Abstract
S5a/Rpn10 is a ubiquitin (Ub)-binding protein that is a subunit of the 26S proteasome but also exists free in the cytosol. It binds poly-Ub chains through its two Ub-interacting motifs (UIMs). We discovered that, unlike typical substrates of Ub ligases (E3s), S5a can be ubiquitinated by all E3s tested including multimeric and monomeric Ring finger E3s (MuRF1, Siah2, Parkin, APC, and SCFβTRCP1), the U-box E3, CHIP, and HECT domain E3s (E6AP and Nedd4) when assayed with UbcH5 or related Ub-conjugating enzymes. However, the E2s, UbcH1 and UbcH13/Uev1a, which function by distinct mechanisms, do not support S5a ubiquitination. Thus, S5a can be used for assay of probably all E3s with UbcH5. Ubiquitination of S5a results from its binding to Ub chains on the E3 (after self-ubiquitination) or on the substrate, as a mutant lacking the UIM domain was not ubiquitinated. Furthermore, if the S5a UIM domains were fused to GST, the protein was rapidly ubiquitinated by MuRF1 and CHIP. In addition, polyubiquitination (but not monoubiquitination) of MuRF1 allowed S5a to bind to MuRF1 and accelerated S5a ubiquitination. This tendency of S5a to associate with the growing Ub chain can explain how S5a, unlike typical substrates, which are recognized by certain E3s through specific motifs, is ubiquitinated by all E3s tested and is rapidly degraded in vivo.
In eukaryotes, ubiquitination of proteins plays a key role in the regulation of many cellular processes ranging from cell cycle and antigen presentation to gene transcription (1, 2). A primary biochemical function of ubiquitination is to serve as a substrate recognition mechanism that targets specific proteins for degradation by the 26S proteasome. This large complex degrades most cellular proteins in an ATP hydrolysis-dependent manner (1). Ubiquitination of protein requires the sequential action of three types of enzymes. First, the Ub-activating enzyme (E1)3 activates the ubiquitin (Ub) in an ATP-dependent reaction and forms a thioester bond between the C-terminal carboxyl group of the Ub and a cysteine on the E1. The activated Ub is then transferred to a cysteine residue on a Ub-conjugating enzyme (E2), and finally, a Ub-protein ligase (E3) facilitates formation of an isopeptide linkage between the C terminus of the Ub and a lysine residue on the substrate or on the preceding Ub (3).
In the final step, the Ub ligase (E3) binds both the substrate and the Ub∼E2 thioester and thus determines which substrate is ubiquitinated. E3s can be classified into two types based on their mechanism of transferring Ub to the substrate. One type, which comprises most E3s, contains a Ring motif (or the related U-box motif) and facilitates transfer of the activated Ub directly from the E2 to the substrate (3). The second type contains a HECT domain, which forms a thioester intermediate with Ub and then transfers the Ub to the substrate (3). Eukaryotic cells contain a large number of different E3s (probably hundreds), and each E3 is believed to recognize only a limited number of proteins as substrates (1). In most cases the E3 recognizes a specific structure or amino acid sequence in the substrate (often termed the “degron”) (3). For example, the “N-end rule” E3, Ubr-1, recognizes particular types of amino acids at the N terminus of the substrates (4), whereas the E3 SCFCdc4 complex recognizes a specific phosphorylated amino acid sequence in its substrate, Sic1 (5).
Cells contain a variety of proteins that non-covalently bind Ub or Ub chains through several distinct Ub binding domains (6). Many of these proteins have been reported to become ubiquitinated in cells or in vitro, and deletion or mutation of Ub binding domains abolishes their ubiquitination (6–10). Interestingly, ubiquitination of some of these proteins reportedly blocks their binding to other ubiquitinated proteins and seems to reduce the activities of these Ub-binding proteins (9, 11). However, despite an increasing number of reports about ubiquitination of Ub-binding proteins, only a few studies have identified E2s and E3s responsible for this process (8, 12), and the mechanisms for their ubiquitination remain uncertain.
S5a/Rpn10 is a major Ub-binding protein that binds preferentially to poly-Ub chains (13). It is found as a subunit of the 26S proteasome, but unlike other proteasome subunits, S5a exists predominantly as a free protein in the cytosol (i.e. not incorporated into the proteasome) (14, 15). S5a contains two stretches of about 15 amino acids called the ubiquitin interacting motif (UIM) which is responsible for its affinity for the Ub chains (16, 17). In yeast, the homolog of S5a, Rpn10, is required for degradation of a subset of cellular proteins by the proteasome (14, 18–20). While investigating how the presence of free S5a affects ubiquitination of typical substrates of E3s, we observed that S5a can be ubiquitinated by two very different E3s (MuRF1 and CHIP) with the E2, UbcH5. Because a substrate is usually ubiquitinated by only a particular E3 and because these two enzymes recognize substrates by distinct mechanisms, this observation suggests that S5a interacts with E3s in an atypical manner. The present study demonstrates systematically that a large variety of E3s, which differ widely in enzymatic mechanism, size, structure, and specificity, can ubiquitinate S5a provided they function with UbcH5 and related E2s. S5a, thus, can be considered as a new type of substrate that is ubiquitinated by a novel mechanism involving the association of S5a with growing poly-Ub chains.
EXPERIMENTAL PROCEDURES
Plasmids—The bacterial expression plasmids for His10-S5a and its mutants (16) were kindly provided by Dr. Patrick Young (Stockholm University), GST fusion Siah2 (21) by Dr. Ze'ev Ronai (Burnham Institute), GST fusion MuRF1 (22) by Dr. David J. Glass (Novartis Institute), GST fusion E6AP and Nedd4 (23) by Dr. Allan M. Weissman (National Institutes of Health), and His6-CHIP (24) by Dr. Cam Patterson (University of North Carolina Chapel Hill). The GST fusion UIM domain of S5a, MJD1, KIAA1594, KIAA1386, Eps15, and Epsin were subcloned from yellow fluorescent protein-UIMs (7). Expression plasmids for His6-Parkin and GST-Mdm2 were generated by using cDNAs inserted into pGEX-6X expression plasmids.
Expression and Purification of Proteins—Most proteins were expressed in Escherichia coli (BL21(DE3)STAR) by induction with 0.2 mm isopropyl 1-thio-β-d-galactopyranoside at 16 or 25 °C for 14 h. Cells were lysed in phosphate-buffered saline with a French press, and the crude lysate was cleared by centrifugation at 100,000 × g for 1 h. All GST fusion proteins were purified using glutathione 4B-Sepharose (GE Healthcare), and all His-tagged proteins were purified using nickel-nitrilotriacetic acid-agarose (Qiagen). His6-E1 was purified with Ub-agarose (Sigma) in lysis buffer (50 mm Tris-Cl, pH 7.5, 1 mm dithiothreitol, 5 mm MgCl2, 20 mm KCl, and 2 mm ATP). UbcH5 was purified with Ub-agarose in lysis buffer containing 10 nm E1. The SCFβTRCP1 complex was purified from SF9 insect cells expressing His-Cul1, myc-Rbx1, myc-Skp1, and FLAG-βTRCP1 isolated as previously described (25). APCCdc20 was immunoprecipitated from Xenopus oocytes, as described by Kirkpatrick et al. (26).
Radiolabeling of S5a—To radiolabel S5a by 125I, 150 μl of 70 μm purified His10-S5a in phosphate-buffered saline was first incubated with IODO-BEADS (Pierce) for 5 min at room temperature. Then 250 μl of 0.5 mCi of 125I in phosphate-buffered saline was added, and the mixture was incubated for an additional 10 min at room temperature. Free iodine was removed with a PD10 desalting column (GE Healthcare). To metabolically label S5a, His10-S5a was expressed in E. coli (BL21(DE3)STAR) in media containing [35S]methionine, and the 35S-labeled S5a was purified with a nickel-nitrilotriacetic acid column as described above.
Ubiquitination Assays—To assay the ubiquitination of S5a, 400 nm His10-S5a was incubated at 37 °C for 30–60 min with 20 nm E1, 350–700 nm E2, 150–700 nm E3, and 58 μm Ub in conjugation buffer A (20 mm Tris-Cl, pH 7.5, 20 mm KCl, 5 mm MgCl2, 2 mm ATP, and 1 mm dithiothreitol). The same procedure was performed for the ubiquitination of GST-UIMs. To ubiquitinate S5a by SCFβTRCP1, 200 nm His10-S5a was incubated at 30 °C for 60 min with 230 nm E1, 1.5 μm UbcH5c, 58 μm Ub, 1 μm Ub-aldehyde, 6 nm SCFβTRCP1, 12 nm Nedd8, 6 nm UbcH12, and 6 nm Nedd8-activating enzyme in conjugation buffer A supplemented with 5 mm NaF, 20 mm creatine phosphate, and 5 μm creatine kinase. To measure ubiquitination of S5a by Parkin, 200 nm His10-S5a was incubated at 37 °C for 30–60 min with 400 nm E1, 5 μm UbcH7, 1.4 μm Parkin, and 200 μm Ub in conjugation buffer B (50 mm HEPES-KCl, pH 8.8, 50 mm NaCl, 10 mm MgCl2, 1 mm ATP). To study ubiquitination of S5a by Mdm2, 1 μm of His10-S5a was incubated at 37 °C for 30–60 min with 400 nm E1, 5 μm UbcH5c, 900 nm GST-Mdm2, and 200 μm Ub in the conjugation buffer B. To measure ubiquitination of troponin I, 400 nm troponin I was incubated at 37 °C for 30–120 min with 20 nm E1, 350 nm E2, 150 nm GST-MuRF1, and 58 μm Ub in conjugation buffer A. Ubiquitination of S5a by APC was assayed as described by Kirkpatrick et al. (26). Ubiquitinated proteins were analyzed by SDS-PAGE followed by autoradiography or immunoblotting using specific antibodies.
Binding of S5a to Immobilized MuRF1—GST-MuRF1 from 1 liter of E. coli culture was immobilized on 400 μl (bed volume) of glutathione resin and washed with phosphate-buffered saline. The resin was equilibrated with conjugation buffer A. Then E1 was added to reach final concentrations of 20 nm, 350 nm UbcH5c, 58 μm Ub, and 2 mm ATP. The mixture was incubated at 37 °C for 1 h with gentle rotation. A control sample was incubated in the same manner without E1, UbcH5c, or Ub. After incubation, the resin was washed 3 times with 40 bed volumes of Ub binding buffer (25 mm HEPES-KCl, pH 7.0, 125 mm potassium acetate, 5 mm EGTA, 0.5% (v/v) Triton X-100, and 1 mm dithiothreitol). The resin was then equilibrated with Ub binding buffer containing 1 mg/ml bovine serum albumin. 5 μl of the resin was mixed with 125I-S5a at a final concentration of 400 nm and rotated at 4 °C for 2 h. The resin was then transferred to a spin column (Bio-Rad) and washed twice with 500 μl of Ub binding buffer. The bound proteins were eluted with SDS-PAGE sample loading buffer, and the radioactive 125I-S5a in the eluate was measured using a gamma counter.
A similar experiment was performed under conditions where GST-MuRF1 and S5a were ubiquitinated in the same test tube. GST-MuRF1 immobilized on the glutathione resin was prepared as above, and 125I-S5a (400 nm) was added to reach final concentrations of 20 nm E1, 350 nm UbcH5c, 58 μm Ub, and 2 mm ATP. In the control reaction, Ub was not included in the mixture. The mixtures were rotated at 37 °C, and aliquots were taken every 15 min. The resin was washed, and the bound S5a was measured as described above.
Mass Spectrometry Analysis—His10-S5a was ubiquitinated by GST-MuRF1 or GST-E6AP as described above. The sample was resolved on SDS-PAGE and visualized with Coomassie Blue. The upper portion of the gel, corresponding to ubiquitinated S5a, was excised and subjected to in-gel trypsin digestion. The tryptic peptides were extracted from the gel and analyzed by liquid chromatography MS/MS. Peptides were separated across a 50-min gradient ranging from 7 to 30% (v/v) acetonitrile in 0.1% (v/v) trifluoroacetic acid in a microcapillary (125 μm × 18 cm) column packed with C18 reverse-phase material (Magic C18AQ, 5-μm particles, 200-Å pore size, Michrom Bioresources) and analyzed on-line on a hybrid linear ion trap-Orbitrap mass spectrometer (LTQ-Orbitrap, Thermo-Electron). For each cycle, one full MS scan acquired on the Orbitrap at high mass resolution was followed by 10 MS/MS spectra on the linear ion trap from the 10 most abundant ions. MS/MS spectra were searched using the Sequest algorithm against the human IPI protein data base. Dynamic modifications of 114.0429275 Da on lysine was allowed for ubiquitination. All peptide matches were initially filtered based on enzyme specificity, mass measurement error, Xcorr and dCorr scores and further manually validated for peptide identification and site localization.
RESULTS
All E3s Tested Can Ubiquitinate S5a—During recent studies, we observed that S5a was rapidly ubiquitinated by the E2, UbcH5, functioning with two different ubiquitin ligases, the muscle specific Ring finger E3, MuRF1, which is induced in atrophying muscles (27), and the U-box E3, CHIP, which ubiquitinates unfolded proteins bound to Hsp70 (28, 29). We, therefore, speculated that S5a, unlike typical substrates, can be ubiquitinated by many different types of E3s. To test this possibility, we initially examined whether S5a could be a substrate of the E2, UbcH5, with several types of monomeric Ring E3s, including MuRF1, Siah2 (which is involved in apoptosis induced by tumor necrosis factor α (21)), and Mdm2 (which catalyzes the degradation of p53 (30)) (Fig. 1). We also examined its ubiquitination by an oligomeric Ring E3, SCFβTRCP1 (which catalyzes the degradation of various phosphoproteins including I-κBα and β-catenin (31)), a U-box E3, CHIP, and the HECT domain E3s, E6AP (which is responsible for degradation of p53 upon infection by human papillomaviruses (32)) and Nedd4 (which is involved in endocytosis of membrane proteins (33)) (Fig. 1). Ubiquitination of S5a and its detection were performed as described under “Experimental Procedures.” In a typical reaction with MuRF1, S5a (400 nm) was incubated with MuRF1 (700 nm), E1 (20 nm), UbcH5 (700 nm), Ub (58 μm), and ATP (2 mm). All these E3s formed long Ub chains on S5a with UbcH5, and no ubiquitination was observed without an E3 added. Furthermore, to demonstrate that S5a is also ubiquitinated when it is in molar excess over E3, we performed the ubiquitination reaction with 2000 nm S5a and 400, 200, or 100 nm GST-MuRF1. Even at 100 nm GST-MuRF1, the majority of S5a in the reaction mixture was ubiquitinated within 1 h (supplemental Fig. S2). This finding indicates that during these reactions MuRF1 (and presumably other E3s) ubiquitinates S5a in a catalytic (not a stoichiometric) manner.
FIGURE 1.
S5a can be ubiquitinated by various types of E3s with UbcH5. His10-S5a was incubated with various types of E3s for 60min as described under “Experimental Procedures.” A, ubiquitination by MuRF1 requires E1, E2, Ub, and ATP. S5a was probed by immunoblotting using an anti-His6 antibody. B, Siah2, MuRF1, CHIP, E6AP, and Nedd4 can all polyubiquitinate S5a. 35S-Labeled S5a (700cpm/pmol) was detected by autoradiography. C, ubiquitination of S5a by SCFβTRCP1 was assayed by immunoblotting using an anti-S5a antibody. D, ubiquitination of S5a by GST-MDM2 was assayed by immunoblotting with an anti-S5a antibody.
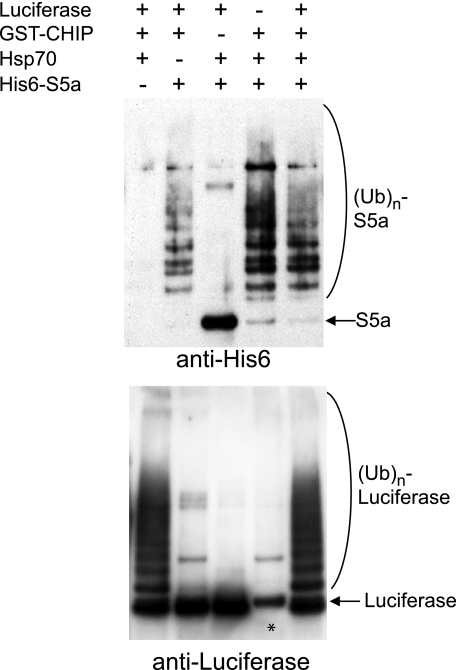
It is noteworthy that ubiquitination of S5a by CHIP does not require the presence of a molecular chaperone for substrate recognition (Fig. 2). Typical substrates of CHIP are proteins recognized and bound to the molecular chaperones Hsp70 or Hsp90 (28). Accordingly, CHIP ubiquitinates the model substrate, luciferase, only when it is denatured and bound to Hsp70 (Fig. 2). By contrast, in the absence of chaperones, CHIP ubiquitinates itself, which seems to be important in the ubiquitination of S5a without Hsp70 or Hsp90 present (see below).
FIGURE 2.
S5a ubiquitination by CHIP does not require Hsp70 unlike ubiquitination of a typical substrate, denatured luciferase. 200 nm His10-S5a or luciferase were ubiquitinated in the presence or absence of 200 nm Hsp70 for 60 min (before the ubiquitination reaction, 2 μm luciferase was denatured by preincubation with 2 μm Hsp70 at 43 °C for 10 min). S5a was ubiquitinated by CHIP even in the absence of Hsp70 or without heat-denaturation, whereas luciferase was ubiquitinated only in the presence of Hsp70. Ubiquitination of S5a and luciferase was assayed by immunoblotting with an anti-His6 or anti-luciferase antibody, respectively. The asterisk indicates that the anti-luciferase antibody cross-reacted with GST-MuRF1.
Ubiquitination of S5a Results from Binding to the Growing Ub Chain—These findings raised the possibility that S5a is promiscuously ubiquitinated by many E3s because of a mechanism that does not require a substrate recognition site of E3. Several studies have reported the ubiquitination of Ub-binding proteins in vivo, which has been proposed to occur because of their high affinity for Ub, as deletion or mutation in their Ub binding domains diminishes their ubiquitination in vivo (7, 11, 34, 35) and in vitro (10, 36). Therefore, we tested systematically if the ubiquitination of S5a by various E3s requires the UIMs using mutant S5a in which the critical hydrophobic residues within the UIM1 and/or UIM2 region are replaced with alanines (16). These mutations abolish the affinity of the UIMs for Ub (supplemental Fig. S1 and Ref. 16). Mutations in both the UIM1 and UIM2 domains of S5a abolished ubiquitination of S5a by MuRF1, CHIP, and E6AP, with UbcH5 as the E2 (Fig. 3 and supplemental Fig. S1). However, S5a lacking just one functional UIM (UIM1 or UIM2) domain was still rapidly ubiquitinated. Thus, UIMs are required for the ubiquitination of S5a, but either of these UIMs is sufficient to allow S5a ubiquitination (Fig. 3, supplemental Fig. S1).
FIGURE 3.
The rapid ubiquitination of S5a by MuRF1 or E6AP requires its UIM domains. His10-S5a and the mutants, which lack hydrophobic residues in either or both UIM domains (16), were incubated with MuRF1 (A) or E6AP (B) and UbcH5 for 60 min. S5a was probed by immunoblotting using an anti-His6 antibody. The asterisk shows His6-E1 present in the reaction.
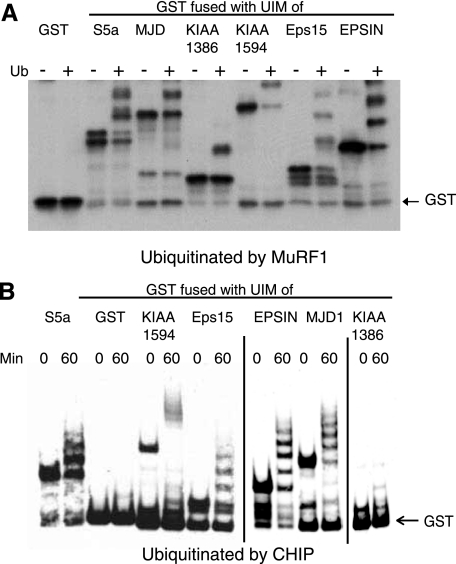
To test whether the UIM regions of S5a are sufficient to promote ubiquitination of a protein, we assayed ubiquitination of GST fused at its C terminus to residues 203–329 of S5a that contain both UIM1 and UIM2. This fusion protein, GST-UIMS5a, was rapidly ubiquitinated by MuRF1 or CHIP with UbcH5, whereas GST without the UIM was not ubiquitinated (Fig. 4). To determine whether the UIM of S5a is unique in this regard, we also assayed the ubiquitination of GST fusion proteins containing UIMs of various other proteins (EPSIN, Eps15, MJD1, KIAA1594, or KIAA1386). These GST fusions were all ubiquitinated by GST-MuRF1 with UbcH5 (Fig. 4A). Also, His6-CHIP with UbcH5 ubiquitinated all these GST fusions, but not GST itself, and did so even in the absence of a molecular chaperone (Fig. 4B). Because these two E3s have very different specificities for substrates and CHIP does not directly bind substrates, these UIM proteins must function as substrates due to their tendency to bind to Ub chains. Accordingly, all these GST fusions bound to free poly-Ub chains (not attached to a protein substrate) containing 2–7 Ub molecules (supplemental Fig. S1).
FIGURE 4.
The attachments of a UIM domain from various UIM proteins to GST is sufficient to cause its ubiquitination by multiple E3s. GST or GST fused with UIM domains from S5a, Eps15, EPSIN, MJD1, KIAA1594, or KIAA1386 were subjected to ubiquitination by MuRF1 (A) or CHIP in the absence of molecular chaperones (B) with UbcH5 for 60 min. Ubiquitination of GST-UIMs was assayed by immunoblotting using an anti-GST antibody.
Self-ubiquitination of MuRF1 Specifically Accelerated the Ubiquitination of S5a—These observations make it very likely that binding to the growing Ub chains on the E3s or on the substrate proteins leads to the ubiquitination of the UIM proteins. To determine whether this self-ubiquitination of a E3 promotes the ubiquitination of S5a, MuRF1 was first incubated with E1, UbcH5, Ub, and ATP for 25 min to allow self-ubiquitination of MuRF1, and then S5a was added to the reaction. In a control reaction, MuRF1 was first incubated with E1, UbcH5, and ATP but without Ub, and then the capacity to ubiquitinate S5a was assayed by adding S5a and Ub. The ubiquitination of S5a was significantly faster when MuRF1 had been preubiquitinated before the addition of S5a (Fig. 5A). For example, ubiquitination of S5a by ubiquitinated MuRF1 was clearly evident 4 min after S5a addition, whereas in the reaction with control MuRF1, ubiquitination of S5a was not observed until 8 min.
FIGURE 5.
Self-ubiquitination of MuRF1 with UbcH5 accelerates ubiquitination of S5a but inhibits ubiquitination of troponin I. A, GST-MuRF1 was first self-ubiquitinated by incubation with UbcH5, ATP, E1, and E2 at 37 °C for 25 min, and then S5a was added. For comparison, MuRF1 with E1 and UbcH5 was incubated first without Ub, and then S5a and Ub were added to the reaction mixture. GST-MuRF1 (i) in the reaction was assayed using an anti-GST antibody and S5a (ii) with an anti His6 antibody. The asterisk shows His6-E1 present in the reaction. iii, unmodified S5a was quantified by densitometric analysis of the immunoblot image shown in Aii. B, a similar experiment was conducted with troponin I (TnI) in place of S5a, and GST-MuRF1 (i) or troponin I (ii) was probed by immunoblotting using an anti-GST or an anti-troponin I antibody respectively. iii, unmodified troponin I was quantified by densitometric analysis of the immunoblot image shown in Bii.
In contrast to S5a, troponin I was ubiquitinated more slowly by self-ubiquitinated MuRF1 than by unmodified MuRF1 (Fig. 5B). Ubiquitination of a typical substrate, thus, does not require and seems to be rather reduced by the presence of Ub chains on the E3. Furthermore, the self-ubiquitination of MuRF1 appeared to precede the ubiquitination of S5a in the control reaction, whereas S5a ubiquitination was slow in onset but accelerated later when MuRF1 had been self-ubiquitinated (Fig. 5A). By contrast, ubiquitination of troponin I and of MuRF1 appeared to start simultaneously (Fig. 5B). These observations further support our hypothesis that self-ubiquitination of MuRF1 is a prerequisite for the ubiquitination of S5a but not for ubiquitination of a typical substrate.
If ubiquitination of S5a is caused by binding of S5a to the growing Ub chains on the E3, S5a should bind to ubiquitinated MuRF1 but not to unmodified MuRF1. To test this prediction, we compared the binding of the 125I-S5a to ubiquitinated or unubiquitinated GST-MuRF1 immobilized on a glutathione resin. A much greater amount of 125I-S5a associated with the ubiquitinated GST-MuRF1 than with GST alone or unmodified GST-MuRF1 (Fig. 6A). In addition, as MuRF1 was being ubiquitinated (i.e. during incubation of immobilized GST-MuRF1 with 125I-S5a, E1, UbcH5, Ub, and ATP), the binding of 125I-S5a to the immobilized MuRF1 increased markedly with time (Fig. 6B). However, when ubiquitination was prevented by omission of Ub, no binding of S5a occurred with time.
FIGURE 6.
S5a binds to ubiquitinated MuRF1 but not unmodified MuRF1. A, GST-MuRF1 was immobilized on a glutathione resin and allowed to self-ubiquitinate for 30 min by incubation with UbcH5. The resulting Ubn-GST-MuRF1, GST-MuRF1 (unmodified), and GST on the glutathione resin were then incubated with 125I-labeled S5a (7500 cpm/pmol). i, S5a bound to the resin was measured using a gamma counter (the data represent averages of three replicates, and the error bar represents S.E.). ii, autoradiograph of the samples corresponding to Ai. iii, self-ubiquitination of the immobilized GST-MuRF1 corresponding to Ai is shown by immunoblotting with an anti-GST antibody. B, 125I-labeled S5a was incubated with E1, UbcH5, ATP, Ub, and GST-MuRF1 immobilized on glutathione resin. Aliquots were taken at different times, and radioactivity bound to the resin was analyzed by gamma counting (i) or autoradiography (ii). iii, self-ubiquitination of GST-MuRF1 in the same aliquots is shown by immunoblotting using an anti-GST antibody. As a control, the same procedure was performed with the reaction mixture lacking Ub.
S5a has a high affinity for poly-Ub chains but only a low affinity for monoubiquitinated proteins (37). To test if monoubiquitination of the E3s is sufficient to cause ubiquitination of S5a, we assayed ubiquitination of S5a using methylated Ub (MeUb) in place of Ub to prevent formation of poly-Ub chains. Although troponin I and MuRF1 were monoubiquitinated with MeUb, no ubiquitination of S5a with MeUb by MuRF1 and UbcH5 was observed (Fig. 7A). To confirm that this result was because of a low affinity of S5a for monoubiquitinated MuRF1, we tested whether polyubiquitinated GST-MuRF1 can ubiquitinate S5a with MeUb. After self-ubiquitination of GST-MuRF1 and removal of unconjugated Ub from the reaction mixture, S5a was added with MeUb, E1, E2, and ATP. As expected, MeUb was conjugated to S5a when MuRF1 was polyubiquitinated before the reaction with MeUb (Fig. 7B, lane 8). These findings together demonstrate that S5a is ubiquitinated by binding to the growing Ub chains and, thus, in a very different manner from typical substrates.
FIGURE 7.
Methylated Ub, although supporting monoubiquitination of MuRF1 and troponin I, does not support ubiquitination of S5a unless the E3, MuRF1, is first polyubiquitinated. A, His10-S5a and troponin I were ubiquitinated by GST-MuRF1 with methylated Ub (MeUb) or Ub for 60 min. Ubiquitination of His10-S5a was assayed by immunoblotting with anti-His6, troponin I (TnI) with anti-troponin I, and MuRF1 with anti-GST antibodies. B, to test if S5a can be conjugated to MeUb when MuRF1 is polyubiquitinated, GST-MuRF1 immobilized on the glutathione resin was first polyubiquitinated by incubation with E1, UbcH5, Ub, and ATP at 37 °C for 30 min. As a control, the immobilized GST-MuRF1 was preincubated similarly but without E1 and E2. The resin containing GST-MuRF1 was washed extensively and then incubated with S5a, E1, UbcH5, ATP, and MeUb at 37 °C for 60 min (lanes 4 and 8). No Ub was added to a set of reaction mixtures (lanes 2 and 6) to show that the washing after the first incubation was complete. In parallel, Ub was added to another set of reaction mixtures (lanes 3 and 7) to confirm that the GST-MuRF1 retained its activity after the first incubation. The asterisk shows His6-E1 present in the reaction.
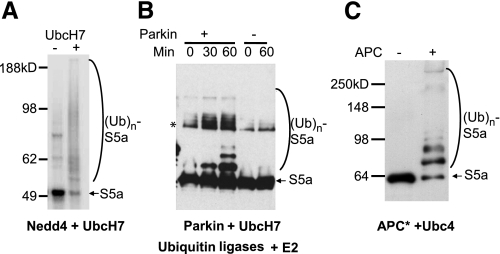
Only UbcH5 and Related Class I E2s Support Ubiquitination of S5a—UbcH5 belongs to the Class I family of E2s which contains a catalytic core (UBC domain) without a distinct Ub binding domain (38). To test whether other Class I E2s can also support ubiquitination of S5a, we assayed the ubiquitination of S5a with UbcH7 and the E3s, Nedd4, or Parkin. With either of these E3s, UbcH7 supported ubiquitination of S5a (Fig. 8, A and B). In addition, another Class I E2, Ubc4, a close homolog of UbcH5, supported ubiquitination of S5a by the APC, a multimeric Ring finger E3 responsible for cell cycle progression through mitosis (39) (Fig. 8C). Thus, multiple Class I E2s can support ubiquitination of S5a by various types of E3s (Table 1).
FIGURE 8.
Other Class I E2s, UbcH7, and Ubc4 can support ubiquitination of S5a. S5a was ubiquitinated as described in Fig. 1 using UbcH7 or Ubc4 as the E2 for 60 min. A, ubiquitination by Nedd4 with UbcH7. 125I-Labeled S5a (7500 cpm/pmol) was detected by autoradiography. B, ubiquitination by Parkin with UbcH7. S5a was probed by immunoblotting using an anti-S5a antibody. C, ubiquitination by Xenopus APC with Ubc4. S5a was probed by immunoblotting using an anti-S5a antibody.
TABLE 1.
Summary for ubiquitination of S5a by E2/E3 pairs tested in this study
“Yes” indicates that rapid ubiquitination of S5a was observed. “No” indicates that little or no ubiquitination of S5a was observed.
|
E3 type
|
E3
|
E2 type
|
Class I
|
Class II
|
Hetero-dimeric E2
|
||
|---|---|---|---|---|---|---|---|
| E2 | UbcH5 | Ubc4 | UbcH7 | UbcH1 | UbcH13/Uev1a | ||
| U-box | CHIP | Yes | No | ||||
| Monomeric Ring | MuRF1 | Yes | No | No | |||
| Siah2 | Yes | ||||||
| Mdm2 | Yes | ||||||
| Parkin | Yes | ||||||
| Oligomeric Ring | SCFβTRCP1 | Yes | |||||
| APC | Yes | ||||||
| HECT | E6AP | Yes | |||||
| Nedd4 | Yes | Yes | |||||
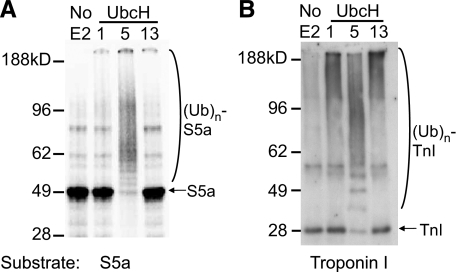
In addition, we tested whether two very different types of E2s, UbcH1 and UbcH13, can support ubiquitination of S5a. UbcH1 belongs to the Class II family of E2s, which are characterized by a C-terminal Ub-associated (UBA) domain (40). UbcH13 functions as a heterodimer with its non-catalytic homologous protein, Uev1a, which contains Ub binding sites (41). Unlike UbcH5, which forms Ub chains containing all possible isopeptide linkages (29), UbcH1 with a Ring finger E3 synthesizes processively Ub chains containing homogeneous Lys-48 isopeptide linkages, whereas UbcH13/Uev1a forms homogeneous Lys-63 chains (29). We used MuRF1 as the E3 as it functions with all these E2s to ubiquitinate one of its typical substrates, troponin I (29). Unlike UbcH5, neither UbcH1 nor UbcH13 supported ubiquitination of S5a by MuRF1 (Fig. 9A and Table 1) even though these E2s did support ubiquitination of troponin I (Fig. 9B). These findings provide further evidence that the mechanisms for ubiquitination of a typical substrate and for S5a must differ.
FIGURE 9.
Unlike UbcH5, UbcH1 and UbcH13 cannot support ubiquitination of S5a by MuRF1, although they support ubiquitination of a typical MuRF1 substrate, troponin I. A, 125I-labeled S5a (7500 cpm/pmol) was incubated with MuRF1 and UbcH1, UbcH5a, or UbcH13/Uev1a for 120 min. S5a was detected by autoradiography. B, troponin I (TnI) was also ubiquitinated under the same conditions and probed by immunoblotting using an anti-troponin I antibody.
In the absence of a distinct substrate, ubiquitination of S5a seems to require the self-ubiquitination of the E3 after which S5a binds to the ubiquitinated E3. Because self-ubiquitination of E3 appeared to be a prerequisite for ubiquitination of S5a, we tested if UbcH1 and UbcH13/Uev1a can support self-ubiquitination of MuRF1. As shown in supplemental Fig. S5A, UbcH1 can support self-ubiquitination of MuRF1, but UbcH13/Uev1a cannot do so. Thus, the failure of UbcH13/Uevla to support S5a ubiquitination may be because of its inability to catalyze auto-ubiquitination of the E3, but this explanation cannot apply to UbcH1. We then tested whether UbcH1 or UbcH13/Uevla could catalyze ubiquitination of S5a if MuRF1 was first self-ubiquitinated with UbcH5, so that the S5a had a Ub chain on the E3 to which it could bind. Under these conditions neither of these E2s allowed ubiquitination of S5a (supplemental Fig. S5B). Therefore, the inability of UbcH1 and UbcH13/Uevla to support S5a ubiquitination is most likely because of their distinct catalytic mechanisms, which differs from that of the UbcH5 family (see below).
The Same Sites on S5a Are Ubiquitinated by Two Different Types of E3s—As demonstrated above, various types of E3s (e.g. Ring finger and HECT domain E3s) ubiquitinate S5a when functioning together with UbcH5 or other Class I E2s. This result was surprising as they function by very different mechanisms; for example, the monomeric Ring finger E3s facilitate a direct transfer of an activated Ub from Ub∼E2 thioester to any lysine on the preceding ubiquitin, whereas the HECT domain E3s form an intermediate Ub thioester before the activated Ub is transferred to either Lys-48 or Lys-63 on the preceding Ub. Because of this difference in conjugation mechanism, we expected that these enzymes would ubiquitinate S5a at very different sites. Therefore, after incubation of S5a with UbcH5 and MuRF1 or E6AP, we identified the ubiquitinated lysine residues on S5a using mass spectrometry.
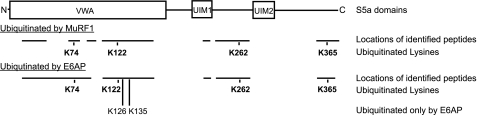
Both E3s ubiquitinated S5a at multiple sites that are located at distant sites from one another. Surprisingly, the same four Lys residues on S5a, Lys-74, Lys-122, Lys-262, and Lys-365 were ubiquitinated by MuRF1 and E6AP (Fig. 10). Two of the common ubiquitination sites, Lys-74 and Lys-122, are located within the S5a von Willebrand factor A domain. Another common site, Lys-262, is located between UIM1 and UIM2, and the last, Lys-365, is located close to the C terminus. Two additional lysines, Lys-126 and Lys-122, were ubiquitinated only by E6AP. In repeated experiments with these E3s, we identified more than 30 peptides from S5a that accounted for 70–80% of its 24 lysines. Although some peptides and lysines were never recovered, these data indicate that these very different E3s ubiquitinate a similar set of Lys residues on S5a.
FIGURE 10.
MuRF1 and E6AP attach ubiquitins to the same four lysines in S5a. S5a was ubiquitinated by MuRF1 or E6AP, and the proteins in the sample were resolved on SDS-PAGE and digested by trypsin. The tryptic peptides were analyzed by liquid chromatography-MS/MS using a LTQ-Orbitrap hybrid mass spectrometer, and the ubiquitination sites were identified by searching a data base using SEQUEAST. An increase in the mass of the Lys residues by 114.0429275 Da was considered as the ubiquitination signature. Twenty (with MuRF1) or 17 (by E6AP) Lys residues of a total 24 Lys residues in S5a were identified. Four Lys residues (Lys-74, -122, -262, and -365) were ubiquitinated by both E3s. Two additional Lys residues (Lys-126 and -135) were ubiquitinated by E6AP. The data represent two independent experiments for each E3, which yielded similar results.
DISCUSSION
Ubiquitin ligases are highly specific enzymes that ubiquitinate a small number of proteins by recognizing specific structural features of their substrates, such as the presence of N-terminal basic amino acids (42), or short sequences containing phosphoserines (43) or hydroxyproline residues (44). Nevertheless, we demonstrated here that a large variety of ligases, including monomeric Ring finger E3s (MuRF1, Mdm2, Siah2, Parkin), large multimeric Ring finger E3s (SCF and APC), the U-box E3, CHIP, and the HECT domain E3s (E6AP and Nedd4), can all ubiquitinate S5a. The narrow substrate specificities of these E3s are well documented. For example, we have confirmed that CHIP does not ubiquitinate native globular proteins (e.g. luciferase) but does so after heat denaturation provided Hsp70 was present (28). Also as reported previously (27), MuRF1 was found to ubiquitinate troponin I, but it could not ubiquitinate a variety of control proteins (e.g. GST, ovalbumin, luciferase). Because all E3s tested ubiquitinated S5a when functioning with UbcH5 (or a closely related class I E2), modification of S5a appears to be a universal property of Ub ligases. Moreover, because S5 is an abundant intracellular protein, ubiquitination of S5a (through its UIM domains) probably occurs in vivo (see below). In fact, a yellow fluorescent protein fused with the UIM regions of S5a was previously shown to be ubiquitinated in cultured mammalian cells (7).
The ability of virtually all E3s together with UbcH5 to ubiquitinate S5a and GST-UIMS5a suggests that this molecule may be useful as a universal substrate for the assay of ubiquitin ligases. Assay of these enzymes is often difficult because the substrates are unknown or not readily available. Although assaying self-ubiquitination of the E3 is a possible alternative approach, measuring Ub conjugation to S5a may offer several advantages. As shown here, ubiquitination of S5a is catalytic and, thus, is likely to allow more sensitive assays than stoichiometric self-ubiquitination. Also, measuring E3 self-ubiquitination may require sensitive immunoassays, which may not be available, and such reagents are available for following S5a ubiquitination. Also for high throughput assays, it is most convenient to have a formatted general assay (e.g. with S5a) that is amenable for routine use.
The mechanism of S5a ubiquitination clearly differs from that of typical E3 substrates. For example, we found that CHIP ubiquitinates S5a without a molecular chaperone present (Fig. 2). The high affinity of S5a for poly-Ub chains is critical for its ubiquitination by diverse E3s. For example, S5a binds to MuRF1 only after MuRF1 undergoes self-ubiquitination. Also, mutations in S5a UIM domains that abolish the binding of S5a to poly-Ub chains greatly reduce its ubiquitination by MuRF1, CHIP, and E6AP (Fig. 3 and supplemental Fig. S1). Furthermore, GST-UIMS5a was rapidly ubiquitinated by MuRF1 and CHIP (Fig. 4). Thus, the binding of a protein to a Ub chain on an E3 appears to be sufficient to make it a good substrate. Although S5a (or other UIM proteins) are ubiquitinated by various E3s, unlike typical substrates, S5a shows no inherent affinity for the unubiquitinated E3s.
In fact, in the absence of a substrate, formation of poly-Ub chains on the E3 seems to be required for (and must precede) the ubiquitination of S5a. Also, pre-ubiquitination of MuRF1 accelerated S5a ubiquitination. Together, these observations support our conclusion that S5a is ubiquitinated because it binds through its UIM domains to the growing poly-Ub chains on the E3s (during self-ubiquitination) (Fig. 6) or on the substrate bound to the E3. In the reaction, when S5a and the typical substrate, troponin I, were both present with MuRF1, S5a was also rapidly ubiquitinated (supplemental Fig. S4), and it reduced slightly the apparent size of the Ub chains on troponin I (data not shown) (presumably because of competition for Ub as S5a is ubiquitinated when it binds to the growing Ub chains on the substrate or on the E3s).
Another UIM protein, Eps15, which plays an important role in endocytosis, has been shown by Woelk et al. (10) to be monoubiquitinated by Nedd4 by a very similar mechanism to that proposed in Fig. 11 for S5a. This ubiquitination of Eps15 is a consequence of its binding to the self-ubiquitinated Nedd4 through the Eps15 UIM domain. In our study we found that S5a can be ubiquitinated by perhaps all E3s (when functioning with UbcH5 or a related E2), as would be expected if S5a ubiquitination is dependent on its binding to a ubiquitinated E3 or substrate (45). At least in vitro it is possible that a large variety of E3s can also ubiquitinate Eps15. We showed that GST fused with UIM of several proteins (including Eps15) can be ubiquitinated by two different E3s, MuRF1 and CHIP (Fig. 4). Therefore, not only S5a but also other UIM proteins (and perhaps other types of ubiquitin-binding proteins) can be ubiquitinated by several different E3s.
FIGURE 11.
The proposed mechanism of ubiquitination of S5a. S5a binds to growing Ub chain on the E3 and is ubiquitinated because of its proximity to the highly reactive Ub thioester. After multiple round of ubiquitination, S5a is supposedly released as our data (supplemental Fig. S2) showed that ubiquitination of S5a is a catalytic process.
Alternatively, there might be an additional mechanism that allows only specific E3s to ubiquitinate Eps15 or other Ub-binding proteins in cells. For example, Eps15 is also ubiquitinated in cells by the Ring finger E3, Parkin, in a mechanism dependent on a direct binding of the Eps15 UIM to the Parkin Ub-like domain (46). In such a case, the E3 should recognize specific structures in the Ub-binding proteins. Based on our data obtained using in vitro systems, we cannot exclude the latter possibility. Nevertheless, in cells Ub-binding proteins are likely to encounter and bind to self-ubiquitinated E3s as self-ubiquitination of several E3s has been reported in vivo (10, 47–49). We also showed that ubiquitination of S5a is not an artifact that occurs only in the absence of other substrates. In fact, the presence of equimolar concentrations of the MuRF1 substrate, troponin I, did not reduce the rate of S5a ubiquitination (supplemental Fig. S4). Furthermore, S5a was ubiquitinated even in the presence of a 4-fold excess of troponin I (supplemental Fig. S4). It will be interesting to test if other types of Ub-binding proteins, such as Sts1 and Vps9, which contain UBA or CUE domains, are also ubiquitinated by a large variety of E3s, as most of these proteins have been shown to be ubiquitinated in cells (11).
In the present experiments we never observed ubiquitination of S5a unless an E3 was present (Figs. 1 and 8). However, recently Hoeller et al. (50) demonstrated that several Ub-binding proteins can be ubiquitinated by E1–E2 mixtures (including UbcH5 and UbcH7) even without an E3 present. Their ubiquitination results from their association with the highly reactive Ub∼E2 thioester. A possible explanation of these opposite findings is that S5a, which has only a weak affinity for monomeric Ub (37), does not bind to a single Ub∼E2 thioester sufficiently to allow its ubiquitination. Our finding that MeUb also does not support ubiquitination of S5a (unless the E3 is previously polyubiquitinated) demonstrates that S5a must first bind to a poly-Ub chain to be ubiquitinated.
Although UbcH1 and UbcH13/Uev1a support ubiquitination of troponin I by MuRF1, these E2s do not support ubiquitination of S5a, unlike Class I E2s. One possible explanation for the inability of UbcH13 to support ubiquitination of S5a seems to be its inability to catalyze self-ubiquitination of E3. On the other hand, UbcH1 supported self-ubiquitination of MuRF1 but not S5a ubiquitination. The most likely explanation for the inability of UbcH1 and also UbcH13/Uevla to ubiquitinate S5a is that these E2s (unlike UbcH5) conjugate Ub in a sterically specific manner. It is noteworthy that UbcH1 as well as UbcH13/Uev1a could not support ubiquitination of S5a even when MuRF1 was self-ubiquitinated before the addition of these E2s. Therefore, UbcH1 and UbcH13 do not simply conjugate Ub to proteins in their proximity. As shown previously, UbcH1 synthesizes homogeneous Ub chains containing only Lys-48 linkages, whereas UbcH13/Uev1a forms only Lys-63 chains (29). In contrast, UbcH5 forms highly heterogeneous chains containing all seven types of isopeptide linkages as well as forked chains in which two Ub moieties are attached to the proximal Ub (29). Our observations here and in a previous report by Kim et al. (29) agree with a recent report that self-ubiquitination of CHIP and ubiquitination of the chaperone, Hsp90, can be supported by UbcH5a but not by UbcH13/Uev1a (51). Similarly, when functioning with APC, the Class I E2, Ubc4, conjugates Ub at multiple sites on the substrates (securin or cyclin B), whereas the Class II E2, Ubc1 (the yeast homolog of UbcH1), links a Ub to only one site (52). Thus, the inability of UbcH1 and UbcH13/Uev1a, in contrast to UbcH5, to ubiquitinate S5a seems to be because of their distinct mechanisms of ubiquitination, which ensure that Ub is conjugated only to a specific Lys residue on the preceding Ub.
The tendency of Ring finger/U-box E3s with UbcH5 to form forked Ub chains composed of all possible isopeptide linkages (29) indicates that these UbcH5/E3 pairs conjugate Ub in a relatively nonspecific manner in which the activated Ub released by the E2 becomes covalently attached to any nearby Lys residue (51). It seems quite likely that ubiquitination of S5a also occurs because of this nonspecific conjugation of the activated Ub from the E2. Elsewhere we will show that the presence of S5a during this ubiquitination reaction with UbcH5 prevents the formation of forked Ub chains on the substrate probably because the S5a on the growing Ub chain shields lysine residues on the proximal Ub from reacting with the Ub released from the E2.4 By contrast, the HECT domain E3s together with UbcH5 form homogeneous chains composed of a single type of isopeptide linkage (Lys-48 by E6AP and Lys-63 by Nedd4). Thus, these enzymes must transfer the activated Ub in a rather precise, stereo-specific manner (29). Nevertheless, both E6AP and Nedd4 extensively ubiquitinated S5a, and E6AP ubiquitinates S5a at multiple Lys residues. This finding suggests that HECT domain E3s require a less specific orientation of the Ub-acceptor Lys residues in the substrate than in the proximal Ub during Ub chain synthesis. The mechanism of Ub conjugation to the substrate and that of Ub-chain elongation by the HECT domain E3s differ markedly. For example, the HECT domain E3s, E6AP and KIAA10, appear to pre-synthesize Ub chains before transferring them to their substrates (53). Thus, chain synthesis and Ub conjugation to the substrate might be two distinct processes.
These observations suggest that Ub-chain formation by E3s involves two kinds of specificity that are dependent on types of the E2/E3 pairs; the first specificity defines the Lys residue of the substrate to which the first Ub is attached, and the second concerns the types of isopeptide linkages formed within the Ub chain. These two types of specificity may not necessarily correlate with each other. Accordingly, MuRF1 and E6AP, despite their contrasting mechanisms, conjugated multiple Ub residues to the same four sites on S5a. These findings further argue that the conjugation of the initial Ub to a substrate protein is quite a different process from the elongation of Ub chains.
The rapid ubiquitination of S5a by various E3s in vitro would predict that this protein, at least its free form present in the cytosol (not in the proteasome), would be rather short-lived. In fact, in yeast, the S5a homolog, Rpn10 (54), is extensively ubiquitinated and rapidly degraded by the proteasome in vivo (54), and in C2C12 (mouse) myoblasts we observed that S5a has a half-life of about 30 min (supplemental Fig. S3). The short half-life of S5a presumably is because of the presence of the UIM domain and reflects the ubiquitination of free S5a by many E3s. At first glance this rapid turnover would appear to be quite wasteful for the cell. Although ubiquitination of S5a may perhaps serve some important regulatory purpose, it seems more likely from the present findings to represent an untoward side effect of substrate ubiquitination that would require cells to continually synthesize and destroy (or deubiquitinate) S5a molecules.
Supplementary Material
Acknowledgments
We are grateful to Mary Dethavong for valuable assistance in the preparation of this manuscript. We thank Lulu Ang for providing materials for the reaction SCFβTRCP1 and Dr. Nathaniel Hathaway for performing the ubiquitination reaction with APC.
This work was supported, in whole or in part, by National Institutes of Health Grants GM51923 and AR055255 (to A. L. G.) and Grant GM67945 (to S. P. G.). This work was also supported by the Fund for Innovation from Elan Pharmaceuticals (to A. L. G.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
Footnotes
The abbreviations used are: E1, ubiquitin-activating enzyme; Ub, ubiquitin; E2, ubiquitin-conjugating enzyme; E3, ubiquitin-protein ligase; UIM, ubiquitin interacting motif; APC, anaphase-promoting complex; CHIP, C terminus of Hsc70-interacting protein; SCF, Skp1-Cullin-F-box ubiquitin ligase; MeUb, methylated Ub; GST, glutathione S-transferase; MS, mass spectrometry.
Kim, H. T., Kim, K. P., Uchiki, T., Gygi, S. P., and Goldberg, A. L. (2009) EMBO J., in press.
References
- 1.Glickman, M. H., and Ciechanover, A. (2002) Physiol. Rev. 82 373–428 [DOI] [PubMed] [Google Scholar]
- 2.Goldberg, A. L. (2003) Nature 426 895–899 [DOI] [PubMed] [Google Scholar]
- 3.Pickart, C. M. (2001) Annu. Rev. Biochem. 70 503–533 [DOI] [PubMed] [Google Scholar]
- 4.Varshavsky, A. (1997) Genes Cells 2 13–28 [DOI] [PubMed] [Google Scholar]
- 5.Nash, P., Tang, X., Orlicky, S., Chen, Q., Gertler, F. B., Mendenhall, M. D., Sicheri, F., Pawson, T., and Tyers, M. (2001) Nature 414 514–521 [DOI] [PubMed] [Google Scholar]
- 6.Hicke, L., Schubert, H. L., and Hill, C. P. (2005) Nat. Rev. Mol. Cell Biol. 6 610–621 [DOI] [PubMed] [Google Scholar]
- 7.Miller, S. L., Malotky, E., and O'Bryan, J. P. (2004) J. Biol. Chem. 279 33528–33537 [DOI] [PubMed] [Google Scholar]
- 8.Polo, S., Sigismund, S., Faretta, M., Guidi, M., Capua, M. R., Bossi, G., Chen, H., De Camilli, P., and Di Fiore, P. P. (2002) Nature 416 451–455 [DOI] [PubMed] [Google Scholar]
- 9.Meray, R. K., and Lansbury, P. T., Jr. (2007) J. Biol. Chem. 282 10567–10575 [DOI] [PubMed] [Google Scholar]
- 10.Woelk, T., Oldrini, B., Maspero, E., Confalonieri, S., Cavallaro, E., Di Fiore, P. P., and Polo, S. (2006) Nat. Cell Biol. 8 1246–1254 [DOI] [PubMed] [Google Scholar]
- 11.Hoeller, D., Crosetto, N., Blagoev, B., Raiborg, C., Tikkanen, R., Wagner, S., Kowanetz, K., Breitling, R., Mann, M., Stenmark, H., and Dikic, I. (2006) Nat. Cell Biol. 8 163–169 [DOI] [PubMed] [Google Scholar]
- 12.Timsit, Y. E., Miller, S. L., Mohney, R. P., and O'Bryan, J. P. (2005) Biochem. Biophys. Res. Commun. 328 550–559 [DOI] [PubMed] [Google Scholar]
- 13.Deveraux, Q., Ustrell, V., Pickart, C., and Rechsteiner, M. (1994) J. Biol. Chem. 269 7059–7061 [PubMed] [Google Scholar]
- 14.van Nocker, S., Sadis, S., Rubin, D. M., Glickman, M., Fu, H., Coux, O., Wefes, I., Finley, D., and Vierstra, R. D. (1996) Mol. Cell. Biol. 16 6020–6028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rubin, D. M., van Nocker, S., Glickman, M., Coux, O., Wefes, I., Sadis, S., Fu, H., Goldberg, A., Vierstra, R., and Finley, D. (1997) Mol. Biol. Rep. 24 17–26 [DOI] [PubMed] [Google Scholar]
- 16.Young, P., Deveraux, Q., Beal, R. E., Pickart, C. M., and Rechsteiner, M. (1998) J. Biol. Chem. 273 5461–5467 [DOI] [PubMed] [Google Scholar]
- 17.Fu, H., Sadis, S., Rubin, D. M., Glickman, M., van Nocker, S., Finley, D., and Vierstra, R. D. (1998) J. Biol. Chem. 273 1970–1981 [DOI] [PubMed] [Google Scholar]
- 18.Elsasser, S., Chandler-Militello, D., Muller, B., Hanna, J., and Finley, D. (2004) J. Biol. Chem. 279 26817–26822 [DOI] [PubMed] [Google Scholar]
- 19.Mayor, T., Lipford, J. R., Graumann, J., Smith, G. T., and Deshaies, R. J. (2005) Mol. Cell. Proteomics 4 741–751 [DOI] [PubMed] [Google Scholar]
- 20.Verma, R., Oania, R., Graumann, J., and Deshaies, R. J. (2004) Cell 118 99–110 [DOI] [PubMed] [Google Scholar]
- 21.Habelhah, H., Frew, I. J., Laine, A., Janes, P. W., Relaix, F., Sassoon, D., Bowtell, D. D., and Ronai, Z. (2002) EMBO J. 21 5756–5765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bodine, S. C., Latres, E., Baumhueter, S., Lai, V. K., Nunez, L., Clarke, B. A., Poueymirou, W. T., Panaro, F. J., Na, E., Dharmarajan, K., Pan, Z. Q., Valenzuela, D. M., DeChiara, T. M., Stitt, T. N., Yancopoulos, G. D., and Glass, D. J. (2001) Science 294 1704–1708 [DOI] [PubMed] [Google Scholar]
- 23.Hatakeyama, S., Jensen, J. P., and Weissman, A. M. (1997) J. Biol. Chem. 272 15085–15092 [DOI] [PubMed] [Google Scholar]
- 24.Ballinger, C. A., Connell, P., Wu, Y., Hu, Z., Thompson, L. J., Yin, L. Y., and Patterson, C. (1999) Mol. Cell. Biol. 19 4535–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skowyra, D., Koepp, D. M., Kamura, T., Conrad, M. N., Conaway, R. C., Conaway, J. W., Elledge, S. J., and Harper, J. W. (1999) Science 284 662–665 [DOI] [PubMed] [Google Scholar]
- 26.Kirkpatrick, D. S., Hathaway, N. A., Hanna, J., Elsasser, S., Rush, J., Finley, D., King, R. W., and Gygi, S. P. (2006) Nat. Cell Biol. 8 700–710 [DOI] [PubMed] [Google Scholar]
- 27.Kedar, V., McDonough, H., Arya, R., Li, H. H., Rockman, H. A., and Patterson, C. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 18135–18140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murata, S., Minami, Y., Minami, M., Chiba, T., and Tanaka, K. (2001) EMBO Rep. 2 1133–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim, H. T., Kim, K. P., Lledias, F., Kisselev, A. F., Scaglione, K. M., Skowyra, D., Gygi, S. P., and Goldberg, A. L. (2007) J. Biol. Chem. 282 17375–17386 [DOI] [PubMed] [Google Scholar]
- 30.Oliner, J. D., Kinzler, K. W., Meltzer, P. S., George, D. L., and Vogelstein, B. (1992) Nature 358 80–83 [DOI] [PubMed] [Google Scholar]
- 31.Nakayama, K. I., and Nakayama, K. (2005) Semin. Cell Dev. Biol. 16 323–333 [DOI] [PubMed] [Google Scholar]
- 32.Huibregtse, J. M., Scheffner, M., and Howley, P. M. (1991) EMBO J. 10 4129–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rotin, D., Staub, O., and Haguenauer-Tsapis, R. (2000) J. Membr. Biol. 176 1–17 [DOI] [PubMed] [Google Scholar]
- 34.Shih, S. C., Prag, G., Francis, S. A., Sutanto, M. A., Hurley, J. H., and Hicke, L. (2003) EMBO J. 22 1273–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Delft, S., Govers, R., Strous, G. J., Verkleij, A. J., and van Bergen en Henegouwen, P. M. (1997) J. Biol. Chem. 272 14013–14016 [DOI] [PubMed] [Google Scholar]
- 36.Oldham, C. E., Mohney, R. P., Miller, S. L., Hanes, R. N., and O'Bryan, J. P. (2002) Curr. Biol. 12 1112–1116 [DOI] [PubMed] [Google Scholar]
- 37.Wang, Q., Young, P., and Walters, K. J. (2005) J. Mol. Biol. 348 727–739 [DOI] [PubMed] [Google Scholar]
- 38.Jentsch, S. (1992) Annu. Rev. Genet 26 179–207 [DOI] [PubMed] [Google Scholar]
- 39.Baker, D. J., Dawlaty, M. M., Galardy, P., and van Deursen, J. M. (2007) Cell. Mol. Life Sci. 64 589–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haldeman, M. T., Xia, G., Kasperek, E. M., and Pickart, C. M. (1997) Biochemistry 36 10526–10537 [DOI] [PubMed] [Google Scholar]
- 41.McKenna, S., Spyracopoulos, L., Moraes, T., Pastushok, L., Ptak, C., Xiao, W., and Ellison, M. J. (2001) J. Biol. Chem. 276 40120–40126 [DOI] [PubMed] [Google Scholar]
- 42.Bartel, B., Wunning, I., and Varshavsky, A. (1990) EMBO J. 9 3179–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hao, B., Oehlmann, S., Sowa, M. E., Harper, J. W., and Pavletich, N. P. (2007) Mol. Cell 26 131–143 [DOI] [PubMed] [Google Scholar]
- 44.Kaelin, W. G. (2005) Cold Spring Harbor. Symp. Quant. Biol. 70 159–166 [DOI] [PubMed] [Google Scholar]
- 45.Haglund, K., and Stenmark, H. (2006) Nat. Cell Biol. 8 1218–1219 [DOI] [PubMed] [Google Scholar]
- 46.Fallon, L., Belanger, C. M., Corera, A. T., Kontogiannea, M., Regan-Klapisz, E., Moreau, F., Voortman, J., Haber, M., Rouleau, G., Thorarinsdottir, T., Brice, A., van Bergen En Henegouwen, P. M., and Fon, E. A. (2006) Nat. Cell Biol. 8 834–842 [DOI] [PubMed] [Google Scholar]
- 47.Sato, S., Aoyama, H., Miyachi, H., Naito, M., and Hashimoto, Y. (2008) Bioorg. Med. Chem. Lett. 18 3354–3358 [DOI] [PubMed] [Google Scholar]
- 48.Sasiela, C. A., Stewart, D. H., Kitagaki, J., Safiran, Y. J., Yang, Y., Weissman, A. M., Oberoi, P., Davydov, I. V., Goncharova, E., Beutler, J. A., McMahon, J. B., and O'Keefe, B. R. (2008) J. Biomol. Screen 13 229–237 [DOI] [PubMed] [Google Scholar]
- 49.Nuber, U., Schwarz, S. E., and Scheffner, M. (1998) Eur. J. Biochem. 254 643–649 [DOI] [PubMed] [Google Scholar]
- 50.Hoeller, D., Hecker, C. M., Wagner, S., Rogov, V., Dotsch, V., and Dikic, I. (2007) Mol. Cell 26 891–898 [DOI] [PubMed] [Google Scholar]
- 51.Windheim, M., Peggie, M., and Cohen, P. (2008) Biochem. J. 409 723–729 [DOI] [PubMed] [Google Scholar]
- 52.Rodrigo-Brenni, M. C., and Morgan, D. O. (2007) Cell 130 127–139 [DOI] [PubMed] [Google Scholar]
- 53.Wang, M., and Pickart, C. M. (2005) EMBO J. 24 4324–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Crosas, B., Hanna, J., Kirkpatrick, D. S., Zhang, D. P., Tone, Y., Hathaway, N. A., Buecker, C., Leggett, D. S., Schmidt, M., King, R. W., Gygi, S. P., and Finley, D. (2006) Cell 127 1401–1413 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.