laity, J. H., and Andrews, G. K. (2006) Mol. Cell. Biol. 26 5580–5587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang, B., Georgiev, O., Hagmann, M., Gunes, C., Cramer, M., Faller, P., Vasak, M., and Schaffner, W. (2003) Mol. Cell. Biol. 23 8471–8485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Potter, B. M., Feng, L. S., Parasuram, P., Matskevich, V. A., Wilson, J. A., Andrews, G. K., and Laity, J. H. (2005) J. Biol. Chem. 280 28529–28540 [DOI] [PubMed] [Google Scholar]
- 27.Westin, G., and Schaffner, W. (1988) EMBO J. 7 3763–3770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang, H., Daniels, P. J., and Andrews, G. K. (2003) J. Biol. Chem. 278 30394–30402 [DOI] [PubMed] [Google Scholar]
- 29.Giedroc, D. P., Chen, X., Pennella, M. A., and LiWang, A. C. (2001) J. Biol. Chem. 276 42322–42332 [DOI] [PubMed] [Google Scholar]
- 30.Bi, Y., Palmiter, R. D., Wood, K. M., and Ma, Q. (2004) Biochem. J. 380 695–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bi, Y., Lin, G. X., Millecchia, L., and Ma, Q. (2006) J. Biochem. Mol. Toxicol. 20 57–68 [DOI] [PubMed] [Google Scholar]
- 32.Saydam, N., Steiner, F., Georgiev, O., and Schaffner, W. (2003) J. Biol. Chem. 278 31879–31883 [DOI] [PubMed] [Google Scholar]
- 33.Saydam, N., Adams, T. K., Steiner, F., Schaffner, W., and Freedman, J. H. (2002) J. Biol. Chem. 277 20438–20445 [DOI] [PubMed] [Google Scholar]
- 34.Adams, T. K., Saydam, N., Steiner, F., Schaffner, W., and Freedman, J. H. (2002) Environ. Health Perspect. 110 Suppl. 5, 813–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.LaRochelle, O., Gagne, V., Charron, J., Soh, J. W., and Seguin, C. (2001) J. Biol. Chem. 276 41879–41888 [DOI] [PubMed] [Google Scholar]
- 36.Gomez-Caminero, A., Howe, P., Hughes, M. F., Kenyon, E., Lewis, D. R., Moore, M., Ng, J., Aitio, A., and Becking, G. (2001) Environmental Health Criteria 224. Arsenic and Arsenic Compounding, pp. 1–521, World Health Organization, Geneva, Switzerland
- 37.Duker, A. A., Carranza, E. J., and Hale, M. (2005) Environ. Int. 31 631–641 [DOI] [PubMed] [Google Scholar]
- 38.Tseng, C. H. (2005) J. Environ. Sci. Health C. Environ. Carcinog. Ecotoxicol. Rev. 23 55–74 [DOI] [PubMed] [Google Scholar]
- 39.Taylor, P. R., Qiao, Y. L., Schatzkin, A., Yao, S. X., Lubin, J., Mao, B. L., Rao, J. Y., McAdams, M., Xuan, X. Z., and Li, J. Y. (1989) Br. J. Ind. Med. 46 881–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leonard, A., and Lauwerys, R. R. (1980) Mutat. Res. 75 49–62 [DOI] [PubMed] [Google Scholar]
- 41.Golub, M. S., Macintosh, M. S., and Baumrind, N. (1998) J. Toxicol. Environ. Health B Crit. Rev. 1 199–241 [DOI] [PubMed] [Google Scholar]
- 42.Mastin, J. P. (2005) Cardiovasc. Toxicol. 5 91–94 [DOI] [PubMed] [Google Scholar]
- 43.Chappell, W. R., Beck, B. D., Brown, K. G., Chaney, R., Cothern, R., Cothern, C. R., Irgolic, K. J., North, D. W., Thornton, I., and Tsongas, T. A. (1997) Environ. Health Perspect. 105 1060–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, Z. Y., and Chen, Z. (2008) Blood 111 2505–2515 [DOI] [PubMed] [Google Scholar]
- 45.Ma, Q., Kinneer, K., Bi, Y., Chan, J. Y., and Kan, Y. W. (2004) Biochem. J. 377 205–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nishikimi, A., Kira, Y., Kasahara, E., Sato, E. F., Kanno, T., Utsumi, K., and Inoue, M. (2001) Biochem. J. 356 621–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aitken, A., and Learmonth. (1996) in Protein Protocol Handbook (Walker, J. M., ed) pp. 487–494, Humana Press, Totowa, NJ
- 48.Bianco, N. R., Chaplin, L. J., and Montano, M. M. (2005) Biochem. J. 385 279–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stuart, G. W., Searle, P. F., and Palmiter, R. D. (1985) Nature 317 828–831 [DOI] [PubMed] [Google Scholar]
- 50.He, X., Chen, M. G., Lin, G. X., and Ma, Q. (2006) J. Biol. Chem. 281 23620–23631 [DOI] [PubMed] [Google Scholar]
- 51.He, X., Chen, M. G., and Ma, Q. (2008) Chem. Res. Toxicol. 21 1375–1383 [DOI] [PubMed] [Google Scholar]
- 52.He, X., Lin, G. X., Chen, M. G., Zhang, J. X., and Ma, Q. (2007) Toxicol. Sci. 98 298–309 [DOI] [PubMed] [Google Scholar]
- 53.Chen, X., Zhang, B., Harmon, P. M., Schaffner, W., Peterson, D. O., and Giedroc, D. P. (2004) J. Biol. Chem. 279 4515–4522 [DOI] [PubMed] [Google Scholar]